Abstract
As the need for new and more effective antibiotics increases, untapped sources of biodiversity are being explored in an effort to provide lead structures for drug discovery. Endophytic fungi from marine macroalgae have been identified as a potential source of biologically active natural products, although data to support this is limited. To assess the antibiotic potential of temperate macroalgal endophytes we isolated endophytic fungi from algae collected in the Bay of Fundy, Canada and screened fungal extracts for the presence of antimicrobial compounds. A total of 79 endophytes were isolated from 7 species of red, 4 species of brown, and 3 species of green algae. Twenty of the endophytes were identified to the genus or species level, with the remaining isolates designated codes according to their morphology. Bioactivity screening assays performed on extracts of the fermentation broths and mycelia of the isolates revealed that 43 endophytes exhibited antibacterial activity, with 32 displaying antifungal activity. Endophytic fungi from Bay of Fundy macroalgae therefore represent a significant source of antibiotic natural products and warrant further detailed investigation.
Keywords: marine macroalgae, endophyte, antibacterial, antifungal
1. Introduction
The threat to human health as a result of the growing emergence of microbial resistance to antibiotic agents is both real and significant; current forecasts predict that broad-scale antimicrobial ineffectiveness is imminent and we may soon once again face the problems that challenged medicine in the “pre-antibiotic” era [1,2,3]. There is, therefore, an urgent need to accelerate the discovery of antibiotic molecules to facilitate the development of new therapeutic agents with novel modes-of-action to combat infectious disease [4,5]. Endophytes of macroalgae have recently gained attention as an untapped source of biodiversity with potential to yield novel bioactive metabolites [6,7,8] and high proportions of isolates from both tropical [9] and temperate [10,11] environments have exhibited significant antibiotic activities. However, data relating to the bioactivity of endophyte assemblages obtained from a given algal species is limited; evaluating the true potential of macroalgal endophytes as a source of antibiotic compounds is therefore problematic. Recent reports also suggest that there may be significant differences in the endophyte assemblages found within tropical and temperate marine macroalgal hosts [9,11], although further work is required to confirm these preliminary observations. The objectives of this study were to perform a preliminary investigation of the endophytes from macroalgae from the Atlantic coast of Canada and evaluate their potential for the production of antimicrobial compounds.
2. Experimental Section
2.1. Algal Collection
Fourteen species of algae were collected from L’Etete, New Brunswick, Canada (45° 02.372′ N, 66° 53.424′ W) from August to November 2009. Seven species of red algae (Porphyra umbilicalis, Porphyra purpurea, Palmaria palmata, Chondrus crispus, Devaleraea ramentacea, Mastocarpus stellatus and Polysiphonia lanosa), four species of brown algae (Fucus spiralis, Fucus vesiculosus, Saccharina latissima and Ascophyllum nodosum) and three species of green algae (Spongomorpha arcta, Ulva intestinalis and Ulva lactuca) were investigated for the presence of endophytic fungi. In all cases algae with no obvious signs or symptoms of disease were used.
2.2. Surface Sterilization of Algae and Culture Techniques
The surfaces of the algal samples were sterilized by immersion in various sterilant solutions [11]. Prior to the isolation of endophytic fungi from marine algae, an optimal surface sterilization method was developed for each algal species (Table 1). Portions (5 cm2) of algal tissue were individually surface sterilized using the appropriate optimized technique, blotted dry on autoclaved paper towel and rubbed across the surface of 2 plates of 2% malt extract agar (MEA, Becton Dickinson, Sparks, MD, USA) prepared with artificial seawater (MEA-SW, 24.4 g·L−1; Instant Ocean® Sea Salt, Cincinnati, OH, USA) to verify surface sterilization had been effective. Sterilized algal species were cut using a sterile cork borer and placed on Petri plates of 2% MEA and 2% MEA with seawater. Petri plates were sealed with Parafilm™ (Pechiney Plastic Packaging Company, Chicago, IL, USA) prior to incubation.
Table 1.
Surface sterilization techniques used on marine algae collected from the Bay of Fundy, Canada. 1
| Algal Species | Sterilant Immersion Duration (Seconds) | |
|---|---|---|
| Bleach (5.25%) | Ethanol (70%) | |
| Chondrus crispus | 5 | 10 |
| Devaleraea ramentacea | 5 | 10 |
| Mastocarpus stellatus | 5 | 15 |
| Palmaria palmata | 0 | 15 |
| Polysiphonia lanosa | 0 | 10 |
| Porphyra purpurea | 0 | 10 |
| Porphyra umbilicalis | 0 | 10 |
| Ascophyllum nodosum | 0 | 15 |
| Fucus spiralis | 0 | 15 |
| Fucus vesiculosus | 0 | 15 |
| Saccharina latissima | 5 | 15 |
| Spongomorpha arcta | 0 | 20 |
| Ulva intestinalis | 0 | 20 |
| Ulva lactuca | 0 | 20 |
1 All surface sterilizations were confirmed by the absence of microbial colony formation on 2 surface verification plates (2% malt extract with artificial seawater).
2.3. Isolation of Endophytes
Petri plates containing surface sterilized macroalgal pieces were incubated for 14 days at room temperature (approximately 25 °C) under ambient light conditions and monitored daily for the presence of hyphae growing from the cut edges of the algal segments. The isolation frequency (IF) of emerging hyphae was determined for each species of algae collected [12]:
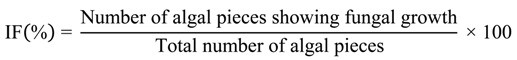 |
(1) |
Endophytes growing from the cut edges of the segments were subcultured onto fresh media (2% MEA) to obtain pure isolates. Pure isolates were also grown on Czapek, potato dextrose, cornmeal and marine agars (Becton Dickinson, Sparks, MD, USA) to induce sporulation and differentiate between colony morphologies. Individual isolates from each algal species were then sorted into groups of homogeneous morphotypes, and a representative colony of each distinct isolate was used for the fungal identification, fermentation, and extraction.
2.4. Identification of Fungi
Fungal isolates were identified taxonomically through examination of colony and spore morphology with taxonomic classifications being confirmed by comparison of the internal transcribed spacer and 5.8S rRNA gene (ITS) DNA regions [13] with corresponding sequences available in the GenBank database (National Center for Biotechnology Information, U.S. National Library of Medicine, Bethesda, MD, USA).
The genomic DNA of all distinct fungal isolates was extracted using a DNeasy® plant mini kit (Qiagen, Toronto, Ontario, Canada) and amplified by PCR as previously described [11]. Samples of amplified ITS DNA were submitted for sequencing (Génome-Québec; Montreal, Québec, Canada) along with the ITS 1 and ITS 4 primers. The sequences obtained were checked for ambiguity and submitted to the GenBank database and compared with existing GenBank sequence data using BLAST. In cases where electrophoresis indicated that the DNA extraction/ PCR procedure had been unsuccessful, the procedure was repeated on a fresh sample of the fungal isolate.
Isolates identified taxonomically to species level had ≥99% sequence similarities to entries for conspecifics in GenBank, isolates identified to the genus level had ≥96% sequence similarities with congeneric species, and isolates identified to class level had sequence similarities ≥82% to entries from the corresponding taxonomic rank. Isolates that could not be unequivocally identified to species level based on morphological observations were only classified to the corresponding genus even when the sequence similarities ≥98% were obtained with sequence data for congeneric species in GenBank. If DNA from the ITS region was not isolated after three extraction/amplification attempts, the corresponding isolate was identified on morphological observations alone. Sterile isolates that did not provide sequence data upon repeated attempts were given codes according to their morphology in plate culture [9,14,15]. All distinct fungal isolates have been archived in the UNB Saint John fungal repository (Saint John, New Brunswick, Canada).
2.5. Preparation of Extracts
For each fungal isolate, a portion (5 mm2) of the fungal colony on solid medium was transferred to a 250 mL Erlenmeyer flask containing 2% Bacto™ malt extract broth (100 mL). Flasks were shaken (150 rpm) at room temperature under ambient light conditions for two weeks.
After incubation, the fungal mycelia from each culture were separated from the spent culture broth using vacuum filtration. Mycelia were extracted once with methanol (50 mL; Fisher Scientific, Ottawa, Ontario, Canada) in the dark for 24 h at 4 °C, solid residue and cell debris were removed by vacuum filtration, and the resulting solution was concentrated in vacuo to give a crude fungal extract. The spent culture broths were extracted 3 times with 50 mL ethyl acetate (Fisher Scientific) and the combined organic extracts were concentrated in vacuo to give a crude extract of the growth media. All crude extracts were stored at −20 °C until required.
2.6. Antibacterial and Antifungal Activity Assay
Antibacterial and antifungal activity against Pseudomonas aeruginosa (ATCC 10145), Staphylococcus aureus (ATCC 29213) and Candida albicans (ATCC 14053) was evaluated using a microbroth dilution antimicrobial susceptibility assay as previously described [11] with extracts being tested at a concentration of 200 μg·mL−1.
2.7. Statistical Analyses
Antimicrobial activities of extracts were compared to the negative control using the Kruskal-Wallis non-parametric test as the data was not normally distributed (Shapiro-Wilk, p < 0.05) and the variances were not equal (Levene’s test, p < 0.05). Post hoc analysis (stepwise-stepdown) was performed to determine which extracts differed from the negative control. Crude extracts were defined as biologically active if their effect on the growth of the test organism was significantly different (p < 0.05) to the negative control (0% inhibition). All statistical tests were performed using SPSS (PASW Statistics 18, IBM Corporation, Armonk, NY, USA).
3. Results and Discussion
3.1. Isolation of Endophytic Fungi from Marine Algae
The Bay of Fundy, with its extreme tidal range, presents a unique temperate habitat and supports a high diversity of marine macroalgal species [16,17], many of which have not been investigated for the presence of endophytes. Our results indicate that these algae support a sizeable and diverse assemblage of endophytes: fungi were isolated from 632 out of a total 3081 algal pieces resulting in an overall isolation frequency of 26% (Table 2). The isolation frequencies of endophytic fungi varied by host, with the highest isolation frequencies obtained from the red algae P. palmata and P. umbilicalis, at rates of 87% and 72% respectively (Table 2). The lowest isolation frequencies were from the red alga C. crispus at a rate of 0.08% (Table 2).
Table 2.
Isolation frequencies and distinct number of isolates of endophytic fungi from marine algae collected from the Bay of Fundy, Canada.
| Seaweed Species | Total Algae Segments | Pieces with Endophytic Growth | Isolation Frequency (%) | Distinct Fungal Taxa |
|---|---|---|---|---|
| Chondrus crispus | 246 | 2 | 0.08 | 2 |
| Devaleraea ramentacea | 108 | 46 | 43 | 13 |
| Mastocarpus stellatus | 98 | 32 | 33 | 9 |
| Palmaria palmata | 167 | 146 | 87 | 7 |
| Polysiphonia lanosa | 67 | 12 | 18 | 5 |
| Porphyra purpurea | 85 | 17 | 20 | 2 |
| Porphyra umbilicalis | 217 | 156 | 72 | 3 |
| Ascophyllum nodosum | 152 | 16 | 11 | 1 |
| Fucus spiralis | 192 | 16 | 8 | 4 |
| Fucus vesiculosus | 109 | 6 | 6 | 1 |
| Saccharina latissima | 1138 | 95 | 8 | 13 |
| Spongomorpha arcta | 184 | 49 | 27 | 11 |
| Ulva intestinalis | 117 | 24 | 21 | 5 |
| Ulva lactuca | 201 | 15 | 8 | 3 |
| Total | 3081 | 632 | 26 | 79 |
Seventy-nine distinct endophyte species were isolated from the ten algal hosts (Table 3). The number of distinct fungal isolates obtained varied by host with a maximum of 13 endophytic fungal species from D. ramentacea and S. latissima to a minimum of one from A. nodosum and F. vesiculosus (Table 3). Penicillium spp. were isolated from seven algae: C. crispus, D. ramentacea, M. stellatus, P. palmata, S. latissima, S. arcta and U. lactuca with a total of 11 isolates representing the six species isolated (Table 3). Six distinct Aspergillus spp. were isolated from five algae species: C. crispus, P. umbilicalis, A. nodosum, S. latissima and U. intestinalis (Table 3). Botrytis sp. was isolated from two algal hosts (D. ramentacea and P. palmata; Table 3) as was Aureobasidium pullulans (D. ramentacea and P. lanosa; Table 3). Cladosporium sp., Trametes versicolor, Coniothyrium sp., Coelomycete I, Hypoxylon sp., Helicomyces sp. and Botryotinia fuckeliana were only isolated from one host (Table 3).
Table 3.
Identification of endophytic fungi isolated from marine algae collected from the Bay of Fundy.
| Host species | Isolate | Species | Accession number |
|---|---|---|---|
| Chondrus crispus | AF8-053 | Aspergillus sp. I | KF572134 |
| AF8-129 | Penicillium crustosum I | KF572146 | |
| Devaleraea ramentacea | AF8-079 | Cladosporium sp. | KF572136 |
| AF8-081 | Septate Pigmented I | - | |
| AF8-083 | Septate Pigmented II | - | |
| AF8-085 | Sterile Hyaline I | - | |
| AF8-087 | Sterile Hyaline II | - | |
| AF8-089 | Botrytis sp. I | - | |
| AF8-091 | Penicillium decumbens I | KF572138 | |
| AF8-093 | Septate Pigmented III | - | |
| AF8-095 | Aureobasidium pullulans I | - | |
| Devaleraea ramentacea | AF8-109 | Trametes versicolor | KF572133 |
| AF8-111 | White Fluffy II | - | |
| AF8-113 | Coniothyrium sp. | KF572141 | |
| AF8-115 | Botrytis sp. II | - | |
| Mastocarpus stellatus | AF8-055 | Penicillium sp. | - |
| AF8-057 | Red Yeast I | - | |
| AF8-059 | Septate Pigmented IV | - | |
| AF8-061 | Red Yeast II | - | |
| AF8-063 | Sterile Hyaline III | - | |
| AF8-065 | Sterile Hyaline IV | - | |
| AF8-067 | Coelomycete I | - | |
| AF8-101 | Penicillium decumbens II | KF572139 | |
| Palmaria palmata | AF8-069 | Sterile Hyaline V | - |
| AF8-071 | Hypoxylon sp. | KF572135 | |
| AF8-073 | Penicillium decumbens III | KF572149 | |
| AF8-075 | Penicillium chrysogenum I | KF572137 | |
| AF8-077 | Helicomyces sp. | - | |
| AF8-103 | Penicillium crustosum II | KF572147 | |
| AF8-105 | Botrytis sp. III | - | |
| AF8-107 | Sterile Hyaline VI | - | |
| AF8-123 | Sterile Hyaline VII | - | |
| Polysiphonia lanosa | AF8-097 | Sterile Hyaline VIII | - |
| AF8-099 | Aureobasidium pullulans II | KF572140 | |
| AF8-117 | Sterile Hyaline IX | - | |
| AF8-119 | Botryotinia fuckeliana | KF572148 | |
| AF8-121 | Sterile Hyaline X | - | |
| Porphyra purpurea | AF8-125 | Sterile Hyaline XI | - |
| AF8-127 | Sterile Hyaline XII | - | |
| Porphyra umbilicalis | AF8-049 | Sterile Hyaline XIII | - |
| AF8-051 | Sterile Hyaline XIV | - | |
| AF8-131 | Aspergillus sydowii | KF572145 | |
| Ascophyllum nodosum | AF1-141B | Aspergillus sp. II | - |
| Fucus spiralis | AF1-021A | Black Hyaline I | - |
| AF1-021B | Septate Pigmented V | - | |
| AF1-021D | Pigmented Hyaline I | - | |
| AF1-021G | Sterile Hyaline XV | - | |
| Fucus vesiculosus | AF2-059A | Sterile Hyaline XXI | - |
| Saccharina latissima | AF1-029A2 | White Hyaline IV | - |
| AF1-029B2 | Black Hyaline II | - | |
| AF1-073C | Penicillium chrysogenum II | KF572144 | |
| AF1-073D | Aspergillus sp. III | - | |
| AF1-073E | White Hyaline I | - | |
| AF1-073G | Sterile Beige I | - | |
| AF1-073N | Sterile Beige II | - | |
| AF2-055B | Penicillium soppii I | KF572151 | |
| AF2-055F | Sterile Hyaline XVI | - | |
| AF2-055G | White Hyaline II | - | |
| AF2-055O | Penicillium chrysogenum III | KF572150 | |
| AF2-055P | Pigmented Hyaline IV(Red) | - | |
| Spongomorpha arcta | AF1-037C2 | Sterile Hyaline XVII | - |
| AF1-037F | Sterile Beige III | - | |
| AF1-153A | Pigmented Hyaline VII | - | |
| AF1-153B | Sterile Hyaline XVIII | - | |
| AF1-153C | White Hyaline III | - | |
| AF1-153D | Penicillium spinulosum | KF572143 | |
| AF1-153H | Black Hyaline III | - | |
| AF1-153J | Pigmented Hyaline VI | - | |
| AF1-153L | Pigmented Hyaline V | - | |
| AF1-153M | Penicillium sp. | KF572152 | |
| AF1-153N | Penicillium soppii II | - | |
| Ulva intestinalis | AF2-063C | Sterile Hyaline XIX | - |
| AF2-063E | Septate Pigmented VIII | - | |
| AF2-063F | Pigmented Hyaline VIII | - | |
| AF2-063G | Aspergillus sp. V | - | |
| AF2-063H | Septate Pigmented VI | - | |
| Ulva lactuca | AF1-033A2 | Septate Pigmented VII | - |
| AF1-033B | Sterile Hyaline XX | - | |
| AF1-033C | Penicillium chrysogenum IV | KF572142 |
The majority of isolates (74%), however, could neither be identified morphologically nor through the use of molecular genetic techniques. These isolates were designated codes according to their plate morphology (Table 3). This proportion of sterile mycelia (74%) is high in comparison to other marine macroalgal endophytes from the North Atlantic (45%) [11] and may have contributed to a correspondingly lower success rate observed for the molecular identification of isolates in this study (26% compared with 55% for endophytes isolated from macroalgae of the Shetland Islands, UK) [11]. The magnitude of these discrepancies is surprising and may be due to the particular assemblage of fungal species isolated from the Bay of Fundy algae. Further work will be required to optimize procedures used for molecular identification of sterile algal endophytes in an effort to increase our ability to identify fungi isolated from this source.
Penicillium spp. and Aspergillus spp. are common endophytes of marine macroalgae [9,11], and represent nearly one quarter (24%, 19/79 isolates) of the distinct isolates obtained from macroalgae of the Bay of Fundy. The majority of fungi obtained from Bay of Fundy macroalgae were isolated as sterile mycelia and several of the endophytes, i.e., Botrytis spp., Hypoxylon sp., and Helicomyces sp., have not been previously isolated from marine algal hosts. Many of the macroalgae investigated in this study, S. latissima, S. arcta, P. purpurea, P. umbilicalis, P. palmata, M. stellatus and D. ramentacea, have not been previously investigated for their endophytic fungi. Endophytes have previously been reported from A. nodosum, F. spiralis, F. vesiculosus, P. lanosa, U. intestinalis and U. lactuca [9,11,18,19,20,21,22,23,24,25,26,27,28,29,30]. However, there was a high degree of variability in the diversity of fungi obtained in these studies, which suggests limited host specificity of these fungal endophytes to their hosts.
A point of particular interest is the fact that none of the fungi isolated from the brown alga A. nodosum were identified as Mycosphaerella ascophylli, despite it being a well documented endophyte of that host [18,19,20,21,22,24,27,28,30]. Whilst it is possible that M. ascophylli may not have been isolated due to the particular sterilization and culture conditions used or the low growth rate of the fungus [19], it is more probable that M. ascophylli is represented in the mycelia sterilia obtained from A. nodosum as it is known to display a sterile morphotype of fine white septate hyphae [19,31].
3.2. Antimicrobial Screening Results of Algal-Derived Fungal Endophytes
Crude extracts of the endophyte isolates were screened against three pathogenic microorganisms, the Gram positive bacterium Staphylococcus aureus, the Gram negative bacterium Pseudomonas aeruginosa and the fungus Candida albicans. Extracts were defined as bioactive if they inhibited growth of the test organism in comparison to the negative control as indicated by heterogeneous subsets (p < 0.05) identified through post hoc testing of Kruskal-Wallis analyses. Seventy-eight crude extracts, tested at 200.0 μg·mL−1, inhibited at least one of the test microorganisms. Forty-four crude extracts significantly inhibited the growth of S. aureus, whereas, 18 and 36 extracts inhibited the growth of P. aeruginosa and C. albicans, respectively. These extracts were derived from 57 individual fungal isolates (Table 4) and comprised 39 mycelia-derived and 39 medium-derived crude extracts. Of the 156 crude extracts tested, 59 showed activity against only 1 test microorganism, 18 inhibited 2 microorganisms, and 1 showed activity against all 3 test microorganisms (Table 4). In this study, 73% of the isolates (57/78) were found to have antimicrobial activity against P. aeruginosa, S. aureus, and C. albicans (43/78, 55% antibacterial; 32/78, 41%, antifungal). The antimicrobial screening results of this study are comparable to those obtained from marine algae of the Shetland Islands, UK where 61% of the isolates (39/64) had activity against at least one of the same three test organisms (36/64 isolates antibacterial and 24/64 isolates antifungal) [11]. However, these antimicrobial screening results are in contrast to those obtained from endophytic fungi of macroalgae of the Southern Indian coast (82%, 31/38 isolates) [9] and the coasts of the North and Baltic Seas (83%, 249/300) [10]. The crude extracts showing strong antimicrobial bioactivity in this work should be investigated to identify the biologically active constituents responsible for the observed bioactivity.
Table 4.
Activity of crude extracts showing significant inhibition to S. aureus, P. aeruginosa and C. albicans, obtained from endophytic fungi isolated from marine algae collected from the Bay of Fundy.
| Fungal Taxa | Host Alga | Extract 1 Source | Inhibition (%) 2 | ||
|---|---|---|---|---|---|
| SA 3 | PA 3 | CA 3 | |||
| Aspergillus sp. I | C. crispus | Mycelia | - | 18 ± 1 | - |
| Aspergillus sp. I | C. crispus | Media | 28 ± 3 | - | 15 ± 3 |
| Penicillium crustosum I | C. crispus | Media | 43 ± 1 | - | - |
| Cladosporium sp. | D. ramentacea | Mycelia | 36 ± 1 | 9 ± 1 | - |
| Septate Pigmented I | D. ramentacea | Mycelia | - | - | 17 ± 2 |
| Sterile Hyaline I | D. ramentacea | Mycelia | - | 20 ± 1 | - |
| Sterile Hyaline I | D. ramentacea | Media | 29 ± 2 | - | - |
| Botrytis sp. I | D. ramentacea | Mycelia | 94 ± 1 | - | - |
| P. decumbens I | D. ramentacea | Mycelia | 36 ± 4 | - | 98 ± 1 |
| P. decumbens I | D. ramentacea | Media | - | - | 99 ± 1 |
| Septate Pigmented III | D. ramentacea | Mycelia | 47 ± 4 | - | - |
| Aureobasidium pullulans I | D. ramentacea | Media | 82 ± 4 | 51 ± 2 | 13 ± 2 |
| Trametes versicolor | D. ramentacea | Mycelia | - | 17 ± 2 | - |
| White Fluffy II | D. ramentacea | Media | - | - | 12 ± 1 |
| Coniothyrium sp. | D. ramentacea | Mycelia | 31 ± 1 | - | - |
| Coniothyrium sp. | D. ramentacea | Media | 25 ± 2 | 28 ± 2 | - |
| Botrytis sp. II | D. ramentacea | Mycelia | - | - | 14 ± 1 |
| Botrytis sp. II | D. ramentacea | Media | 97 ± 1 | - | - |
| Red Yeast I | M. stellatus | Mycelia | 31 ± 2 | - | - |
| Septate Pigmented IV | M. stellatus | Mycelia | - | 16 ± 3 | - |
| Septate Pigmented IV | M. stellatus | Media | 23 ± 1 | - | 70 ± 3 |
| P. decumbens II | M. stellatus | Media | - | - | 12 ± 1 |
| Sterile Hyaline V | P. palmata | Mycelia | - | 11 ± 1 | 15 ± 4 |
| Sterile Hyaline V | P. palmata | Media | - | - | 16 ± 1 |
| Hypoxylon sp. | P. palmata | Mycelia | - | 13 ± 3 | 30 ± 3 |
| Hypoxylon sp. | P. palmata | Media | - | - | 18 ± 2 |
| P. decumbens III | P. palmata | Mycelia | - | - | 16 ± 5 |
| P. chrysogenum I | P. palmata | Media | 58 ± 3 | - | - |
| Helicomyces sp. | P. palmata | Mycelia | 21 ± 1 | - | - |
| Helicomyces sp. | P. palmata | Media | - | - | 21 ± 5 |
| P. crustosum II | P. palmata | Mycelia | 82 ± 5 | - | 22 ± 3 |
| P. crustosum II | P. palmata | Media | 39 ± 2 | - | - |
| Botrytis sp. III | P. palmata | Media | 92 ± 1 | - | - |
| Sterile Hyaline VI | P. palmata | Mycelia | - | - | 13 ± 2 |
| Sterile Hyaline VI | P. palmata | Media | 19 ± 6 | - | - |
| Sterile Hyaline VII | P. palmata | Media | 90 ± 2 | - | 24 ± 2 |
| Aureobasidium pullulans II | P. lanosa | Mycelia | 49 ± 1 | 34 ± 3 | - |
| Sterile Hyaline IX | P. lanosa | Media | - | - | 40 ± 5 |
| Botryotinia fuckeliana | P. lanosa | Mycelia | 96 ± 1 | - | 30 ± 2 |
| Botryotinia fuckeliana | P. lanosa | Media | 41 ± 2 | - | - |
| Sterile Hyaline X | P. lanosa | Mycelia | - | - | 20 ± 11 |
| Sterile Hyaline XI | P. purpurea | Mycelia | 22 ± 1 | - | - |
| Sterile Hyaline XI | P. purpurea | Media | 34 ± 9 | - | - |
| Sterile Hyaline XII | P. purpurea | Mycelia | 50 ± 1 | - | - |
| Sterile Hyaline XIII | P. umbilicalis | Media | - | - | 33 ± 1 |
| Sterile Hyaline XIV | P. umbilicalis | Mycelia | - | 14 ± 1 | 27 ± 6 |
| Sterile Hyaline XIV | P. umbilicalis | Media | 22 ± 5 | - | - |
| Aspergillus sydowii | P. umbilicalis | Media | 51 ± 3 | - | - |
| Aspergillus sp. II | A. nodosum | Mycelia | 66 ± 4 | - | - |
| Aspergillus sp. II | A. nodosum | Media | 85 ± 1 | - | 15 ± 3 |
| Black Hyaline I | F. spiralis | Media | 41 ± 2 | - | - |
| Septate Pigmented V | F. spiralis | Mycelia | 39 ± 3 | 8 ± 1 | - |
| Septate Pigmented V | F. spiralis | Media | 27 ± 3 | - | - |
| Pigmented Hyaline I | F. spiralis | Mycelia | 53 ± 4 | - | - |
| White Hyaline IV | S. latissima | Media | - | - | 30 ± 1 |
| Black Hyaline II | S. latissima | Mycelia | - | - | 13 ± 5 |
| P. chrysogenum II | S. latissima | Mycelia | 25 ± 4 | - | - |
| P. chrysogenum II | S. latissima | Media | 31 ± 4 | - | - |
| Aspergillus sp. III | S. latissima | Mycelia | 25 ± 1 | - | - |
| Sterile Beige II | S. latissima | Media | - | - | 14 ± 3 |
| White Hyaline II | S. latissima | Mycelia | 28 ± 3 | - | 27 ± 5 |
| White Hyaline II | S. latissima | Media | 21 ± 1 | - | 18 ± 9 |
| Pigmented Hyaline IV(Red) | S. latissima | Mycelia | - | 12 ± 1 | - |
| Pigmented Hyaline IV(Red) | S. latissima | Media | - | - | 17 ± 1 |
| Sterile Hyaline XVII | S. arcta | Mycelia | - | - | 13 ± 1 |
| Pigmented Hyaline VII | S. arcta | Media | - | 15 ± 3 | - |
| White Hyaline III | S. arcta | Mycelia | 27 ± 7 | - | - |
| White Hyaline III | S. arcta | Media | - | - | 13 ± 3 |
| Penicillium spinulosum | S. arcta | Mycelia | 65 ± 4 | - | - |
| Penicillium spinulosum | S. arcta | Media | 37 ± 1 | - | - |
| Pigmented Hyaline V | S. arcta | Media | 87 ± 3 | - | 82 ± 1 |
| Septate Pigmented VIII | S. arcta | Mycelia | - | 17 ± 1 | - |
| Septate Pigmented VIII | S. arcta | Media | 36 ± 2 | - | - |
| Penicillium soppii II | S. arcta | Media | - | 9 ± 3 | - |
| Sterile Hyaline XIX | U. intestinalis | Media | - | 9 ± 2 | 22 ± 2 |
| Septate Pigmented VI | U. intestinalis | Mycelia | - | - | 13 ± 2 |
| Septate Pigmented VII | U. lactuca | Mycelia | - | - | 18 ± 2 |
| Sterile Hyaline XX | U. lactuca | Mycelia | - | 24 ± 3 | - |
1 Crude extracts were tested at 200.0 μg·mL−1; activity is defined as microbial inhibition significantly differing from negative control; 2 Percentage inhibition is represented as the mean of 3 readings ± SE; 3 SA: Staphylococcus aureus; PA: Pseudomonas aeruginosa; CA: Candida albicans.
4. Conclusions
Our research has demonstrated that marine macroalgae from the Bay of Fundy, Canada, have the potential to be an excellent source of endophytic fungi. Seventy-eight distinct isolates were obtained from 14 algal hosts, with most of the endophytes that could be taxonomically identified belonging to the genera Penicillium and Aspergillus. The results from the antimicrobial screening on the mycelium and broth extracts of the endophytic fungi suggest that they are a promising source of antimicrobial extracts, with 18 extracts exhibiting >50% inhibition in the screening assays. These crude extracts should be subjected to bioassay-guided fractionation in an attempt to identify the bioactive constituents of the extracts. Further work should also be focused on improving the rate of identification either through the use of molecular techniques, as only 26% of the obtained isolates were identified through this method, or through the induction of conidia or other distinguishing morphological characteristics. The results from this study has led to further work into the endophytic fungi of Atlantic coast marine macroalgae, with a current investigation now focused on the isolation, identification and antimicrobial screening of endophytic fungi from the diverse range of macroalgae present in the Bay of Fundy.
Acknowledgments
The authors would like to thank Christopher Martyniuk and Thierry Chopin (University of New Brunswick, Saint John, Canada) for their assistance in the isolation and amplification of DNA and the identification of the host algae, respectively. Financial support for this research was provided by the Natural Sciences and Engineering Research Council of Canada (Discovery Grant to C.A.G.; USRA to E.F.), the New Brunswick Health Research Foundation (Seed Operating Grant to C.A.G. and J.A.J.), the Harrison McCain Foundation (Young Scholar Award to C.A.G.) and UNB (University Research Fund grants to C.A.G. and J.A.J.; Vaughan Fellowship to A.J.F.) and is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Appelbaum P.C. 2012 and beyond: Potential for the start of a second pre-antibiotic era? J. Antimicrob. Chemother. 2012;67:2062–2068. doi: 10.1093/jac/dks213. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi S. Beginning and possibly the end of the antibiotic era. J. Paediatr. Child Health. 2012;49:179–182. doi: 10.1111/jpc.12032. [DOI] [PubMed] [Google Scholar]
- 3.Spellberg B., Bartlett J.G., Gilbert D.N. The future of antibiotics and resistance. N. Engl. J. Med. 2013;368:299–302. doi: 10.1056/NEJMp1215093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopra I. The 2012 garrod lecture: Discovery of antibacterial drugs in the 21st century. J. Antimicrob. Chemother. 2013;68:496–505. doi: 10.1093/jac/dks436. [DOI] [PubMed] [Google Scholar]
- 5.Butler M.S., Cooper M.A. Screening strategies to identify new antibiotics. Curr. Drug Targets. 2012;13:373–387. doi: 10.2174/138945012799424624. [DOI] [PubMed] [Google Scholar]
- 6.Debbab A., Aly A.H., Proksch P. Endophytes and associated marine derived fungi-ecological and chemical perspectives. Fungal Divers. 2012;57:45–83. doi: 10.1007/s13225-012-0191-8. [DOI] [Google Scholar]
- 7.Suryanarayanan T.S. Fungal Endosymbionts of Seaweeds. In: Raghukumar C., editor. Biology of Marine Fungi. Springer; Berlin, Germany: 2012. pp. 53–69. [DOI] [PubMed] [Google Scholar]
- 8.Jones E.B.G., Pang K.-L., Stanley S.J. Fungi from Marine Algae. In: Jones E.B.G., Pang K.-L., editors. Marine Fungi and Fungal-Like Organisms. Walter de Gruyter; Berlin, Germany: 2012. pp. 329–344. [Google Scholar]
- 9.Suryanarayanan T.S., Venkatachalam A., Thirunavukkarasu N., Ravishankar J.P., Doble M., Geetha V. Internal mycobiota of marine macroalgae from the tamilnadu coast: Distribution, diversity and biotechnological potential. Bot. Mar. 2010;53:457–468. [Google Scholar]
- 10.Schulz B., Boyle C., Draeger S., Rommert A.K., Rohn K. Endophytic fungi: A source of novel biologically active secondary metabolites. Mycol. Res. 2002;106:996–1004. doi: 10.1017/S0953756202006342. [DOI] [Google Scholar]
- 11.Flewelling A.J., Johnson J.A., Gray C.A. Isolation and bioassay screening of fungal endophytes from north atlantic marine macroalgae. Bot. Mar. 2013;56:287–297. [Google Scholar]
- 12.Petrini O., Fisher P., Petrini L. Fungal endophytes of bracken (Pteridium aquilinum), with some reflections on their use in biological control. Sydowia. 1992;44:282–293. [Google Scholar]
- 13.White T.J., Bruns T.D., Lee S.B., Taylor J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. pp. 315–322. [Google Scholar]
- 14.Dobranic J.K., Johnson J.A., Alikhan Q.R. Isolation of endophytic fungi from eastern larch (Larix laricina) leaves from new-brunswick, Canada. Can. J. Microbiol. 1995;41:194–198. doi: 10.1139/m95-026. [DOI] [Google Scholar]
- 15.Suryanarayanan T.S., Kumaresan V., Johnson J.A. Foliar fungal endophytes from two species of the mangrove Rhizophora. Can. J. Microbiol. 1998;44:1003–1006. [Google Scholar]
- 16.Bates C.R., Saunders G., Chopin T. Historical versus contemporary measures of seaweed biodiversity in the bay of fundy. Botany. 2009;87:1066–1076. doi: 10.1139/B09-067. [DOI] [Google Scholar]
- 17.South G.R. A checklist of marine algae of eastern canada, 2nd revision. Can. J. Bot. 1984;62:680–704. doi: 10.1139/b84-102. [DOI] [Google Scholar]
- 18.Fries N., Thorentolling K. Identity of fungal endophyte of Ascophyllum with Mycosphaerella ascophylli established by means of fluorescent-antibody technique. Bot. Mar. 1978;21:409–411. [Google Scholar]
- 19.Fries N. Physiological characteristics of Mycosphaerella ascophylli, a fungal endophyte of the marine brown alga Ascophyllum nodosum. Physiol. Plant. 1979;45:117–121. doi: 10.1111/j.1399-3054.1979.tb01674.x. [DOI] [Google Scholar]
- 20.Fries L. Ascophyllum nodosum (Phaeophyta) in Axenic culture and its response to the endophytic fungus Mycosphaerella ascophylli and epiphytic bacteria. J. Phycol. 1988;24:333–337. [Google Scholar]
- 21.Garbary D.J., Gautam A. The Ascophyllum/Polysiphonia/Mycosphaerella symbiosis. I. Population ecology of Mycosphaerella from Nova Scotia. Bot. Mar. 1989;32:181–186. [Google Scholar]
- 22.Garbary D.J., Burke J., Tian L.N. The Ascophyllum/Polysiphonia/Mycosphaerella symbiosis. II. Aspects of the ecology and distribution of Polysiphonia lanosa in Nova Scotia. Bot. Mar. 1991;34:391–401. [Google Scholar]
- 23.Stanley S.J. Observations on the seasonal occurance of marine endophytic and parasitic fungi. Can. J. Bot. 1992;70:2089–2096. doi: 10.1139/b92-259. [DOI] [Google Scholar]
- 24.Garbary D.J., London J.F. The Ascophyllum/Polysiphonial/Mycosphaerella symbiosis. V. Fungal infection protects A. nosodum from desiccation. Bot. Mar. 1995;38:529–533. [Google Scholar]
- 25.Osterhage C., Kaminsky R., Konig G.M., Wright A.D. Ascosalipyrrolidinone A, an antimicrobial alkaloid, from the obligate marine fungus Ascochyta salicorniae. J. Org. Chem. 2000;65:6412–6417. doi: 10.1021/jo000307g. [DOI] [PubMed] [Google Scholar]
- 26.Abdel-Lateff A., Fisch K.M., Wright A.D., Konig G.M. A new antioxidant isobenzofuranone derivative from the algicolous marine fungus Epicoccum sp. Planta Med. 2003;69:831–834. doi: 10.1055/s-2003-43209. [DOI] [PubMed] [Google Scholar]
- 27.Deckert R., Garbary D. Ascophyllum and its symbionts. VI. Microscopic characterization of the Ascophyllum nodosum (Phaeophyceae), Mycophycias ascophylli (Ascomycetes) symbiotum. Algae. 2005;20:225–232. doi: 10.4490/ALGAE.2005.20.3.225. [DOI] [Google Scholar]
- 28.Garbary D.J., Jamieson M.M., Fraser S.J., Ferguson C.A., Cranston P.S. Ascophyllum (Phaeophyceae) and its symbionts. IX. A novel symbiosis between Halocladius variabilis (Chironomidae, Insecta) and Elachista fucicola (Elachistaceae, Phaeophyceae) from marine rocky shores of Nova Scotia. Symbiosis. 2005;40:61–68. [Google Scholar]
- 29.Schulz B., Draeger S., dela Cruz T.E., Rheinheimer J., Siems K., Loesgen S., Bitzer J., Schloerke O., Zeeck A., Kock I., et al. Screening strategies for obtaining novel, biologically active, fungal secondary metabolites from marine habitats. Bot. Mar. 2008;51:219–234. [Google Scholar]
- 30.Xu H., Deckert R.J., Garbary D.J. Ascophyllum and its symbionts. X. Ultrastructure of the interaction between A. nodosum (Phaeophyceae) and Mycophycias ascophylli (Ascomycetes) Botany. 2008;86:185–193. doi: 10.1139/B07-122. [DOI] [Google Scholar]
- 31.Webber F.C. Observations on the structure, life history and biology of Mycosphaerella ascophylli. Trans. Br. Mycol. Soc. 1967;50:583–601. doi: 10.1016/S0007-1536(67)80090-1. [DOI] [Google Scholar]


