Abstract
Gld2, a noncanonical cytoplasmic poly(A) polymerase, interacts with the RNA binding protein CPEB1 to mediate polyadenylation-induced translation in dendrites of cultured hippocampal neurons. Depletion of Gld2 from the hippocampus leads to a deficit in long-term potentiation evoked by theta burst stimulation. At least in mouse liver and human primary fibroblasts, Gld2 also 3′ monoadenylates and thereby stabilizes specific miRNAs, which enhance mRNA translational silencing and eventual destruction. These results suggest that Gld2 would be likely to monoadenylate and stabilize miRNAs in the hippocampus, which would produce measurable changes in animal behavior. We now report that using Gld2 knockout mice, there are detectable alterations in specific miRNA monoadenylation in the hippocampus when compared to wild type, but that these modifications produce no detectable effect on miRNA stability. Moreover, we surprisingly find no overt change in animal behavior when comparing Gld2 knockout to wild-type mice. These data indicate that miRNA monoadenylation-mediated stability is cell type-specific and that monoadenylation has no measurable effect on higher cognitive function.
Keywords: Gld2, miRNA, monoadenylation, hippocampus, behavior
INTRODUCTION
The cytoplasmic polyadenylation element binding protein (CPEB1) is an RNA binding protein that promotes cytoplasmic polyadenylation and translation. Through its regulation of mRNA translation, CPEB1 influences a number of biological events including gamete development and early embryogenesis, cell cycle progression, cellular senescence, glucose homeostasis, neuronal synaptic plasticity, and learning and memory (Ivshina et al. 2014). CPEB1 recognizes the cytoplasmic polyadenylation element (CPE) that resides in mRNA 3′ UTRs and nucleates several factors to promote polyadenylation among which are PARN [poly(A) ribonuclease], a deadenylating enzyme (Kim and Richter 2006), and Gld2 [germline development 2; also referred to as poly(A) polymerase containing domain 4, PAPD4, or terminal uridylyl transferase 2, TUT2], a noncanonical poly(A) polymerase (Barnard et al. 2004; Kim and Richter 2006). CPEB1 also binds a second noncanonical poly(A) polymerase, Gld4 (PAPD5), which promotes polyadenylation (Burns et al. 2011).
In addition to mRNA polyadenylation, Gld2 catalyzes 3′ monoadenylation of specific miRNAs (Katoh et al. 2009, 2015; Burns et al. 2011; D'Ambrogio et al. 2012). miR122, the most abundant miRNA in liver, is post-transcriptionally 3′ monoadenylated in wild-type (WT) but not in Gld2 knockout (KO) mice; the lack of this modification leads to miRNA instability. PARN appears to be the key enzyme that deadenylates and destabilizes miR122 (Katoh et al. 2015). In human primary fibroblasts, miR122 and let-7 family members are also monoadenylated, and, in concordance with the observations of Katoh et al. (2009), Gld2 depletion leads to miRNA deadenylation and destabilization (Burns et al. 2011). A third activity of Gld2 is monouridylation of pre-miRNAs, which promotes their maturation to miRNAs (Heo et al. 2012; Kim et al. 2015). Gld2 also uridylates mature miRNAs in vitro, albeit with less efficiency compared to other terminal uridylases (D'Ambrogio et al. 2012).
In the rodent brain, Gld2, PARN, and CPEB1 are present in the hippocampus, particularly in dendrites and at post-synaptic sites (Udagawa et al. 2012). In response to synaptic stimulation, Gld2 catalyzes cytoplasmic polyadenylation, which leads to the translation of GluN2A mRNA (Swanger et al. 2013; Udagawa et al. 2013). GluN2A is a subunit of the post-synaptic N-methyl-d-aspartate receptor (NMDAR) and is a key component that mediates synaptic plasticity. Consistent with these observations, stereotactic injection of lentivirus expressing Gld2 shRNA into the rat hippocampus followed by theta burst stimulation (TBS) evoked a deficit in long-term potentiation (LTP), a form of synaptic plasticity strongly linked to learning and memory (Malinow et al. 2000; Kelleher et al. 2004). These observations therefore suggest that mice lacking Gld2 would have altered learning and memory and perhaps other behavioral anomalies as well.
Based on the foregoing, we addressed two questions: Does Gld2 monoadenylate or monouridylate miRNAs in the mouse hippocampus and do Gld2 knockout (KO) mice exhibit impaired learning and memory or other behavioral deficits? We find that Gld2 mediates monoadenylation but has no observable effect on miRNA abundance. We find only a single miRNA with a statistically significant increase and no miRNAs with a statistically significant decrease in monouridylation in Gld2 KO mouse hippocampus. In addition, we detect no observable change in the behavior of Gld2 KO mice compared to WT. Thus, although we can attribute several molecular and cellular events to Gld2 in the mouse brain, they have no apparent influence on higher cognitive function.
RESULTS AND DISCUSSION
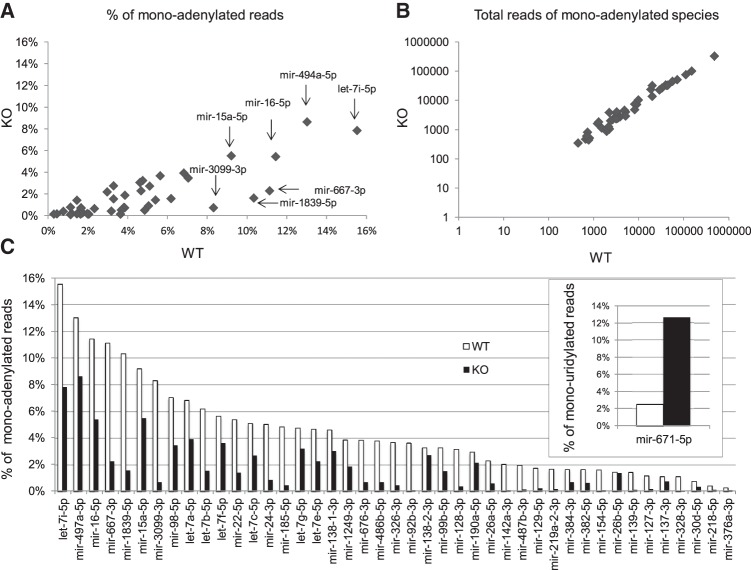
We obtained global Gld2 KO mice (Nakanishi et al. 2007) and confirmed that in contrast to wild type (WT), Gld2 is not expressed in the hippocampus in these animals (Supplemental Fig. 1A). To assess the involvement of Gld2 in nontemplated nucleotide addition to miRNAs, hippocampi from six WT and six Gld2 KO male mice were isolated and cDNA libraries were constructed from gel-isolated RNAs between 18 and 32 bases in length, which primarily include miRNAs. The libraries had an average length of 22 base pairs (bp) with a general size distribution of 19–25 bp (Supplemental Fig. 1B). The libraries had 3–5.3 million total sequences although the Gld2 KO libraries contained ∼23% fewer sequences (Supplemental Fig. 1C). However, the total miRNA reads were similar in the libraries from both genotypes (Supplemental Fig. 1D,E). Of 1149 miRNAs detected in the hippocampus, 97 showed changes in 3′ monoadenylate residues between the genotypes; however, only 43 of these had a statistically significant reduction in the amount of monoadenylation in the Gld2 KO compared to the WT hippocampus (Fig. 1A; Supplemental Table 1). However, there were no statistically significant differences in the steady-state levels of these miRNAs between the two genotypes (Fig. 1B; Supplemental Fig. 1). We confirmed the levels of 14 miRNAs by qPCR (Supplemental Fig. 2).
FIGURE 1.
Gld2 controls miRNA monoadenylation. (A) Scatter plot of mean percentages of 3′ monoadenylated counts for 43 miRNAs in six WT and six Gld2 KO hippocampi. (B) Scatter plot on log scale of the steady-state levels (average read counts) of 43 specific hippocampal miRNAs that are differentially 3′ monoadenylated in WT and Gld2 KO mice as identified in panel A. (C) Bar plot representation of panel A showing mean percentages of 43 specific miRNAs that are differentially monoadenylated in WT and Gld2 KO hippocampus (all have a P-value <0.05; Mann-Whitney U-test). (C, inset) Mean percentages of mir-671-5p that are monouridylated in Gld2 KO hippocampus (P-value <0.05).
Figure 1C quantifies the changes in monoadenylation of specific miRNAs. The miRNAs with the highest proportion of monoadenylation include members of the let-7 family, which were also the most abundant miRNAs in the hippocampus. For example, let-7i-5p was ∼16% monoadenylated in WT hippocampus but ∼8% monoadenylated in Gld2 KO hippocampus. In the majority of cases, monoadenylation was reduced in the Gld2 KO hippocampus versus WT, but as noted above, there were no detectable changes in overall miRNA abundance regardless of miRNA modifications between the genotypes (Fig. 1B; Supplemental Fig. 1E). An alignment of the hippocampal miRNAs that are monoadenylated does not reveal any particular motif or conserved nucleotides at the 3′ end (Supplemental Fig. 3). These results are in contrast to those of Katoh et al. (2009) and Burns et al. (2011), who demonstrated that Gld2 controls monoadenylation and miRNA stability in liver and primary human fibroblasts, respectively. Our sequencing data also showed that only mir-671-5p had a statistically significant increase in its 3′ uridylation in Gld2 KO versus WT hippocampus (Fig. 1C, inset; Supplemental Table 2).
We have found that nontemplated 3′ monoadenylation of miRNAs is widespread in the mouse hippocampus but that the proportion of any given miRNA that is modified in this manner is modest. For example, let-7i-5p, at ∼16%, is the most monoadenylated of any miRNA in this tissue; most miRNAs, however, are <5% monoadenylated. In human primary fibroblasts, miR122 is the most monoadenylated at ∼40%; ∼9% of miRNAs overall are monoadenylated (Burns et al. 2011; D'Ambrogio et al. 2012). In this regard, the hippocampus and primary human fibroblasts are roughly similar. However, these values vary substantially from those in human embryonic stem cells and several cancer cell lines in which ∼50% or more of miRNAs are monoadenylated (Burroughs et al. 2010; Wyman et al. 2011), among the most prevalent of which is miR21 (Boele et al. 2014). In mouse liver, miR122, the most abundant miRNA in that tissue, is ∼75% monoadenylated (Katoh et al. 2009). In many of these cases, miRNA monoadenylation is correlated with enhanced stability.
In contrast to what occurs in human fibroblasts, mouse liver, or other cell types, we detected no loss of any miRNA in Gld2 KO hippocampus, indicating tissue-specific destabilization when specific miRNAs are not normally monoadenylated. It might be argued that because PARN is so important for miRNA destruction (Boele et al. 2014; Katoh et al. 2015), this enzyme might be absent from the hippocampus and thus nonadenylated miRNAs are stable. However, we have shown that PARN is quite abundant in that tissue, that it resides in a complex containing Gld2, and that these two enzymes colocalize in dendritic spines (Udagawa et al. 2012). We therefore surmise that the lack of miRNA destabilization is not due to the absence of PARN. It might also be argued that miRNA monoadenylation might have some effect on, say, the formation or stability of miRNA–mRNA duplexes. If true, then one predicted outcome would be altered animal behavior, which is not the case (see below).
Gld2 KO mouse behavior is normal
Although Gld2 had no effect on miRNA abundance in the hippocampus, it was still possible that Gld2-catalyzed monoadenylation might alter miRNA metabolism such as loading into RISC, which could have an effect on mRNA translation and/or stability. This possibility, as well as the observations that Gld2 regulates polyadenylation-induced translation in the hippocampus and synaptic plasticity in that brain region, suggested that this enzyme would be likely to mediate animal behavior. Accordingly, we performed a battery of behavioral tests to determine whether Gld2 is involved in learning and memory, anxiety, or repetitive disorders.
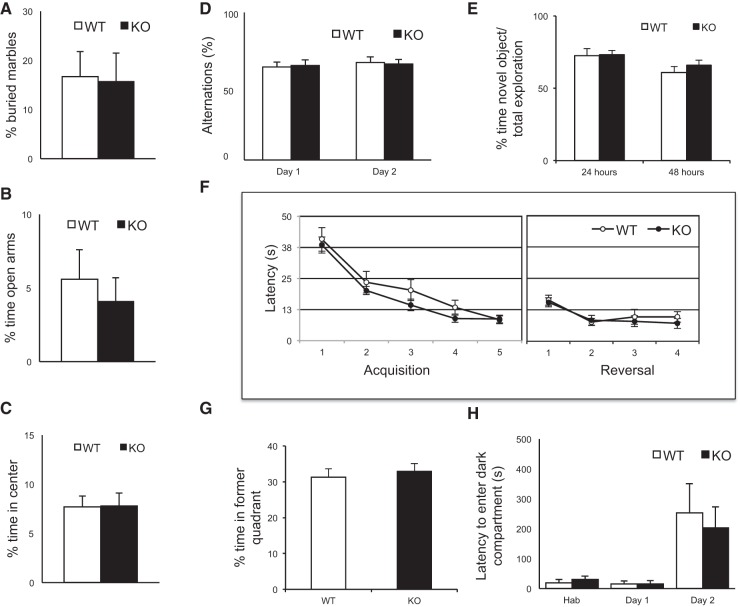
Figure 2A demonstrates results of a marble burying assay, which measures anxiety and obsessive-compulsive behavior. In this test, the rate at which WT and Gld2 KO mice bury 15 marbles in their bedding was measured over 30 min. There was no difference between genotypes. Figure 2B depicts results from an elevated plus maze, an apparatus commonly used to assess anxiety. Here, a mouse must choose between closed or open arms on an elevated platform; anxiety is correlated with increased time in the closed arms. Again, there was no statistical difference between WT and KO animals. Figure 2C shows results of an open field test, which is a measure of an animal's willingness to explore as well as anxiety. In this assay, the time an animal spends in the center of a plexiglass box is measured relative to the time at the periphery; no difference between the genotypes was observed. Figure 2D shows results from the T-maze assay, which reflects working or short-term memory. Here, the animal has a left–right choice as it explores a maze, and the number of spontaneous alternations was determined. No difference between the genotypes was observed. Figure 2E displays results from a novel object recognition assay. Mice usually spend more time with a new object than with a familiar one. There was no difference between genotypes at either 24 or 48 h with this assay. Figure 2F (right panel) shows results from a Morris water maze, which measures spatial learning and memory. With this test, an animal is placed in a tank of water and the time it takes for it to find a submerged platform is measured. As the animal learns the position of the platform (spatial learning), the faster it swims to it. There was no difference in latency times between genotypes. In a reversal experiment, the platform is moved to a different quadrant of the water maze and the time the animal takes to learn the new position of the new platform is measured. As before, there was no difference between genotypes (Fig. 2F, left panel). In a further measure of learning, the platform is removed completely from the water maze and the time the animal swims in the quadrant where the platform was previously is measured. As before, there was no distinction between the genotypes (Fig. 2G). Finally, Figure 2H shows results from a passive avoidance test. In this test, an animal is placed in a lighted chamber; a door between this chamber and a dark chamber is then opened and the animal immediately moves to the dark. Once in the dark chamber, the animal receives a mild foot shock; 24 h later, it is then placed back into the light chamber and the time it takes to re-enter the dark chamber is measured (this is referred to as latency). In this passive avoidance assay, which is a measure of learning and memory, there was no difference in the latency between the genotypes.
FIGURE 2.
WT and Gld2 KO mice exhibit similar behaviors. (A) Marble burying test to measure obsessive-compulsive behavior and aversion to new objects (WT = 12 mice; Gld2 KO = 14 mice; P > 0.05, unpaired t-test). All error bars in the figure refer to SEM. (B) Elevated plus maze used to assess anxiety objects (WT = 11 mice; Gld2 KO = 11 mice; P < 0.05, unpaired t-test). (C) Open field test to measure anxiety (WT = 12 mice; Gld2 KO = 12 mice; P < 0.05, unpaired t-test). (D) T-maze assay of spontaneous alternation used to assess working memory (WT = 11 mice; Gld2 KO = 12 mice; P < 0.05, unpaired t-test). (E) Novel object recognition as a memory and object recognition test (WT = 12 mice; Gld2 KO = 14 mice; P < 0.05, unpaired t-test). (F) Morris water maze to investigate spatial learning and memory; left panel depicts acquisition (WT = 12 mice; Gld2 KO = 13 mice; P < 0.05, unpaired t-test) and right panel depicts reversal (WT = 12 mice; Gld2 KO = 13 mice; P < 0.05, unpaired t-test). (G) Morris water maze probe trial after acquisition phase (WT = 12 mice; Gld2 KO = 13 mice; P < 0.05, unpaired t-test). (H) Passive avoidance test used to assess learning and memory (WT = 3 mice; Gld2 KO = 3 mice; P < 0.05, unpaired t-test). (Hab) Habituation.
The behaviors we have assessed are linked to many brain regions but particularly the hippocampus. Learning and memory assays such as the Morris water maze, passive avoidance, novel object recognition, and the T-maze all require hippocampal activity (Berger-Sweeney et al. 2006; Tort et al. 2008). The elevated plus maze measure of anxiety generally requires the amygdala but the hippocampus as well. Although there are of course many other tests to assess higher cognitive function, the assays we used are widely used to identify defects in learning and memory.
The observation that Gld2 KO mice display behaviors indistinguishable from WT is surprising. First, Gld2 mediates hippocampal LTP, which is strongly correlated with, if not causative for, memory formation and processing (Malinow and Malenka 2002). Second, Gld2 mediates both basal and synaptic stimulation of mRNA polyadenylation in hippocampal dendrites and thereby controls Gln2A (NR2A) mRNA translation (Udagawa et al. 2012; Swanger et al. 2013). Moreover, this translation results in the insertion into membranes of Gln2A, a component of N-methyl-d-aspartate receptors (NMDARs) that regulate synaptic plasticity (Swanger et al. 2013). Thus, the loss of this Gld2-regulated Gln2A mRNA translation would be expected to result in impaired synaptic efficacy and deficits in animal behavior. Third, Gld2 is important for long-term memory in Drosophila (Kwak et al. 2008). However, we cannot exclude the possibility that other enzymes such as Gld4 (PAPD5) can compensate for the lack of Gld2 vis-à-vis miRNA monoadenylation or animal behavior (but not for LTP because there is a deficit in this measure following Gld2 depletion in rats [Udagawa et al. 2013]). Indeed, Wyman et al. (2011) demonstrated that both Gld2 and Gld4 can monoadenylate miRNAs. However, these in vitro experiments are not obviously reflected in in vivo experiments showing that loss of Gld2 alone induces miRNA instability (Katoh et al. 2009; Burns et al. 2011). Animal behavior of course is much more complex than in vitro monoadenylation assays, and thus it is possible that Gld4 (or other enzyme) substitution for Gld2 cannot be ruled out. If such is the case, it is likely to involve cytoplasmic mRNA polyadenylation rather than miRNA monoadenylation.
MATERIALS AND METHODS
Animals and library construction
All experiments with mice were conducted in accordance with approved institutional IACUC protocols for the treatment and handling of animals. Total RNA was TRIzol extracted from the hippocampi of six 3-mo-old wild-type and six Gld2 KO male mice on C57BL/6 backgrounds (Nakanishi et al. 2007) from littermates derived from Gld2 heterozygous matings. RNA was resolved by gel electrophoresis and the species migrating with a size of 28–32 bases was excised and used for cDNA library construction, as described by Gu et al. (2009). The barcoded samples were sequenced on an Illumina Hi-Seq 2500 instrument.
miRNA validation by RT-qPCR
To confirm the expression of miRNAs in WT and Gld2 KO hippocampus, the levels of 14 mature miRNAs were measured by RT-qPCR using a miScript Sybr Green PCR kit (Qiagen) in combination with miScript primer assays (Qiagen) and universal primer (Qiagen) according to the manufacturer's recommended conditions in three independent RNA samples with two technical replicates. The input cDNA was synthesized using 1 µg of total hippocampal RNA using HiSpec buffer. Real-time RT-PCR detection was performed on an Applied Biosystems 7300 real-time PCR system in 25 µL reactions. Candidate targets were normalized to the reference gene (snoRD6) expression, and the fold difference in Gld2 KO samples relative to the WT were calculated using the 2−ΔΔCT method.
Data preprocessing
The total number of 50-bp single-end reads sequenced was 86.9 million. The data preprocessing has three steps: (i) The fastq reads were demultiplexed using the fastx barcode_splitter of the FASTX-toolkit (v0.0.14) (http://hannonlab.cshl.edu/fastx_toolkit/index.html) allowing 1-bp mismatch in the barcode (that is 4 bp long). (ii) Poor quality reads and the barcodes were trimmed out using Trimmomatic (v0.32) (Bolger et al. 2014). (iii) Adapter sequence and reads <12 bp were removed using Cutadapt (v1.3) (http://cutadapt.readthedocs.org/en/stable/guide.html). FastQC v0.10.1 (http://www.bioinformatics.bbsrc.ac.uk/projects/fasQCc/) was used to generate sequence quality reports at the end of each step. The preprocessing step resulted in 12 fastq sequences, each ranging between 3.0 and 5.3 million reads (Supplemental Fig. 1C), and the sequence length distribution peaks between 21 and 23 bp (Supplemental Fig. 1B), indicative of miRNA enrichment.
Quantification of miRNA gene expression
Three pipelines were used to quantify miRNA gene expression: MiRDeep2 (v2.0.0.5) (Friedländer et al. 2008), Omiras (March 2015) (Müller et al. 2013), and Kraken (v12.164) (Davis et al. 2013). miRDeep2 (Friedländer et al. 2008) is a software package to identify novel and known miRNAs in deep sequencing data. mirDeep2 analyzes the compatibility of sequenced RNAs with miRNA biogenesis. mirDeep2 incorporates third-party tools such as Bowtie v0.12.9 for read mapping (Langmead et al. 2009); RNAFold v2.0 for RNA folding (Gruber et al. 2008); and Randfold v2.0 for calculating significance of free energies (Bonnet et al. 2004). The 12 preprocessed fastq sequences were used as input for the mirDeep2 analysis. In this analysis, the mouse genome (Ensembl GRCm38) and mirBase annotation (version 21) are used. Omiras (Müller et al. 2013) is a web server for differential expression analysis of miRNAs derived from small RNA-seq experiments. Omiras incorporates third-party tools: Bowtie for read mapping (Langmead et al. 2009); mirDeep2 for finding novel miRNA (Friedländer et al. 2008); and DESeq for normalizing the data and calculating differential expression (Anders and Huber 2010). The 12 preprocessed fastq sequences were submitted to the Omiras web server. The results generated were annotations including length distribution, mapping statistics, alignments, and gene quantification tables for each library plus lists of differentially expressed ncRNAs (http://tools.genxpro.net/omiras/85f099c7f02c). Kraken (Davis et al. 2013) is a set of tools for quality control and analysis of high-throughput sequence data. The raw (unprocessed) sequence data were entered into the Kraken pipeline. Sequence Imp, the key Kraken module, incorporates other Kraken modules and streamlines the miRNA sequence analysis. The fastq files were demultiplexed. The adapter sequences were trimmed and filtered for quality and contamination. The filtered reads were aligned using Bowtie (Langmead et al. 2009) against the mouse genome (Ensembl GRCm37) (Flicek et al. 2011). The genomic alignments were compared to genomic coordinates for miRBase mature miRNAs (version 18) (Kozomara and Griffiths-Jones 2011), resulting in a matrix of redundant and nonredundant read depths from each known mature miRNA.
Differential expression calculations
The nonredundant read depths for each miRNA generated from mirDeep2 and Kraken were used for differential expression analysis using DESeq2 (Love et al. 2014). Omiras incorporates DESeq (Anders and Huber 2010) for the differential expression calculation.
Detection of post-transcriptional modifications
The post-transcriptional modification profiles for the miRNAs from the small RNA sequences were detected using the web-based system Chimira (Vitsios and Enright 2015). The preprocessed sequences were uploaded to Chimira (Vitsios and Enright 2015) and mapped against the mouse miRBase (version 21) (Griffiths-Jones et al. 2008) using BLASTn (Boratyn et al. 2013), allowing up to two mismatches for each sequence. miRNAs with modifications at the 3p end with at least 10 mapped reads and at the +1 position were extracted to quantify additional Us, As, Cs, and Gs. Counts for monoadenylation and monouridylation were compiled for the wild-type and the GLD2 knockout samples. Two additional post-transcriptional analysis pipelines were used: a workflow incorporating HAMR (Ryvkin et al. 2013) and an in-house pipeline used previously (D'Ambrogio et al. 2012). Shapiro–Wilk and Kolmogorov–Smirnov normality tests were used to assess normal distribution. Nonparametric Mann-Whitney U-tests were used to test between conditions and determine P-values (Supplemental Tables 1, 2).
Behavioral assays
Adult male wild-type and Gld2 knockout mice (Nakanishi et al. 2007), 8–14 wk of age and generated from Gld2 heterozygous matings were used for all behavioral tests. Mice were group-housed (2–5 animals) and maintained on a 12-h light/dark cycle. Experiments were performed sequentially in the following order: marble burying, elevated plus maze, T-maze, open field test, novel object recognition, and Morris water maze. Passive avoidance was performed with a separate cohort of animals.
Marble burying
Marble burying is one indicator of obsessive-compulsive behavior or anxiety (Njung'e and Handley 1991). Polycarbonate cages (19 × 30 × 12 cm) filled with a 6-cm layer of bedding were used for the tests. Animals were habituated to the cages 2 d for ∼1 h each day. Fifteen evenly spaced marbles were placed on the bedding on the third day. The mice were then placed into the cage and allowed to bury the marbles for 30 min. The number of buried marbles was determined.
Elevated plus maze
The elevated plus maze, which is used to determine anxiety, has two open and two closed arms. Mice were placed at the intersection of the open and closed arms, and mouse behavior was recorded during a 5-min test interval. The time spent in the open and closed arms as well as the number of entries into each arm was measured.
T-maze
This assay tests spatial working or short-term memory (Udagawa et al. 2013). The maze consists of three arms, a start arm and two T arms. Mice placed in the start arm will explore one of the other two arms when it makes a left–right choice. The percentage of time the mice chose left–right alternatives 15 times was recorded for two consecutive days. Chance level of alternation is 50%.
Open field test
This is an assay that measures exploratory behavior as well as anxiety. Animals are placed in the center of a plexiglass box; the distance they traveled and time they spent in the center versus the periphery of the box were recorded for 10 min total duration. The time spent in the periphery is correlated with heightened anxiety.
Novel object recognition
This test is an indicator of memory. Animals are subjected to a 10-min training period when they are able to explore two novel objects. Twenty-four and 48 h later, the animals are placed back in the same space with one of the original objects but also with a new object. The percentage of time the animals spent with the novel object relative to total time spent with the objects was calculated.
Morris water maze
This test, which was performed as described by Berger-Sweeney et al. (2006), measures learning and memory. A circular water-filled pool is hypothetically divided into four quadrants with different visual cues above each quadrant. A platform just underneath the surface is placed in one of the quadrants, and the mice use the visual cues to navigate to the platform. Testing consists of an acquisition phase and a reversal of learning phase. The acquisition phase took place over 5 d with four trials per day as the animals learned the position of the platform. The probe trial took place 24 h after the last day of acquisition. Here, the platform was removed, and the time the animal spent in the quadrant that formerly contained the platform was determined. For the reversal phase, the platform was placed in the opposite quadrant, and the time required to find this newly positioned platform was measured as it was in the acquisition phase.
Passive avoidance
This assay uses an aversive stimulus to measure learning and memory. The test was performed over three consecutive days and was composed of three phases: habituation, acquisition, and retention. For habituation, mice were placed in a brightly lit chamber, in which they can access a dark chamber through a door. The time the animals took to enter the dark chamber once the door was opened was measured. The next day, the experiment was repeated except that when the animals entered the dark chamber, they received a 0.25 mA foot shock for 2 sec (acquisition). On the third day (retention), the animals were again placed in the lighted chamber and the door opened; the latency for the animals to enter the dark chamber was measured. If the animals did not enter the dark chamber within 600 sec, the experiment was terminated.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Baba and G. Gao for providing Gld2 KO mouse strain. We thank Andrew Tapper and Jennifer Ngolab for use of their behavioral mouse facilities and for initial help with the elevated plus maze and marble burying assays and Douglas T. Golenbock and Techen Tzeng for use and initial help with the water maze. F.M. was supported by a Science Without Borders Fellowship, CNPq, Brazil. This work was supported by National Institutes of Health grants NS079415 and GM46779.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.056937.116.
REFERENCES
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651. [DOI] [PubMed] [Google Scholar]
- Berger-Sweeney J, Zearfoss NR, Richter JD. 2006. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem 13: 4–7. [DOI] [PubMed] [Google Scholar]
- Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Søkilde R, Hawkins SM, Coarfa C, Ikeda K, Takayama K, et al. 2014. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci 111: 11467–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. 2004. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20: 2911–2917. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41: 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, D'Ambrogio A, Nottrott S, Richter JD. 2011. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrogio A, Gu W, Udagawa T, Mello CC, Richter JD. 2012. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep 2: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MP, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. 2013. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y. 2011. Ensembl 2012. Nucleic Acids Res 39: D800–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36 Suppl 2: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. 2012. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151: 521–532. [DOI] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD. 2014. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol 30: 393–415. [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Hojo H, Suzuki T. 2015. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res 43: 7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ III, Govindarajan A, Tonegawa S. 2004. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron 44: 59–73. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183. [DOI] [PubMed] [Google Scholar]
- Kim B, Ha M, Loeff L, Chang H, Simanshu DK, Li S, Fareh M, Patel DJ, Joo C, Kim VN. 2015. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J 34: 1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. 2008. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci 105: 14644–14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. 2002. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25: 103–126. [DOI] [PubMed] [Google Scholar]
- Malinow R, Mainen ZF, Hayashi Y. 2000. LTP mechanisms: from silence to four-lane traffic. Curr Opin Neurobiol 10: 352–357. [DOI] [PubMed] [Google Scholar]
- Müller S, Rycak L, Winter P, Kahl G, Koch I, Rotter B. 2013. omiRas: a Web server for differential expression analysis of miRNAs derived from small RNA-Seq data. Bioinformatic 29: 2651–2652. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Kumagai S, Kimura M, Watanabe H, Sakurai T, Kimura M, Kashiwabara S, Baba T. 2007. Disruption of mouse poly(A) polymerase mGLD-2 does not alter polyadenylation status in oocytes and somatic cells. Biochem Biophys Res Commun 364: 14–19. [DOI] [PubMed] [Google Scholar]
- Njung'e K, Handley SL. 1991. Effects of 5-HT uptake inhibitors, agonists and antagonists on the burying of harmless objects by mice; a putative test for anxiolytic agents. Br J Pharmacol 104: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, Wang LS. 2013. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19: 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SA, He YA, Richter JD, Bassell GJ. 2013. Dendritic GluN2A synthesis mediates activity-induced NMDA receptor insertion. J Neurosci 33: 8898–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tort AB, Kramer MA, Thorn C, Gibson DJ, Kubota Y, Graybiel AM, Kopell NJ. 2008. Dynamic cross-frequency couplings of local field potential oscillations in rat striatum and hippocampus during performance of a T-maze task. Proc Natl Acad Sci 105: 20517–20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. 2012. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell 47: 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, et al. 2013. Genetic and acute CPEB depletion ameliorate Fragile X pathophysiology. Nat Med 19: 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitsios DM, Enright AJ. 2015. Chimira: analysis of small RNA sequencing data and microRNA modifications. Bioinformatics 31: 3365–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21: 1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.