Abstract
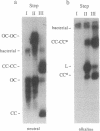
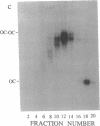
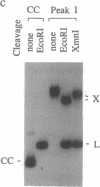
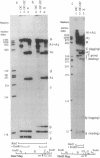
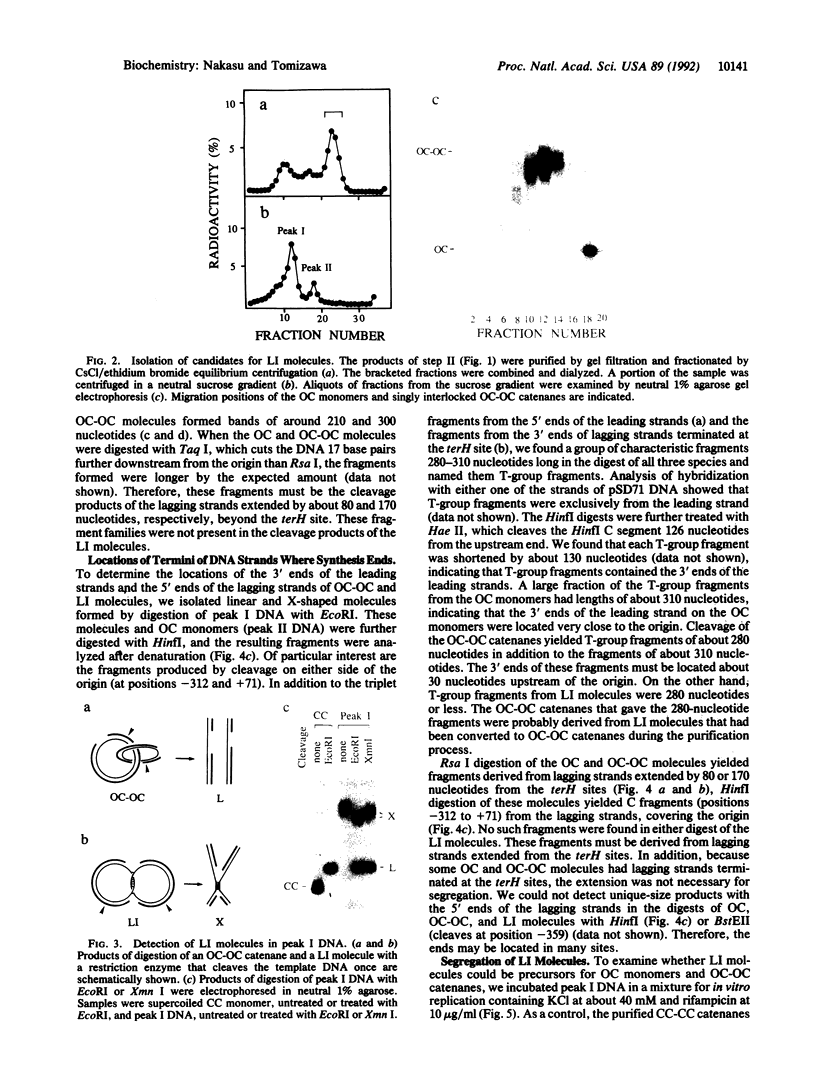
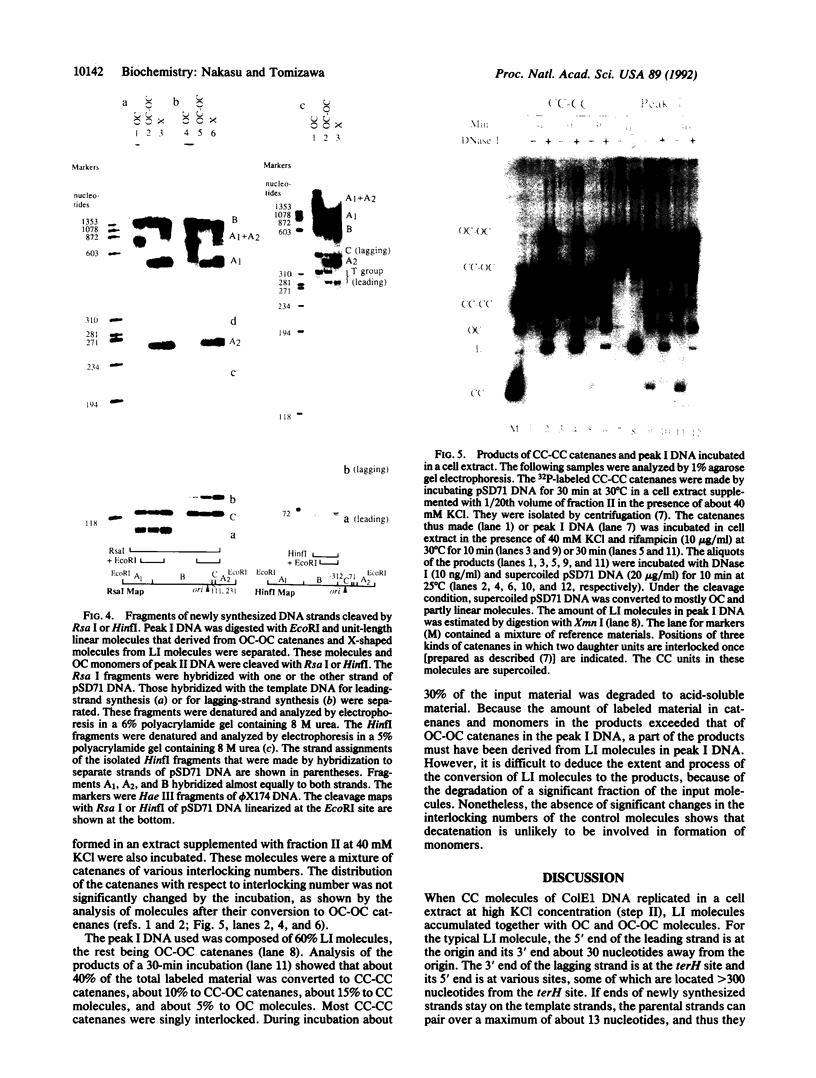
The segregation of daughter DNA molecules at the end stage of replication of plasmid ColE1 was examined. When circular ColE1 DNA replicates in a cell extract at a high KCl concentration (140 mM), a unique class of molecules accumulates. When the molecule is cleaved by a restriction enzyme that cuts the ColE1 DNA at a single site, an X-shaped molecule in which two linear components are held together around the origin of DNA replication is made. For a large fraction of these molecules, the 5' end of the leading strand remains at the origin and the 3' end of the strand is about 30 nucleotides upstream of the origin. The 3' end of the lagging strand is located at the terH site (17 nucleotides upstream of the origin) and the 5' end of the strand is a few hundred nucleotides upstream of the terH site. Thus the parental strands of the molecule intertwine with each other only once. When the KCl concentration is lowered to 70 mM, practically all of these molecules are converted to daughter circular monomers or to catenanes consisting of two singly interlocked circular units.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dasgupta S., Masukata H., Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987 Dec 24;51(6):1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- Dean F., Krasnow M. A., Otter R., Matzuk M. M., Spengler S. J., Cozzarelli N. R. Escherichia coli type-1 topoisomerases: identification, mechanism, and role in recombination. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):769–777. doi: 10.1101/sqb.1983.047.01.088. [DOI] [PubMed] [Google Scholar]
- DiGate R. J., Marians K. J. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988 Sep 15;263(26):13366–13373. [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990 Oct 19;63(2):393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978 Oct 25;253(20):7149–7157. [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985 Aug 5;260(16):9316–9325. [PubMed] [Google Scholar]
- Sakakibara Y., Suzuki K., Tomizawa J. I. Formation of catenated molecules by replication of colicin E1 plasmid DNA in cell extracts. J Mol Biol. 1976 Dec 15;108(3):569–582. doi: 10.1016/s0022-2836(76)80137-4. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y., Tomizawa J. I. Replication of colicin E1 plasmid DNA in cell extracts. Proc Natl Acad Sci U S A. 1974 Mar;71(3):802–806. doi: 10.1073/pnas.71.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y., Tomizawa J. Replication of colicin E1 plasmid DNA in cell extracts. II. Selective synthesis of early replicative intermediates. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1403–1407. doi: 10.1073/pnas.71.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivenugopal K. S., Lockshon D., Morris D. R. Escherichia coli DNA topoisomerase III: purification and characterization of a new type I enzyme. Biochemistry. 1984 Apr 24;23(9):1899–1906. doi: 10.1021/bi00304a002. [DOI] [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984 Apr;36(4):1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980 Aug;21(1):103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Sakakibara Y., Kakefuda T. Replication of colicin E1 plasmid DNA added to cell extracts. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1050–1054. doi: 10.1073/pnas.72.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Masukata H. Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Cell. 1987 Nov 20;51(4):623–630. doi: 10.1016/0092-8674(87)90131-0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Sakakibara Y., Kakefuda T. Replication of colicin E1 plasmid DNA in cell extracts. Origin and direction of replication. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2260–2264. doi: 10.1073/pnas.71.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Tanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986 May;5(5):1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases: why so many? J Biol Chem. 1991 Apr 15;266(11):6659–6662. [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- Weaver D. T., Fields-Berry S. C., DePamphilis M. L. The termination region for SV40 DNA replication directs the mode of separation for the two sibling molecules. Cell. 1985 Jun;41(2):565–575. doi: 10.1016/s0092-8674(85)80029-5. [DOI] [PubMed] [Google Scholar]
- Yang L., Wold M. S., Li J. J., Kelly T. J., Liu L. F. Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A. 1987 Feb;84(4):950–954. doi: 10.1073/pnas.84.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]