Abstract
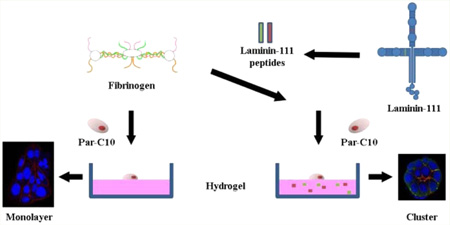
Previous studies showed that mouse submandibular gland cells form three-dimensional structures when grown on Laminin-111 gels. The use of Laminin-111 for tissue bioengineering is complicated due to its lack of purity. By contrast, the use of synthetic peptides derived from Laminin-111 is beneficial due to their high purity and easy manipulation. Two Laminin-111 peptides have been identified for salivary cells: the A99 peptide corresponding to the α1 chain from Laminin-111 and the YIGSR peptide corresponding to the β1 chain from Laminin-111, which are important for cell adhesion and migration. We created three-dimensional salivary cell clusters using a modified fibrin hydrogel matrix containing immobilized Laminin-111 peptides. Results indicate that the YIGSR peptide improved morphology and lumen formation in rat parotid Par-C10 cells as compared to cells grown on unmodified fibrin hydrogel. Moreover, a combination of both peptides not only allowed the formation of functional three-dimensional salivary cell clusters but also increased attachment and number of cell clusters. In summary, we demonstrated that fibrin hydrogel decorated with Laminin-111 peptides supports attachment and differentiation of salivary gland cell clusters with mature lumens.
Graphical abstract
INTRODUCTION
Proper salivary gland function is critical for oral health. Autoimmune disorders (such as Sjögren’s syndrome), genetic diseases (such as ectodermal dysplasia), and γ-irradiation therapies (for head and neck cancers) cause salivary secretory dysfunction and lead to severe dryness of the oral cavity.1–3 Dry mouth can lead to oral infections, sleep disturbances, oral pain, and difficulty in chewing or swallowing food.4–6 However, the current treatments for dry mouth only provide temporary relief, and no tissue engineering approaches are currently available for patients suffering with dry mouth.7 Therefore, it is important to design a clinically safe matrix capable of supporting the growth of a functional salivary gland structure first in vitro and then in vivo applications.
Previous studies showed that rat parotid gland (Par-C10) cells and mouse parotid cells form three-dimensional (3D) cell clusters displaying tight junctions (TJ) and agonist-induced secretory responses when grown on Growth Factor-Reduced Matrigel (GFR-MG).8,9 Matrigel is rich in extracellular matrix (ECM) proteins, with the major components being Laminin-111 (L1) (60%) and collagen IV (30%). Although Matrigel allows the formation of 3D salivary constructs from single acinar cells, clinical application may be hindered by reports of basement membrane matrices supporting tumor growth. Also, it is derived from mouse cancer cells, which could trigger possible immune reactions in humans.10,11
Hydrogels are widely used as a tissue engineering scaffold.12 Among the hydrogels, fibrin hydrogel (FH) is the most extensively studied hydrogel. FHs are water-swollen, polymeric structures that form scaffolds capable of supporting cell viability and differentiation for long periods of time by interaction of cells with fibrin. Fibrin forms a hydrogel at physiological temperatures and contains native arginine-glycine-aspartic acid (RGD) sites that promote cell attachment.13 In addition, several studies demonstrated engineering of FH with conjugated growth factors, genes and recombinant viruses for multiple applications ranging from wound healing, vascular tissue engineering and lentiviral arrays. These include delivery of keratinocyte growth factor (KGF) to promote wound healing,14 a peptide-TGF-β1 fusion protein to improve the contractile function, extracellular matrix synthesis and mechanical properties of vascular grafts,15,16 and plasmid DNA and recombinant lentivirus for engineering gene delivery microarray platforms.17–19
Previous studies also showed that a combination of GFR-MG and FH allowed Par-C10 cells to form 3D structures with a hollow lumen resembling salivary glands. Additionally, critical components within GFR-MG (i.e., a combination of EGF andIGF-1) enhanced salivary gland differentiation when polymerized within FH.8 However, EGF and IGF-1 were found to be incapable of independently producing organized cell clusters.8 More recently, another critical component of GFRMG, L1, was able to produce organized salivary cell clusters using primary mouse submandibular (mSMG) cells.20 Nonetheless, the full L1 sequence may not be suitable for clinical applications, as some protein domains are known to promote tumorigenesis or immunogenic response that may outweigh the potential benefits provided by the whole molecule.21,22 Conversely, the use of synthetic peptides is relatively simple and inexpensive as compared to animal-derived proteins. In addition, controlled densities of peptides can be conjugated to the target material surfaces. Moreover, it is possible to minimize the immune reactivity or pathogen transfer.23 In 1999, Yoshida et al. reported that the YIGSR peptide has an inhibitory effect on tumor growth and an antiproliferative effect.24 Moreover, the cell attachment and proliferation of mouse fibroblasts were improved on RGD-modified films in 2012.25 Therefore, the use of a combination of peptides (RGD and YIGSR) within the L1 sequence may provide the synergistic effects while minimizing the risks: (a) the A99 (RGD) peptide corresponding to the α1 chain from L1 (important for cell adhesion),25–27 and (b) the YIGSR peptide corresponding to the β1 chain from L1 (also important for cell adhesion, inhibitory effect on tumor growth, and migration).24,26,28
The goal of this study was to test whether L1 peptides (corresponding to regions that promote intact salivary gland formation, see Table 1) were able to induce lumen formation in Par-C10 salivary cell clusters. Our results indicate that cell clusters were formed on the FH modified with YIGSR peptide. Specifically, it improved morphology and lumen formation in rat parotid Par-C10 cells as compared to cells grown on unmodified FH. Moreover, a combination of YIGSR and A99 peptides not only allowed the formation of functional 3D salivary cell clusters, but also increased attachment and cell cluster numbers. In summary, FH decorated with Laminin-111 peptides supports attachment and differentiation of salivary gland cell clusters with mature lumens.
Table 1.
Sequence of Synthetic Peptides
| peptide | sequence | molecular mass | L1 sequence |
|---|---|---|---|
| A99 (RGD) | CGGALRGDN-amide | 860.9 | laminin α1 chain (1145–1150) |
| YIGSR | CGGADPGYIGSRGAA-amide | 1350.5 | laminin β1 chain (925–936) |
| RAD | CGGALRADN-amide | 875.0 | scrambled peptide for A99 |
| SGIYR | CGGADPGSGIYRGAA-amide | 1350.5 | scrambled peptide for YIGSR |
MATERIALS AND METHODS
Materials
Lyophilized fibrinogen from human plasma was purchased from EMD Millipore (Billerica, MA). Spectra/Por 7 dialysis membrane (MWCO = 3.5 kDa) was purchased from Spectrum Laboratories (Rancho Dominguez, CA). A Whatman syringe filter (0.8 µm) was purchased from GE Healthcare Life Sciences (Pittsburgh, PA). A Millex syringe filter (0.22 µm) was purchased from Merck Millipore (Billerica, MA, USA). Paraformaldehyde (PFA) was purchased from Baker (Phillipsburg, NJ). Insulin-transferrin-sodium selenite media supplement, retinoic acid, hydrocortisone, gentamicin, epidermal growth factor (EGF) from murine submaxillary gland, dl-dithiothreitol (DTT), and ε-aminocaproic acid (εACA) were purchased from Sigma-Aldrich (St. Louis, MO). Rabbit antizonula occludens-1 (ZO-1) antibody was purchased from Invitrogen (Carlsband, CA). Dulbecco’s modified eagle medium/nutrient mixture F-12 (1:1) (DMEM/F12 (1:1)), fetal bovine serum (FBS), glutamine, Lab-Tek chambered coverglass (8-well), sulfosuccinimidyl 6-(3′-(2-pyridyldithio)propionamido)hexanoate (Sulfo-LC-SPDP), Alexa Fluor 488-conjugated goat antirabbit secondary antibody, Alexa Fluor 568-conjugated phalloidin, TO-PRO-3 iodide nuclear stain, and Fura-2-acetoxymethylester were purchased from Thermo Fisher Scientific (Newington, NH). Peptides were synthesized by University of Utah DNA/Peptide synthesis core facility.
Peptide Synthesis
Two biologically active peptides derived from L1 were synthesized on an ABI431 or ABI433 peptide synthesizer using a standard Fmoc solid-phase peptide synthesis as follows: Amino acids were protected at their amino terminus by the Fmoc (9-fluorenylmethoxycarbonyl) group and coupled to the growing chain after activation of the carboxylic acid terminus. Then, the Fmoc group was removed by piperidine treatment and the process repeated. After the peptide was assembled, it was removed from the resin by treatment with trifluoroacetic acid (TFA). At the same time, protecting groups on amino acid side chains were removed yielding the crude linear peptide. Finally, one-step purification by reverse-phase HPLC yielded peptides with >95% purity. Two scrambled peptides were synthesized as a control. The overall synthesis scheme of scrambled peptides was the same as described above. All peptides were synthesized with a cysteine and two glycine residues (Cys–Gly–Gly, CGG) at the N-terminus. A cysteine free thiol group was used for coupling with thiol-reactive fibrinogen and the two glycine residues used as a spacer, respectively. A list of these peptides is shown in Table 1.
Peptide-Conjugated Fibrinogen
Lyophilized fibrinogen was dissolved in 0.1 M phosphate-buffered saline (PBS, pH 7.2, 0.15 M NaCl, 1 mM EDTA) and dialyzed using a disposable cellulose membrane (MWCO = 3.5 kDa) overnight. Then, the fibrinogen solution was purified using a 0.8 µm filter. In order to produce a thiol-reactive fibrinogen, 7.2 equiv of Sulfo-LC-SPDP was added to the purified fibrinogen solution and incubated for 1 h at room temperature. Subsequently, the excess Sulfo-LC-SPDP and its hydrolysis products (N-hydroxysulfosuccinimide, Sulfo-NHS) were removed by dialysis.
The level of LC-SPDP-modification was determined by measuring the absorbance of pyridine-2-thione at 343 nm. Briefly, 10 µL of DTT (15 mg/mL) was added to 1 mL of modified fibrinogen. After 15 min of incubation, absorbance at 343 nm was measured, and the change in absorbance was calculated: ΔA343 = (A343 after DTT) – (A343 before DTT). The level of SPDP modification was calculated using the following equation:
| (1) |
where 341 kDa reflects the molecular weight of fibrinogen, and the value 8080 reflects the extinction coefficient for pyridine-2-thione at 343 nm: 8.08 × 103 M−1 cm−1.29,30
For peptide conjugation, LC-SPDP activated fibrinogen was dissolved in 50 mM PBS (pH 7.2, 0.15 M NaCl, 10 mM EDTA). Two equivalents of peptide per 2-pyridyldithiol groups of LC-SPDP-fibrinogen was added to the solution, and the mixture was reacted for 18 h at room temperature. The reaction was monitored by thin-layer chromatography (TLC). Finally, the product was dialyzed against ultrapure water using a dialysis membrane (MWCO = 3.5 kDa) as described above and products were filtered using a 0.22 µm syringe filter from Merck Millipore. Then, peptide-conjugated fibrinogen was lyophilized and stored at −80 °C until use.
The concentration of fibrinogen was calculated using the following equation:
| (2) |
where εFib, the extinction coefficient at 280 nm for human fibrinogen, is 1.51 mL mg−1 cm−1.31
Molecular Weight Determination
Static light scattering has been used to determine the size and molecular weight of macromolecules since the 20th century.32 When the light hits a macromolecule (e.g., a polymer or protein), some of the light is absorbed and re-emitted in all directions. The Rayleigh eq 3 describes the relationship between molecular weight and scattered light. By using this equation, the molecular weight of the modified fibrinogen can be determined.
| (3) |
where K is an optical constant, C is the sample concentration, θ is the measurement angle, Rθ is the Rayleigh ratio, M is the molecular weight, and A2 is the second virial coefficient.
Fibrin Hydrogel Preparation
The cross-linked FH was generated by mixing plasma-derived bovine thrombin (2.5 U/mL) and fibrinogen (2.5 mg/mL) in Tris-buffered saline (TBS) with CaCl2 (2.5 mM) and εACA (2 mg/mL) as previously described.18 One hundred microliters of mixture per well in eight-well chambers was allowed to solidify in the incubator at 37 °C overnight. The overall preparation scheme of YIGSR (50%)- and A99 (50%)-conjugated FH was the same as described above. YIGSR-conjugated fibrinogen (1.25 mg/mL) and A99-conjugated fibrinogen (1.25 mg/mL) were used as monomers.
Rheological Parameters
Rheological measurements of fibrin hydrogel were performed on a stress-controlled rheometer (TA Instruments, AR 2000ex). All tests were performed using the cone plate geometry (4°/20 mm) with a truncation height of 114 µm at 37 °C. Human fibrinogen (2.5 mg/mL) and thrombin (2.5 U/mL) solutions were rapidly mixed in TBS buffer (2.5 mM CaCl2, 2 mg/mL εACA) and then applied to the bottom of the rheometer plate. To prevent evaporation, the shear gap was covered with a solvent trap cover. The modulus of elasticity (G′) and the strain (%) were recorded 5 min after FH addition. Data were analyzed by two-way ANOVA with pairwise comparisons where p < 0.05 represents significant differences between experimental groups.
Par-C10 Cell Culture
The polarized rat parotid cell line (Par-C10) was derived from freshly isolated rat parotid gland acinar cells by transformation with simian virus 40 and exhibits morphological, biochemical, and functional characteristics of freshly isolated acinar cells.33,34 Par-C10 cells (5 × 105 at passages 40–60) were grown to confluence in DMEM/F12 (1:1) containing 2.5% (v/v) FBS and the following supplements: 0.1 µM retinoic acid, 80 ng/mL EGF, 2 nM triiodothyronine, 5 mM glutamine, 0.4 µg/mL hydrocortisone, 5 µg/mL insulin, 5 µg/mL transferrin, 5 ng/mL sodium selenite, and 50 µg/mL gentamicin. Two thousand cells were plated on top of different hydrogels as a two-dimensional (2D) culture and incubated at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
Par-C10 Cell Morphometric Analysis
After 3 days of incubation (shown in previous studies to be optimal for sphere formation),8,35 cells were fixed in 2% PFA for 20 min at room temperature and stained for 10 min using 200 µL of PBS containing 0.1% Triton X-100 with 30 µM DAPI. After washing three times with PBS, cell morphology was observed under an inverted microscope (Leica DMI6000B, Germany) at 10× magnification. Then, the DAPI stained cells in three randomly selected fields were counted using ImageJ software.36,37 All experiments were performed in triplicate and repeated three times. All data are presented as means ± SD. Statistical analysis was performed using GraphPad Prism software. Data were analyzed by one-way ANOVA followed by pairwise post hoc Tukey’s t-test where p < 0.05 represents significant differences between experimental groups.
Intracellular Free Calcium Levels
The intracellular free calcium levels of Par-C10 salivary cell clusters on FH were determined using a Leica DMI6000B imaging system. After 3 days of incubation, cells were treated with 4 µM Fura-2-acetoxymethylester (Fura-2 AM) for 20 min at 37 °C in cell culture medium (as described above) and washed with cell culture medium. The cells were stimulated with 100 µM carbachol (Cch). Then, images were recorded and analyzed using Leica Application Suite X software. To determine statistical significance, the fluorescence intensity was measured by a Tecan Infinite M200 Pro spectrophotometer (Tecan Group Ltd., Männedorf, Switzerland) at room temperature. Dual excitation measurements at 340 and 380 nm were performed, and the emission intensity was recorded at 510 nm. All experiments were performed in sextuplicate. Data were analyzed by one-way ANOVA followed by pairwise post hoc Tukey’s t-test where p < 0.01 represents significant differences between experimental groups.
Immunofluorescence (IF)
After 3 days of incubation, Par-C10 cells were fixed in 2% PFA for 10 min, incubated with 0.1% Triton X-100 in PBS for 10 min and washed three times with PBS for 5 min at room temperature. For ZO-1 staining, Par-C10 cells were blocked for 2 h in 5% goat serum at room temperature and incubated with a rabbit anti-ZO-1 antibody (1:50) in 5% goat serum overnight at 4 °C. The following day, cells were warmed to room temperature for 20 min and washed three times for 5 min with PBS. Cells were incubated for 1 h with Alexa Fluor 488-conjugated goat antirabbit secondary antibody (1:500) in 5% goat serum then washed three times with PBS. For the immunofluorescent staining of F-actin, cells were stained with Alexa Fluor 568-conjugated phalloidin (1:400, PBS) for 1 h at room temperature and washed three times for 5 min with PBS. For nuclear staining, cells were incubated with TO-PRO-3 iodide (1:1,000, PBS) for 15 min at room temperature and washed three times for 5 min with PBS. Cells were visualized using a Carl Zeiss 700 LSM confocal microscope. The average lumen diameter was calculated using the ZEN software (Carl Zeiss, Thornwood, NY). Apical ZO-1 stained cells in randomly selected fields were counted as a cluster. However, cell aggregates were counted as beehive-like pattern structures lacking apical ZO-1. All data are presented as means ± SD (n = 9). Statistical analysis was performed using GraphPad Prism software. Data were analyzed by one-way ANOVA followed by pairwise post hoc Tukey’s t-test where p < 0.05 represents significant differences between experimental groups. Microscope settings were kept consistent for all samples.
RESULTS AND DISCUSSION
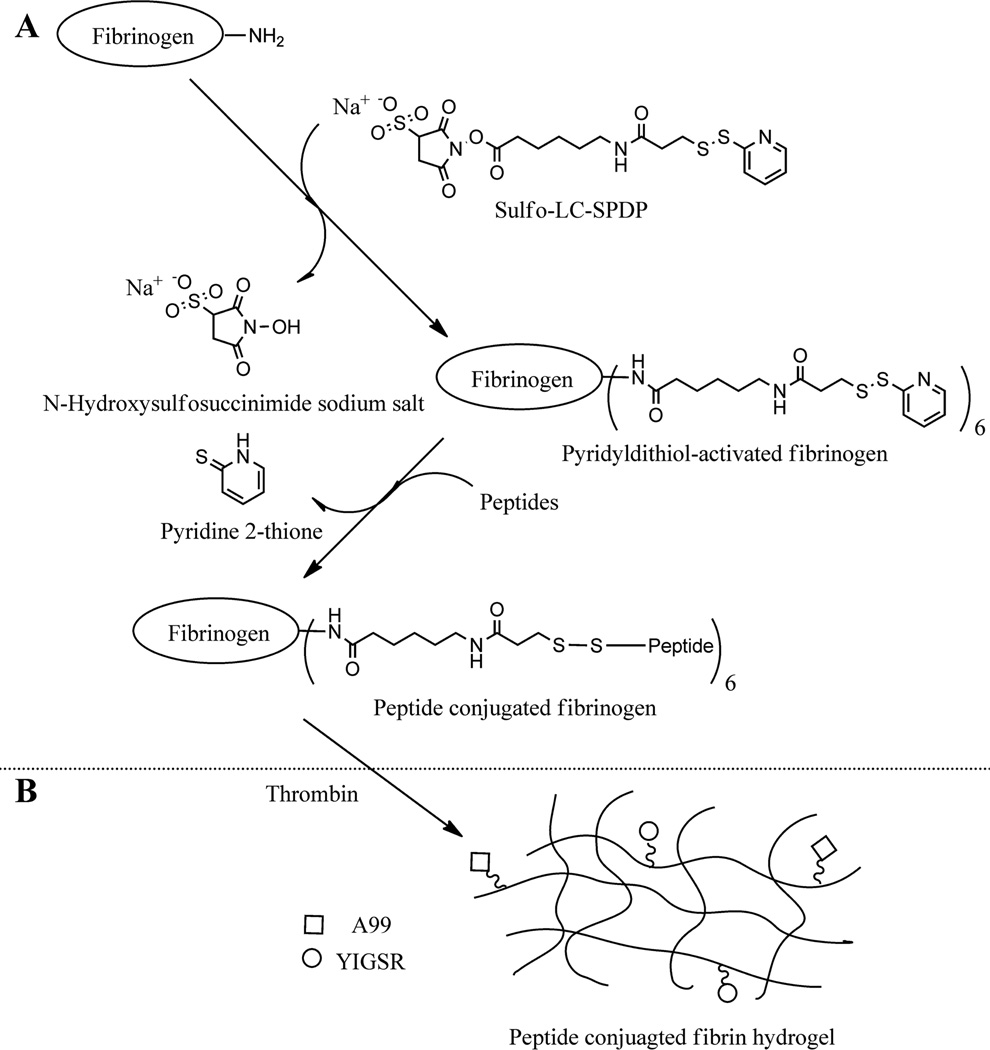
L1-derived peptide-conjugated fibrinogen was prepared to induce formation of Par-C10 salivary cell clusters. Peptide-conjugated fibrinogen was prepared following the synthetic procedure illustrated in Figure 1. Briefly, the primary amine groups of fibrinogen were reacted with Sulfo-LC-SPDP. The cross-linker is able to react with both the side chain of lysine (ε-amino group) and the α-amine at the N-terminus. However, the coupling efficiency of the α-amine and the ε-amine is highly dependent on pH. At a neutral pH, ε-amino of lysine is rapidly protonated. Therefore, coupling of the cross-linker through the α-amine of N-terminus is more efficient than the ε-amino of lysine.38,39 The reaction was monitored by thin layer chromatography (TLC) and Ultraviolet–visible (UV) spectroscopy. The level of LC-SPDP-modification was calculated using eq 1 as described in the Materials and Methods section. Based on the result of the UV measurements (Supplementary Figure S1), six cross-linkers were conjugated to fibrinogen. Then, L1-derived peptide-conjugated fibrinogen was obtained by reaction between a free thiol group of cysteine terminated peptide (A99 or YIGSR) and a pyridyldithiol-activated fibrinogen. The products were dialyzed against ultrapure water using a dialysis membrane (MWCO = 3.5 kDa) and filtered using a 0.22 µm syringe filter. Then, the peptide-conjugated fibrinogen was freeze-dried. The percent yields for the products were 79.47% (A99), 90.04% (YIGSR), 83.17% (RAD), and 80.05% (SGIYR), respectively. Finally, lyophilized fibrinogen was stored at −80 °C until further use.
Figure 1.
(A) Synthetic scheme of L1-peptide-conjugated fibrinogen. (B) Preparation of fibrin hydrogel.
Peptide conjugation was confirmed using and UV–vis spectrum data (Supplementary Figure S1) and static light scattering data (Supplementary Figure S2). The molecular weight of L1 derived peptide-conjugated fibrinogens was slightly increased. Based on the UV–vis spectrum data we estimated that six peptides were conjugated to a single fibrinogen molecule (Table 2).
Table 2.
Molecular Weights of the Unmodified and Modified Fibrinogena
| kDa | |
|---|---|
| fibrinogen | 330 ± 7.87 |
| YIGSR-conjugated fibrinogen | 350 ± 3.70 |
| A99-conjugated fibrinogen | 347 ± 2.98 |
| SGIYR-conjugated fibrinogen | 349 ± 4.78 |
| RAD-conjugated fibrinogen | 345 ± 5.12 |
Results are expressed as mean ± SD (n = 3).
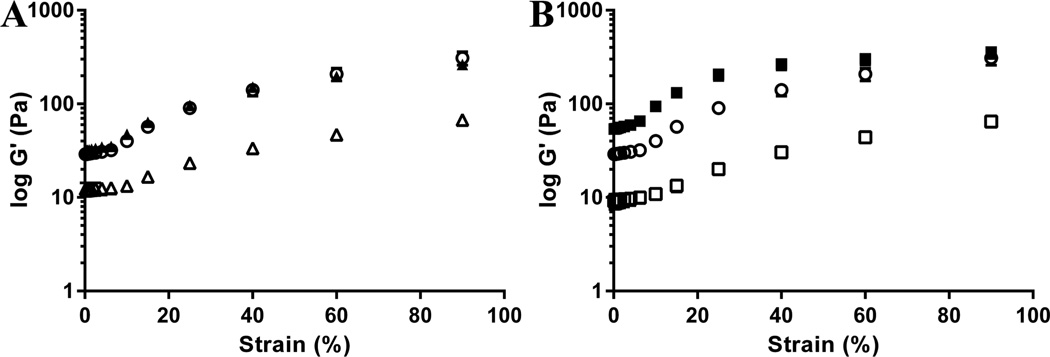
FH has both elastic and viscous properties, and these properties are highly sensitive to changes in polymerization.13,40 In addition, the rheological parameters can provide information about the structural changes.41 Therefore, the peptide-conjugated FHs were characterized using rheological techniques. As shown in Figure 2, the elasticity of YIGSR-conjugated FH and A99-conjugated FH was slightly less than unmodified FH, and RAD-conjugated FH was slightly greater than unmodified FH. These results were significantly different from the control (FH alone) except SGIYR-conjugated FH. These results indicate that peptide conjugation affect the overall physical structure of the FH.
Figure 2.
Rheological Parameters of FH. (A) YIGSR measurements and (B) A99 measurements. Data represent the elasticity of unmodified FH (○), YIGSR-conjugated FH (△), A99-conjugated FH (□), SGIYR-conjugated FH (▲), and RAD-conjugated FH (■). Each data point represents the mean ± SD (n = 3, p < 0.05).
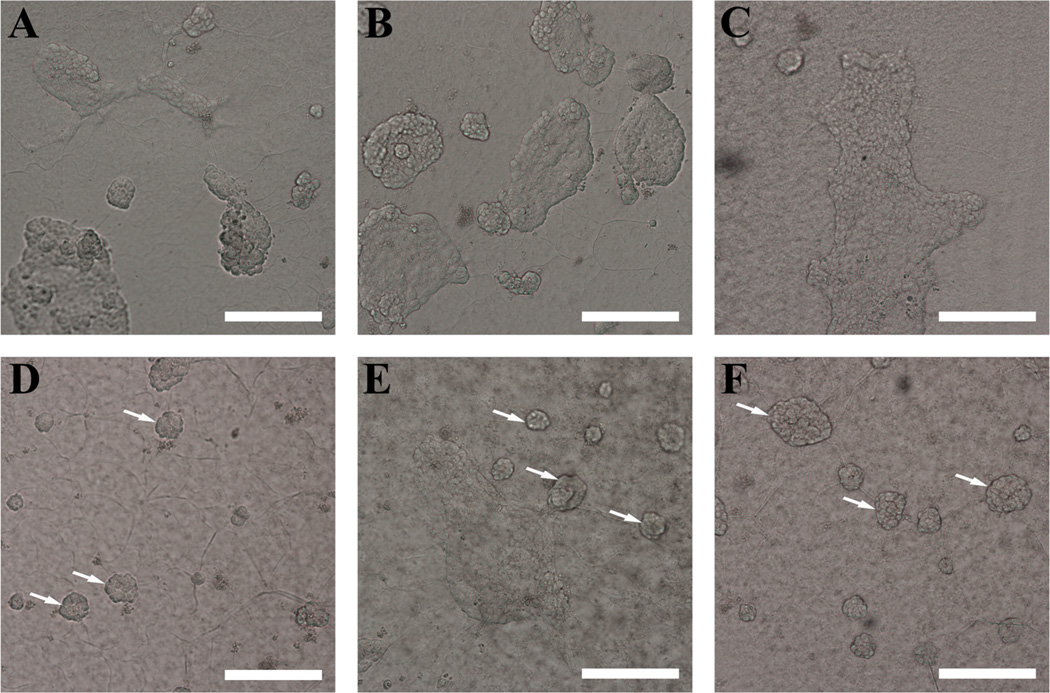
Par-C10 cells were plated on FH as described in the Materials and Methods section. After 3 days in culture, Par-C10 cells formed fibroblast-like monolayers when grown on unmodified FH (Figure 3A). In addition, Par-C10 cells displayed fibroblast-like monolayers when grown on scrambled peptide-conjugated FH (Figure 3B; SGIYR, Figure 3C; RAD). This result suggests that both unmodified FH and scrambled peptide-conjugated FH are not suitable for formation of Par-C10 salivary cell clusters.
Figure 3.
Par-C10 salivary cell cluster formation organization on peptide-conjugated FH. (A) Unmodified FH, (B) SGIYR-conjugated FH, (C) RAD-conjugated FH, (D) YIGSR-conjugated FH, (E) A99-conjugated FH, and (F) a combination of YIGSR (50%)- and A99 (50%)-conjugated FH. Par-C10 cells grown on YIGSR and/or A99 peptide-conjugated FH formed round organized structures (white arrows). Scale bars represent 200 µm.
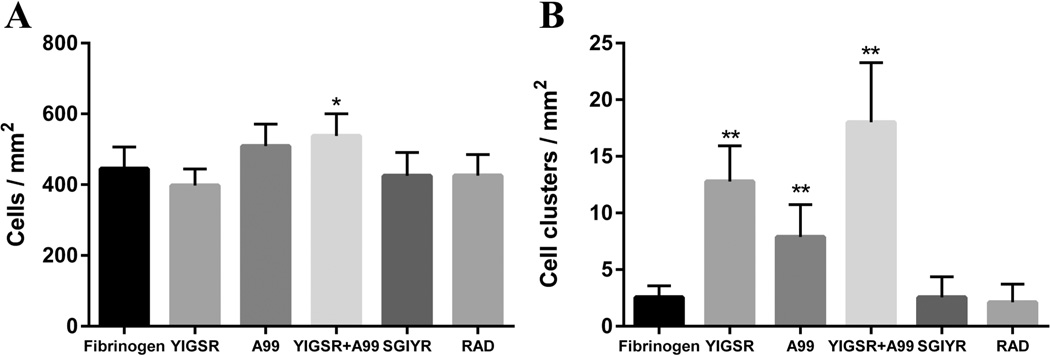
However, Par-C10 cells grown on YIGSR and/or A99 peptide-conjugated FH formed round organized structures, with an average cell cluster diameter of approximately 70 µm when grown on a combination of YIGSR (50%)- and A99 (50%)-conjugated FH (Figure 3F). Moreover, a combination of the peptides (YIGSR 50% with A99 50%) showed an increase in cell attachment (537.78 ± 62.61 cells/mm2) and Par-C10 cell cluster formation (18.00 ± 5.29 clusters/mm2) as compared to the unmodified FH (444.78 ± 61.65 cells/mm2, 2.56 ± 1.01 clusters/mm2) (Figure 4). These results are relevant to both human and mouse studies, as previous studies showed that both of these cell types form 3D salivary cell clusters when grown on different scaffolds, such as Matrigel or L1.20,42,43
Figure 4.
(A) Total cell number and (B) Par-C10 salivary cell cluster formation were calculated. A combination of the peptides (YIGSR 50% with A99 50%) showed an increase in cell attachment and Par-C10 cell cluster formation as compared to the unmodified FH. Each data point represents the mean ± SD (n = 9, *p < 0.05, **p < 0.01).
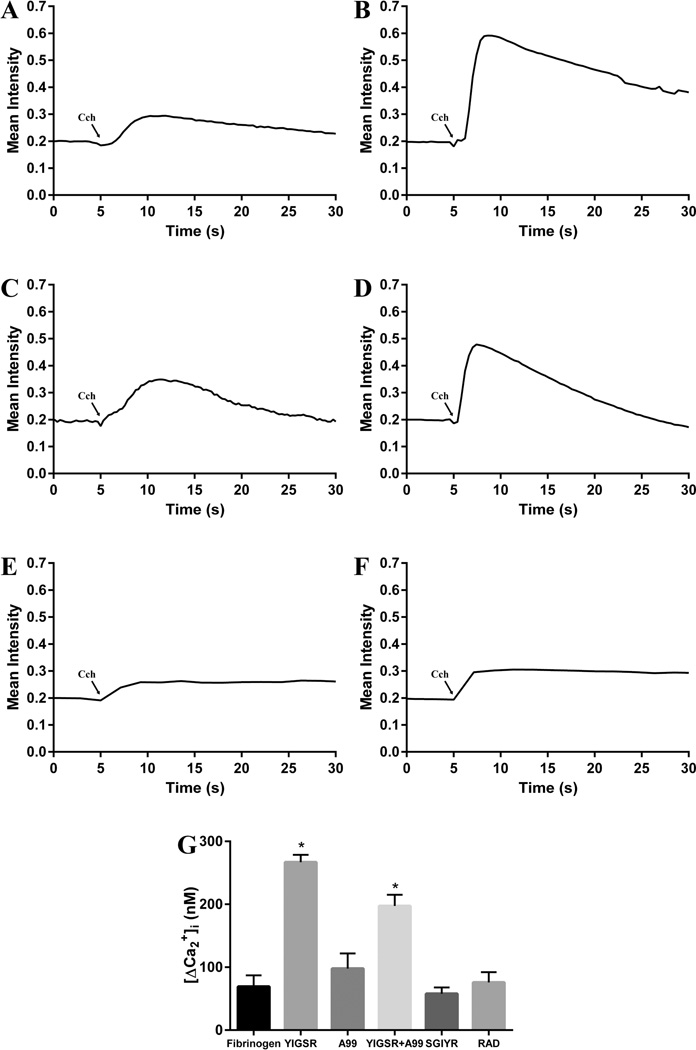
Carbachol (Cch) is a cholinergic agonist that stimulates the M3 muscarinic acetylcholine receptor in salivary glands, leading to increased intracellular free calcium concentration ([Ca2+]i).44 Cch (100 µM) induced an increase in [Ca2+]i in Par-C10 cells cultured under all the conditions studied (i.e., FH, SGIYR, RAD, YIGSR, A99 alone and in combination, Figure 5A–F), consistent with results from previous studies using rat parotid gland and Par-C10 cells.8,20 However, Par-C10 cells cultured on YIGSR-modified FH (Figure 5B,G) displayed a significantly higher increase of [Ca2+]i as compared to unmodified FH (Figure 5A,G) and A99-modified FH (Figure 5C,G). Furthermore, increases of [Ca2+]i in Par-C10 cells on FH containing both YIGSR and A99 peptides (Figure 5D,G) was significantly different from the [Ca2+]i response observed on FH modified with YIGSR alone. The greater [Ca2+]i response is important because increases in [Ca2+]i are critical for eliciting the physiological secretory function in salivary glands.
Figure 5.
Intracellular calcium concentration measurements. The cells were stimulated with 100 µM carbachol (Cch). Then, images were recorded and analyzed using Leica Application Suite X software. Par-C10 cells plated on unmodified FH (A), YIGSR-conjugated FH (B), A99-conjugated FH (C), a combination of YIGSR (50%)- and A99 (50%)-conjugated FH (D), SGIYR-conjugated FH (E) and RAD-conjugated FH (F). (G) Par-C10 cells cultured on YIGSR-modified FH displayed increased [Ca2+]i. Data are expressed as means ± SD, where *p < 0.01 indicates a significant difference from control (unmodified FH).
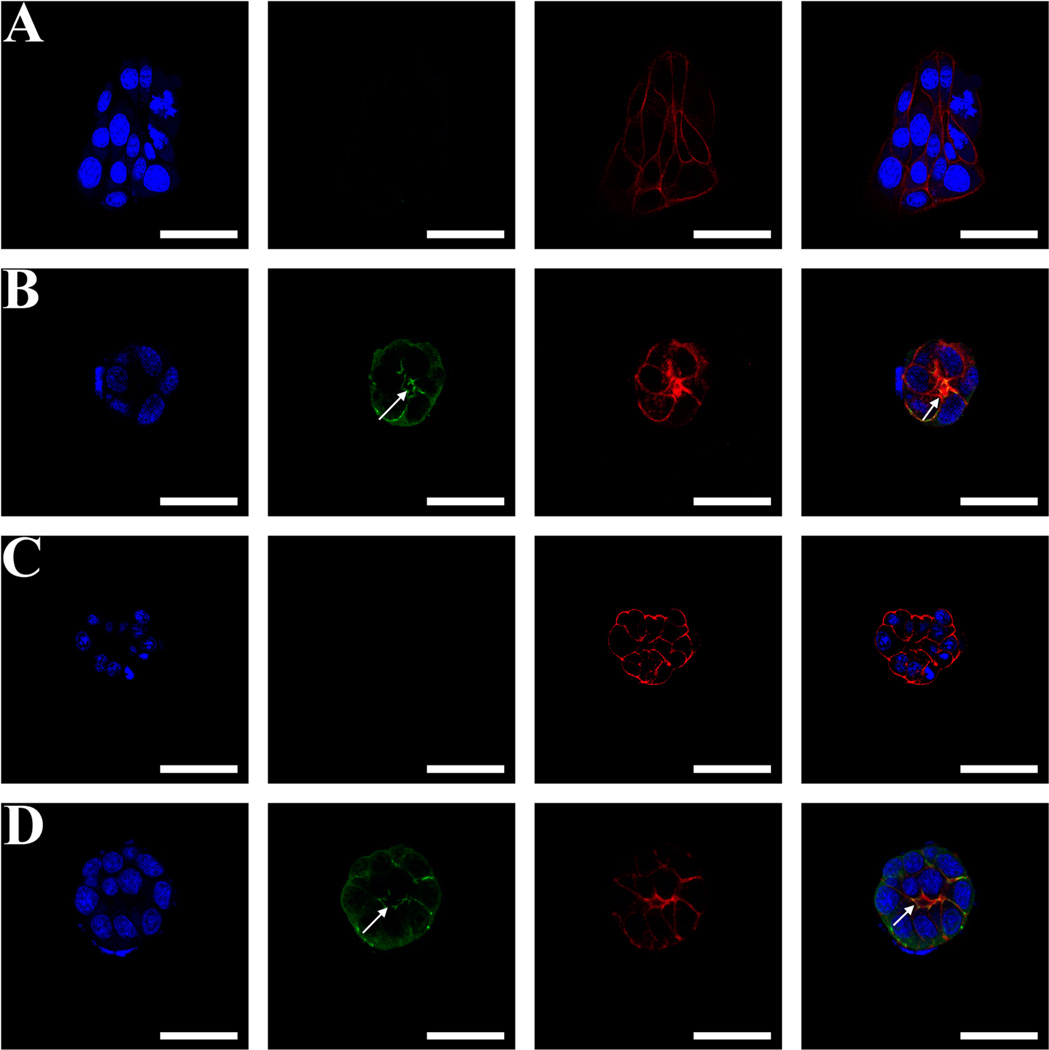
Par-C10 cells formed monolayers when cultured on unmodified FH (Figure 6A) and scrambled peptide-conjugated FH (Supplementary Figure S3). On the other hand, Par-C10 cells grown on A99-modified FH formed salivary cell clusters but with no lumens (Figure 6C). Notably, cells grown in the presence of YIGSR peptide (i.e., YIGSR-modified FH or YIGSR combined with A99-modified FH) were able to form lumens, as indicated by the intense F-actin and ZO-1 staining on the apical region (Figure 6B,D). As shown in Figure 6, a combination of YIGSR (50%)- and A99 (50%)-conjugated FH exhibited a higher level of ZO-1 polarization and a well-defined lumen structure (16.48 ± 3.95 µm) as compared to unmodified FH (form monolayers) and A99-conjugated FH (2.64 ± 1.60 µm).
Figure 6.
Confocal microscopy images show ZO-1 (green) as stained by Alexa Fluor 488-conjugated goat antirabbit secondary antibody, F-actin as stained by Alexa Fluor 568-conjugated phalloidin (red), and nuclei (blue) as stained by TO-PRO-3 iodide. Right images are merged images. All images were obtained and analyzed using a Carl Zeiss 700 LSM confocal microscope. Par-C10 cells plated on unmodified FH (A), YIGSR-conjugated FH (B), A99-conjugated FH (C), and a combination of YIGSR (50%)- and A99 (50%)-conjugated FH (D). Scale bars represent 50 µm. White arrows indicate lumen formation.
Saliva is a mixture of water, electrolytes, and proteins. However, it mostly consists of water. Lumen formation is an essential morphological feature of salivary glands to allow saliva production into the lumen, ductal system, and the oral cavity.45 Saliva production and function are highly sensitive to structural changes in salivary glands.46 Developing natural tissue-like structures is very important to achieve a functional organ that could be used for bioengineering purposes. Three-dimensional cultures allow a more realistic and controllable system to mimic tissue structures when compared with 2D culture systems;47 this is because cells can communicate more effectively in 3D than 2D culture systems.48 Furthermore, both human and mouse salivary gland cells form 3D structures similar to Par-C10 cell clusters.9 Therefore, 3D culture systems are very useful when studying cell signaling, morphology, gene expression and differentiation.49 Over the past two decades, hydrogels have been used as 3D culture and tissue engineering matrices in various tissues such as cornea, cartilage, cardiac muscle and valves, skin wound healing, bone, liver, urethra and urinary bladder and nerve.50,51 Our previous studies showed that GFR-MG and L1 allow formation of 3D structures with hollow lumens resembling acinar lumens in Par-C10 cells. However, animal derived hydrogel systems are restricted for clinical applications and medical research because of concerns for immunogenic reactions and infection from animal tissue.52
In comparison with other hydrogel systems, FH can be produced from the patient’s fibrinogen, minimizing potential immune reactions.53 Moreover, FH has other advantages such as high cell-adhesion efficiency, uniform cell distribution, flexibility, biocompatibility, and biodegradability.54 Despite these advantages, the use of FH itself for tissue engineering of salivary gland is still far from ideal because salivary cells only form monolayers when cultured on FH (Figures 3A and 6A). In order to solve this problem, we designed a L1 peptide-modified FH using L1 peptides that were previously demonstrated to be useful for salivary gland cell attachment and differentiation.26–28 We found that L1-modified FHs are indeed useful for achieving formation of 3D structures with lumens (Figures 3D and 6D). Our results are consistent with previous studies demonstrating that L1 peptides enhance the properties of other bioengineering materials such as chitosan and PLLA.55,56 Through the use of L1, we have developed a consistent method through which the biological character of FH can be augmented with an active factor such as L1 peptides, creating a material that can be optimized for clinical applications. When studying the physical properties of L1-conjugated FH, we were able to demonstrate that the use of YIGSR and A99 together did not significantly modify the rheological properties of the L1 peptide-conjugated FH. A fundamental understanding of peptide-conjugated hydrogel mechanical properties and underlying formation and deformation mechanisms is crucial for determining whether these biomaterials are potentially suitable for bioengineering uses.57 Here, we demonstrated that L1-modified FHs have optimal properties for use with soft tissues such as the salivary glands, which are unlikely affected by shear flow, although future studies will be required to confirm this notion.
Regarding cell attachment, our results indicate that Par-C10 cells cultured on a combination of YIGSR (50%) and A99 (50%) peptide-conjugated FH allowed for successful cell attachment as compared with single peptides. Previous studies have demonstrated that putting mixtures of peptides together in groups can improve their function (e.g., chitosan scaffolds conjugated with a mixture of L1 peptides promote cell attachment, spreading, and neurite outgrowth).58 Interestingly, peptides in tandem, connected by specific domains, have also demonstrated a greater impact (e.g., assisted gene transfer is significantly increased when multivalent fusion peptides are used instead of a mixture of two peptides).59 As demonstrated in Figure 6, Par-C10 cells cultured on a combination of YIGSR (50%) and A99 (50%) peptide-conjugated FH were able to differentiate into 3D structures with a central lumen without affecting cell viability (Figure 5).
CONCLUSIONS
We were able to determine that combinations of L1 peptides offer superior outcomes as compared to single peptides. As demonstrated in Figure 6, Par-C10 cells cultured on a combination of YIGSR (50%) and A99 (50%) peptide-conjugated FH were able to respond to the salivary secretory agonist, carbachol (Figure 5), indicating that salivary cells maintain viability. These results are consistent with our previous studies showing that Par-C10 cells maintain their secretory responses on different extracellular matrices.8,9 These results suggest that FH decorated with both YIGSR and A99 peptides has great potential as a 3D culture matrix for engineering salivary glands.
Supplementary Material
Acknowledgments
The authors would like to thank Ms. Monika Sima (University of Utah, Department of Bioengineering) for her assistance in performing the rheological experiments. This study is supported by the National Institutes of Health-National Institute of Dental and Craniofacial Research Grants R01DE022971 (to O.B. and S.A.) and R01DE021697 (to O.B.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Supplementary Figure S1: The UV–vis absorption spectra of LC-SPDP activated fibrinogen (A) and YIGSR-conjugated fibrinogen (B) after incubation in the presence (red dots) or absence (blue dots) of DTT for 15 min. Supplementary Figure S2: Debye plots for fibrinogen (●), YIGSR-conjugated fibrinogen (▲) and A99-conjugated fibrinogen (■). Supplementary Figure S3: Maximum intensity projection of confocal images data at low magnification (10×). Par-C10 cells plated on unmodified FH (A), SGIYR-conjugated FH (B), RAD-conjugated FH (C), YIGSR-conjugated FH (D), A99-conjugated FH (E), and a combination of YIGSR (50%)-and A99 (50%)-conjugated FH (F). Scale bars represent 200 µm. (PDF)
The authors declare no competing financial interest.
REFERENCE
- 1.Almståhl A, Wikström M, Fagerberg-Mohlin B. Oral Dis. 2008;14:541–549. doi: 10.1111/j.1601-0825.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- 2.Castro I, Sepulveda D, Cortes J, Quest AF, Barrera MJ, Bahamondes V, Aguilera S, Urzua U, Alliende C, Molina C, Gonzalez S, Hermoso MA, Leyton C, Gonzalez MJ. Autoimmun. Rev. 2013;12:567–574. doi: 10.1016/j.autrev.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Callea M, Teggi R, Yavuz I, Tadini G, Priolo M, Crovella S, Clarich G, Grasso DL. Int. J. Pediatr Otorhinolaryngol. 2013;77:1801–1804. doi: 10.1016/j.ijporl.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Turner MD, Ship JA. J. Am. Dent. Assoc., JADA. 2007;138:S15–S20. doi: 10.14219/jada.archive.2007.0358. [DOI] [PubMed] [Google Scholar]
- 5.Kałużny J, Wierzbicka M, Nogala H, Milecki P, Kopeć T. Otolaryngol. Pol. 2014;68:1–14. doi: 10.1016/j.otpol.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Lin C-Y, Ju S-S, Chia J-S, Chang C-H, Chang C-W, Chen M-H. J. Dent. Sci. 2015;10:253–262. [Google Scholar]
- 7.Han P, Suarez-Durall P, Mulligan R. J. Prosthodont Res. 2015;59:6–19. doi: 10.1016/j.jpor.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 8.McCall AD, Nelson JW, Leigh NJ, Duffey ME, Lei P, Andreadis ST, Baker OJ. Tissue Eng., Part A. 2013;19:2215–2225. doi: 10.1089/ten.tea.2012.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker OJ, Schulz DJ, Camden JM, Liao Z, Peterson TS, Seye CI, Petris MJ, Weisman GA. Tissue Eng., Part C. 2010;16:1135–1144. doi: 10.1089/ten.tec.2009.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridman R, Giaccone G, Kanemoto T, Martin GR, Gazdar AF, Mulshine JL. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6698–6702. doi: 10.1073/pnas.87.17.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen P, Ritchie A, Langdon SP, Miller WR. Int. J. Cancer. 1996;67:816–820. doi: 10.1002/(SICI)1097-0215(19960917)67:6<816::AID-IJC10>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 12.El-Sherbiny IM, Yacoub MH. Glob Cardiol Sci. Pract. 2013;2013:38. doi: 10.5339/gcsp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janmey PA, Winer JP, Weisel JW. J. R. Soc., Interface. 2009;6:1–10. doi: 10.1098/rsif.2008.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geer DJ, Swartz DD, Andreadis ST. Am. J. Pathol. 2005;167:1575–1586. doi: 10.1016/S0002-9440(10)61242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang MS, Andreadis ST. Biomaterials. 2011;32:8684–8693. doi: 10.1016/j.biomaterials.2011.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang MS, Koobatian M, Lei P, Swartz DD, Andreadis ST. Biomaterials. 2013;34:7281–7291. doi: 10.1016/j.biomaterials.2013.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei P, Padmashali RM, Andreadis ST. Biomaterials. 2009;30:3790–3799. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raut SD, Lei P, Padmashali RM, Andreadis ST. J. Controlled Release. 2010;144:213–220. doi: 10.1016/j.jconrel.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmashali RM, Andreadis ST. Biomaterials. 2011;32:3330–3339. doi: 10.1016/j.biomaterials.2011.01.035. [DOI] [PubMed] [Google Scholar]
- 20.Maruyama CL, Leigh NJ, Nelson JW, McCall AD, Mellas RE, Lei P, Andreadis ST, Baker OJ. J. Dent. Res. 2015;94:1610–1617. doi: 10.1177/0022034515600157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topley P, Jenkins DC, Jessup EA, Stables JN. Br. J. Cancer. 1993;67:953–958. doi: 10.1038/bjc.1993.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beliveau A, Mott JD, Lo A, Chen EI, Koller AA, Yaswen P, Muschler J, Bissell MJ. Genes Dev. 2010;24:2800–2811. doi: 10.1101/gad.1990410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellis SL. Biomaterials. 2011;32:4205–4210. doi: 10.1016/j.biomaterials.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida N, Ishii E, Nomizu M, Yamada Y, Mohri S, Kinukawa N, Matsuzaki A, Oshima K, Hara T, Miyazaki S. Br. J. Cancer. 1999;80:1898–1904. doi: 10.1038/sj.bjc.6690618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlrab S, Müller S, Schmidt A, Neubauer S, Kessler H, Leal-Egaña A, Scheibel T. Biomaterials. 2012;33:6650–6659. doi: 10.1016/j.biomaterials.2012.05.069. [DOI] [PubMed] [Google Scholar]
- 26.Frith JE, Mills RJ, Hudson JE, Cooper-White JJ. Stem Cells Dev. 2012;21:2442–2456. doi: 10.1089/scd.2011.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada Y, Hozumi K, Katagiri F, Kikkawa Y, Nomizu M. Biomaterials. 2013;34:6539–6547. doi: 10.1016/j.biomaterials.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 28.Hosokawa Y, Takahashi Y, Kadoya Y, Yamashina S, Nomizu M, Yamada Y, Nogawa H. Dev., Growth Differ. 1999;41:207–216. doi: 10.1046/j.1440-169x.1999.00419.x. [DOI] [PubMed] [Google Scholar]
- 29.Stuchbury T, Shipton M, Norris R, Malthouse JP, Brocklehurst K, Herbert JA, Suschitzky H. Biochem. J. 1975;151:417–432. doi: 10.1042/bj1510417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlsson J, Drevin H, Axén R. Biochem. J. 1978;173:723–737. doi: 10.1042/bj1730723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marder VJ, Shulman NR, Carroll WR. J. Biol. Chem. 1969;244:2111–2119. [PubMed] [Google Scholar]
- 32.Debye P. J. Phys. Colloid Chem. 1947;51:18–32. doi: 10.1021/j150451a002. [DOI] [PubMed] [Google Scholar]
- 33.Quissell DO, Turner JT, Redman RS. Eur. J. Morphol. 1998;36:50–54. [PubMed] [Google Scholar]
- 34.Turner JT, Redman RS, Camden JM, Landon LA, Quissell DO. Am. J. Physiol. 1998;275:C367–C374. doi: 10.1152/ajpcell.1998.275.2.C367. [DOI] [PubMed] [Google Scholar]
- 35.Odusanwo O, Chinthamani S, McCall A, Duffey ME, Baker OJ. Am. J. Physiol Cell Physiol. 2012;302:C1331–C1345. doi: 10.1152/ajpcell.00207.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner MD, Ship JA. J. Am. Dent. Assoc., JADA. 2007;138:S15–S20. doi: 10.14219/jada.archive.2007.0358. [DOI] [PubMed] [Google Scholar]
- 38.Kinstler O, Molineux G, Treuheit M, Ladd D, Gegg C. Adv. Drug Delivery Rev. 2002;54:477–485. doi: 10.1016/s0169-409x(02)00023-6. [DOI] [PubMed] [Google Scholar]
- 39.Gauthier MA, Klok H-A. Chem. Commun. 2008:2591–2611. doi: 10.1039/b719689j. [DOI] [PubMed] [Google Scholar]
- 40.Weisel JW. Biophys. Chem. 2004;112:267–276. doi: 10.1016/j.bpc.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 41.Wedgwood J, Freemont AJ, Tirelli N. Macromol. Symp. 2013;334:117–125. [Google Scholar]
- 42.Feng J, van der Zwaag M, Stokman MA, van Os R, Coppes RP. Radiother. Oncol. 2009;92:466–471. doi: 10.1016/j.radonc.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 43.Leigh NJ, Nelson JW, Mellas RE, McCall AD, Baker OJ. J. Tissue Eng. Regener. Med. 2014 doi: 10.1002/term.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foskett JK, Melvin JE. Science. 1989;244:1582–1585. doi: 10.1126/science.2500708. [DOI] [PubMed] [Google Scholar]
- 45.Wells KL, Patel N. Front Oral Biol. 2010;14:78–89. doi: 10.1159/000313708. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Shan Z, Ou G, Liu X, Zhang C, Baum BJ, Wang S. Int. J. Radiat. Oncol., Biol., Phys. 2005;62:1510–1516. doi: 10.1016/j.ijrobp.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 47.Goodman TT, Ng CP, Pun SH. Bioconjugate Chem. 2008;19:1951–1959. doi: 10.1021/bc800233a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FD. J. Cell. Physiol. 2015;230:16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 49.Edmondson R, Broglie JJ, Adcock AF, Yang L. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaikh FM, Callanan A, Kavanagh EG, Burke PE, Grace PA, McGloughlin TM. Cells Tissues Organs. 2008;188:333–346. doi: 10.1159/000139772. [DOI] [PubMed] [Google Scholar]
- 51.Dhandayuthapani B, Yoshida Y, Maekawa T, Kumar DS. Int. J. Polym. Sci. 2011;2011:1–19. [Google Scholar]
- 52.Zhu J, Marchant RE. Expert Rev. Med. Devices. 2011;8:607–626. doi: 10.1586/erd.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alston SM, Solen KA, Broderick AH, Sukavaneshvar S, Mohammad SF. Transl Res. 2007;149:187–195. doi: 10.1016/j.trsl.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Swartz DD, Russell JA, Andreadis ST. Am. J. Physiol Heart Circ Physiol. 2005;288:H1451–H1460. doi: 10.1152/ajpheart.00479.2004. [DOI] [PubMed] [Google Scholar]
- 55.Mochizuki M, Kadoya Y, Wakabayashi Y, Kato K, Okazaki I, Yamada M, Sato T, Sakairi N, Nishi N, Nomizu M. FASEB J. 2003;17:875–877. doi: 10.1096/fj.02-0564fje. [DOI] [PubMed] [Google Scholar]
- 56.He L, Liao S, Quan D, Ngiam M, Chan CK, Ramakrishna S, Lu J. Biomaterials. 2009;30:1578–1586. doi: 10.1016/j.biomaterials.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 57.Yan C, Pochan DJ. Chem. Soc. Rev. 2010;39:3528–3540. doi: 10.1039/b919449p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hozumi K, Sasaki A, Yamada Y, Otagiri D, Kobayashi K, Fujimori C, Katagiri F, Kikkawa Y, Nomizu M. Biomaterials. 2012;33:4241–4250. doi: 10.1016/j.biomaterials.2012.02.055. [DOI] [PubMed] [Google Scholar]
- 59.Hanenberg H, Xiao XL, Dilloo D, Hashino K, Kato I, Williams DA. Nat. Med. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.