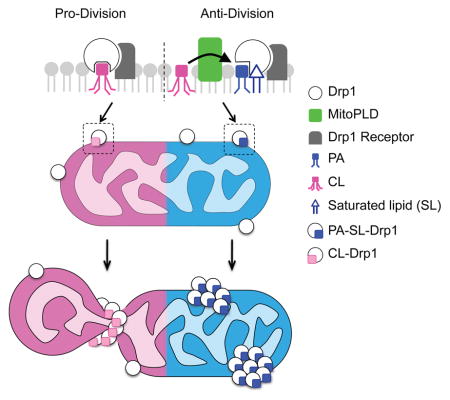
Summary
Mitochondria divide to control their size, distribution, turnover and function. Dynamin-related protein (Drp1) is a critical mechanochemical GTPase that drives constriction during mitochondrial division. It is generally believed that mitochondrial division is regulated during recruitment of Drp1 to mitochondria and its oligomerization into a division apparatus. Here, we report an unforeseen mechanism that regulates mitochondrial division by coincident interactions of Drp1 with the headgroup and acyl chains of phospholipids. Drp1 recognizes the headgroup of phosphatidic acid (PA) and two saturated acyl chains of another phospholipid by penetrating into the hydrophobic core of the membrane. The dual phospholipid interactions restrain Drp1 via inhibition of oligomerization-stimulated GTP hydrolysis that promotes membrane constriction. Moreover, a PA-producing phospholipase, MitoPLD, binds Drp1, creating a PA-rich microenvironment in the vicinity of a division apparatus. Thus, PA controls the activation of Drp1 after the formation of the division apparatus.
Graphical Abstract
Introduction
Mitochondria are dynamic organelles that undergo regulated cycles of division and fusion. Precise control over these cycles of division and fusion ensures an appropriate subcellular mitochondrial distribution and their turnover when damaged (Shutt and McBride, 2012; Youle and van der Bliek, 2012). Hyper- or hypo-activation of mitochondrial division has been linked to many neurological and metabolic disorders (Roy et al., 2015). The conserved dynamin-related protein 1 (Drp1) plays an essential role in mitochondrial division (Bui and Shaw, 2013; Tamura et al., 2011). A loss of Drp1 results in mitochondrial elongation and enlargement due to unopposed mitochondrial fusion. Excessively large mitochondria escape engulfment by autophagosomes and become severely defective in mitophagy (Tanaka et al., 2010; Twig et al., 2008). Therefore, Drp1 loss results in the accumulation of damaged, dysfunctional mitochondria, leading to progressive neurodegeneration and fatal cardiac failure in Drp1 knockout (KO) mice (Ishihara et al., 2014; Kageyama et al., 2014; Kageyama et al., 2012).
Drp1 is a soluble protein that forms small foci on the mitochondria in cells that function as division machinery. A current model suggests three key mechanistic steps for Drp1 in mitochondrial division: 1) the recruitment of Drp1 to the mitochondria; 2) the oligomerization of Drp1; and 3) the constriction of Drp1 oligomers (Bleazard et al., 1999; Koirala et al., 2013; Lackner et al., 2009; Sesaki and Jensen, 1999; Smirnova et al., 2001; Yoon et al., 2001). The recruitment of Drp1 to mitochondria is mediated by interactions with outer membrane proteins such as Mff, Mid49, Mid51 and Fis1. This step is regulated via post-translational modifications of Drp1 and these outer membrane proteins (Chang and Blackstone, 2007; Cribbs and Strack, 2007; Kashatus et al., 2015; Serasinghe et al., 2015; Toyama et al., 2016). Oligomerization of Drp1 is stimulated by GTP binding, and actin filaments facilitate Drp1 oligomerization at ER-mitochondria contact sites (Friedman et al., 2011; Ji et al., 2015; Korobova et al., 2013; Manor et al., 2015). The constriction of Drp1 oligomers initiates mitochondrial division and is driven by GTP hydrolysis. However, it is unknown how the constriction of Drp1 oligomers is regulated. There are many oligomerized Drp1 foci on mitochondria that appear not to engage in mitochondrial division (Roy et al., 2015), suggesting the presence of an inhibitory signal that coordinates via positive cues to precisely control the execution of mitochondrial division.
The founding member of the dynamin superfamily, endocytic dynamin, binds PI(4,5)P2, which anchors this protein to the plasma membrane via a pleckstrin homology domain (Ramachandran, 2011). In contrast, Drp1 lacks a known lipid-binding domain, and the exact role that lipids play in the regulation of mitochondrial division is largely unknown. Here we describe that Drp1 recognizes the headgroup of PA and the saturated acyl chains of another phospholipid. This dual lipid interaction restrains Drp1 in mitochondrial division by suppressing oligomerization-stimulated GTP hydrolysis. These data reveal that PA, together with saturated phospholipids, functions as a key negative regulator of constriction of Drp1 oligomers in mitochondrial division.
Results and Discussion
Drp1 binds saturated PA
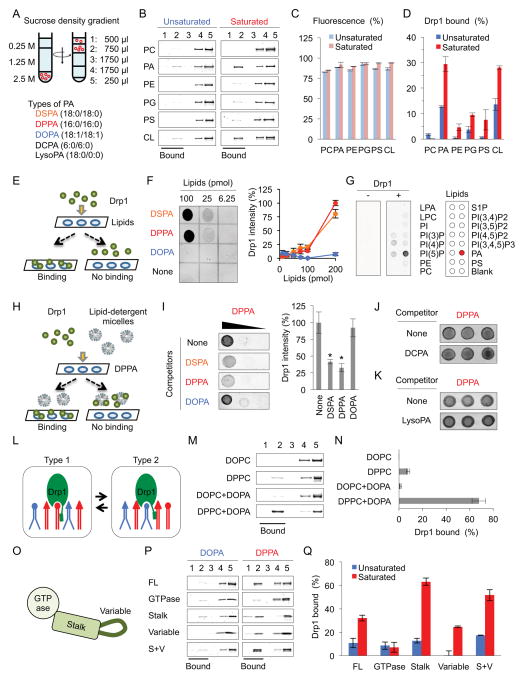
The structure of lipids is determined by their headgroup and acyl chains (Osman et al., 2011). To systematically analyze the interactions of Drp1 with phospholipids, we examined interactions of purified His6-Drp1 with saturated and unsaturated forms of major mitochondrial phospholipids, including phosphatidylcholine (PC), phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylserine (PS), and cardiolipin (CL) using liposome flotation assays (Fig. 1A). Liposomes comprised of palmitoyl-oleoylphosphatidylcholine (POPC, an unsaturated PC; 84%), fluorescent rhodamine-PE (1%), and one of six distinct lipids with saturated or unsaturated acyl chains (15%) were incubated with His6-Drp1 and placed at the bottom of a sucrose gradient. After ultracentrifugation, we collected five fractions from the top of the tube and analyzed them using SDS-PAGE and silver staining (Fig. 1B). Greater than 80% of the liposomes floated to the top two fractions based on rhodamine fluorescence (Fig. 1C). Drp1 was preferentially associated with liposomes-containing PA with saturated acyl chains compared to unsaturated acyl chains (Fig. 1D). As expected from previous studies (Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2014; Montessuit et al., 2010; Stepanyants et al., 2015), Drp1 also preferentially bound liposomes containing a saturated CL. Although the exact role of this mitochondria specific phospholipid CL remains to be determined, binding to CL both stimulates the Drp1 GTPase activity that is required for mitochondrial division, and in a non-GTPase activity-dependent manner, enables cytochrome c release from mitochondria during apoptosis.
Figure 1. Coincident interaction of Drp1 with saturated PA.
(A) Liposome floatation assays. After ultracentrifugation, we collected five fractions from the top and then analyzed the Drp1-liposome association in (B). Rhodamine-PE fluorescence in (C) and Drp1 band intensity in (D) were normalized to the volume of fractions (Mean ± SEM; n = 3). (E) Lipid dot-blot assays. (F and G) His6-Drp1 was incubated with PVDF membranes spotted for the indicated lipids in (F) or PIP strip (Echelon P-6001) in (G), and then detected Drp1-lipid interactions using anti-Drp1 antibodies (Mean ± SEM; n = 3). (H) Lipid competition assays. (I–K) Drp1 was incubated with membranes spotted for DPPA in the presence of the indicated lipids. (L) Model for Drp1-phospholipid interactions. (M and N) We analyzed interactions of His6-Drp1 with the indicated liposomes using floatation assays. (O) Domain structure of Drp1. (P and Q) Different domains were analyzed in the liposome floatation assay (Mean ± SEM; n = 3). “n” indicates the number of independent experiments. Student’s t-test: *p< 0.05. See also Figure S1.
To determine how Drp1 recognizes saturated PA, we used a lipid dot-blot assay (Fig. 1E–G) and a competition assay (Fig. 1H–K). In the dot-blot assay, we incubated His6-Drp1 with PDVF membranes spotted with different lipids (Fig. 1E). We detected the Drp1-lipid interactions with anti-Drp1 antibodies and fluorescently-labeled secondary antibodies. The dot-blot assays revealed that Drp1 binds saturated PA with C16:0 acyl chains (DPPA) or C18:0 acyl chains (DSPA) but not unsaturated PA with C18:1 acyl chains (DOPA) (Fig. 1F) and other saturated lipids (Fig. 1G) with this method of lipid presentation. In a competition assay, saturated PA (DPPA) was spotted onto PVDF membranes and incubated with His6-Drp1 and competitor lipids in the presence of a detergent (Fig. 1H). Saturated PA (DPPA or DSPA) successfully competed for the spotted DPPA, but the unsaturated PA (DOPA) did not (Fig. 1I). Additionally, saturated PA with short acyl chains (DCPA) (Fig. 1J) and lysoPA with only one saturated acyl chain (stearoyl-lysoPA) (Fig. 1K) failed to compete for DPPA. Thus, Drp1 binds saturated PA via a coincident mechanism by recognizing the headgroup as well as the saturation, number, and length of the acyl chains (Fig. S1A).
The phosphorylation of Drp1 regulates its recruitment to mitochondria. To determine whether phosphorylation controls the interaction of Drp1 with saturated PA, we purified a phosphomimetic Drp1 mutant carrying the S600D mutation. Equivalent phosphomimetic mutations have been demonstrated to inhibit the recruitment of Drp1 to mitochondria and mitochondrial division (Chang and Blackstone, 2007; Cribbs and Strack, 2007). In our lipid dot blot assay, we found that wildtype (WT) Drp1 and Drp1S600D similarly bind to saturated PA (DPPA) (Fig. S1B). In addition, another phosphomimetic mutant Drp1S579D also bound saturated PA normally. Phosphorylation at these sites does not appear to modulate the association of Drp1 with saturated PA.
Drp1 separately recognizes the headgroup of PA and saturated acyl chains
To define the mechanism of the coincident interaction between Drp1 and saturated PA, we asked whether binding to the headgroup and acyl chains were separable by testing whether Drp1 can simultaneously interact with the headgroup of unsaturated PA and the acyl chains of saturated non-PA phospholipids. If the regions of Drp1 that interact with the headgroup and acyl chains are spatially separated sufficiently to reach two phospholipids, Drp1 may interact with the PA headgroup and saturated acyl chains of an adjacent phospholipid (Fig. 1L, type 2). To test this model, we performed flotation assays with Drp1 and liposomes containing unsaturated PA (DOPA) and saturated PC (DPPC). Drp1 was associated with liposomes containing both the phospholipids, but not control liposomes only containing the saturated PC (Fig. 1M, N). Saturated acyl chains of DPPC were essential for this interaction, as liposomes containing DOPA and unsaturated PC (DOPC) did not bind Drp1 (Fig. 1M, N). Therefore, Drp1 can separately recognize the headgroup and saturated acyl chains in two phospholipid molecules. This unique Drp1-saturated PA interaction also suggests that Drp1 penetrates the hydrophobic core of the lipid bilayer to reach acyl chains (Fig. 1L).
Drp1 contains two saturated PA binding regions: the stalk and variable domains
Drp1, which lacks a known lipid-binding motif, consists of three major domains: the GTPase domain, the stalk domain, and the variable domain (also called the B-insert) (Frohlich et al., 2013) (Fig. 1O). To identify the saturated PA-binding regions in Drp1, we purified the three domains from E. coli and performed liposome flotation assays (Fig. 1P, Q). We found that the His6-stalk domain strongly bound saturated PA over unsaturated PA. The variable domain also preferentially interacted with saturated PA, but to a lesser extent than the stalk domain. Conversely, the His6-GTPase domain exhibited negligible interaction levels with PA, regardless of acyl chain saturation. The combination of the stalk and variable domains did not increase the lipid interactions in comparison with the stalk domain alone, suggesting that these two domains mediate saturated-PA binding in an overlapping manner. The stalk domain contains alpha-helices and unstructured loops with hydrophobic amino acids; the variable domain of ~100 residues is largely disordered. These unstructured regions may mediate the insertion of Drp1 into the membrane.
PA inhibits Drp1-mediated mitochondrial division
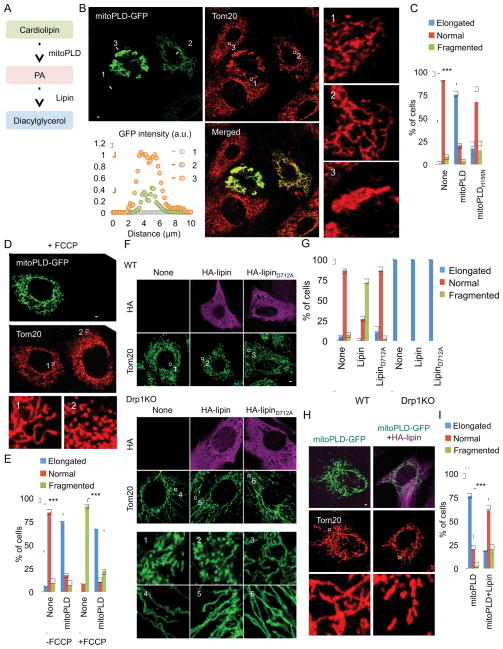
To determine the impact of Drp1-phospholipid interactions on mitochondrial division in cells, we altered levels of PA and acyl chain saturation and examined mitochondrial morphology and dynamics in mouse embryonic fibroblasts (MEFs). MitoPLD, a phospholipase that is tail-anchored to the cytoplasmic surface of the mitochondrial outer membrane, converts CL to PA there (Fig. 2A). Immunofluorescence confocal microscopy with antibodies to the mitochondrial protein Tom20 demonstrated that mitochondria undergo peri-nuclear aggregation upon increased expression of MitoPLD (Fig. 2B–#3) as previously reported (Baba et al., 2014; Choi et al., 2006; Huang et al., 2011; Zhang et al., 2016). This morphology is most likely a non-specific consequence of outer membrane protein overexpression, as observed for catalytically inactive MitoPLDH156N-GFP (4–#3). In cells in which MitoPLD is modestly expressed (~50% of high expression; Fig. 2B–#2 and 2C), the mitochondria are elongated compared with controls (untransfected; Fig. 2B–#1 and S2–#1 and MitoPLDH156N-GFP; Fig. S2–#2); hence the elongation results from a MitoPLD activity-dependent pathway. To determine if mitochondrial elongation resulted from decreased division, we treated MitoPLD-GFP-expressing cells with carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), which induces Drp1-mediated mitochondrial division (Li et al., 2015). FCCP fragmented mitochondria in untransfected cells, as expected (Fig. 2D, E). Cells containing MitoPLD-GFP were resistant to FCCP-induced mitochondrial fragmentation, indicating that MitoPLD overexpression inhibits mitochondrial division. We showed that FCCP decreases the membrane potential similarly in both control and MitoPLD-GFP-expressing cells using the membrane potential-dependent fluorescent dye tetramethylrhodamine ethyl ester (Fig. S3).
Figure 2. Suppression of Drp1-mediated mitochondrial division by PA.
(A) Conversion of lipids mediated by MitoPLD and lipin. MEFs were transfected with MitoPLD-GFP in (B) or catalytically inactive MitoPLDH156N-GFP in Supplementary Figure 3. We quantified GFP intensity along the lines #1–3. Mitochondrial shape was visualized using immunofluorescence with anti-Tom20 antibodies. The boxed regions are enlargements. (C) Quantification of mitochondrial morphology in cells modestly expressing MitoPLD-GFP or MitoPLDH156N-GFP. Mean ± SD (n = 3, 90 cells). (D and E) Mitochondria were analyzed in MEFs modestly expressing MitoPLD-GFP or MitoPLDH156N-GFP after FCCP treatment for 30 min. The boxed regions are enlargements. Mean ± SD (n = 3, 90 cells). (F) WT and Drp1KO MEFs were transfected with HA-lipin1b, which converts PA to diacylglycerol, or enzymatically inactive HA-lipinD712A. The cells were subjected to immunofluorescence microscopy with anti-Tom20 antibodies. (G) We quantified mitochondrial shape. Mean ± SEM (n = 3, 90 cells). (H and I) Mitochondrial morphology was examined in MEFs modestly expressing MitoPLD-GFP in the presence or absence of HA-lipin using immunofluorescence microscopy. Mean ± SD (n = 3, 90 cells). “n” indicates the number of independent experiments. Student’s t-test: ***p< 0.001. Bar, 10 μm. See also Figure S2, S3 and S4.
In addition to stimulating mitochondrial division, FCCP also inhibits mitochondrial fusion by inducing the proteolytic cleavage of Opa1 (Fig. S4A). While FCCP affects both mitochondrial division and fusion, FCCP first fragments mitochondria due to increased mitochondrial division. The fragmented mitochondria then remain unfused due to fusion defects. Therefore, FCCP-induced mitochondrial fragmentation has been used extensively to assess mitochondrial division in previous studies (Li et al., 2015; Palmer et al., 2011). Nonetheless, to separate mitochondrial division from fusion, we used another inducer of mitochondrial division, the calcium ionophore 4Br-A23187 (Tan et al., 2011). Unlike FCCP, we found that 4Br-A23187 does not cause Opa1 cleavage (Fig. S4A). The mitochondrial fragmentation induced by 4Br-A23187 was suppressed by MitoPLD-GFP but not by MitoPLDH156N-GFP (Fig. S4B and C). These data further support the conclusion that mitochondrial division is decreased upon MitoPLD-GFP overexpression.
To test whether the reduced mitochondrial division in MitoPLD-GFP-expressing cells was caused by increased PA levels or decreased CL levels, we co-expressed MitoPLD-GFP with HA-tagged lipin (HA-lipin 1b), a PA phosphatase that converts PA to diacylglycerol and therefore reduces PA levels (Csaki et al., 2013; Huang et al., 2011) (Fig. 2A). Single overexpression of HA-lipin 1b fragmented mitochondria in WT MEFs, but not in Drp1KO MEFs (Fig. 2F, G). This Drp1-dependent mitochondrial fragmentation was contingent on the catalytic activity of lipin 1b (Fig. 2F, G). If the effect of MitoPLD was mediated by an increase in PA, the co-expression of HA-lipin 1b should decrease the effect. Indeed, HA-lipin 1b counteracted mitochondrial elongation caused by MitoPLD-GFP (Fig. 2H, I). Given that CL acyl chains are mainly unsaturated and that the PA produced by MitoPLD is likely unsaturated, our data suggest that MitoPLD expression promotes Drp1 interactions with unsaturated PA and saturated non-PA phospholipids (Fig 1L, type 2).
Saturated acyl chains suppress Drp1-mediated mitochondrial division
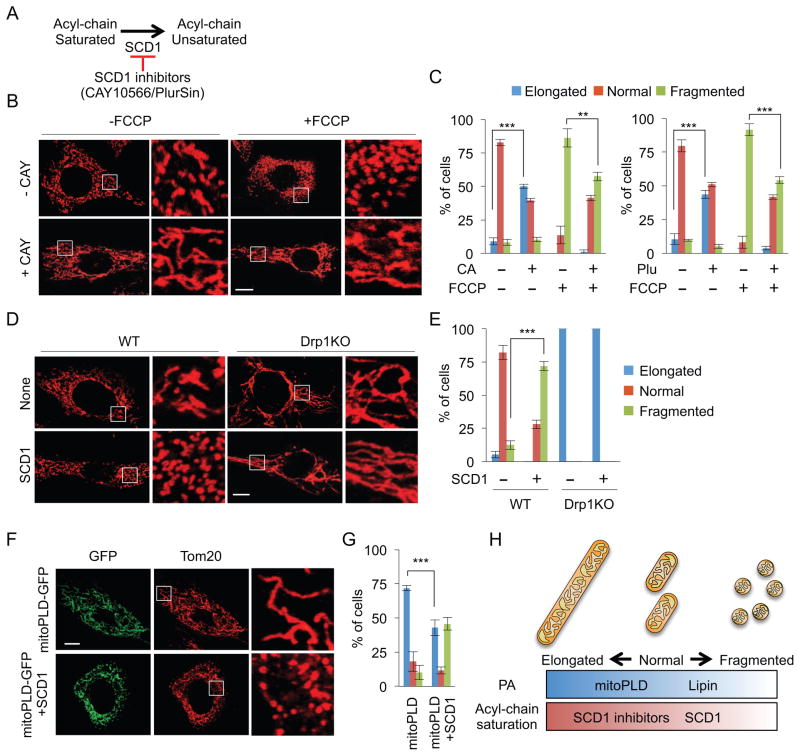
Stearoyl-CoA desaturase-1 (SCD1) converts saturated fatty acids to unsaturated ones (Paton and Ntambi, 2009) (Fig. 3A). The products of SCD1 serve as substrates for the synthesis of different unsaturated lipids. To assess the function of acyl chain saturation in mitochondrial division, we treated WT MEFs with two structurally different SCD1 inhibitors: CAY10566 and PluriSln1 (Ben-David et al., 2013; Liu et al., 2007) (Fig. 3B, C). After incubation with these inhibitors, the mitochondria became elongated, suggesting reduced mitochondrial division. To test this idea, we induced Drp1-mediated mitochondrial division using FCCP. Pretreating with the SCD1 inhibitors made the cells partially resistant to FCCP-induced mitochondrial fragmentation (Fig. 3B, C). We confirmed that FCCP decreases membrane potential in the presence or absence of the SCD1 inhibitor using flow cytometry with the membrane potential dye MitoLite NIR (Fig. S3). Therefore, reducing the formation of unsaturated lipids decreases mitochondrial division (Fig. 3H). We next asked whether overexpression of SCD1 and a concomitant increase in unsaturated lipids simulates mitochondrial division. In WT MEFs, SCD1 overexpression induced mitochondrial fragmentation in a Drp1 dependent manner (Fig. 3D, E). We next tested how acyl chain saturation and levels of PA impinge on mitochondrial morphology by co-expressing SCD1 (increases unsaturated acyl chains) with MitoPLD-GFP (increases PA). The mitochondria became less elongated in the SCD1/MitoPLD-expressing cells compared with the mitochondria in the MitoPLD-expressing cells (Fig. 3F, G). Therefore, the saturation of acyl chains is required for PA to inhibit mitochondrial division (Fig. 3H).
Figure 3. Reduction of mitochondrial division via saturation of acyl chains.
(A) SCD1 mediates the desaturation of fatty acid acyl chains. (B and C) After incubation with SCD1 inhibitors, CAY10566 (CAY) and PluriSln1 (Plu), WT MEFs were treated with 10 μM FCCP for 30 min. Mitochondrial shape was analyzed using immunofluorescence microscopy with anti-Tom20 antibodies. Mean ± SD (n = 3, 90 cells). (D and E) WT and Drp1KO MEFs were transfected with SCD1 and analyzed for mitochondrial morphology using immunofluorescence microscopy. Mean ± SD (n = 3, 90 cells). (F and G) We examined mitochondria in WT MEFs mildly expressing MitoPLD-GFP in the presence or absence of SCD1. Mean ± SD (n = 3, 90 cells). (H) Summary of the effects of PA and saturated acyl chain on mitochondrial shape. “n” indicates the number of independent experiments. Student’s t-test: **p< 0.01; ***p< 0.001. Bar, 10 μm.
MitoPLD interacts with Drp1 in vitro and in vivo
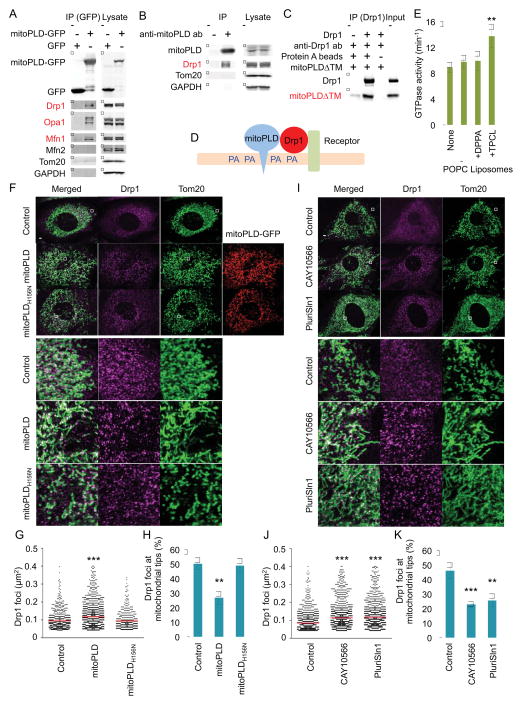
The mitochondrial membrane contains relatively low levels of PA (less than 5%) (Potting et al., 2013; Zinser et al., 1991). How does Drp1 find PA in the mitochondrial membrane? One possibility is that PA is produced near Drp1. To test this model, we asked whether Drp1 binds the PA-producing enzyme MitoPLD. We expressed GFP or MitoPLD-GFP in HEK293T cell and found that Drp1 specifically co-immunoprecipitated with the MitoPLD-GFP, but not with GFP alone (Fig. 4A). To determine whether endogenous Drp1 and MitoPLD interact in vivo, we isolated testes from mice and immunoprecipitated MitoPLD using anti-MitoPLD antibodies. Again, Drp1 co-precipitated specifically with MitoPLD (Fig. 4B). To ask if this interaction was direct, we purified a FLAG-tagged cytoplasmic domain of MitoPLD (mitoPLDΔTM) and incubated it with His6-Drp1. We immunoprecipitated His6-Drp1 using anti-Drp1 antibodies. As a negative control, we incubated FLAG-mitoPLDΔTM with anti-Drp1 antibody without His6-Drp1. FLAG-mitoPLDΔTM specifically bound His6-Drp1 (Fig. 4C). These data suggest that MitoPLD directly associates with Drp1 on the mitochondria and produces PA in the vicinity of Drp1, thereby locally increasing PA concentrations near the mitochondrial division apparatus (Fig. 4D).
Figure 4. Mechanism of phospholipid-mediated suppression of mitochondrial division.
(A) Cells expressing MitoPLD-GFP or GFP were subjected to immunoprecipitation using anti-GFP antibodies. Drp1, Mfn1 and Opa1 specifically bind mitoPLD-GFP. (B) Mouse testes were subjected to immunoprecipitation using anti-MitoPLD antibodies. Drp1 specifically bind MitoPLD. (C) A FLAG-tagged cytoplasmic domain of MitoPLD (mitoPLDΔTM) was incubated with His6-Drp1, and His6-Drp1 was pulled down with anti-Drp1 antibodies. Drp1 directly binds mitoPLDΔTM. (D) Model for inhibitory lipid microenvironment created by the Drp1-MitoPLD interaction. (E) The GTPase activity of His6-Drp1 was measured in the presence of the indicated liposomes. Saturated PA containing liposomes does not stimulate the GTPase activity of Drp1. Mean ± SEM (n = 5). (F–K), WT MEFs expressing MitoPLD-GFP or MitoPLDH156N-GFP (F, G and H) or treated with SCD1 inhibitors (I, J and K) were subjected to immunofluorescence microscopy with antibodies specific for Tom20 and Drp1. The sizes of the Drp1 foci in the mitochondria were quantified (G and J). Red lines correspond to means (more than 300 Drp1 foci were analyzed). The relative abundance of Drp1 foci associated with the tips of mitochondrial tubules was determined (H and K). Mean ± SEM (n = 10 cells). Student’s t-test: **p< 0.01, ***p< 0.01. Bar, 10 μm.
In addition to Drp1, two mitochondrial fusion proteins, the outer membrane protein mitofusin 1 and the inner membrane protein Opa1, were associated with MitoPLD-GFP in our co-immunoprecipitation study (Fig. 4A). These interactions are interesting because PA stimulates mitofusin-mediated mitochondrial fusion (Choi et al., 2006; Huang et al., 2011). Our data suggest that PA is also produced near mitofusin 1 to promote mitochondrial fusion. Because Opa1 functions together with mitofusin 1 but not with mitofusin 2 during mitochondrial fusion (Cipolat et al., 2004), MitoPLD may stimulate a specific type of fusion mechanism.
PA and saturated acyl chains restrain oligomerization-stimulated GTPase activity of Drp1
To gain mechanistic insights into how saturated PA suppresses mitochondrial division, we measured the GTPase activity of Drp1 in the presence of saturated PA. We incubated His6-Drp1 with unsaturated PC (POPC) liposomes containing either saturated PA (DPPA) or CL (TPCL). It has been previously shown that CL interactions facilitate Drp1 oligomerization and lead to oligomerization-stimulated GTP hydrolysis (Macdonald et al., 2014). As expected, TPCL/POPC liposomes stimulated the GTPase activity of Drp1 compared with POPC liposomes (Fig. 4E). In contrast, DPPA/POPC liposomes did not enhance GTPase activity. Our data suggest that interactions with saturated PA do not stimulate the enzymatic activity of Drp1 and instead keep this protein inert on the surface of the membrane. To test this model, we examined Drp1 foci that were formed by Drp1 oligomerization on mitochondria in MitoPLD-overexpressing cells (Fig. 4F, G, H) or SCD1-inhibitor-treated cells (Fig. 4I, J, K) using immunofluorescence microscopy with antibodies to Drp1 and Tom20. MitoPLD and SCD1 inhibitors significantly increased the size of the Drp1 foci (Fig. 4G, J). Furthermore, we found significant decreases in the relative number of Dpr1 foci associated with the tips of mitochondrial tubules, which are indicative of recent division events, in MitoPLD-overexpressing cells (Fig. 4H) and SCD1 inhibitor-treated cells (Fig. 4K). These data suggest that the PA headgroup and acyl chains of saturated phospholipids restrain oligomerized Drp1 from constricting mitochondria by suppressing the activation of GTP hydrolysis, enabling continuous growth of the Drp1 foci.
In this study, we reveal a previously unrecognized mechanism for mitochondrial division via coincident Drp1-PA interactions. We propose that this protein-lipid interaction suppresses the activation of GTP hydrolysis that is required for constriction of mitochondria after Drp1 oligomerization. Our data also suggest that MitoPLD creates an inhibitory microenvironment in the vicinity of the mitochondrial division apparatus. Previous studies have shown that PA stimulates mitofusin-mediated mitochondrial fusion by changing the biophysical property of the outer membrane (Choi et al., 2006; Huang et al., 2011; Zhang et al., 2016). Considering the crucial roles of mitochondrial shape in the control of function, distribution and turnover of this essential organelle, we propose that synthesis of PA coordinates division and fusion to orchestrate dynamic changes in mitochondrial shape. Such control provides a robust intrinsic mechanism to maintain the integrity of mitochondria that continuously grow by importing proteins and lipids. In addition, PA may also function as a signaling molecule that regulates mitochondrial shape in response to stress to maintain bioenergetic status in cells or to remove dysfunctional mitochondria producing excess reactive oxygen species. Since mitochondrial division and saturated lipids are linked to many human diseases, our findings may contribute to understanding and treating these diseases.
Experimental Procedures
Plasmids
Drp1 and its domains and the cytoplasmic domain of MitoPLD were cloned into pET15b. SCD1 was cloned into the pcDNA3.1.
Protein purification
Recombinant His6-tagged proteins were purified using Ni-NTA beads from Rosetta 2(DE3) pLysS cells. The purified proteins were stored at −80°C.
Liposome floatation assay
Recombinant proteins were incubated with unilamellar liposomes and analyzed by a sucrose density gradient. Each fraction was analyzed by SDS-PAGE and silver staining.
Detailed experimental procedures, plasmids, protein purification, lipid binding assays, immunofluorescence microscopy, GTPase assay, immunoprecipitation, and protein binding assay are available in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank members of the Iijima and Sesaki labs for helpful discussion. This work was supported by grants to MI (NIH, GM084015), HS (NIH, GM089853 and NS084154; M. J. Fox Foundation, 11143; AHA, 15GRNT25380005), and MAF (NIH, GM084251).
Footnotes
Author Contributions
YA, MI, and H.S. designed the study. YA, KI, YT, KC, and PM performed experiments. TLS, MAF, and RR assisted with experiments or provided valuable reagents. HS, MI, and YA wrote the manuscript and all authors contributed to writing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baba T, Kashiwagi Y, Arimitsu N, Kogure T, Edo A, Maruyama T, Nakao K, Nakanishi H, Kinoshita M, Frohman MA, et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. The Journal of biological chemistry. 2014;289:11497–11511. doi: 10.1074/jbc.M113.531921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167–179. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui HT, Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Current biology : CB. 2013;23:R891–899. doi: 10.1016/j.cub.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One. 2014;9:e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO reports. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaki LS, Dwyer JR, Fong LG, Tontonoz P, Young SG, Reue K. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog Lipid Res. 2013;52:305–316. doi: 10.1016/j.plipres.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. The EMBO journal. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Developmental cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, et al. Dynamics of mtDNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Molecular and cellular biology. 2014 doi: 10.1128/MCB.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015:4. doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. The EMBO journal. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. The Journal of cell biology. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Molecular cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1342–1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu S, Roelofs BA, Boyman L, Lederer WJ, Sesaki H, Karbowski M. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. The Journal of cell biology. 2015;208:109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Lynch JK, Freeman J, Liu B, Xin Z, Zhao H, Serby MD, Kym PR, Suhar TS, Smith HT, et al. Discovery of potent, selective, orally bioavailable stearoyl-CoA desaturase 1 inhibitors. J Med Chem. 2007;50:3086–3100. doi: 10.1021/jm070219p. [DOI] [PubMed] [Google Scholar]
- Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Molecular biology of the cell. 2014;25:1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015:4. doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. The Journal of cell biology. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO reports. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. American journal of physiology Endocrinology and metabolism. 2009;297:E28–37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potting C, Tatsuta T, Konig T, Haag M, Wai T, Aaltonen MJ, Langer T. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 2013;18:287–295. doi: 10.1016/j.cmet.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Ramachandran R. Vesicle scission: dynamin. Seminars in cell & developmental biology. 2011;22:10–17. doi: 10.1016/j.semcdb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Roy M, Reddy PH, Iijima M, Sesaki H. Mitochondrial division and fusion in metabolism. Current opinion in cell biology. 2015;33C:111–118. doi: 10.1016/j.ceb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, Jabado O, Hoehn K, Kageyama Y, Sesaki H, et al. Mitochondrial Division Is Requisite to RAS-Induced Transformation and Targeted by Oncogenic MAPK Pathway Inhibitors. Molecular cell. 2015;57:521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, McBride HM. Staying cool in difficult times: Mitochondrial dynamics, quality control and the stress response. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandran R. Cardiolipin’s propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Molecular biology of the cell. 2015;26:3104–3116. doi: 10.1091/mbc.E15-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Itoh K, Sesaki H. SnapShot: Mitochondrial dynamics. Cell. 2011;145:1158, 1158 e1151. doi: 10.1016/j.cell.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AR, Cai AY, Deheshi S, Rintoul GL. Elevated intracellular calcium causes distinct mitochondrial remodelling and calcineurin-dependent fission in astrocytes. Cell Calcium. 2011;49:108–114. doi: 10.1016/j.ceca.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. The Journal of cell biology. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu X, Bai J, Tian X, Zhao X, Liu W, Duan X, Shang W, Fan HY, Tong C. Mitoguardin Regulates Mitochondrial Fusion through MitoPLD and Is Required for Neuronal Homeostasis. Molecular cell. 2016;61:111–124. doi: 10.1016/j.molcel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.