Abstract
Water samples of the Drinking Water Supply System (DWSS) of the city of Braunschweig were analysed for its Legionella species composition using genus-specific PCR amplicons and single-strand conformation polymorphism (SSCP) fingerprint analyses based on 16S rRNA genes. These analyses comprised the whole supply chain including raw water, treatment process and large-scale storage, and a seasonal study of finished drinking water sampled monthly from cold and hot tap water. Treatment of raw water had a major impact on Legionella species by reducing their diversity and abundances. The Legionella species composition of the tap water was highly distinct from that of both source waters. In cold water, 8–14 different phylotypes of Legionella (PTLs) were observed per sample with relative abundances ranging from >1% to 53%. In hot water, L. pneumophila was present during all seasons at high relative abundances (8–40%) accompanied by 5–14 other PTLs of which 6 PTLs were in common with cold water. This thermophilic Legionella community, including L. pneumophila, was able to grow in the hot water above 50 °C. Such thermophilic Legionella populations are of general relevance for drinking water management and public health, but also for the ecology and evolution of the genus Legionella.
Introduction
The genus Legionella comprises currently 59 species of which about half have been demonstrated to be pathogenic to humans, and the majority is considered as facultative pathogenic (Mercante and Winchell, 2015). The most well-known and best-studied Legionella species is L. pneumophila because it is the aetiological agent of legionellosis (primarily lung infections) in >90% of the cases -in most countries where appropriate surveillance is in place (Bartram et al., 2007; European Centre for Disease Prevention and Control, 2015). Non-pneumophila Legionella species account usually for <10% of human infections, especially L. micdadei, L. bozmanii, L. longbeachae, L. dumoffii and L. feeleii are isolated repeatedly from hospitalized patients whereas L. anisa, L. wadsworthii and L. cincinnatiensis are only rarely found (Yu et al., 2002; Beauté et al., 2013; European Centre for Disease Prevention and Control, 2015). A special case is L. longbeachae that accounts for about 30% of the human Legionella infections in Australia (Fields et al., 2002; Whiley and Bentham, 2011). The genus Legionella has the highest human health relevance for water-based bacterial infections with respect to fatality in the developed countries (Phin et al., 2014).
In Europe, local legionellosis outbreaks due to aerosols distributed by cooling towers and spas regularly affect hundreds of people despite strict control and surveillance measures. Most cases, however, are caused by sporadic infections and contaminated water systems still pose the greatest threat to public health (European Centre for Disease Prevention and Control, 2015). Despite a substantial amount of research on the ecology, the physiology and the molecular virulence mechanisms of Legionella since its discovery annual incidence rates remained on a high level (Fields et al., 2002; Phin et al., 2014; European Centre for Disease Prevention and Control, 2015).
Most Legionella species are of aquatic origin including L. pneumophila; a few are also isolated from non-aquatic habitats such as potting soil and compost (especially L. longbeachae) (Fields et al., 2002; Borella et al., 2005a; Casati et al., 2010; Lück et al., 2010; Whiley and Bentham, 2011; Travis et al., 2012; Currie et al., 2014). Many Legionella species are considered well adapted to low-nutrient conditions in water, and therefore pre-adapted to conditions in drinking water (Vervaeren et al., 2006). Best studied are the environmental reservoirs of L. pneumophila, which has been found in natural freshwater environments such as lakes, streams and Drinking Water Supply Systems (DWSS) (Fields, 1996). In DWSS, L. pneumophila survives planktonically, grows in biofilms and infects and replicates within protozoa (Steinert et al., 2002; Molmeret et al., 2005; Declerck et al., 2007b; Steinert et al., 2007). These bacteria can survive in moist places for a long time at relatively high temperatures and in the presence of disinfectants such as chlorine (Dupuy et al., 2011).
Temperature is an important factor for the abundance and growth of Legionella in natural and man-made aquatic systems. Legionella are mesophilic with a temperature range for growth ranging from 12 °C to 42 °C and an optimum from 25 °C to 40 °C (Ohno et al., 2003). This is the reason why warm water systems (showers, cooling towers and air conditioners) were often shown to be of special health relevance, and heating above 50 °C is highly recommended to decrease the risk of legionellosis (Stout et al., 1986). Temperature might also be a key factor for the annual summer peak of Legionella cases observed in Europe (Joseph et al., 2010; Beauté et al., 2013). On the average, in late winter (February and March) a minimum in the number of cases is observed which is followed by a 3- to 4-fold increase in summer with a maximum in August. Due to this seasonality of Legionella infections, it might be worthwhile studying seasonal effects on their abundances in drinking water to understand their environmental control factors. Seasonal variations of Legionella spp. communities have already been detected in natural waters and reservoirs, but these studies did not include the connected DWSS (Parthuisot et al., 2010; Kao et al., 2014).
Next to temperature, the abundances of Legionella species in aquatic ecosystems are controlled by a variety of factors such as available nutrients, grazing by protozoa and eventually by lysis of bacteriophages in a regulatory circuit called microbial loop (Lammertyn et al., 2007; Pomeroy et al., 2007; Abdel-Nour et al., 2013). In addition to these classical ecological parameters, concentrations of disinfectants have an important role in many DWSS as essential abiotic regulators. In man-made freshwater supply systems, treatment procedures further remove particles from the source water as well as components of the original food web and the subsequent preparation of hot water applies substantial temperature stress on the remaining microbiota. Some of these processes were shown to have a major impact on the bacterial community and have not yet been assessed for Legionella in surface water-derived drinking water (Eichler et al., 2006; Henne et al., 2013). Eichler et al. (2006) demonstrated for a DWSS that the overall bacterial community of the surface water (deep water from two reservoirs) changed very little due to processing by sand filtration and flocculation. However, chlorination had a major impact on the species composition and increased the abundance of presumably nitrifying taxa. The present bacterial community in the finished drinking water, on the other hand, was still composed to a large extent of taxa derived from the two reservoirs while only the active fraction of the community was substantially altered. In contrast, an almost complete change of the bacterial community was observed during hot water preparation by heating cold drinking water to about 63 °C (Henne et al., 2013). The majority of the cold water bacteria decayed during heating with their nutrients being used by a rapidly growing hot water community reaching finally cell numbers of about 80% of the cold water community. Most bacterial members of the hot water community were phylogenetically closely affiliated with thermophilic species.
Currently, there is a very restricted set of studies on Legionella species diversity in DWSSs; most studies have concentrated on L. pneumophila. Wullings and van der Kooij (2006) provided a first molecular survey on Legionella species occurring in surface- and groundwater-based source waters and the respective finished drinking water in the Netherlands, showing that surface waters tend to lead to a higher Legionella load in drinking water. In addition, Wullings et al. (2011) described the Legionella diversity of two groundwater-based DWSS, showing that Legionella abundance and diversity is influenced by the dissolved organic carbon in source and drinking water. So far, there is neither a study following the seasonal diversity and dynamics of the genus Legionella from source water via cold to the hot drinking water nor a study that sees the Legionella dynamics in the frame of the overall bacterial community.
Our study aims to understand the composition and dynamics of Legionella communities in drinking water as a result of the typical treatments for drinking water processing such as raw water flocculation, sand filtration, chlorination, storage and hot water preparation. Based on our previous studies and the overall understanding of the total bacterial community of a DWSS (Eichler et al., 2006; Kahlisch et al., 2012; Henne et al., 2013), we will provide detailed insights into the community ecology of a highly health relevant genus, that is, Legionella, its changes and their triggering factors. In our DWSS, the following approach was pursued to accomplish our objectives: (i) genus-specific single-strand conformation polymorphism (SSCP) fingerprints were analysed to illustrate changes in the Legionella community structure along the drinking water production line and during a seasonal cycle of the finished drinking water before and after the heating process; (ii) single phylotypes were identified by sequencing of major bands to assess phylogenetic community compositions; (iii) community structures and compositions were compared with respect to the impact of drinking water processing, hot water production and season; (iv) the Legionella community and its dynamics were compared with the total bacterial community; (v) the contribution of the genus Legionella to the total bacterial community in terms of cell numbers was quantified by real-time PCR; and (vi) responses to water temperature of the whole Legionella community and abundant phylotypes were evaluated. To this end, we approached a better understanding of the long-term community ecology of the genus Legionella that is urgently needed for freshwater management and public health prevention measures.
Materials and methods
Study sites and sampling
Cold drinking water was sampled monthly from May 2008 to October 2009. It was taken from the tap of laboratory D0.04 on the campus of the Helmholtz Centre for Infection Research (HZI), Braunschweig, Germany, with 5-min flushing to prevent stagnant water to be sampled. In former studies, it was shown that the HZI campus drinking water is representative for the whole city area of Braunschweig with respect to physicochemical parameters and the bacterial community (Eichler et al., 2006; Henne et al., 2012).
Hot drinking water was sampled monthly from September 2008 to November 2009. It was taken from a shower next to lab D0.04 after several minutes flushing. The hot drinking water was made from the cold drinking water on the HZI campus by heating it up to 63 °C (range: 60–63 °C) in two 600-litre heating vessels. The cold drinking water was heated in the heating vessels together with the hot water of the circular systems at variable mixing ratios from about 1:1 to 1:10 (cold to hot water) during work hours, that is, our sampling time. More details about the cold and hot water supply system are given elsewhere (Eichler et al., 2006; Henne et al., 2013).
The drinking water originates from two surface water reservoirs (Grane reservoir, oligotrophic water, average dissolved organic carbon 2.2 mg C l−1, 51° 45' 44” N, 10° 22' 38” E; Ecker reservoir, dystrophic water, average dissolved organic carbon 6.2 mg C l−1, 51° 50' 27” N, 10° 34' 45” E) situated in the Harz Mountains 40 km south of Braunschweig. Processing of the drinking water by the local supplier Harzwasserwerke GmbH included flocculation/coagulation, sand filtration and chlorination (0.2–0.7 mg chlorine l−1). At both reservoirs, the obtained raw water is processed similarly (sampling sites G=Grane and E=Ecker reservoir, samples for raw water are called GR1 and ER1, processed raw water is called GR2 and ER2, respectively). After treatment of the raw water by physical and chemical means, pipe systems lead from both reservoirs to the Lewerberg (Lb) storage container where both waters are mixed. This mixed water consisted of about 77% water from the Grane reservoir and 23% water from the Ecker reservoir. From the Lb container, the mixed water is transported to the Lindenberg (Li) storage container, directly located at the southern outskirts of Braunschweig, to which the local drinking water supply net is connected, including the sampled tap water source at the institute. The finished drinking water had an average dissolved organic carbon content of 1.6 mg C l−1, with a maximum of 2.7 mg C l−1 in early June. Samples along the water processing line from source water to the tap were collected on 3 May 2004. More details on the respective DWSS and sampling are given elsewhere (Eichler et al., 2006).
Drinking water microorganisms from cold and hot drinking water were sampled by filtration according to Eichler et al. (2006). In brief, 5 litres of drinking water were filtered through a filter sandwich consisting of a 0.2-μm pore size polycarbonate filter (90 mm diameter; Nucleopore; Whatman, Maidstone, UK) with a precombusted glass fibre filter on top (90 mm diameter; GF/F; Whatman). Biomass harvested on filter sandwiches was stored at −70 °C until further analysis.
Cell counts of drinking water bacteria
For total bacterial cell counts, formaldehyde-fixed samples (2% final concentration) of drinking water were stained with Sybr Green I dye (1:10 000 final dilution; Molecular Probes, Invitrogen, Carlsbad, CA, USA) and analysed directly by epifluorescence microscopy or stored frozen (−20 °C) until examination as described in more detail by Henne et al. (2013).
DNA extraction and PCR
For extraction of DNA from the filter sandwiches, a modified DNeasy protocol (Qiagen, Hilden, Germany) was used as described in detail by Henne et al. (2013). Quantification of DNA was carried out using Picogreen (dsDNA quantification, Molecular Probes; Invitrogen) according to the manufacturer.
For the SSCP fingerprints, a Legionella genus-specific PCR was performed using the biotinylated primer pair Lgsp17Fb (5′-bio-GGCCTACCAAGGCGACGATCG-3′) and Lgsp28R (5′-CACCGGAAATTCCACTACCCTCTC-3′) targeting the 16S rRNA gene (Kahlisch et al., 2010). For L. pneumophila, the amplicon size is 421 bp spanning the V2 and V3 regions of the 16S rRNA (positions 264–661 E. coli numbering). Each amplification was carried out using 0.2–2.0 ng of DNA template in a final volume of 50 μl under the following conditions: Initial denaturation was at 95 °C for 15 min; followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 66.5 °C for 30 s, elongation at 72 °C for 30 s and final elongation for 10 min at 72 °C. Amplification was achieved using HotStarTaq DNA polymerase at 1 U μl−1, 2 mm dNTPs, 2 mm MgCl2 (all from Qiagen) and primers at a final concentration of 0.2 μm (Eurofins MWG, Ebersberg, Germany) in the reaction buffer provided by the manufacturer of the DNA polymerase.
SSCP electrophoresis and sequencing of single-stranded DNA bands from SSCP gels
For the preparation of single-stranded DNA and the SSCP electrophoresis, the protocol described by Henne et al. (2013) was applied. After electrophoresis, the gel was silver stained according to Bassam et al. (1991). Dried SSCP gels were digitized using an Epson Expression 1600 Pro scanner (Epson, Long Beach, CA, USA). Sequence information of individual bands was obtained following the protocol of Eichler et al. (2006). The partial 16S rRNA gene sequences obtained from the genus-specific fingerprints are accessible under the GenBank/EMBL/DDBJ accession numbers KP822824 to KP822921 and KP863802 to KP863862. Individual accession numbers of representative sequences are listed in Supplementary Tables S1 and S2.
Sequence comparison and statistical analyses
Forward and reverse sequences obtained from each excised band were assembled to contigs, analysed and manually checked using the Sequencher software (v. 5.2, Gene Codes Cooperation, Ann Arbor, MI, USA). Primer sequences were trimmed; resulting in partial 16S rRNA gene sequences of 376 bp. Fragments were checked for chimeras with the DECIPHER web tool (Wright et al., 2012) and pooled to clusters using the Sequencher's assembling tool for dirty data while applying a 99% sequence similarity threshold in order to group highly similar sequences while still discriminating between well-described species. Consensus sequences of each cluster were designated ‘phylotype of Legionella' (PTL-01 to -35) and ‘phylotype of reservoirs' (PTR-01 to -42), respectively. Phylogenetic identification of PTL and PTR consensus sequences was obtained by comparison with a curated database of type strain isolates using the Seqmatch function of the Ribosomal Database Project (RDP, accessed January 2015) (Cole et al., 2014). RDP Seqmatch results and similarity scores are listed in Supplementary Table S1 (PTRs) and Supplementary Table S2 (PTLs).
Digitized images of dried SSCP gels were analysed with the GelCompare II software (Applied Maths, Kortrijk, Belgium), bands were extracted applying appropriate settings (2.0% minimum profiling, 0.0% ‘grey zone', 0.5% minimum area and shoulder sensitivity=0) and edited manually (Supplementary Figure S3). Relative abundance measures for each band were obtained and assigned to its respective 16S rRNA gene sequence. Similarity matrices and Pearson's correlation coefficients of Legionella community fingerprints were calculated and subsequently, cluster analyses were performed by Neighbor-Joining algorithm.
Phylotype sequences were aligned with reference sequences of all 62 described Legionella type species (including subspecies) listed by the DSMZ (http://www.dsmz.de/bacterial-diversity/prokaryotic-nomenclature-up-to-date, last accessed: December 2014) using the RDP alignment tool. In case sequences of type strains had numerous ambiguous positions, additional high-quality sequences were searched in and extracted from the NCBI database and added to complement the alignment, for example, for L. hackeliae and L. spiritensis. The alignment was then further analysed in MEGA (version 6) (Tamura et al., 2013). Phylogenetic comparison was conducted calculating a Neighbor-Joining tree of 1000 bootstrap replicates using the Kimura 2-parameter model with a discrete Gamma distribution (shape parameter=5) and to allow for invariant sites (46.8%).
Species richness, evenness and abundance-weighted comparisons and other statistical analyses were conducted in R (v. 3.1.2, ‘Pumpkin Helmet') within the package Picante (v. 1.6–2) (Kembel et al., 2010). To compare Legionella community structures phylogenetically, Bray-Curtis distances among all samples were calculated to take phylotype composition and abundances into account. Samples were then hierarchically clustered using UPGMA in order to visualize Bray-Curtis dissimilarity and reveal seasonal clustering and differentiation, respectively. Mean pairwise distance (mpd) and mean nearest taxon distance (mntd) were calculated for the presence–absence data as standardized effect size (of mpd and mntd, respectively) versus null communities as implemented in Picante to investigate community phylogenetic structures.
Quantification of total Legionella cell numbers by real-time PCR
DNA as extracted from the water samples was diluted 10- or 100-fold in 1 × Tris-EDTA (TE) buffer, pH 8.0 (Sigma-Aldrich, Munich, Germany) before all qPCR analyses in a LightCycler 480 System (Roche, Mannheim, Germany). Final DNA content in each reaction ranged from 0.1 to 1.8 ng depending on the sample. We measured bacterial 16S rRNA gene abundance using a set of community primers (‘Com') adapted from Labrenz et al. (2004) amplifying a fragment spanning positions 519–926 of the E. coli 16S rRNA. Briefly, Com qPCRs (20 μl) contained 10 μl of LightCycler 480 SYBR Green I master mix (Roche), 0.8 μm of each primers (Eurofins MWG), PCR grade sterile water (Roth, Karlsruhe, Germany) and 5 μl of sample DNA. An initial activation step of 5 min at 95 °C was followed by 50 amplification cycles (30 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C). Potential formation of primer dimers was determined by melting-point analysis in a range from 65 °C to 97 °C. No peaks indicating primer dimers were observed in all samples analysed. Quantification of Legionella spp. 16S rRNA gene abundance (‘Leg') was performed using primers L4F (5′-agaaccttacctacccttgacatacagt-3′) and L4R (5′-gacagccatgcagcacctgta-3′) amplifying an 89-bp fragment spanning positions 975–1063 of the L. pneumophila strain Philadelphia 1 16S rRNA gene combined with an internal fluorescent reporter probe L4P (5′-LC610-gtgccttcgggaacactg-BHQ1-3′). Results obtained were comparable with a Sybr green assay using the same primers and amplification conditions as for the Legionella spp. SSCP analysis (data not shown). Leg qPCR reactions (20 μl) consisted of 10 μl LightCycler 480 Probes Master (Roche), 0.5 μm of each primer (Eurofins MWG), 0.2 μm of the probe (Eurofins MWG), PCR grade sterile water (Roth) and 5 μl of sample DNA. Amplification conditions were 5 min at 95 °C followed by 50 cycles of 10 s at 95 °C and 20 s at 60 °C. Each run contained a 10-fold serial dilution (5 × 101–5 × 105 genome units per reaction) of a commercial quantification standard for genomic L. pneumophila DNA (Minerva Biolabs, Berlin, Germany) for calibration and a non-template control (TE buffer).
We calculated the ratio of Legionella spp. cells to all bacterial cells (RLeg) of each sample by assuming an average 16S copy number of 2.3 cell−1 for freshwater communities and 3 cell−1 for Legionella spp. (Equation 1) with 16Scom 16SLeg and 16Scom16SLeg being the numbers of 16S copies measured by qPCR of Com and Legionella spp., respectively. The value for Legionella spp. cells per litre (NLeg) in each sample was obtained by multiplying total bacterial cell counts per litre (TCC) by the ratio RLeg, assuming further an equal extraction efficiency for the bacterial community and the Legionella community (Equation 2).
Results
Total bacterial cell counts and environmental conditions
During the treatment process, total cell numbers were reduced by about 75% due to flocculation and sand filtration (see decrease from samples GR1 to GR2 and samples ER1 to ER2 in Supplementary Figure S2). This reduction in cell numbers did not affect the community structure as demonstrated previously (Eichler et al., 2006). After chlorination, the cell counts increased to about a level that can be considered typical for finished drinking water, that is, ranging from 2.0 × 108 to 4.0 × 108 cells l−1. This increase was accompanied by a change in the active bacterial community as indicated by RNA-based fingerprints (Eichler et al., 2006).
Seasonal samples of cold and hot drinking water have been analysed previously for their total bacterial communities (Henne et al., 2013). For a comprehensive understanding of the environmental conditions of these samples, the conditions are reiterated here. Monthly samples of cold drinking water comprised 18 samples from May 2008 to October 2009. The mean temperature of the cold water was 12.5 °C and varied, depending on the season, between 7.6 °C in January and 16.9 °C in September (Supplementary Figure S1A). The pH of the cold drinking water varied between 8.2 and 8.8 with a mean of 8.5. During the sampling period, no chlorine was detected (detection limit of 0.1 mg l−1) and conductivity had a mean of 142.8±7.5 μS cm−1. In parallel, hot drinking water was sampled 15 times from September 2008 onwards. The water entered the circular hot water system with 60 °C yet, its temperature at the sampling point fluctuated between 49.2 °C and 57.9 °C with an annual mean of 53.1 °C (Supplementary Figure S1B); the pH varied between 7.9 and 8.5 (mean of 8.3); the mean conductivity was 146.4±8.3 μS cm−1. The number of total bacterial counts of the cold drinking water varied between 0.8 × 108 and 4.8 × 108 cells l−1 with a mean of 2.4 × 108 cells l−1 showing no clear seasonal trend, that is, no maxima or minima with respect to seasons. The dynamics of the bacterial cell counts in hot water followed the cold water dynamics at a lower level (on average by 20%) (Henne et al., 2013).
Effects of treatment and source water on the distribution of Legionella species in drinking water
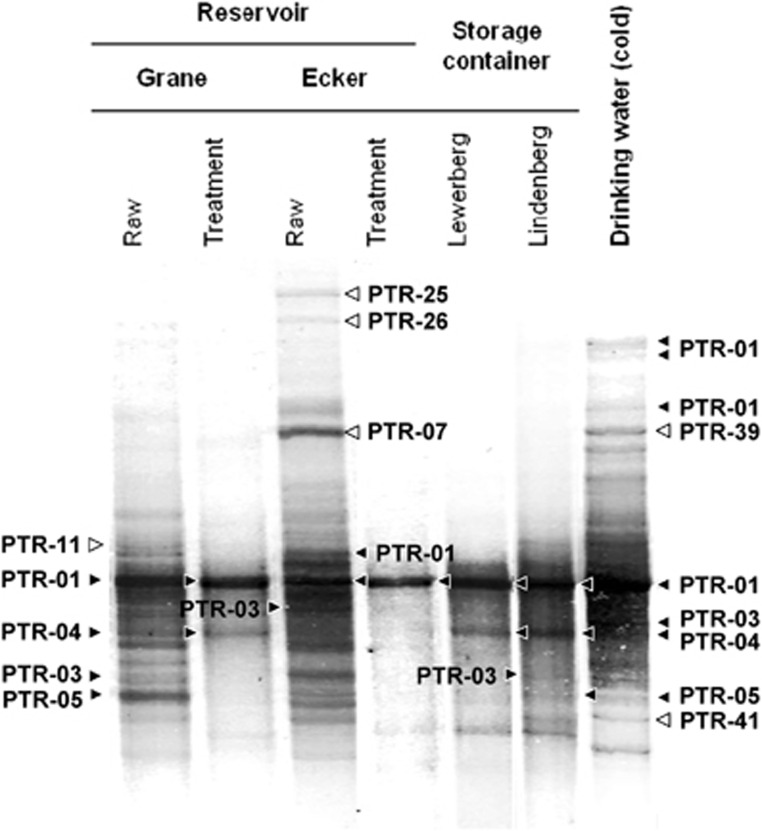
SSCP fingerprint analysis of Legionella genus-specific amplicons was done on seven bulk water samples of the DWSS of the city of Braunschweig to understand the effects of treatment on the Legionella species composition in drinking water (Figure 1). Fingerprints of raw water from the two drinking water reservoirs (Grane and Ecker reservoir) displayed complex and distinct patterns, indicating a unique composition of diverse Legionella phylotypes in the different raw waters. Complexity of fingerprints of both raw waters was markedly reduced after treatment, recovered in the storage containers (Lewerberg and Lindenberg) and was eventually completely restored to higher levels in the distribution system. Sequence analysis of excised bands confirmed the high diversity of Legionella spp. phylotypes and uniqueness of Legionella communities in the reservoirs and the drinking water at the HZI (Supplementary Table S1). Overall, we identified more than 40 environmental phylotypes of which only two were non-Legionella species (see Figure 2 and asterisks in Supplementary Table S1). These sequencing results demonstrated the validity and specificity of the selected primers for the molecular assessment of Legionella species in freshwater environments.
Figure 1.
Effect of raw water treatment by chlorination and drinking water distribution on Legionella communities as observed by genus-specific and 16S rRNA gene-based SSCP fingerprints. Distinct bands representing PTRs are indicated by filled arrow heads when found in several points along the DWSS and open arrow heads indicate PTRs detected solely in the marked samples. Details of all PTRs are summarized in Supplementary Table S1.
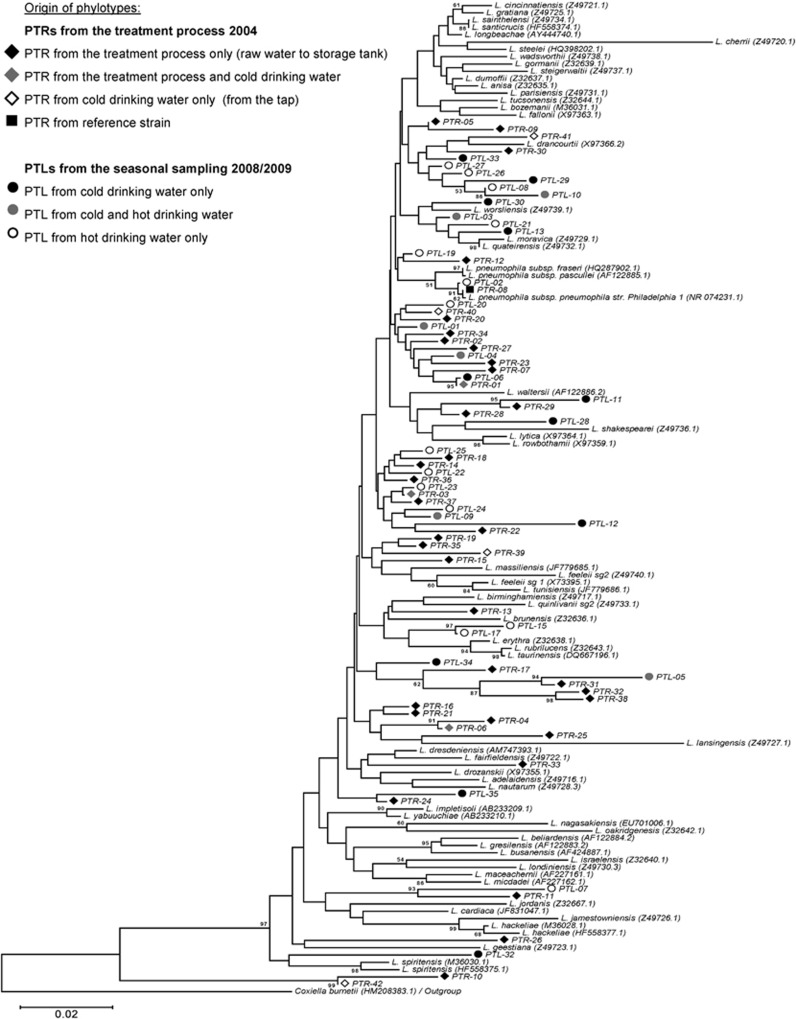
Figure 2.
Phylogenetic comparison 16S rRNA gene sequences of Legionella reference species and environmental phylotypes (PTLs, fragment size 376 bp) obtained in this study. For individual PTL designation, see Supplementary Tables S1 and S2. The phylogenetic tree compares the phylotypes obtained from the treatment process (PTRs=diamonds) with the ones from the seasonal sampling of finished drinking water (PTLs=dots). Scale bar indicates the base substitutions for each position and Coxiella burnetii was used as an outgroup.
Phylogenetic analysis of the 16S rRNA gene sequences (Figure 2) revealed the following: (i) a certain phylotype dominated the communities throughout all treatment steps (PTR-01); (ii) some phylotypes from both reservoirs re-emerged at the end of the distribution system (PTR-04 and -05) whereas some others vanished completely (PTR-11, -25 and -26) and (iii) the finished drinking water harboured phylotypes that were not found in either of the reservoirs (PTR-39 and -41) (Supplementary Table S1).
Some of the closest cultured neighbours of the phylotypes found in the raw water samples were related to pathogenic Legionella species according to the Seqmatch results (Supplementary Table S1,Figure 2). In the two raw waters only phylotypes related to L. dumoffii (PTR-01) were in common, whereas PTR-13 (L. erythra), -18 (L. drancourtii) and -11 (L. impletisoli) only occurred in raw water from the Grane reservoir and PTR-36 (L. tucsonensis) and -34 (L. longbeachae) were detected only in the Ecker reservoir. Relative to the other pathogenic Legionella species, L. longbeachae from the Grane reservoir occurred in several bands (PTR-03) in the finished drinking water (Figure 1). The majority of the PTRs listed in Supplementary Table S1 were distributed across the whole phylogenetic diversity of the Legionella genus as shown in Figure 2.
Seasonal dynamics of Legionella species in cold drinking water
A set of 18 cold drinking water samples, taken in monthly intervals, was analysed by SSCP fingerprints (Supplementary Figure S3A). For a better understanding of the banding patterns on the SSCP gels, a cluster analysis was performed using Pearson correlations (Supplementary Figure S4A). Single seasonal clusters occurred with durations of 3–4 months if an overall similarity of >70% was used.
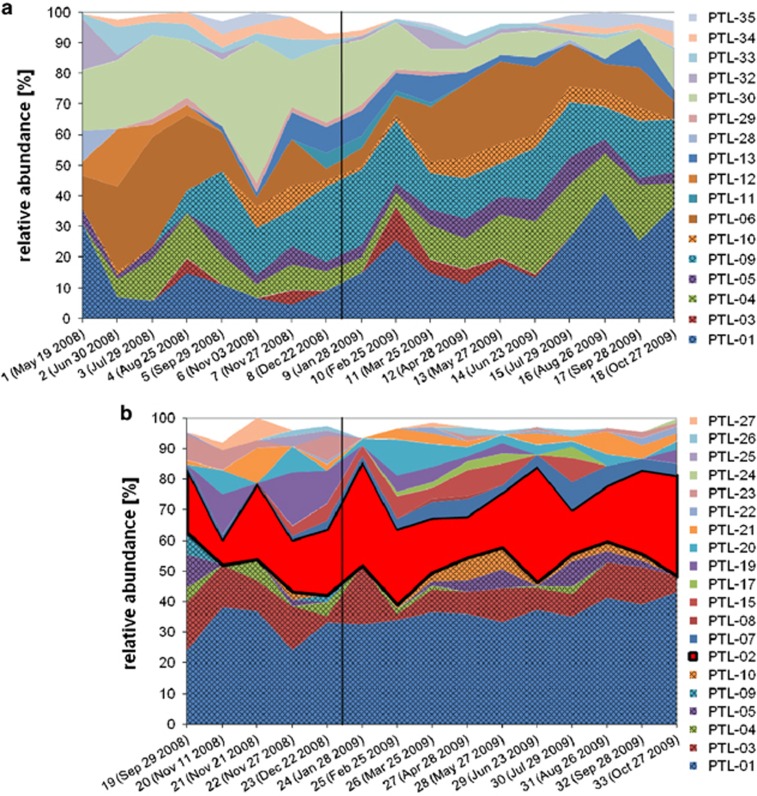
The abundances of individual PTLs were quantified with the help of GelCompare using the relative intensity of individual bands (Figure 3a). The seasonal dynamics of the relative abundances of Legionella phylotypes defined at about the species level showed 8–14 different PTLs (average=11±1.4) per sample in cold drinking water with abundances ranging from >1% to 53% (Figure 3a). This overall species richness for Legionella, that is, the number of phylotypes above a certain threshold per sample (here: 1.0%), did not show a significant seasonal pattern in synchrony with temperature, precipitation or total bacterial cell counts. Only PTL-01, -06 and -30 were observed to be present above this threshold each month. Although PTL-30 consistently accounted for a major portion (20–50%) of all Legionella phylotypes, changes in abundance of PTL-06 (3–40%) were more pronounced with minima in winter 2008/09 and October 2009 and maxima in July 2008 and May/June 2009. Interestingly, these two major PTLs (-06 and -30) were only detected in cold drinking water. Six other PTLs (-01, -03, -04, -05, -09 and -10) were also found in the hot drinking water during the sampling period (marked with gridded areas in Figures 3a and b and grey dots in Figure 2). Of those, one (PTL-09) emerged in August 2008 and became a major part of the cold drinking water Legionella community, ranging from 7.5% to 24.5% in relative abundance. Only one sequence (PTL-02.5, Figure 4) was identical to L. pneumophila but, it was only observed in a single faint band that occurred in January 2008.
Figure 3.
Overview of the seasonal dynamics of Legionella species communities in cold (a) and hot (b) drinking water based on the relative abundances of the individual PTLs. For individual PTL designation, see Supplementary Table S2. Only six PTLs occurred in both systems (PTL-01, -03, -04, -05, -09 and -10, gridded areas) accounting for a fraction of approximately 50% in each community. PTL-02, a phylotype identical in sequence with the most pathogenic species L. pneumophila (red fraction in b), was only detected above the limit of relative abundance of 1% in the hot water community where it constantly constituted a substantial fraction (22.15±7.94%).
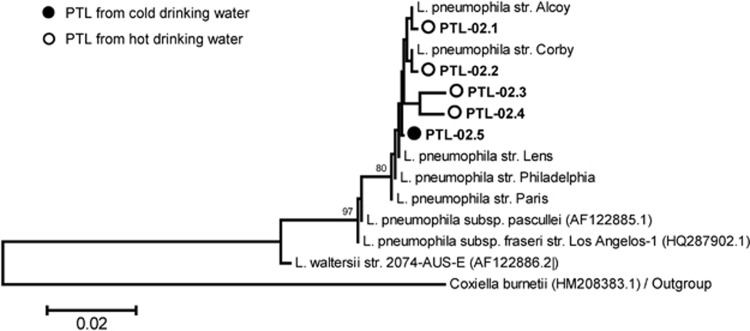
Figure 4.
Phylogenetic comparison 16S rRNA gene sequences from L. pneumophila reference strains and from the environmental phylotype PTL-02, the PTL most similar to L. pneumophila. The phylotype is represented by five sample-specific sequences, designated PTL-02.1 to -02.5, obtained from cold (filled circles) and hot (open circles) drinking water. Scale bar indicates the base substitutions for each position.
A detailed analysis of partial 16S rRNA gene sequences resulted in a total of 19 Legionella phylotypes in cold water summarized in Supplementary Table S2. The most relevant Legionella species concerning human health is L. pneumophila and therefore, we compared our environmental sequences in detail with the best-known L. pneumophila strains and subspecies (Figure 4). The amplicon of a size of 376 bp used in our analyses includes three positions that can vary intragenomically in the genomes of the L. pneumophila strains Philadelphia, Alcoy, Corby, Lens and Paris (positions 187, 261 and 284 of the amplicon). Therefore, the 16S rRNA gene fragment used allowed distinguishing these strains from the two subspecies, L. pneumophila fraseri and pasculleri.
Seasonal dynamics of Legionella species in hot drinking water
Fifteen samples from hot drinking water were analysed by SSCP fingerprints (Supplementary Figure S3B). The cluster analysis revealed only three clusters ranging from 3 to 8 months (Supplementary Figure S4B). Several sequences of strong SSCP bands present in every hot water sample were identical to L. pneumophila and defined as PTL-02.
In hot drinking water, we observed 6–15 PTLs (average=10.4±2.5) per sample with abundances ranging from >1 to 40% (Figure 3b). PTL-01, -02 and -03 represented a stable core community, which was present each month in a similar composition. These three PTLs comprised always >50% of all phylotypes ranging in summary from 50% to 82% of the total abundances. Strikingly, the PTL-02 (red area in Figure 3b) representing L. pneumophila-like sequences (PTL-02.1 to PTL-02.4, Figure 4) consistently accounted for 8–40% of all Legionella phylotypes in the hot drinking water with an exceptional minimum of 8% in November 2008 and maxima of 40% and 38% in January and June 2009, respectively. In cold drinking water, PTL-02 (sequence PTL-02.5) was observed only at one sampling date (28th of January 2008) and seemed to be slightly different from the PTL-02 sequences derived from hot water (Figure 4). PTL-01, however, displayed a similar abundance as in cold water varying from 24.2% to 44.7% in hot water (compared with 3.5–30%). Remarkably, common PTLs found in both, cold and hot water systems (marked with grey dots in Figure 2 and gridded areas in Figures 3a and b), were generally accounting for 50% of the Legionella communities but this ratio varied more drastically in cold than in hot water.
In hot water, the detailed analysis of the 16S rRNA gene sequences resulted in a total of 24 Legionella phylotypes (Supplementary Table S2). Six of the hot water PTLs were in common with cold water and one PTL (-02) clustered within L. pneumophila (Figures 3 and 4). Based on these high sequence similarities, we think that both types of finished drinking water contained bacteria belonging to the species L. pneumophila, but they are substantially more abundant in hot than in cold drinking water.
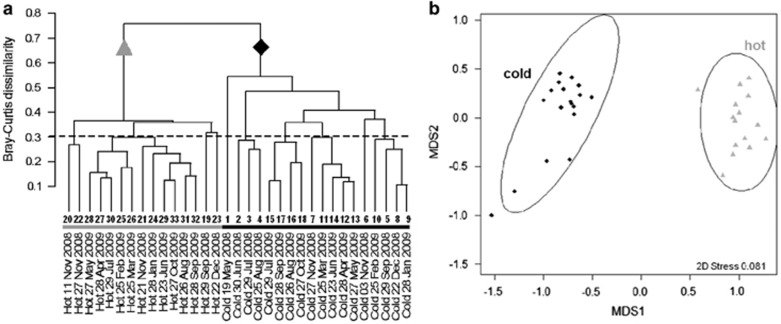
Comparison of Legionella community compositions in hot and cold drinking water was done by detailed cluster analysis (Figure 5). These analyses confirmed the general pattern of the fingerprints derived from SSCP gels. Cold water communities were substantially different from the hot water communities as demonstrated by the dendrogram using a Bray-Curtis distance matrix of untransformed abundance values (Figure 5a). Ordination by non-metric multidimensional scaling (NMDS) of PTL abundance and phylogenetic distance between phylotypes showed, like the SSCP fingerprints, more and smaller clusters in cold than in hot water communities when applying a similarity threshold of 70% (Figure 5b).
Figure 5.
Comparison of Legionella community composition in hot and cold drinking water by using different statistical approaches. (a) Dendrogram based on a Bray-Curtis distance matrix of untransformed abundance values of individual PTLs as shown in Figure 3. Cold water communities (black diamonds) are substantially different from the hot water communities (grey triangles). (b) Ordination plot obtained by NMDS of PTL abundance and phylogenetic distances between phylotypes (here: mean nearest taxon distance, mntd, see Materials and methods).
Comparison of Legionella community with total bacterial community
The Legionella community comprised on average 0.67% (±0.58) of the cold water bacterial community corresponding to an average cell number of 1.7 × 106 cells l−1 (Supplementary Figure S6). These genus-specific cell numbers were based on quantification by real-time PCR with primers targeting the genus Legionella and the total bacterial cell number of the water sample as detailed in Materials and methods. The Legionella community of the hot water comprised a significantly larger fraction, that is, on average 2.4% (±1.5), corresponding to 3.8 × 106 cells l−1. These numbers indicated a substantial increase of the Legionella fraction of the total bacterial community with respect to the relative abundance and the absolute cell numbers from cold to hot water.
Rank-abundance curves of Legionella communities from cold and hot water did not differ substantially and variation for both Legionella regression analyses was high as indicated by their relatively low coefficients for determination (Supplementary Figure S5). Additionally, mean Legionella species richness per sample did not significantly differ between both environments (Table 1) although total Legionella species richness over the whole sampling period was slightly higher in hot water (cold water: n=17, hot water: n=20). Despite these similarities of mean species richness, both environments differed in other metrics of alpha diversity. Shannon and (inverse) Simpson index of the Legionella communities were significantly higher for cold water samples (Table 1) as observed also for the bacterial communities. Common indices for beta diversity and evenness coherently showed significantly higher values for the Legionella community in cold water, which is in line with its more dynamic community structure (Table 1, Figure 3) (Henne et al., 2013). Phylogenetic distance (pd) was slightly higher for cold water Legionella, and also the negative values for mean nearest taxon distance (mntd) and mean pairwise distance (mpd) indicated that PTLs in cold water were more distantly related to each other (Table 1) (Webb, 2000). In other words, hot water PTLs were clustering closer to each other.
Table 1. Biodiversity indices of the Legionella communities and whole bacterial communities in hot and cold drinking water (values displaying significant differences are given in bold; P-value⩽0.05).
|
Legionella community |
Bacterial community |
|||||
|---|---|---|---|---|---|---|
| Cold mean (s.d.) | Hot mean (s.d.) | P-value | Cold mean (s.d.) | Hot mean (s.d.) | P-value | |
| Alpha diversitya | ||||||
| Observed species richness | 11.00 (±1.37) | 10.40 (±2.47) | 0.104 | 18.72 (±1.32) | 9.27 (±1.49) | <0.001 |
| Shannon index | 2.02 (±0.17) | 1.81 (±0.24) | 0.009 | 2.43 (±0.16) | 1.56 (±0.14) | <0.001 |
| Simpson index | 0.83 (±0.04) | 0.77 (±0.05) | 0.002 | 0.88 (±0.03) | 0.73 (±0.04) | <0.001 |
| Inverse Simpson index | 6.12 (±1.48) | 4.62 (±0.97) | 0.001 | 8.88 (±2.09) | 3.82 (±0.53) | <0.001 |
| DHill | 9.33 (±1.26) | 8.33 (±1.99) | 0.102 | 14.68 (±1.50) | 6.30 (±0.94) | <0.001 |
| Evennessa | ||||||
| Shannon-based | 0.84 (±0.05) | 0.78 (±0.06) | 0.003 | 0.83 (±0.05) | 0.71 (±0.06) | <0.001 |
| Inverse Simpson-based | 0.56 (±0.11) | 0.46 (±0.10) | 0.008 | 0.47 (±0.10) | 0.42 (±0.07) | 0.078 |
| DHill | 0.85 (±0.04) | 0.80 (±0.05) | 0.009 | 0.78 (±0.05) | 0.68 (±0.07) | <0.001 |
| Phylogenetic diversity | ||||||
| Faith's phylogenetic distance (pd) | 0.24 (±0.04) | 0.19 (±0.05) | 0.010 | 1.34 (±0.12) | 1.10 (±0.07) | <0.001 |
| Mean phylogenetic distance (mpd) | 0.01 (±0.73) | −0.53 (±0.91) | 0.073 | −1.11 (±0.75) | 1.48 (±0.49) | <0.001 |
| Mean nearest taxon distance (mntd) | 0.10 (±0.60) | −0.94 (±0.71) | <0.001 | −2.15 (±0.42) | 0.77 (±0.52) | <0.001 |
| Beta diversity a | ||||||
| Sørensens index | 0.76 (±0.11) | 0.66 (±0.11) | <0.001 | 0.92 (±0.04) | 0.84 (±0.07) | <0.001 |
| Jaccard index | 0.68 (±0.15) | 0.51 (±0.13) | <0.001 | 0.85 (±0.06) | 0.73 (±0.10) | <0.001 |
| Bray-Curtis dissimilarity | 0.59 (±0.12) | 0.69 (±0.08) | <0.001 | 0.68 (±0.10) | 0.78 (±0.10) | <0.001 |
Indices were calculated for communities of single samples and average values are given; indices for the whole site differ since species accumulate over extended periods of sampling.
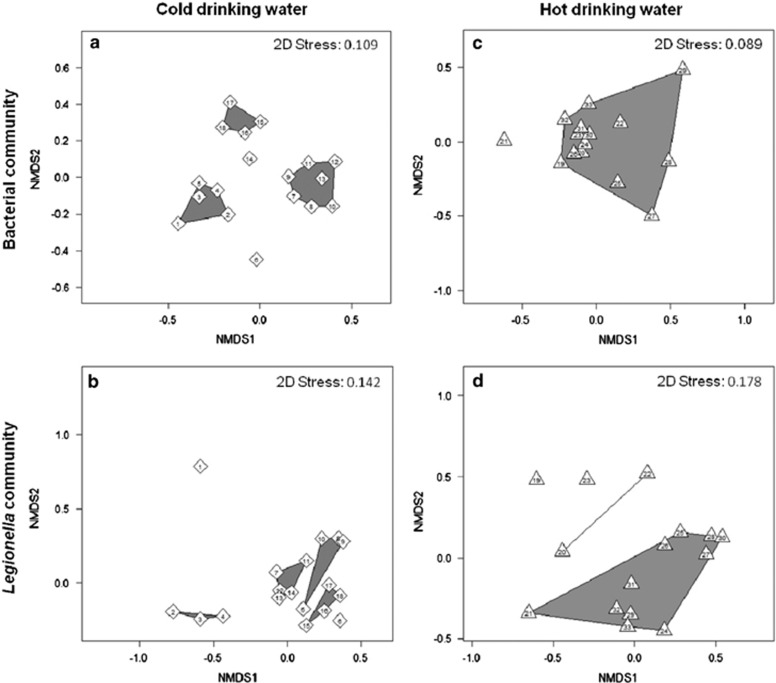
Taking phylogenetic relationships and relative abundances of single phylotypes into account NMDS ordination revealed dynamics of bacterial and Legionella communities (Figure 6). For better illustration of clusters, ordination plots are shown separately for both types of drinking water since their communities shared only few phylotypes. Cold water bacterial communities clustered into three groups of four to seven sampling dates (Figure 6a). Clusters were aggregations of subsequent dates with samples 6 and 14 being outliers. These outliers had been identified as turning points between winter and summer phases of the total bacterial community (Henne et al., 2013). Similarly, Legionella communities from the same samples clustered into four groups of three to four sampling dates with two outliers (Figure 6b). Although clusters contained mainly subsequent dates, this pattern was not as strictly followed here, while samples 1 and 6 were outliers. Clusters were mainly characterized by the constant decrease in the abundance of PTL-30 and the constant increase in PTL-09 during the sampling period (Figure 3a).
Figure 6.
Comparison of the whole bacterial communities and Legionella communities using NMDS ordination plots. In cold water (diamonds), both the bacterial communities (a) and the Legionella communities (b) are divided into three to four clusters consisting of three to seven successive dates. In hot water (triangles), the bacterial communities (c) and the Legionella communities (d) cluster mainly in one large single group. Similarity thresholds of the clusters are 75% and 70% for whole bacterial and Legionella communities, respectively. Numbers of the single data points represent the sampling dates indicated in Figure 3 at the x axes.
Clusters of hot water samples looked very different from the cold water clusters. Concerning the bacterial communities all sample dates clustered into one single group, except for one outlier (Figure 6c). This reflected the rather stable community composition dominated by single phylotypes from five different taxonomic groups (Henne et al., 2013). Similarly, most of the Legionella communities from hot water clustered into one single group, but here two outliers were identified next to one smaller additional cluster of samples 20 and 22 (Figure 6d). This clustering also reflected a more constant species composition, mainly due to steadily high abundances of the major PTLs (Figure 3b). Overall, dynamics of Legionella communities in drinking water behave similar to the one of the most abundant bacterial phylotypes as deduced from diversity indices and clustering patterns in cold and hot drinking water. However, differences in the sample composition of single clusters and phylogenetic structure could indicate a selection pressure specific for Legionella species.
Relationship of Legionella abundances with temperature regime in cold and hot water
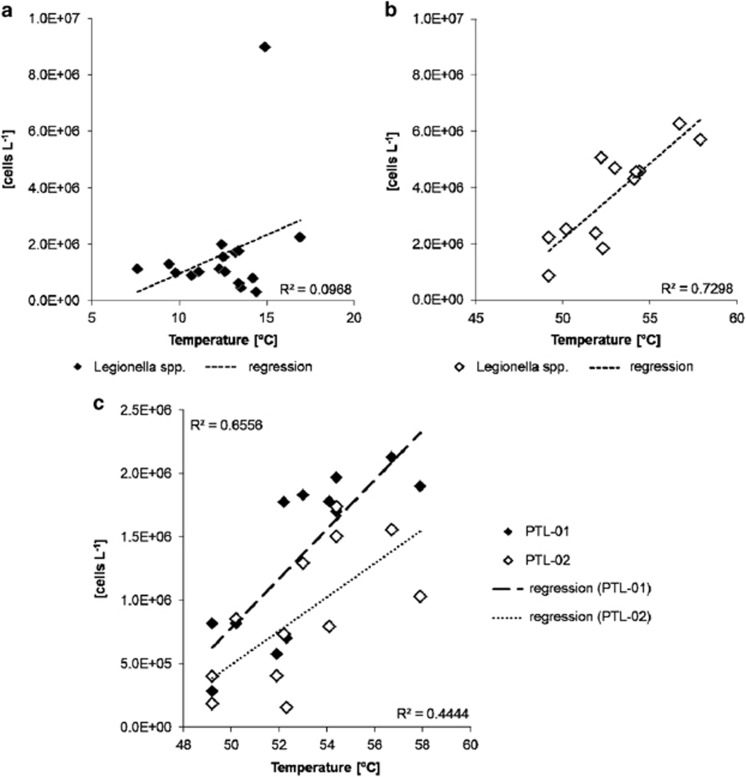
Since the temperature regime is an important controlling factor of Legionella growth and survival, the abundance of the Legionella cell numbers per litre was compared with the temperature observed in the respective sample. For cold water, the Legionella cell numbers did not show a clear trend within the range of the observed in situ temperatures from 7.6 °C to 16.9 °C (Figure 7a). In addition, specific cell numbers of single phylotypes were calculated using the fraction of the respective phylotype derived from SSCP analyses and the respective total Legionella cell number of a given sample. For single phylotypes of the total Legionella community also no correlation to temperature was observed (Supplementary Table S3). By contrast, for hot water, there was a significant increase in the total Legionella abundances from 49.2 °C to 57.9 °C by a factor of about three (Figure 7b). A positive response of cell numbers to temperature was also observed for the two most abundant phylotypes (Supplementary Table S3). This increase with temperature is shown in detail for the most abundant phylotypes PTL-01 and PTL-02 closely affiliated with L. longbeachae and L. pneumophila, respectively (Figure 7c). For these phylotypes, an increase in cell numbers by an order of magnitude from 49.2 °C to 57.9 °C was observed, indicating a preference for these high temperatures and the thermophilic nature of these phylotypes.
Figure 7.
Relationship between abundance of Legionella spp. cells and drinking water temperature as determined by real-time PCR. (a) In cold drinking water, Legionella cell numbers did not show any dependence on water temperature (R2=0.097). (b) In hot water, cell numbers of Legionella spp. increased with higher temperatures (R2=0.730). (c) Relationship between abundance of the two major Legionella phylotypes in hot drinking water (PTL-01 and PTL-02) and drinking water temperature as calculated from their relative abundances in the SSCP fingerprints. Phylotype PTL-02 represents L. pneumophila.
Discussion
Identification and quantification of individual Legionella species by fingerprinting
The taxonomic resolution and the affiliation of the single 16S rRNA sequences with cultured Legionella species has to be considered to understand the relevance of the obtained results for the ecology of this genus. SSCP fingerprints provide phylogenetic and abundance information of single taxa using partial 16S rRNA sequence data and relative peak intensities of individual bands of the fingerprint. Our consensus phylotypes, summarized in Supplementary Tables S1 and S2, can be considered to cover a broad set of different species of the genus Legionella. Figure 2 provides an overview about the phylogenetic positions of all 76 PTLs observed in a total of 40 samples from three different years of the studied DWSS and demonstrates the enormous phylogenetic breadth and depth of the Legionella species present. This finding corroborates with other studies of the environmental occurrence of Legionella species in freshwater environments using cultivation-independent approaches (Calvo-Bado et al., 2003; Riffard et al., 2001; Brooks et al., 2004). How the observed PTLs relate to individual cultured Legionella species is only clear for L. pneumophila (Figure 4). The other PTLs could be divided into two different types: (i) PTLs closely related to cultured species (for example, PTR-41 and PTL-13) and (ii) PTLs belonging to deeper branches with no phylogenetic relationship to cultured species (for example, the PTLs clustered around PTL-012 and PTL-05). The first type of PTLs might stem from strains of the closely related species and the observed sequence variation can be considered to be due to micro-heterogeneity of the 16S rRNA gene sequences as demonstrated for L. pneumophila (Figure 4). The second type of PTLs could relate to the many environmental Legionella species that have not yet been cultured and identified (Costa et al., 2005; Wullings and van der Kooij, 2006; Declerck et al., 2007a; Wullings et al., 2011).
Influence of treatment on the Legionella species composition
In our DWSS, treatment of the raw water reduced substantially the Legionella species diversity to only one resilient and dominant phylotype (PTL-01) occurring after treatment and in the storage containers (Figure 1). However, in concordance with the whole bacterial communities (Eichler et al., 2006), diversity of Legionella spp. was restored along the DWSS. A similar reduction and regrowth of Legionella spp. has been observed in other chlorinated DWSS (Whiley et al., 2014). Interestingly, the occurrence of phylotypes not detected in the raw waters suggests the selection of Legionella species well adapted to our DWSS. This selection pressure could result from low-nutrient content, presence/absence of certain host protozoa or bacterial competitors and effects of chlorination. Furthermore, invasion of Legionella species from mature biofilms of the DWSS has to be taken into consideration (Lau and Ashbolt, 2009; Abdel-Nour et al., 2013). This invasion from biofilm might be a key mechanism for the increase in Legionella species in bulk water because the ratio of biofilm surface to water volume increases on its way from the storage container to the tap at the HZI where the highest number of Legionella species occurred (Figure 1). Notably, no phylotype identical or similar to L. pneumophila could be detected in raw water and along the water treatment process. In contrast, the phylotype related to L. pneumophila (PTL-02) was only detected once in cold drinking water, but was always present at substantially higher abundances in hot drinking water (Figure 3).
Dynamics of Legionella species in cold and hot drinking water
The seasonal dynamics of bacterial community composition in cold and hot water of our DWSS have been recently described in detail at the level of the most dominant bacterial phylotypes (Henne et al., 2013). This study showed by comparing rank-abundance curves that on average the bacterial communities in cold water had a more even species composition than in hot water (see Figure 4 in Henne et al., 2013). Accordingly, total bacterial species richness was significantly higher in cold water (n=55) than in hot water (n=44) (Henne et al., 2013). Here we analysed the same set of samples but focusing only on members of the genus Legionella.
Seasonal dynamics of the Legionella community were observed neither in cold nor in hot drinking water. Statistical analyses showed a closer relatedness of Legionella communities of subsequent months. This is consistent with observations for the total bacteria community of the hot water in our previous study, while for the cold water seasonal dynamics were observed for the whole bacterial community (Henne et al., 2013).
The Legionella community composition of cold and hot drinking water showed that both systems harbour phylogenetically quite distinct communities, but 6 of the major 35 PTLs were shared (Figure 3, Supplementary Table S2). The observed minute differences between the individual sequences of these common PTLs of cold and hot water (Figure 4) could indicate that there were different strains in the different types of water which were better adapted to the respective temperature. High-resolution genotyping analyses of these water samples as performed for L. pneumophila could elucidate such differences on the strain level indicated by the high degree of micro-heterogeneity observed in the 16S rRNA sequences (Kahlisch et al., 2010).
The structure of the Legionella communities in both types of water was dissimilar as suggested by the corresponding statistical analysis (Figure 5). Such a pronounced discrepancy has also been shown for the total bacterial community of cold and hot drinking water (Henne et al., 2013). The Legionella communities of the hot water showed a reduced overall diversity (for example, Shannon index) and less phylogenetic distance among PTLs in comparison with cold water, despite comparable richness (Table 1, Supplementary Figure S5). The dynamics of the Legionella communities of both types of water were also substantially different as revealed by the NMDS ordination plots (Figures 6b and d). These differences in the community dynamics could stem from the different environmental conditions and selection pressure and their seasonal variability in the two types of water.
Transformation of the cold water community to a hot water community favouring thermophilic Legionella species
Hot water production in our DWSS is a rather rapid process: hot water returning from the distribution system and cold water are co-heated at about 63 °C for about 1 h. During this short time span, the cold water bacterial community decays by about 95% thereby providing nutrients for regrowth (Henne et al., 2013). Heating-induced changes of the Legionella community were well pronounced, but still less substantial than those observed for the whole bacterial community where different thermotolerant and thermophilic species dominated the overall hot water community (Henne et al., 2013). This indicates an intrinsic potential to adapt to a wide range of temperature regimes within the genus Legionella and we assume that thermophilic phylotypes have an important role for the following reasons. First, the total abundance of members of the genus Legionella increased relatively to the total bacterial community but also in absolute terms from 1.7 × 106 to 3.8 × 106 cells l−1 from cold to hot water (Supplementary Figure S6). Second, the cell numbers for the genus Legionella increased by a factor of 3 along the temperature range observed for the analysed samples from 49.2 °C to 57.9 °C (Figure 7b). Third, this positive correlation with temperature was confirmed among others (PTL-21, -22 and -24, data not shown) for the most abundant phylotypes PTL-01 and PTL-02 that were most closely affiliated with L. longbeachae and L. pneumophila, respectively (Figure 7c, Supplementary Table S3). In contrast, no clear temperature effect was seen for the cold water Legionella community. Thus we could demonstrate that a set of Legionella species, including L. pneumophila, were able to grow at temperatures above 50 °C, and survive or grow until 63 °C.
Currently, no Legionella species are known that are able to grow at temperature above 50 °C (Stout et al., 1986; Steinert et al., 1998; Vervaeren et al., 2006). Especially L. pneumophila and L. longbeachae have been extensively studied, but although numerous isolates have been obtained among others from hydrothermal springs and spas at temperatures between 40 °C and 60 °C, indicating thermotolerant characteristics of Legionella species, to our knowledge growth above 45 °C has not been documented until now (Veríssimo et al., 1991; Ghrairi et al., 2013). Better survival and viability traits have been described for L. pneumophila strains isolated from a shock heated (70 °C for 30 min) hospital DWSS and a Tunisian therapeutic spa, but growth above 45 °C was not tested (Allegra et al., 2011; Ghrairi et al., 2013). Adaptations, spontaneous mutations or horizontal gene transfer from thermophilic Legionella species or other thermophilic bacteria of the hot water community could have supported this development (Lindsay, 1995). It also appeared that thermophilic traits could readily be gained and lost as shown for Bacillus species (Hobbs et al., 2012). High levels of horizontal gene transfer have been shown for members of the genus Legionella within species and at the genus level by inter- and intra-species genome comparisons rendering it a conceivable means for acquisition of thermophilic traits (Gomez-Valero et al., 2011, 2014). Therefore, the development of thermophilic genotypes could be considered for L. pneumophila but also for other Legionella species.
Potential public health relevance of thermophilic L. pneumophila
Though the whole genus of Legionella is considered as having a potential to cause disease, only a very restricted set of species was shown to cause major health problems with L. pneumophila and L. longbeachae being the most important candidates (Phin et al., 2014; Mercante and Winchell, 2015). In this respect, the hot water system could be considered as a higher health risk than the cold water due to the relatively high abundance of L. pneumophila PTL-02. The use of hot water is in general perceived as a potential public health risk (Borella et al., 2005b; Leoni et al., 2005; Mouchtouri et al., 2007; Phin et al., 2014). However, all these hot water studies did not assume the growth of pathogenic Legionella species in hot water systems above 50 °C, though some of their observations may indicate it (Borella et al., 2005b; Leoni et al., 2005).
The general perception of a reduced risk of legionellosis when water is continuously heated between 50 °C and 60 °C may suggest that thermophilic Legionella do not pose a major health threat. However, it cannot be ruled out that the thermophilic genotypes of pathogenic species, like L. pneumophila, as demonstrated for our hot water systems are of relevance for public health. Therefore, the aspect of high temperature adapted genotypes may deserve further detailed studies with respect to their clonal nature, physiology and virulence traits including the attempt to isolate these genotypes.
Only a restricted set of genotypes of L. pneumophila is virulent and able to cause disease (Sobral et al., 2011; Khan et al., 2013; Phin et al., 2014). On the other hand, DWSS can be considered as highly diverse ecosystems providing a wealth of niches for selection and potentially adaptation or even evolution of specific genotypes as shown by the genotype pattern in large and small water supply systems (Sobral et al., 2011; Khan et al., 2013; Rodríguez-Martínez et al., 2015). Specific genotypes may get established in DWSS that show distinct ecological preferences, for example, for temperature, and a distinct abundance as shown by Rodríguez-Martínez et al. (2015). The relevance of the genotypes for health can only be finally assessed by isolate-based studies in macrophage assays (Epalle et al., 2015; Mercante and Winchell, 2015). To analyse the genotype distribution in a DWSS, high-resolution genotyping methods have to be applied to isolates and/or environmental DNA (Gaia et al., 2005; Ratzow et al., 2007; Kahlisch et al., 2010; Sobral et al., 2011). Strain-based investigations applying genotyping, physiological and virulence assays accompanying the molecular study may link the Legionella diversity to the genotype pattern of L. pneumophila, enabling assessment of their ecotype and potential health risk.
In conclusion, we demonstrated in our DWSS that the water treatment of two raw waters reduced the complexity of their Legionella communities' strongly but it did not fully eradicate the genus. Consequently, Legionella communities gradually re-established complexity along the DWSS resulting in a changed species composition better adapted to the respective conditions. Interestingly, Legionella community dynamics did not display a true seasonal trend yet, its species composition changed gradually due to factors that could not yet be fully identified. Although relative abundance of Legionella spp. stayed below 1% of the total bacterial community, cell numbers of more than 1.5 × 106 cells l−1 were reached exceeding the currently tolerated levels of Legionella colony forming units per litre by two orders of magnitude. Yet, both methods cannot provide a true risk assessment due to the complex lifestyle of Legionella spp. (Robertson et al., 2014). During their parasitic and planktonical life phases, Legionella cells pass through a variety of developmental stages and not all of them allow the bacteria to be cultured on agar plates (for example, VBNC states) or to be infectious. Molecular analyses on the other hand detect theoretically all target genes above a detection limit, but do not discriminate between live and dead cells. The heating to 63 °C further altered the drinking water Legionella community substantially. As for the whole bacterial community heating caused a shift to a thermophilic species composition and up to a threefold increase in Legionella cell numbers. Our findings document an adaptation of the community to the elevated temperatures and active growth of Legionella spp. at temperatures above 50 °C. Notably, cell numbers of L. pneumophila phylotypes increased with temperature, possibly a major concern for drinking water safety and public health. These thermophilic L. pneumophila populations deserve extensive future research using the full array of cellular and molecular methods available in microbiology.
Acknowledgments
This work was supported by funds from the Deutsche Forschungsgemeinschaft (DFG-project HO-930/5-1) and the EU project (No. 311846) AQUAVALENS. We thank Julia Strömpl, Josefin Koch and Verena Maiberg for their skilful technical support. Axel Mohr and Monika Pape are acknowledged for support with the technical devices of the DWSS of the HZI. Karsten Henne and Leila Kahlisch are acknowledged for valuable support with respect to total bacterial cell counts, primer design and SSCP analysis. We are very grateful to the constructive comments of two anonymous reviewers.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abdel-Nour M, Duncan C, Low DE, Guyard C. (2013). Biofilms: the stronghold of Legionella pneumophila. Int J Mol Sci 14: 21660–21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra S, Grattard F, Girardot F, Riffard S, Pozzetto B, Berthelot P. (2011). Longitudinal evaluation of the efficacy of heat treatment procedures against Legionella spp. in Hospital water systems by using a flow cytometric assay. Appl Environ Microbiol 77: 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram J, Chartier Y, Lee JV, Pond K, Surman-Lee S. (eds). (2007) Legionella and the Prevention of Legionellosis. WHO: Geneva. [Google Scholar]
- Bassam BJ, Caetano-Anollés G, Gresshoff PM. (1991). Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196: 80–83. [DOI] [PubMed] [Google Scholar]
- Beauté J, Zucs P, de Jong B, European Legionnaires' Disease Surveillance Network (2013). Legionnaires' disease in Europe, 2009-2010. Euro Surveill 18: 20417. [DOI] [PubMed] [Google Scholar]
- Borella P, Guerrieri E, Marchesi I, Bondi M, Messi P. (2005. a). Water ecology of Legionella and protozoan: environmental and public health perspectives. El-Gewely MR (ed). Biotechnology Annual Review, (Vol. 11)Elsevier: Amsterdam, Netherlands, pp 355–380. [DOI] [PubMed] [Google Scholar]
- Borella P, Montagna MT, Stampi S, Stancanelli G, Romano-Spica V, Triassi M et al. (2005. b). Legionella contamination in hot water of Italian hotels. Appl Environ Microbiol 71: 5805–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks T, Osicki RA, Springthorpe VS, Sattar SA, Filion L, Abrial D et al. (2004). Detection and identification of Legionella species from groundwaters. J Toxicol Environ Health A 67: 1845–1859. [DOI] [PubMed] [Google Scholar]
- Calvo-Bado LA, Morgan JAW, Sergeant M, Pettitt TR, Whipps JM. (2003). Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops. Appl Environ Microbiol 69: 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati S, Conza L, Bruin J, Gaia V. (2010). Compost facilities as a reservoir of Legionella pneumophila and other Legionella species. Clin Microbiol Infect 16: 945–947. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42: D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J, Tiago I, da Costa MS, Veríssimo A. (2005). Presence and persistence of Legionella spp. in groundwater. Appl Environ Microbiol 71: 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie SL, Beattie TK, Knapp CW, Lindsay DSJ. (2014). Legionella spp. in UK composts—a potential public health issue? Clin Microbiol Infect 20: O224–O229. [DOI] [PubMed] [Google Scholar]
- Declerck P, Behets J, van Hoef V, Ollevier F. (2007. a). Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res 41: 3159–3167. [DOI] [PubMed] [Google Scholar]
- Declerck P, Behets J, van Hoef V, Ollevier F. (2007. b). Replication of Legionella pneumophila in floating biofilms. Curr Microbiol 55: 435–440. [DOI] [PubMed] [Google Scholar]
- Dupuy M, Mazoua S, Berne F, Bodet C, Garrec N, Herbelin P et al. (2011). Efficiency of water disinfectants against Legionella pneumophila and Acanthamoeba. Water Res 45: 1087–1094. [DOI] [PubMed] [Google Scholar]
- Eichler S, Christen R, Höltje C, Westphal P, Bötel J, Brettar I et al. (2006). Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl Environ Microbiol 72: 1858–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epalle T, Girardot F, Allegra S, Maurice-Blanc C, Garraud O, Riffard S. (2015). Viable but not culturable forms of Legionella pneumophila generated after heat shock treatment are infectious for macrophage-like and alveolar epithelial cells after resuscitation on Acanthamoeba polyphaga. Microb Ecol 69: 215–224. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (2015). Legionnaires' Disease in Europe, 2013. ECDC: Stockholm. [Google Scholar]
- Fields BS, Benson RF, Besser RE. (2002). Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS. (1996). The molecular ecology of legionellae. Trends Microbiol 4: 286–290. [DOI] [PubMed] [Google Scholar]
- Gaia V, Fry NK, Afshar B, Lück PC, Meugnier H, Etienne J et al. (2005). Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol 43: 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghrairi T, Chaftar N, Jarraud S, Berjeaud JM, Hani K, Frere J. (2013). Diversity of legionellae strains from Tunisian hot spring water. Res Microbiol 164: 342–350. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Jarraud S, Vacherie B, Rouy Z, Barbe V et al. (2011). Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Rolando M, Neou M, Dervins-Ravault D, Demirtas J et al. (2014). Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires' disease. Genome Biol 15: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne K, Kahlisch L, Brettar I, Höfle MG. (2012). Analysis of structure and composition of bacterial core communities in mature drinking water biofilms and bulk water of a citywide network in Germany. Appl Environ Microbiol 78: 3530–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne K, Kahlisch L, Höfle MG, Brettar I. (2013). Seasonal dynamics of bacterial community structure and composition in cold and hot drinking water derived from surface water reservoirs. Water Res 47: 5614–5630. [DOI] [PubMed] [Google Scholar]
- Hobbs JK, Shepherd C, Saul DJ, Demetras NJ, Haaning S, Monk CR et al. (2012). On the origin and evolution of thermophily: reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol Biol Evol 29: 825–835. [DOI] [PubMed] [Google Scholar]
- Joseph CA, Ricketts KD. European Working Group for Legionella Infections (2010). Legionnaires' disease in Europe 2007-2008. Euro Surveill 15: 19493. [DOI] [PubMed] [Google Scholar]
- Joseph CA, Ricketts KD, Yadav R, Patel S. European Working Group for Legionella Infections (2010). Travel-associated Legionnaires' disease in Europe in 2009. Euro Surveill 15: 19683.20961516 [Google Scholar]
- Kahlisch L, Henne K, Draheim J, Brettar I, Höfle MG. (2010). High-resolution in situ genotyping of Legionella pneumophila populations in drinking water by multiple-locus variable-number tandem-repeat analysis using environmental DNA. Appl Environ Microbiol 76: 6186–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlisch L, Henne K, Gröbe L, Brettar I, Höfle MG. (2012). Assessing the viability of bacterial species in drinking water by combined cellular and molecular analyses. Microb Ecol 63: 383–397. [DOI] [PubMed] [Google Scholar]
- Kao P-M, Hsu B-M, Chang T-Y, Hsu T-K, Tzeng K-J, Huang Y-L. (2014). Seasonal variation of Legionella in Taiwan's reservoir and its relationships with environmental factors. Environ Sci Pollut Res Int 22: 6104–6111. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Khan MA, Knox N, Prashar A, Alexander D, Abdel-Nour M, Duncan C et al. (2013). Comparative genomics reveal that host-innate immune responses influence the clinical prevalence of Legionella pneumophila serogroups. PLoS ONE 8: e67298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrenz M, Brettar I, Christen R, Flavier S, Bötel J, Höfle MG. (2004). Development and application of a real-time PCR approach for quantification of uncultured bacteria in the Central Baltic sea. Appl Environ Microbiol 70: 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammertyn E, Voorde JV, Meyen E, Maes L, Mast J, Anné J. (2007). Evidence for the presence of Legionella bacteriophages in environmental water samples. Microb Ecol 56: 191–197. [DOI] [PubMed] [Google Scholar]
- Lau H y, Ashbolt NJ. (2009). The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107: 368–378. [DOI] [PubMed] [Google Scholar]
- Leoni E, De Luca G, Legnani P, Sacchetti R, Stampi S, Zanetti F. (2005). Legionella waterline colonization: detection of Legionella species in domestic, hotel and hospital hot water systems. J Appl Microbiol 98: 373–379. [DOI] [PubMed] [Google Scholar]
- Lindsay JA. (1995). Is thermophily a transferrable property in bacteria? Crit Rev Microbiol 21: 165–174. [DOI] [PubMed] [Google Scholar]
- Lück PC, Jacobs E, Röske I, Schröter-Bobsin U, Dumke R, Gronow S. (2010). Legionella dresdenensis sp. nov., isolated from river water. Int J Syst Evol Microbiol 60: 2557–2562. [DOI] [PubMed] [Google Scholar]
- Mercante JW, Winchell JM. (2015). Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 28: 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M, Santic M, Kwaik YA. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchtouri V, Velonakis E, Tsakalof A, Kapoula C, Goutziana G, Vatopoulos A et al. (2007). Risk factors for contamination of hotel water distribution systems by Legionella species. Appl Environ Microbiol 73: 1489–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno A, Kato N, Yamada K, Yamaguchi K. (2003). Factors influencing survival of Legionella pneumophila serotype 1 in hot spring water and tap water. Appl Environ Microbiol 69: 2540–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthuisot N, West NJ, Lebaron P, Baudart J. (2010). High diversity and abundance of Legionella spp. in a Pristine river and impact of seasonal and anthropogenic effects. Appl Environ Microbiol 76: 8201–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K et al. (2014). Epidemiology and clinical management of Legionnaires' disease. Lancet Infect Dis 14: 1011–1021. [DOI] [PubMed] [Google Scholar]
- Pomeroy L, Le B, Williams P, Azam F, Hobbie J. (2007). The microbial loop. Oceanography 20: 28–33. [Google Scholar]
- Ratzow S, Gaia V, Helbig JH, Fry NK, Lück PC. (2007). Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila Serogroup 1 Strains. J Clin Microbiol 45: 1965–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffard S, Douglass S, Brooks T, Springthorpe S, Filion LG, Sattar SA. (2001). Occurrence of Legionella in groundwater: an ecological study. Water Sci Technol 43: 99–102. [PubMed] [Google Scholar]
- Robertson P, Abdelhady H, Garduño RA. (2014). The many forms of a pleomorphic bacterial pathogen—the developmental network of Legionella pneumophila. Microb Physiol Metab 5: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martínez S, Sharaby Y, Pecellín M, Brettar I, Höfle M, Halpern M. (2015). Spatial distribution of Legionella pneumophila MLVA-genotypes in a drinking water system. Water Res 77: 119–132. [DOI] [PubMed] [Google Scholar]
- Sobral D, Cann PL, Gerard A, Jarraud S, Lebeau B, Loisy-Hamon F et al. (2011). High-throughput typing method to identify a non-outbreak-involved Legionella pneumophila strain colonizing the entire water supply system in the Town of Rennes, France. Appl Environ Microbiol 77: 6899–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Hentschel U, Hacker J. (2002). Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev 26: 149–162. [DOI] [PubMed] [Google Scholar]
- Steinert M, Heuner K, Buchrieser C, Albert-Weissenberger C, Glöckner G. (2007). Legionella pathogenicity: Genome structure, regulatory networks and the host cell response. Int J Med Microbiol 297: 577–587. [DOI] [PubMed] [Google Scholar]
- Steinert M, Ockert G, Lück C, Hacker J. (1998). Regrowth of Legionella pneumophila in a heat-disinfected plumbing system. Zentralbl Bakteriol 288: 331–342. [DOI] [PubMed] [Google Scholar]
- Stout JE, Best MG, Yu VL. (1986). Susceptibility of members of the family Legionellaceae to thermal stress: implications for heat eradication methods in water distribution systems. Appl Environ Microbiol 52: 396–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis TC, Brown EW, Peruski LF, Siludjai D, Jorakate P, Salika P et al. (2012). Survey of Legionella species found in Thai soil. Int J Microbiol 2012: e218791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervaeren H, Temmerman R, Devos L, Boon N, Verstraete W. (2006). Introduction of a boost of Legionella pneumophila into a stagnant-water model by heat treatment. FEMS Microbiol Ecol 58: 583–592. [DOI] [PubMed] [Google Scholar]
- Veríssimo A, Marrão G, da Silva FG, da Costa MS. (1991). Distribution of Legionella spp. in hydrothermal areas in continental Portugal and the island of São Miguel, Azores. Appl Environ Microbiol 57: 2921–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CO. (2000). Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat 156: 145–155. [DOI] [PubMed] [Google Scholar]
- Whiley H, Bentham R. (2011). Legionella longbeachae and Legionellosis. Emerg Infect Dis 17: 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley H, Keegan A, Fallowfield H, Bentham R. (2014). Detection of LegionellaL. pneumophila and Mycobacterium avium complex (MAC) along potable water distribution pipelines. Int J Environ Res Public Health 11: 7393–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ES, Yilmaz LS, Noguera DR. (2012). DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullings BA, Bakker G, van der Kooij D. (2011). Concentration and diversity of uncultured Legionella spp. in two unchlorinated drinking water supplies with different concentrations of natural organic matter. Appl Environ Microbiol 77: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullings BA, van der Kooij D. (2006). Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15 °C. Appl Environ Microbiol 72: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A et al. (2002). Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired Legionellosis: an International Collaborative Survey. J Infect Dis 186: 127–128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.