Abstract
The HIV-1 replication cycle requires the nucleocytoplasmic export of intron-containing viral RNAs, a process that is ordinarily restricted. HIV overcomes this by means of the viral Rev protein, which binds to an RNA secondary structure called the Rev response element (RRE) present in all unspliced or incompletely spliced viral RNA transcripts. The resulting mRNP complex is exported through interaction with cellular factors. The Rev–RRE binding interaction is increasingly understood to display remarkable structural plasticity, but little is known about how Rev–RRE sequence differences affect functional activity. To study this issue, we utilized a lentiviral vector assay in which vector titer is dependent on the activity of selected Rev–RRE pairs. We found that Rev–RRE functional activity varies significantly (up to 24-fold) between naturally occurring viral isolates. The activity differences of the Rev–RRE cognate pairs track closely with Rev, but not with RRE activity. This variation in Rev activity is not correlated with differences in Rev steady state protein levels. These data suggest that Rev sequence differences drive substantial variation in Rev–RRE functional activity between patients. Such variation may play a role in viral adaptation to different immune milieus within and between patients and may be significant in the establishment of latency. The identification of differences in Rev–RRE functional activity in naturally occurring isolates may also permit more efficient production of lentiviral vectors.
Introduction
The binding of the HIV-1 Rev protein to viral mRNA and its subsequent multimerization on the Rev response element (RRE) are critical steps in HIV replication. These interactions allow the unspliced viral genome, and the incompletely spliced viral mRNAs that are generated from this RNA, to reach the cytoplasm through the formation of ribonucleoprotein (RNP) complexes that are competent for nucleocytoplasmic export. Without the formation of these complexes, these mRNAs, which all retain introns, are blocked from export by cellular mechanisms that restrict the export of incompletely spliced mRNA (see reviews in Refs.1,2). Export through the Rev–RRE pathway is also necessary for efficient entry into polyribosomes and translation.3
Most structural and functional studies to date on Rev and the RRE have involved work with laboratory-adapted viruses.4 Earliar work identified the minimal functional RRE as a highly conserved 234 nucleotide structure consisting of multiple stem loops.5,6 More recently, through secondary structure probing studies of the whole viral genome, investigators have come to realize that the lower stem of this structure extends further by about 60 nucleotides on either side, yielding a structure of about 350 nt. The primary binding of Rev to this structure occurs in a region named stem loop IIb.7,8 Additional molecules of the Rev protein then interact with distal regions of the RRE to form a large multimeric complex consisting of at least six Rev monomers.9 The Rev–RRE RNP forms in conjunction with Ras-related nuclear protein-Guanosine-5′-triphosphate and also recruits the cellular export receptor Crm1 to create a large RNP complex that is competent for export.10
The prototype subtype B Rev protein consists of 116 amino acids. Several important highly conserved domains have been identified within Rev, including regions involved in dimer and oligomer formation, nuclear entry, RNA binding, and nuclear export.11,12 The carboxyl terminal domain (∼25 amino acids) does not have a function ascribed to it and is the most highly variable region.13 Rev has also been shown to be a phosphoprotein, although no function has yet been demonstrated for this modification.14
Recently published data have shown that Rev binds to the RRE with great plasticity due to the slippery hydrophobic nature of its dimerization domain and the flexibility of binding shown by its arginine-rich motif (ARM). After the first Rev monomer makes contact with the primary binding site, the additional Rev molecules that form the multimer contact other regions of the RNA. What is remarkable about the resulting complex is that the ARM of each Rev shows great variability in the way it binds to the RNA with different residues of the identical ARMs making contact with the RRE at each binding site.15,16 Recognition of each binding site depends on how the ARM is oriented, as well as on the three-dimensional structure of the RNA, which participates together with the amino acid sequences of the protein to direct the orientation of each Rev monomer. To be exported, the mature Rev–RRE RNP complex then recruits a dimer of Crm1.17 Surprisingly, Crm1 binds to the Rev–RRE RNP through a different surface than it uses for other substrates, where it binds as a monomer, and the binding seems to tolerate considerable variation in the Rev nuclear export signal (NES) sequence.18
Since the Rev–RRE RNP is intrinsically plastic in nature, it is possible that variability in Rev and RRE sequence and structure leads to the formation of complexes that present their NESs to the Crm1 dimer in slightly different configurations.19,20 If so, it would be expected that different Rev–RRE RNPs recruit the Crm1 dimer with different efficiencies, resulting in different export dynamics.21 This may enable HIV to modulate the rate of viral replication by varying RNA export activity.
In fact, small sequence variations in Rev and/or the RRE structure have been shown to affect overall Rev function and virus replication. In one study, viruses were sequenced in a group of HIV-infected long-term survivors and Rev protein variants were identified that had a twofold to fourfold lower functional activity than a control Rev protein from the NL4-3 laboratory isolate.22,23 Other studies have shown that sequence variation of the RRE yielded marked differences in activity among primary isolates,24, 25 and that increased Rev protein function can be observed in patients with advanced stage disease compared with asymptomatic patients.19 We have recently shown that viruses can evolve with different levels of Rev–RRE functional activity during the course of infection within the same patient.19 This study used single genome sequencing to identify cognate pairs of Rev and RRE from infected patients at different time points. It was shown that sequence variation of the RRE, but not Rev, led to the formation of Rev–RRE RNP complexes that had different rates of migration on native gels, implying structural differences, and this appeared to be the underlying reason for the differences in activity. In another study, we found that structural isomerization of the RRE from NL4-3 leads to different levels of replication activity.20 Rev and/or RRE variation during infection has also been observed with other lentiviruses.26–28
Despite the high level of genetic diversity that exists among HIV-1 isolates worldwide, molecular biological studies that have examined the function of specific regulatory and accessory genes have mostly utilized sequences from Group M, subtype B laboratory strains. However, there are a few studies on Rev and RRE function from nonsubtype B isolates,24,25 and some information also exists on subtype differences in Tat, Vif, Vpr, and Nef.29–32 In examining Rev and RRE sequences from circulating recombinant forms (CRFs), we noticed that among the CRF02_AG viruses, which have emerged as the dominant CRF in West and Central Africa and are now spreading in Russia, Rev sequences from subtype G have often recombined with subtype A envelope sequences, creating viruses that contain a subtype A RRE and a subtype G Rev33–36 (reviewed in Ref.37). Given that the flexible nature of Rev–RRE RNP complex formation might lead to activity differences in such recombinants, we decided to examine Rev and RRE function in selected CRF02_AG recombinants. This was compared with Rev and RRE function in selected subtype A and subtype G isolates and in some of the subtype B primary and laboratory isolates that we have previously characterized.
Materials and Methods
Nucleotide sequence selection
HIV Rev and RRE sequences were obtained from the Los Alamos HIV Database (www.hiv.lanl.gov) by searching for subtype A, CRF02_AG, and G sequences with coverage extending over both exons of rev and the RRE. Rev and RRE sequences from four subtype B viruses were obtained through single genome sequencing as described previously.19 The laboratory strain NL4-3 was used for reference sequences. For each virus, the 234-nt RRE sequence and both exons of rev were identified using Geneious (Biomatters Ltd.).
Determination of Rev–RRE functional activity
All plasmids utilized in this study were given the notation pHRXXXX for easy identification. To measure Rev–RRE function, we utilized an HIV vector system that has been previously described.20,38,39 This vector produces a genomic RNA whose nucleocytoplasmic export and packaging are dependent on Rev–RRE function. Thus the titer of the virus-vector particles produced from the system is a measure of Rev–RRE functional activity. Vector titer can be readily measured since the vector transduces target cells with a hygromycin resistance cassette.20
The system utilized 293T cells transfected with five plasmids and rev and RRE sequences that were chemically synthesized (Integrated DNA Technologies). For testing the function of the different RREs, the original vector (pTR167 pHR1266)38 was modified to contain an inactive RRE in the native position (pHR5096)40 and the different RRE sequences to be tested were cloned into the Nef region.20 The rev sequences were cloned into a CMV expression plasmid.19 HIV structural proteins were provided by the GPV-4xCTE (pHR3296) construct, which produces Gag and Pol in a Rev-independent manner.41,42 Vesicular stomatitis virus G protein was provided by pMD-G (pHR2004)43 and Tat was provided by pCMV–Tat (pHR136). A detailed list of the plasmids utilized in these assays is given in the Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/aid).
The day before transfection, 3 × 106 293T/17 cells were seeded onto a 100 mm plate in medium containing Iscove's modified Dulbecco's medium, 10% bovine calf serum, and gentamicin. Using the calcium phosphate method,44 the cells were transfected using 20 μg pTR167 (RRE−) (nef−) (selected RRE+), 15 μg GPV-4xCTE, 5 μg pMD-G, 1 μg CMV–Tat, and 1 μg CMV–Rev such that each producer cell culture would receive the appropriate combination of Rev and RRE sequences to be tested. Cell-free supernatant was harvested from the producer culture 72 h after transfection and serial dilutions of viral stocks were used to infect Hela cells using DEAE dextran. Beginning 2 days after infection, Hela cell cultures were maintained in hygromycin-containing medium (200 μg/ml) until background cells were killed and colonies were clearly visible. The hygromycin-resistant colonies were fixed and stained with crystal violet, and colonies were counted to determine titer.
The cell-free supernatant from the producer cell culture was also used to determine p24 production using an ELISA.45 To calculate the relative Rev–RRE functional activity, the titer of vector as determined by hygromycin-resistant colony counting was normalized to the p24 production of the producer cell culture to account for variation in transfection efficiency. When necessary to compare functional activity levels between assays, colony counts were also normalized to the NL4-3 Rev–RRE cognate pair functional activity as measured by an assay performed on the same day.
Determination of Rev steady state expression
The steady state expression of selected Rev sequences used in the activity assays mentioned was determined by means of Western blotting. Selected CMV–Rev constructs were altered to express a modified Rev protein with an influenza hemagglutinin (HA) epitope consisting of the amino acid sequence YPYDVPDYA at the N terminal end.46
Using the calcium phosphate transfection method, 3 × 106 freshly plated 293T/17 cells were transfected with 5 μg of CMV–HA–Rev plasmid and 1 μg CMV–SEAP (pHR1831) to serve as a control for transfection efficiency. At 72 h, cells were collected and lysed. A Western blot was performed with extracts from the cells using primary antibodies to the HA epitope (Pierce, Thermo Fisher Scientific) and beta-tubulin (Abcam) and fluorescent secondary antibodies (LI-COR Biosciences). Imaging and analysis were performed using an Odyssey infrared imager (LI-COR Biosciences). The Western blot procedure was replicated using alternative blotting membranes, including nitrocellulose and polyvinylidene fluoride, to ensure that results were not secondary to differential blotting efficiency. Cell-free medium was collected during cell harvest and assayed for secreted embryonic alkaline phosphatase (SEAP) activity (Phospha-Light; Applied Biosystems). Relative steady state Rev expression was calculated by normalizing the intensity of the relevant Rev band to the intensity of the beta-tubulin band to control for lane loading and SEAP expression to control for transfection efficiency.
Results
Viruses selected for study
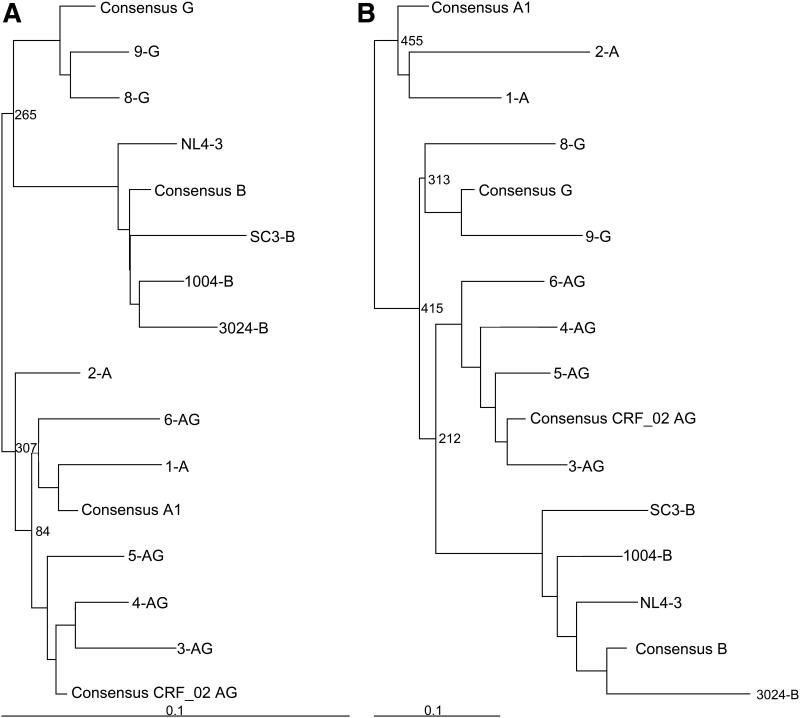
Twelve Rev–RRE pairs from different HIV isolates were selected for functional analysis in this study. Eight of the pairs were from viruses listed in the Los Alamos HIV Database, which included two from subtype A (1-A, accession AB098330 [unpublished data]; 2-A, accession JX20304947), four from subtype CRF02_AG (3-AG, accession AY27169048; 4-AG, accession AY37112449; 5-AG, accession AY37113849; 6-AG, accession AY82921450), and two from subtype G (8-G, accession FJ38936751; 9-G, accession JX14067652). In addition, the Rev–RRE pair from the NL4-3 laboratory strain (7-NL, accession U2694253) and three Rev–RRE pairs from subtype B primary isolates previously tested for function in our laboratory were also included (1004-B [1004-M18A RRE accession KF559150, 1004-M18A Rev accession KF559129], 3024-B [3024-M43A RRE accession KF559153, 3024-M43A Rev accession KF559133], SC3-B [SC3-M57A RRE accession KF559162, SC3-M0A Rev accession KF559145]).19 For each isolate, the 234-nt RRE sequence and the Rev coding exons were identified and phylogenetic trees were constructed using a neighbor joining method with an Simian immunodeficiency virus(cpz) sequence (accession U4272054) as an outgroup [Fig. 1A (RRE), B (Rev)]. As already noted, many of the subtype CRF02_AG viruses have most of the env sequences, including the RRE, contributed by parental subtype A, while both exons of rev are contributed by parental subtype G.55 As expected, the phylogenetic trees show that the CRF02_AG RREs cluster with subtype A sequences, whereas the CRF02_AG Revs are more closely related to the subtype G sequences.
FIG. 1.
Phylogenetic trees of selected RRE and Rev sequences. Phylogenetic trees of the selected RRE nucleotide (A) and Rev amino acid (B) sequences were generated using ClustalX v2.163 and TreeView v1.6.6. Corresponding consensus sequences representing subtypes A, G, B, and CRF02_AG were included to more clearly show subtype clustering. Trees were rooted using an SIV(cpz) sequence as an outgroup. The numbers next to nodes correspond to bootstrap values using 1,000 iterations. RRE, Rev response element.
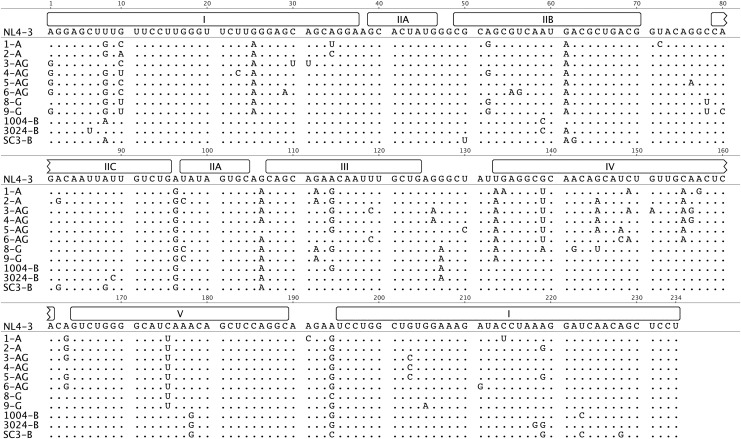
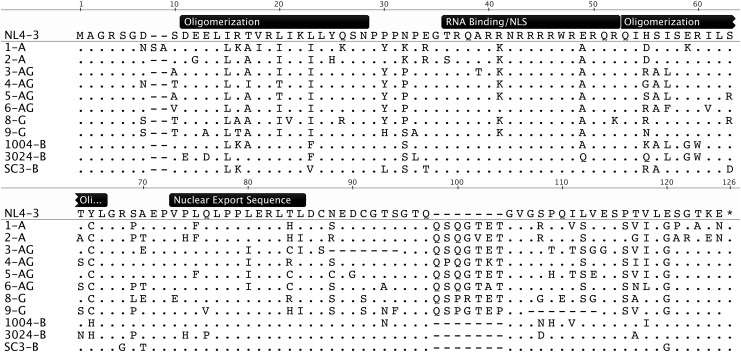
An alignment of these sequences showed substantial nucleotide diversity in the RREs as well as in the Rev coding sequences (Figs. 2 and 3). Interestingly, most of the RRE differences between subtypes occurred in the region corresponding to stem loops III and IV (see also Supplementary Fig. S1 that shows a Highlighter plot of the same alignment that is shown in Fig. 2). In studies using the NL4-3 subtype B laboratory strain, this region of the RRE has previously been shown to adopt alternative secondary structures, which promote different rates of replication.20 In the case of Rev, all of the subtype B isolates had a seven amino acid deletion relative to the A and G isolates. The most variation was observed in the carboxy-terminal region of the protein to which no function has been ascribed. The amino terminus, ARM/RNA binding domain, NLS (nuclear localization signal), and NES domains are highly conserved, as are the key residues Asn26, Arg48, and Gln49 (N28, R50, and Q51 in Fig. 3), which form the critical hydrogen bonding network that aligns the two helices of the monomer in the subtype B Rev crystal structure.56 The oligomerization domains are also conserved, with only conservative amino acid substitutions present, except for the His53 residue present in NL4-3 (H55 in Fig. 3) that is highly variable. In the subtype B crystal structure, hydrogen bonding interactions of this residue with Y23 (Y25 in Fig. 3) add stability to the helical packing of the Rev monomer. This raises the possibility that the observed variations cause changes in monomer stability.56
FIG. 2.
Alignment of selected RRE sequences. The sequences of 234-nt RREs from 12 selected viruses were aligned to the RRE from NL4-3. Dots denote that there is no difference in that position from the NL4-3 sequence. Regions that would fold to form a five stem-loop structure are noted above the NL4-3 sequence. (Modified from Geneious v5.3.64)
FIG. 3.
Alignment of selected Rev amino acid sequences. The amino acid sequences of Rev from 12 selected viruses were aligned to the Rev sequence from NL4-3. The bars above the NL4-3 sequence denote known Rev functional domains. (Modified from Geneious v5.3.64)
The functional activity of naturally occurring Rev–RRE pairs varies greatly
We next tested the relative functional activity of each Rev–RRE pair derived from the different isolates (cognate pairs) in a virus-vector packaging system. In this system, the infectious titer of the virus-vector released from the packaging cells is directly proportional to the activity of the Rev–RRE pair (see Materials and Methods section and Sherpa et al.20), since the virus-vector contains a genomic RNA whose trafficking is dependent on Rev–RRE function. The RNA also contains a hygromycin resistance gene cassette that is expressed in transduced cells. Thus, Rev–RRE function is easily scored by counting hygromycin-resistant colonies after transduction of target cells.
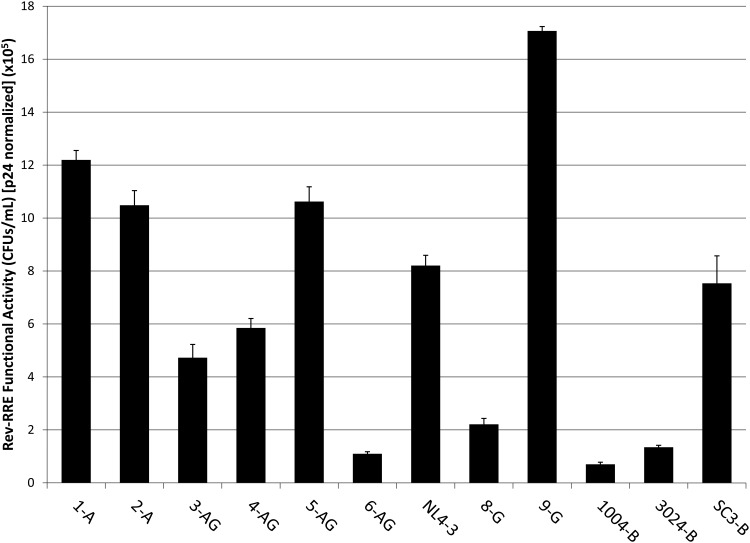
Surprisingly, wide differences in activity of the various pairs were observed with as much as a 24-fold difference in functional activity between the most active (9-G) and least active (1004-B) Rev–RRE cognate pair (Fig. 4). This is considerably of more variation than we previously observed within a patient during the course of an infection.19 Activity levels did not appear to cluster with subtype, although the power to detect such clustering was quite low (data not shown). In fact, subtype G displayed both the most active (9-G) and one of the least active (8-G) Rev–RRE cognate pairs.
FIG. 4.
Functional activity variation of Rev–RRE cognate pairs. An HIV vector assay was used to determine the relative functional activity of Rev–RRE pairs corresponding to each of 12 viruses. The activity of the pair was measured by assaying the efficiency of Rev–RRE-dependent vector production in CFUs per milliliter of production culture medium. This value was normalized to the amount of p24 in each viral stock to control for variation in transfection efficiency. Bars show average deviation from the mean for two replicates. CFUs, colony forming units.
Cognate pair functional activity variation tracks closely with variation in Rev activity
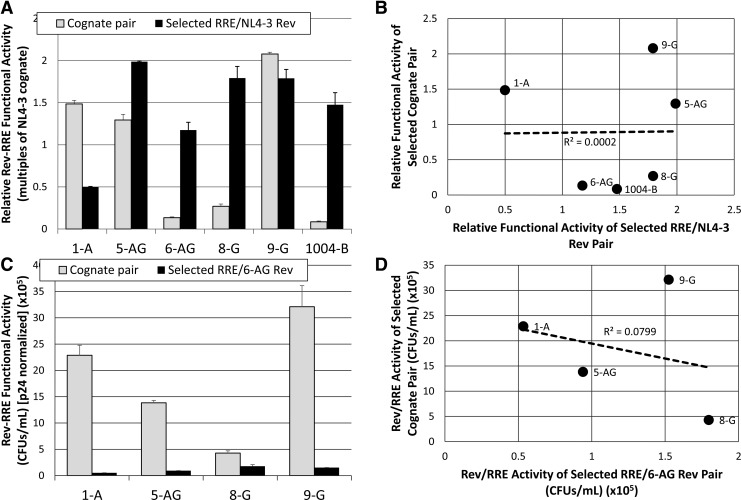
We next sought to determine whether the variation in Rev–RRE cognate pair functional activity was attributable to activity differences in either the RRE or Rev component. To examine this issue, we first paired several selected RREs with a single reference Rev and compared the functional activity of the combination with the functional activity of the cognate pair from which the RRE was derived. In these experiments, either the 7-NL Rev or the 6-AG Rev was used as the reference, since they were derived from different subtypes and their cognate pairs show large activity differences from each other. The SC3-B RRE was specifically excluded from this analysis, as we have previously shown it evolved to be hyperactive over the course of a clinical infection, in conjunction with a Rev protein that did not change sequence.19
These experiments showed that most of the RREs did not promote the same level of functional activity when matched with the 7-NL reference Rev as with their cognate pair (Fig. 5A). In addition, the rank order of activity for each pair was not the same as the rank order of the cognates. Specifically, the RREs of 5-AG, 6-AG, 8-G, and 1004-B showed greater activity with 7-NL Rev than the activity using the cognate pair. In contrast, when paired with the 6-AG Rev, all of the RREs demonstrated lower activity than the cognate pairs, again with a difference in the rank order (Fig. 5C). To analyze these results more fully, we plotted the activity of each cognate pair as a function of the activity of each RRE with the reference Revs. A linear regression performed for both reference Rev experiments (Fig. 5B, D) demonstrated that there was no significant correlation between RRE activity in the context of the reference Rev proteins and cognate pair activity (r2 = 0.0002 for 7-NL Rev pairing and r2 = 0.0799 for 6-AG Rev pairing).
FIG. 5.
Correlation between the RRE component and cognate pair functional activity variation. (A) The relative functional activity of individual RREs was determined by pairing them with NL4-3 Rev (solid bars). The activity of the pair is displayed alongside the functional activity of the corresponding cognate pair (open bars). Rev–RRE functional activity values were normalized to p24 and to the NL4-3 cognate activity level, which was tested as an internal control in each experiment, to permit comparison between experiments. Bars show average deviation from the mean for two replicates. (B) The activity of each Rev–RRE cognate pair is plotted as a function of the activity of each RRE–NL4-3 Rev pair. A linear regression was performed to examine the degree of correlation between variation in RRE activity and cognate pair activity. (C) The functional activity of individual RREs was determined by pairing them with 6-AG Rev (solid bars). The activity of the pair is displayed alongside the functional activity of the corresponding cognate pair (open bars). Activity values are presented as CFUs/ml normalized to p24. Bars show average deviation from the mean for two replicates. (D) The activity of each Rev–RRE cognate pair is plotted as a function of the activity of each RRE–6-AG Rev pair. A linear regression was performed to examine the degree of correlation between variation in RRE activity and cognate pair activity.
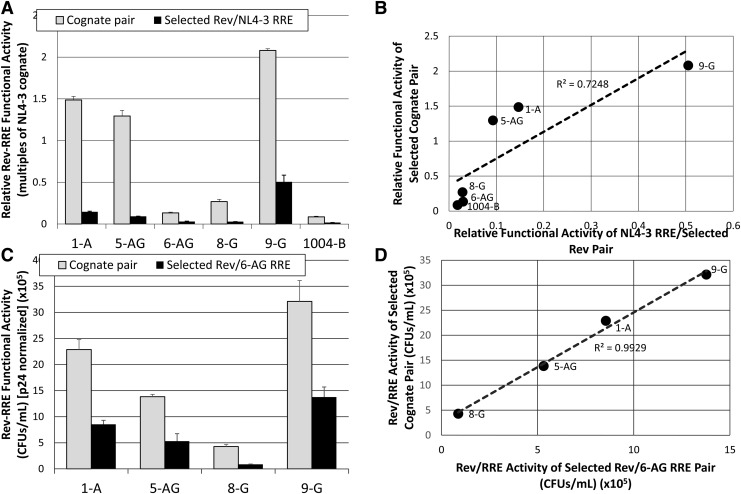
A similar experiment was conducted to assess the relationship between variation in Rev function and variation in cognate pair functional activity. Selected Revs were paired with a reference 7-NL RRE or 6-AG RRE in separate experiments as mentioned. For both reference RREs, the activity of the Rev–reference RRE pairs was less than that of the corresponding cognate pair (Fig. 6A, C). A regression analysis showed that the selected Rev–reference RRE pair functional activities varied linearly with the corresponding cognate pair for both the 7-NL and 6-AG reference (r2 = 0.7248 and 0.9929, respectively; Fig. 6B, D). Thus Rev functional variation, rather than RRE variation, appears to be the key factor that promotes the activity variation observed with the cognate pairs.
FIG. 6.
Correlation between the Rev component and cognate pair functional activity variation. (A) The relative functional activity of individual Revs was determined by pairing them with the NL4-3 RRE (solid bars). The activity of the pair is displayed alongside the functional activity of the corresponding cognate pair (open bars). Rev–RRE functional activity values were normalized to p24 and to the NL4-3 cognate activity level, which was tested as an internal control in each experiment, to permit comparison between experiments. Bars show average deviation from the mean for two replicates. (B) The activity of each Rev–RRE cognate pair is plotted as a function of the activity of each Rev–NL4-3 RRE pair. A linear regression was performed to examine the degree of correlation between variation in Rev activity and cognate pair activity. (C) The functional activity of individual Revs was determined by pairing them with 6-AG RRE (solid bars). The activity of the pair is displayed alongside the functional activity of the corresponding cognate pair (open bars). Activity values are presented as CFUs/ml normalized to p24. Bars show average deviation from the mean for two replicates. (D) The activity of each Rev–RRE cognate pair is plotted as a function of the activity of each Rev–6-AG RRE pair. A linear regression was performed to examine the degree of correlation between variation in Rev activity and cognate pair activity.
Rev–RRE functional activity differences between isolates do not correlate with differences in Rev steady state levels
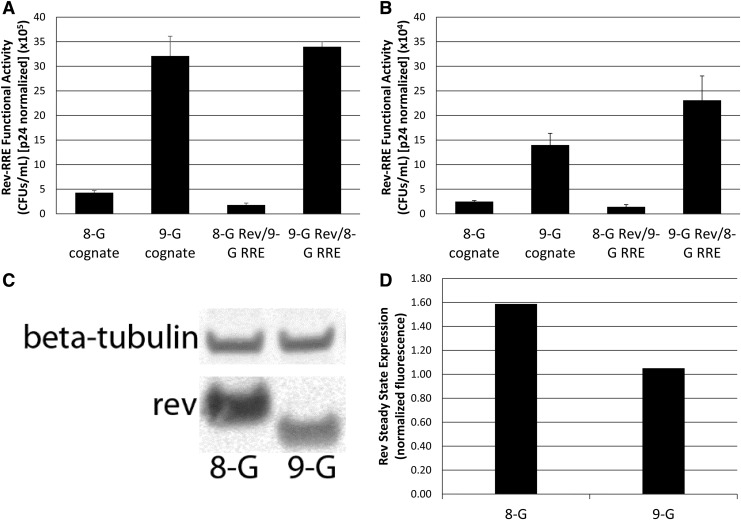
To determine whether expression of Rev from the different isolates led to different steady state levels of Rev protein that might explain the activity differences observed, we created influenza HA epitope-tagged Rev sequences, so that we could detect all of the Rev variants using the same antibody on a Western blot. We then measured the activity of selected HA–Rev and RRE pairs to assess whether the presence of the HA tag changed Rev–RRE activity compared with the pairs with the nontagged Rev. Our results showed that the HA tag did not change the rank order of activity of the different pairs, although lower absolute titers were obtained (data not shown). Figure 7 shows the results of one such comparative activity assay using the 8-G and 9-G cognate pairs, as well as the heterologous combinations. Similar relative activities were observed using the untagged (Fig. 7A) or the HA-tagged (Fig. 7B) Rev sequences and again the function correlated with Rev rather than the RREs. However, surprisingly, the relationship between Rev steady state expression and functional activity was inverse. The pairs containing 9-G Rev were significantly more active than the pairs that contained 8-G Rev, although the Rev from the 8-G isolate displayed 1.5 times greater steady state levels than the Rev from 9-G (Fig. 7C, D).
FIG. 7.
Rev and RRE contributions to functional activity in subtype G viruses. The activity of subtype G virus Rev–RRE cognate pairs as well as Rev–RRE pairs created by transposing the 8-G and 9-G Rev and RRE components were measured using the standard Rev sequences (A) or HA-tagged Rev sequences (B). Bars show average deviation from the mean for two replicates. (C) Western blot showing the level of steady state Rev expression of both subtype G HA-tagged Rev isolates. Rev expression was detected using a fluorescent secondary antibody and infrared imager. (D) Rev intensity values were measured and normalized to beta-tubulin to control for loading variation and to SEAP expression to control for transfection efficiency, yielding arbitrary normalized units. HA, hemagglutinin; SEAP, secreted embryonic alkaline phosphatase.
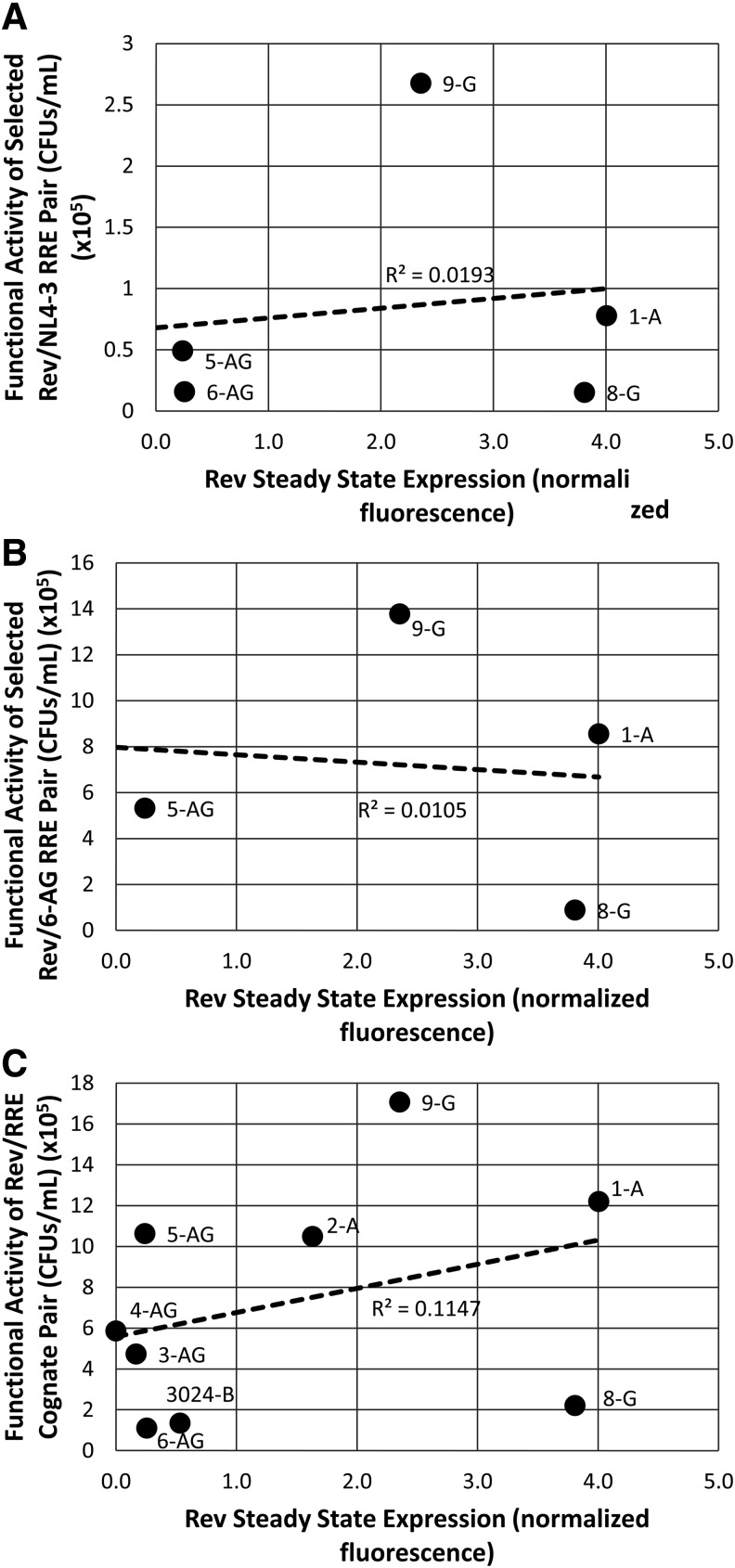
To determine whether the lack of correlation between Rev steady state levels and activity held true for all of the isolates tested, we plotted the activity of each Rev–RRE pair (including the two reference RREs and cognate RREs) as a function of the steady state levels of Rev detected by Western blotting (Fig. 8A–C). Rev protein steady state levels varied considerably among the isolates (x-axis Fig. 8). However, the results of a linear regression analysis show that there was no significant correlation between protein expression and Rev–RRE functional activity (r2 = 0.01913, r2 = 0.0105 and r2 = 0.1147, respectively). Thus Rev sequence variation and not steady state expression level appears to be the determinant of the functional variation we observed.
FIG. 8.
Correlation between Rev steady state protein expression and Rev–RRE functional activity. The functional activity of (A) selected Rev–NL4-3 RRE pairs, (B) selected Rev–6-AG RRE pairs, or (C) cognate pairs were plotted as a function of the Rev steady state levels. The degree of correlation was calculated by performing a linear regression. The quantification of steady state expression for the various Revs was performed by Western blotting as described in the legend of Figure 7. The values for Rev steady state levels represent the average of three different determinations.
Discussion
In this study, we describe a previously unrecognized degree of HIV Rev–RRE functional activity variation between clinical isolates. This activity variation appears to be driven mainly by differences in the activity of the Rev component. Furthermore, the variation in Rev activity is not explainable in terms of differences in Rev steady state protein expression. These results suggest that differences in Rev sequence and structure can result in functional activity differences, potentially by creating Rev–RRE complexes that recruit Crm1, or other cellular factors, with differential efficiencies. This notion is consistent with structural studies that have described the plasticity of the Rev–RRE ribonucleic acid particle.15,16,56 Further experiments are needed to elucidate the mechanisms underlying the differential activities, as well as to clarify the role that functional activity variation may play in pathogenesis.
Although differences in both Rev and the RRE have been demonstrated to be sufficient to cause functional activity variation of the Rev–RRE regulatory axis, few studies to date have attempted to parse the relative contributions of these elements to the activity of the cognate pair. Our laboratory has previously examined Rev–RRE functional activity in five patients at two time points during the course of infection.19 In that study, functional activity variation tracked more closely with variation in RRE activity than with Rev activity. The present study shows the opposite result; there is a close linear relationship between variation in Rev activity and variation in activity of the corresponding Rev–RRE cognate pair, but this does not hold for variation in RRE activity.
One explanation for this discrepancy is that modulation of Rev–RRE functional activity may be accomplished in different ways over the course of infection in a single host (intra-patient variation) compared with longer time scales between hosts (inter-patient variation). Although changes in Rev activity require nonsynonymous mutations that alter protein function, very minor changes in the RRE nucleotide sequence can modify the activity of this element, likely due to the plasticity of RRE secondary structure.20 Thus, viral quasispecies with differential Rev–RRE functional activity might more readily arise due to changes in the RRE rather than in Rev over the course of infection in a single patient.57 However, both Rev and RRE changes would likely be seen over the longer evolutionary time frames that account for interpatient variation.
The mechanisms underlying the observed variation in Rev functional activity were not examined in this study. Although there were substantial differences in Rev steady state protein levels, there was no association between these levels and Rev functional activity. However, it is notable that there were significant differences between the Rev sequences studied, including in residues that are believed to be important for RRE binding, Rev multimerization, and Crm1 interaction. We also observed significant differences in regions of unknown function, particularly at the carboxyterminus. These sequence differences clearly could play a role in mediating the differential activity.56 Further studies using chimeric Rev sequences may be helpful in elucidating which residues are the determinants of differential Rev activity. These data, in turn, may help to inform future structural studies of Rev–RRE interactions.
Although our sample size was small, there does not appear to be any generalizable difference in the functional activity of Rev–RRE between subtypes A, G, and CRF02_AG. This suggests that the recombination of an A-derived RRE and a G-derived Rev sequence is not a primary driving factor in the increasing prevalence of CRF02_AG in western Africa and central Asia. This pattern of recombination may instead yield a competitive advantage due to the Env sequence.58
There is insufficient data about the clinical context in which the viruses used in this study were isolated to permit any correlation with patient outcomes or immune milieu. Only a single patient-derived virus was predicted to utilize CXCR4 (3-AG) (data not shown), preventing any analysis using coreceptor usage as a proxy for disease stage. However, our group has previously shown that the Rev–RRE functional activity of viral quasispecies isolated from a single patient at a single time point during infection tends to cluster together, and that this activity set-point can change over time within the same patient.19 This suggests that different levels of Rev–RRE activity may be selected in the different immune environments in which HIV is replicating. The variation observed in the functional activity of the Rev–RRE cognate pairs used in this study may, therefore, reflect the presumably different clinical contexts in which they were obtained.
Overall, the Rev–RRE regulatory system can be viewed as a rheostat that permits modulation of the production of viral proteins and the rate of viral replication. A cell infected with a virus with higher Rev–RRE functional activity will produce more viral proteins and will permit a faster rate of viral replication. However, it also may be at higher risk for cytotoxic T-lymphocyte-mediated killing, since greater amounts of viral antigens are being produced.59 Thus, one would expect that in the context of an active immune system, selection pressure would favor viruses with lower Rev–RRE functional activity. In contrast, in late stage disease when there is less immune surveillance, viruses with higher Rev–RRE activity would have a selective advantage. Variation in the Rev–RRE regulatory axis could thus be one mechanism by which HIV adapts to the particular immune environment in which it finds itself and fine-tunes the rate of replication to optimize the balance between virus particle production and immune evasion. An alternative or additional role for Rev–RRE modulation could arise out of a proposed oscillatory pattern of viral replication in conjunction with Tat regulation.60 In this model, cyclic viral replication could be tuned by modification of Rev–RRE functional activity to promote immune evasion and minimize direct viral cytotoxicity.
Further investigation into the role of Rev–RRE functional activity variation in pathogenesis is warranted and may have relevance for HIV therapy. Many so-called kick and kill cure strategies rely on the vigorous production of viral proteins after the stimulation of the latent proviral reservoir (reviewed in Ref.61). If lower Rev–RRE functional activity indeed correlates with effective immune evasion in clinical infection, then the latent reservoir may be relatively enriched with viruses that have a less active Rev–RRE. A fuller understanding of this regulatory axis may, therefore, play a key role in developing effective agents for the stimulation of latent cells.
During this study, we identified both naturally occurring and artificial Rev–RRE pairs that yield greater functional activity than that of the laboratory strain NL4-3. High-activity Rev–RRE combinations may have a potential application in the efficient production of lentiviral vectors. The production of high vector titers remains a significant barrier to the commercial use of such vectors,62 and optimization of Rev–RRE function in this context may help to reduce costs for this promising therapeutic modality.
Supplementary Material
Acknowledgments
We thank Laurie Gray for helpful discussions throughout the course of this work and help in developing some of the methods used in this study. Susan Prasad provided valuable assistance in the performance of several of the assays described. Partial salary support for P.E.J. was provided by a Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (5 T32 AI 007046 37). Partial salary support for M.-L.H. and D.R. was provided by the Charles H. Ross, Jr., and Myles H. Thaler Endowments at the University of Virginia. This work was supported by grant number GM110009 from the National Institute of General Medical Sciences and the Myles H. Thaler Endowment.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Frankel AD, Young JA: HIV-1: Fifteen proteins and an RNA. Annu Rev Biochem 1998;67:1–25 [DOI] [PubMed] [Google Scholar]
- 2.Pollard VW, Malim MH: The HIV-1 Rev protein. Annu Rev Microbiol 1998;52:491–532 [DOI] [PubMed] [Google Scholar]
- 3.D'Agostino DM, Felber B, Harrison J, Pavlakis G: The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol 1992;12:1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adachi A, Gendelman HE, Koenig S, et al. : Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 1986;59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malim MH, Hauber J, Le S, Maizel JV, Cullen BR: The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989;338:254–257 [DOI] [PubMed] [Google Scholar]
- 6.Huang XJ, Hope TJ, Bond BL, McDonald D, Grahl K, Parslow TG: Minimal Rev-response element for type 1 human immunodeficiency virus. J Virol 1991;65:2131–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heaphy S, Dingwall C, Ernberg I, et al. : HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the rev response element region. Cell 1990;60:685–693 [DOI] [PubMed] [Google Scholar]
- 8.Tiley LS, Malim MH, Tewary HK, Stockley PG, Cullen BR: Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci U S A 1992;89:758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y, Tambe A, Zhou K, Doudna JA: RNA-guided assembly of Rev-RRE nuclear export complexes. Elife 2014;3:e03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neville M, Stutz F, Lee L, Davis LI, Rosbash M: The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol 1997;7:767–775 [DOI] [PubMed] [Google Scholar]
- 11.Bohnlein E, Berger J, Hauber J: Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: New insights into the domain structure of Rev and Rex. J Virol 1991;65:7051–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zapp ML, Hope TJ, Parslow TG, Green MR: Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: A dual function for an arginine-rich binding motif. Proc Natl Acad Sci U S A 1991;88:7734–7738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malim MH, Böhnlein S, Hauber J, Cullen BR: Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 1989;58:205–214 [DOI] [PubMed] [Google Scholar]
- 14.Hauber J, Bouvier M, Malim MH, Cullen BR: Phosphorylation of the rev gene product of human immunodeficiency virus type 1. J Virol 1988;62:4801–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daugherty MD, D'Orso I, Frankel AD: A solution to limited genomic capacity: Using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell 2008;31:824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaraman B, Mavor D, Gross JD, Frankel AD: Thermodynamics of Rev–RNA interactions in HIV-1 Rev–RRE assembly. Biochemistry 2015;54:6545–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Booth DS, Cheng Y, Frankel AD: The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. Elife 2015;3:e04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fung HYJ, Fu S, Brautigam CA, Chook YM: Structural determinants of nuclear export signal orientation in binding to exportin CRM1. Elife 2015;4:e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloan EA, Kearney MF, Gray LR, et al. : Limited nucleotide changes in the Rev response element (RRE) during HIV-1 infection alter overall Rev-RRE activity and Rev multimerization. J Virol 2013;87:11173–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherpa C, Rausch JW, Le Grice SF, Hammarskjold ML, Rekosh D: The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res 2015;43:4676–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD: RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev–Rev response element complex. Elife 2015;3:e04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Churchill MJ, Chiavaroli L, Wesselingh SL, Gorry PR: Persistence of attenuated HIV-1 rev alleles in an epidemiologically linked cohort of long-term survivors infected with nef-deleted virus. Retrovirology 2007;4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iversen AK, Shpaer EG, Rodrigo AG, et al. : Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol 1995;69:5743–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phuphuakrat A, Auewarakul P: Functional variability of Rev response element in HIV-1 primary isolates. Virus Genes 2005;30:23–29 [DOI] [PubMed] [Google Scholar]
- 25.Phuphuakrat A, Paris RM, Nittayaphan S, Louisirirotchanakul S, Auewarakul P: Functional variation of HIV‐1 Rev response element in a longitudinally studied cohort. J Med Virol 2005;75:367–373 [DOI] [PubMed] [Google Scholar]
- 26.Carpenter S, Chen WC, Dorman KS: Rev variation during persistent lentivirus infection. Viruses 2011;3:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belshan M, Baccam P, Oaks JL, et al. : Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology 2001;279:185–200 [DOI] [PubMed] [Google Scholar]
- 28.Baccam P, Thompson RJ, Li Y, et al. : Subpopulations of equine infectious anemia virus Rev coexist in vivo and differ in phenotype. J Virol 2003;77:12122–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi N, Saiyed Z, Thangavel S, Rodriguez J, Rao K, Nair MP: Differential effects of HIV type 1 clade B and clade C tat protein on expression of proinflammatory and antiinflammatory cytokines by primary monocytes. AIDS Res Hum Retroviruses 2009;25:691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwabu Y, Kinomoto M, Tatsumi M, et al. : Differential anti-APOBEC3G activity of HIV-1 vif proteins derived from different subtypes. J Biol Chem 2010;285:35350–35358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jian H, Zhao LJ: Pro-apoptotic activity of HIV-1 auxiliary regulatory protein vpr is subtype-dependent and potently enhanced by nonconservative changes of the leucine residue at position 64. J Biol Chem 2003;278:44326–44330 [DOI] [PubMed] [Google Scholar]
- 32.Walker PR, Ketunuti M, Choge IA, et al. : Polymorphisms in nef associated with different clinical outcomes in HIV type 1 subtype C-infected children. AIDS Res Hum Retroviruses 2007;23:204–215 [DOI] [PubMed] [Google Scholar]
- 33.Howard TM, Olaylele DO, Rasheed S: Sequence analysis of the glycoprotein 120 coding region of a new HIV type 1 subtype A strain (HIV-1IbNg) from Nigeria. AIDS Res Hum Retroviruses 1994;10:1755–1757 [DOI] [PubMed] [Google Scholar]
- 34.Carr JK, Torimiro JN, Wolfe ND, et al. : The AG recombinant IbNG and novel strains of group M HIV-1 are common in Cameroon. Virology 2001;286:168–181 [DOI] [PubMed] [Google Scholar]
- 35.Ragupathy V, Zhao J, Wood O, et al. : Identification of new, emerging HIV-1 unique recombinant forms and drug resistant viruses circulating in Cameroon. Virol J 2011;8:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baryshev P, Bogachev V, Gashnikova N: Genetic characterization of an isolate of HIV type 1 AG recombinant form circulating in Siberia, Russia. Arch Virol 2012;157:2335–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tebit DM, Arts EJ: Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 2011;11:45–56 [DOI] [PubMed] [Google Scholar]
- 38.Rizvi TA, Panganiban AT: Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol 1993;67:2681–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjold ML, Rekosh D: The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol 1997;71:5841–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zolotukhin AS, Valentin A, Pavlakis GN, Felber BK: Continuous propagation of RRE(-) and Rev(-)RRE(-) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol 1994;68:7944–7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH: Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J 2004;23:2632–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wodrich H, Schambach A, Krausslich HG: Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 gag expression in a context-dependent manner. Nucleic Acids Res 2000;28:901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naldini L, Blömer U, Gallay P, et al. : In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996;272:263–267 [DOI] [PubMed] [Google Scholar]
- 44.Graham FL, van der EB, Alex J: A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 1973;52:456–467 [DOI] [PubMed] [Google Scholar]
- 45.Wehrly K, Chesebro B: P24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 1997;12:288–293 [DOI] [PubMed] [Google Scholar]
- 46.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA: The structure of an antigenic determinant in a protein. Cell 1984;37:767–778 [DOI] [PubMed] [Google Scholar]
- 47.Baalwa J, Wang S, Parrish NF, et al. : Molecular identification, cloning and characterization of transmitted/founder HIV-1 subtype A, D and A/D infectious molecular clones. Virology 2013;436:33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tebit DM, Zekeng L, Kaptué L, Kräusslich H, Herchenröder O: Construction and characterisation of a full-length infectious molecular clone from a fast replicating, X4-tropic HIV-1 CRF02. AG primary isolate. Virology 2003;313:645–652 [DOI] [PubMed] [Google Scholar]
- 49.Kijak GH, Sanders-Buell E, Wolfe ND, et al. : Development and application of a high-throughput HIV type 1 genotyping assay to identify CRF02_AG in West/West Central Africa. AIDS Res Hum Retroviruses 2004;20:521–530 [DOI] [PubMed] [Google Scholar]
- 50.Carr JK, Nadai Y, Eyzaguirre L, et al. : Outbreak of a West African recombinant of HIV-1 in Tashkent, Uzbekistan. J Acquir Immune Defic Syndr 2005;39:570–575 [PubMed] [Google Scholar]
- 51.Yamaguchi J, Ndembi N, Ngansop C, et al. : HIV type 1 group M subtype G in Cameroon: Five genome sequences. AIDS Res Hum Retroviruses 2009;25:469–473 [DOI] [PubMed] [Google Scholar]
- 52.Sanchez AM, DeMarco CT, Hora B, et al. : Development of a contemporary globally diverse HIV viral panel by the EQAPOL program. J Immunol Methods 2014;409:117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salminen MO, Koch C, Sanders-Buell E, et al. : Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology 1995;213:80–86 [DOI] [PubMed] [Google Scholar]
- 54.Haesevelde MMV, Peeters M, Jannes G, et al. : Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 1996;221:346–350 [DOI] [PubMed] [Google Scholar]
- 55.Carr JK, Salminen MO, Albert J, et al. : Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology 1998;247:22–31 [DOI] [PubMed] [Google Scholar]
- 56.Daugherty MD, Liu B, Frankel AD: Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 2010;17:1337–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shuck-Lee D, Chang H, Sloan EA, Hammarskjold ML, Rekosh D: Single-nucleotide changes in the HIV Rev-response element mediate resistance to compounds that inhibit Rev function. J Virol 2011;85:3940–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Njai HF, Gali Y, Vanham G, et al. : The predominance of human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in west central Africa may be related to its replicative fitness. Retrovirology 2006;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bobbitt KR, Addo MM, Altfeld M, et al. : Rev activity determines sensitivity of HIV-1-infected primary T cells to CTL killing. Immunity 2003;18:289–299 [DOI] [PubMed] [Google Scholar]
- 60.Likhoshvai VA, Khlebodarova TM, Bazhan SI, Gainova IA, Chereshnev VA, Bocharov GA: Mathematical model of the tat-rev regulation of HIV-1 replication in an activated cell predicts the existence of oscillatory dynamics in the synthesis of viral components. BMC Genomics 2014;15(Suppl 12):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Margolis DM, Hazuda DJ: Combined approaches for HIV cure. Curr Opin HIV AIDS 2013;8:230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geraerts M, Michiels M, Baekelandt V, Debyser Z, Gijsbers R: Upscaling of lentiviral vector production by tangential flow filtration. J Gene Med 2005;7:1299–1310 [DOI] [PubMed] [Google Scholar]
- 63.Larkin MA, Blackshields G, Brown NP, et al. : Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 64.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. : Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008;105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kearse M, Moir R, Wilson A, et al. : Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.