Abstract
Glioblastoma multiforme (GBM) is the most common malignant brain tumor. This study aimed to identify the hub genes and regulatory factors of GBM subgroups by RNA sequencing (RNA-seq) data analysis, in order to explore the possible mechanisms responsbile for the progression of GBM. The dataset RNASeqV2 was downloaded by TCGA-Assembler, containing 169 GBM and 5 normal samples. Gene expression was calculated by the reads per kilobase per million reads measurement, and nor malized with tag count comparison. Following subgroup classification by the non-negative matrix factorization, the differentially expressed genes (DEGs) were screened in 4 GBM subgroups using the method of significance analysis of microarrays. Functional enrichment analysis was performed by DAVID, and the protein-protein interaction (PPI) network was constructed based on the HPRD database. The subgroup-related microRNAs (miRNAs or miRs), transcription factors (TFs) and small molecule drugs were predicted with predefined criteria. A cohort of 19,515 DEGs between the GBM and control samples was screened, which were predominantly enriched in cell cycle- and immunoreaction-related pathways. In the PPI network, lymphocyte cytosolic protein 2 (LCP2), breast cancer 1 (BRCA1), specificity protein 1 (Sp1) and chromodomain-helicase-DNA-binding protein 3 (CHD3) were the hub nodes in subgroups 1–4, respectively. Paired box 5 (PAX5), adipocyte protein 2 (aP2), E2F transcription factor 1 (E2F1) and cAMP-response element-binding protein-1 (CREB1) were the specific TFs in subgroups 1–4, respectively. miR-147b, miR-770-5p, miR-220a and miR-1247 were the particular miRNAs in subgroups 1–4, respectively. Natalizumab was the predicted small molecule drug in subgroup 2. In conclusion, the molecular regulatory mechanisms of GBM pathogenesis were distinct in the different subgroups. Several crucial genes, TFs, miRNAs and small molecules in the different GBM subgroups were identified, which may be used as potential markers. However, further experimental validations may be required.
Keywords: glioblastoma multiforme, RNA sequencing, immunoreaction, cell cycle, microRNAs, small molecule drugs
Introduction
Glioblastoma multiforme (GBM) is the most frequent and lethal malignant brain tumor (1). Based on the World Health Organization classification, GBM belongs to the grade IV astrocytoma group, which includes diffusely infiltrating astrocytomas that account for half of all primary brain gliomas (2). Despite the advanced detection techniques, such as spectroscopy and perfusion, and improved treatments including surgery and the combination of radiation with chemotherapy, the usual survival time of the majority of patients with GBM after diagnosis is approximately 1 year (3). The major causative factors are the high resistance of GBM to therapeutics and the high variability of the benefits of therapeutic drugs in different GBM patients (4).
To improve the poor prognosis, considerable investigations have been conducted to explore more effective therapeutics at the genetic and protein level. For instance, via the enzyme-linked immunosorbent assay (ELISA), the level of serum glial fibrillary acidic protein (GFAP) was found to be increased in GBM patients, and was thus proposed as a biomarker for the prognosis of GBM (5). Other candidate genes, such as epidermal growth factor receptor (EGFR) (6) and herpes simplex virus (HSV)-thymidine kinase (TK)/interleukin (IL)-2 (7) have been shown to be involved in the prognosis of GBM, and are considered as potential therapeutic markers. MicroRNAs (miRNAs or miRs) are small non-coding RNAs that play central roles in cancer development via the regulation of target genes. Several miRNAs, including miR-21 (8), miR-124 and miR-137 (9) have been verified to play significant roles in anti-apoptosis and in the inhibition of tumor cell proliferation during GBM progression. Several small molecules have been applied in the treatment of GBM, such as temsirolimus (CCI-779), which is an inhibitor of mammalian target of rapamycin (mTOR), and has been identified as a therapeutic target of GBM (10). Additionally, the use of the small molecule drug, vorinostat, has been shown to result in the increased expression of anti-proliferative genes, such as death receptor 5 (DR5), and in the decreased expression of anti-apoptotic genes, such as cyclin-dependent kinase (CDK)2. Via the mediation of these genes, vorinostat serves as another crucial therapeutic target of GBM (11). Recently, a novel small molecule, CG500354, has been reported to target the specific variant, PDE4D, and thus plays a significant role in the suppression of GBM (12).
Previous studies have used the technique of RNA sequencing (RNA-seq), an emerging deep sequencing method for transcriptome profiling and provides more accurate expression measurements (13), to explicate the molecular events in GBM (14). However, most of these studies focused on the function of fusion genes, such as neurofascin (NFASC)-neurotrophic tyrosine kinase receptor type 1 (NTRK1) (14) and RNA splicing-related genes, such as PHD finger protein 5A (PHF5A) (15). The exploration of therapeutic small drugs in GBM patients using RNA-seq, and focusing on different subgroups of individuals in particular, is limited. Considering that different mechanisms may be presented due to different individuals, in this study, we analyzed the RNA-seq data to identify differentially expressed genes (DEGs) in samples between patients with GBM and normal subjects. Furthermore, the non-negative matrix factorization (NMF) method was used to further identify the subgroups of GBM. NMF is a well-known approach that could reduce the dimensionality of a dataset and cluster the data samples (16). The method has been widely used in computational biology. Previous studies have utilized NMF to discover subgroups of patients with complex genetic diseases (16,17).
In this study, by functional enrichment analysis and protein-protein interaction (PPI) network analysis, the altered pathways and the interactions of DEGs in each subgroup were predicted. Furthermore, the miRNAs, transcription factors (TFs) and small molecule drugs related to the DEGs in the subgroups were also explored. All these bioinformatics methods were used in an aim to elucidate the comprehensive molecular mechanisms responsible for the development and progression of GBM, and to provide potential small molecule agents as therapeutic candidates for the treatment of patients with different subgroups of GBM.
Data collection and analysis
Data preprocessing and GBM subgroup classification
The dataset of RNASeqV2 was downloaded using The Cancer Genome Atlas (TCGA)-Assembler (18), consisting of 169 GBM samples and 5 normal samples. The reads per kilobase per million reads (RPKM) measurement were used to calculate gene expression, followed by normalization with the tag count comparison (TCC; http://www.bioconductor.org/packages/release/bioc/html/TCC.html) package (19). The expression value was complemented as '1' for gene loss in several samples, followed by the transformation by log2 to achieve an approximate normal distribution. Following normalization, we applied the NMF method to classify the subgroup of all the GBM samples, and selected the optimal 'k' value based on the cophenetic correlation coefficient (CCC) (20), whose value varies from 0–1. The classification is more stable if the 'k' value is closer to 1. Finally, the highest 'k' value of the CCC was selected as the optimal value.
Identification of DEGs
Following normalization and subgroup classification, the DEGs were screened out using the significance analysis of microarrays (SAM) method (21). The thresholds for DEG selection were a false discovery rate (FDR) <0.05 and |log2 fold change| >1. Top 500 DEGs were clustered using Euclidean distance and the hierarchical clustering method, which were executed by heatmap.2 function in the gplot R package (https://cran.r-project.org/web/packages/gplots/).
Pathway enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html) (22) and Reactome (http://www.reactome.org) (23) pathway enrichment analyses were carried out for the selected DEGs, using the Database for Annotation, Visualization and Integration Discovery (DAVID; http://david.abcc.Ncifcrf.gov/) online software (24), based on the hypergeometric distribution test. The cut-off value for significant terms was p<0.05.
Construction of PPI network and transcriptional regulatory network
The Human Protein Reference Database (HPRD; http://hprd.org/) is a database of curated proteomic information, such as protein annotation, PPI and protein subcellular localization (25). The Pearson correlation coefficient was calculated of two inter-played proteins basing on the miRNA expression profiles, and the proteins with the Pearson correlation coefficient >0.5 were extracted to establish the PPI network. A node in the PPI network serves as a protein, and the degree of a node reflects the number of the interacted proteins with this specific protein. Hub nodes were deemed as the prominent nodes with high degrees.
Prediction of miRNAs, TFs and small molecule drugs
The combinatorial gene regulatory networks (cGRNB; http://www.scbit.org/cgrnb/) is a web server for the establishment of two major network modules such as miRNA-perturbed gene expression (MPGE) datasets and the parallel miRNA/mRNA expression datasets, to provide the combinatorial regulations involving TFs, miRNAs and genes (21). Among these regulations, the miRNA-gene interactions were derived from the starBase (http://starbase.sysu.edu.cn/) database (26), under the criteria of 'microRNA read Num' >0 and 'biological complexity' (BC) >1.
The DrugBank (http://www.drugbank.ca/) database (27) was utilized to recognize potential pharmaceutical molecules and the drug targets. Only the drugs that approved by the Food and Drug Administration (FDA) were downloaded.
GBM subgroup-related miRNAs, TFs and drug selection
Integrating the regulatory information in the cGRNB database, the miRNA-target or TF-target interactions were extracted, while the small molecule drugs and the targets were identified in combination with the information on DrugBank. Subsequently, the overlapping significance between DEGs (gene set N) in a subgroup and target genes (gene set Mi) of selected miRNAs/TFs/drugs (i) were detected based on Fisher's exact test (28). The formula used was as follows:
| (equation 1) |
where 'a', indicates genes included in both Mi and N; 'd', included genes included in neither Mi nor N; 'b', included genes are only included in Mi and 'c', included genes only included in N. The threshold for the selection of eligible miRNAs, TFs or drugs was p<0.05.
Survival curve analysis
By recruiting the survival package in Bioconductor R, we assessed the median survival with the corresponding intervals from Kaplan-Meier (KM) curve (29), to detect the differences in survival time amongst the GBM subgroups.
Results
Pre-processing of the expression profile data and marker genes of GBM subgroups
A set of 11,078 genes was subjected to normalization in the GBM samples. As indicated in the box plot (Fig. 1), the gene expression levels among the different samples were relatively uniform. The top 1,500 genes with variable coefficient in the tumor samples were filtered out for the subgroup classification. The CCC was calculated with the 'k' value varying from 2 to 12, and achieved the highest level when 'k' was 4, which was defined as the final 'k' value for subgroup classification. In all 169 GBM samples, 13 samples were excluded from subsequent analyses for the lack of prognostic information. The remaining 156 samples were classified into 4 subgroups: 52 samples were divided into subgroup 1; 57 in subgroup 2; 26 in subgroup 3 and 21 in subgroup 4.
Figure 1.
Box plot of gene expression among different samples. The x-axis represents samples, while the y-axis represents the expression of genes.
DEGs between GBM and control samples in different subgroups and the disrupted functions and pathways
Based on the aforementioned criteria, a cohort of 19,515 DEGs between the GBM and control samples in the 4 subgroups was identified (subgroup 1: 4,430; subgroup 2: 3,271; subgroup 3: 4,046; subgroup 4: 7,768) (Fig. 2). The Venn diagram indicates substantially overlapping DEGs among the 4 subgroups. The overlapped DEGs in >2 subgroups were considered as the common DEGs, and we identified a set of 7,106 common DEGs, which were mainly enriched in the cell cycle and immunoreaction-related pathways (Table I). The heatmap of the clustering analysis of gene expression is presented in Fig. 3.
Figure 2.
Venn diagram of the differentially expressed genes in the 4 subgroups.
Table I.
Top 15 significantly enriched pathways of the common DEGs.
| Source | Name | q-value Bonferroni |
|---|---|---|
| KEGG | Ribosome | 5.21E-22 |
| REACTOME | Gene expression | 1.71E-14 |
| REACTOME | Cell cycle, mitotic | 5.82E-13 |
| REACTOME | Metabolism of proteins | 4.08E-10 |
| REACTOME | M phase | 6.87E-09 |
| REACTOME | Interferon signaling | 7.66E-08 |
| REACTOME | Disease | 8.23E-08 |
| REACTOME | Cell cycle | 1.01E-07 |
| REACTOME | Cytokine signaling in immune system | 1.57E-07 |
| REACTOME | Class I MHC mediated antigen processing and presentation | 1.88E-06 |
| KEGG | Epstein-Barr virus infection | 4.95E-06 |
| REACTOME | Translation | 1.04E-05 |
| KEGG | Protein processing in endoplasmic reticulum | 1.17E-05 |
| KEGG | Ubiquitin mediated proteolysis | 1.95E-05 |
| REACTOME | Interferon γ signaling | 3.50E-05 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
Figure 3.
Heat map of cluster analysis of differentially expressed genes. The x-axis represents samples, while the y-axis represents genes. Red represents subgroup 1; orange represents subgroup 2; yellow represents subgroup 3; green represents subgroup 4.
The PPI networks of the DEGs in the 4 subgroups
By integrating the information in the HPRD database, a set of 39,240 interactions was obtained, containing 11,390 interactions that involved 7,768 DEGs in the 4 GBM subgroups. Four PPI networks were then constructed based on the Pearson's correlation coefficient. The PPI network for DEGs in subgroup 1 consisted of 790 edges involving 821 nodes, and that for the DEGs in subgroup 2 was comprised of 864 edges and 830 nodes. There were 1,186 edges and 1,174 nodes in the network for subgroup 3, and 2,190 edges and 1,760 nodes in the network for subgroup 4 (data not shown). The top 10 hub nodes in the 4 subgroups are presented in Table II. Among these, CDK1 was the hub node in all 4 subgroups, while several nodes were only remarkable in a specific subgroup, such as lymphocyte cytosolic protein 2 (LCP2), breast cancer 1 (BRCA1), specificity protein 1 (Sp1) and chromodomain-helicase-DNA-binding protein 3 (CHD3) were the hub nodes in subgroups 1, 2, 3 and 4, respectively.
Table II.
Top 10 hub nodes in the 4 PPI networks from the 4 subgroups.
| Subgroup 1
|
Subgroup 2
|
Subgroup 3
|
Subgroup 4
|
||||
|---|---|---|---|---|---|---|---|
| Name | Degree | Name | Degree | Name | Degree | Name | Degree |
| SYK | 20 | CDK1 | 26 | CDK1 | 24 | SRC | 30 |
| CDK1 | 18 | SYK | 22 | SYK | 21 | YWHAG | 26 |
| LYN | 16 | LYN | 16 | PTPN6 | 20 | CDK1 | 26 |
| GRB2 | 16 | PTPN6 | 14 | CREBBP | 18 | CHD3 | 23 |
| CREBBP | 14 | GRB2 | 14 | LYN | 17 | ATN1 | 22 |
| HCK | 13 | BRCA1 | 14 | GRB2 | 17 | CREBBP | 22 |
| PTPN6 | 12 | HCK | 14 | PCNA | 16 | HTT | 21 |
| LCP2 | 11 | PCNA | 13 | Sp1 | 16 | SYK | 20 |
| VAV1 | 10 | CREBBP | 13 | VAV1 | 13 | EP300 | 20 |
| WAS | 10 | MCM7 | 12 | HCK | 13 | EWSR1 | 19 |
The GBM subgroup-related miRNAs, TFs and small molecular drugs
A total of 210,637 pair wise miRNA-target interactions was extracted, containing 207 TFs and 16,862 target genes. A total of 6,108 drug-target interactions was acquired, involving 1,348 small molecule drugs and 1,353 drug targets. According to the formula (equation 1, described above) and the predefined threshold, we screened 8, 17, 6 and 48 miRNAs separately in subgroups 1–4; 34, 50, 45 and 65 TFs respectively in subgroup 1–4; and 2 and 21 small molecule drugs in subgroups 1 and 2, but none in subgroups 3 and 4 (data not shown). The top 5 (ranked by the P-value) significant regulators (miRNAs, TFs and small molecular drugs) are listed in Table III. Among the significant TFs, paired box 5 (PAX5), adipocyte protein 2 (aP2), E2F transcription factor 1 (E2F1) and cAMP-response element-binding protein-1 (CREB1) were the specific regulators in subgroup 1, 2, 3 and 4, respectively. Several miRNAs, such as miR-147b, miR-770-5p, miR-220a and miR-1247 were the specific ones respectively in subgroups 1–4. Furthermore, a spectrum of small molecule drugs was identified in subgroup 2, including natalizumab, trastuzumab, efalizumab, bevacizumab and etanercept.
Table III.
Subgroup-specific miRNAs, TFs and small molecule drugs.
| miRNAs | TFs | Drugs | |
|---|---|---|---|
| Group 1 | miR-147b, miR-1269, miR-744, miR-483-5p, miR-1207-5p | Sp1, DAND5, PSG1, PAX5, Pax-5 | Efalizumab, trastuzumab |
| Group 2 | miR-770-5p, miR-1184, miR-133a, miR-516a-3p, miR-133b | DAND5, PSG1, Sp1, aP2-α, aP2-γ | Natalizumab, trastuzumab, efalizumab, bevacizumab, etanercept |
| Group 3 | miR-220a, miR-492, miR-626, miR-24-1a, miR-489 | Sp1, DAND5, PSG1, E2F1, E2F | NA |
| Group 4 | miR-1247, miR-940,miR-198, miR-1289, miR-214 | DAND5, PSG1, Sp1, Elk-1, CREB1 | NA |
NA, not applicable.
Prognostic analysis of GMB
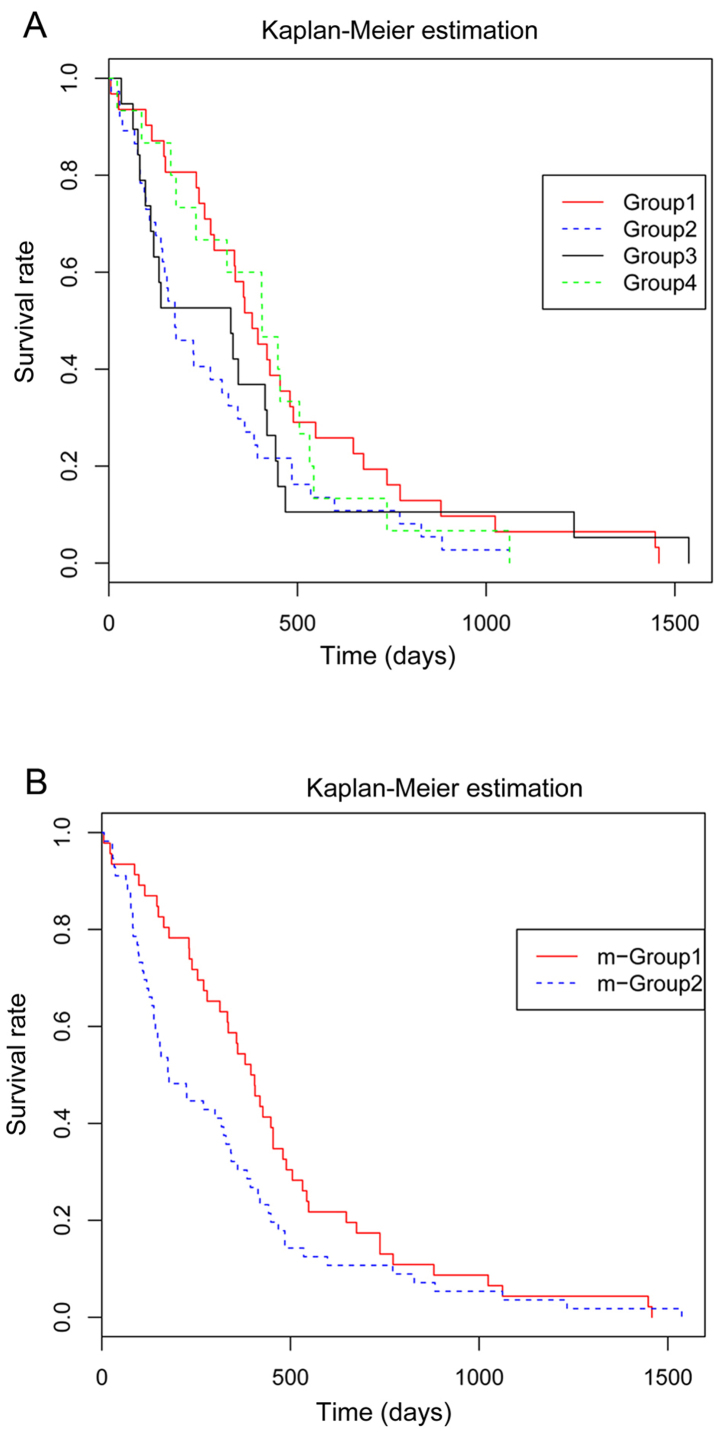
The clinical survival time of the 156 samples was extracted from the TCGA database. With the KM method, the differences among the 4 subgroups were identified. There were 31, 37, 19 and 15 deaths in subgroups 1–4, respectively (data not shown). The KM analysis indicated no significant differences in survival time among the 4 subgroups (p=0.182). However, as shown in Fig. 4A, a similar trend in the survival rate was detected between subgroups 1 and 4, and between subgroups 2 and 3. Therefore, the data in subgroups 1 and 4 were merged into m-group 1, whereas the data in subgroups 2 and 3 were merged into m-group 2. m-group 1 exhibited a pronounced higher survival rate than m-group 2 (p=0.04; Fig. 4B).
Figure 4.
Survival rates of glioblastoma multiforme (GBM) patients by Kaplan-Meier (KM) estimation. (A) Survival rates in the 4 subgroups prior to merging. (B) Survival rates after merging.
Discussion
GBM is the most common malignant brain tumor and is characterized by invasiveness (30). Despite several effective therapeutic agents available, not all patients benefit from these due to individual differences (4). In this study, we utilized the RNA-seq data in the TCGA database to detect gene alterations during the progression of GBM and to further explore the possible mechanisms responsible for its pathogenesis. As a result, 4 subgroups of GBM were classified and a set of 19,515 DEGs between the GBM and control samples was identified; the majority of these genes were enriched in the cell cycle and immunoreaction-related pathways. In the PPI network, LCP2, BRCA1, Sp1 and CHD3 were 4 striking nodes in subgroups 1–4, respectively. PAX5, aP2, E2F1 and CREB1 were the specific TFs in subgroups 1–4, respectively. Several small molecule drugs, such as efalizumab and natalizumab were identified in subgroup 2.
Immunosuppression is the major hallmark of GBM patients, who have to receive radiation therapy in combination with with temozolomide for the restoration of the antitumor immune response (31). LCP2 is known to play significant roles in promoting T cell progression. It was recently reported that a splice variant of LCP2 resulted in severe immune dysregulation (32). The PAX5 encoded transcription factor is a PAX family member which plays important roles in B-cell development (33). Via the chromatin immunoprecipitation (ChIP) technique, PAX5 was found to regulate target genes by directly activating chromatin at the promoter or enhancer region, and PAX5-activated targets are crucial regulators involved in B-cell signaling, migration and immune function (34). Our findings revealed that LCP2 and PAX5 were (separately) the hub nodes in the PPI network and the prominent TFs for subgroup 1 of GBM, and were mainly enriched in the pathway of cytokine signaling in the immune system, suggesting that the LCP2- and PAX5-mediated immune pathway may play important roles in the progression of GBM in subgroup 1. Furthermore, miR-147b was identified as an oncomiR in prostate cancer (PC), but with no detectable targets (35). Our study indicated that miR-147b was the specific miRNA in subgroup 1, suggesting that it may be an oncomiR in this GBM subgroup.
BRCA1 is known to act as a tumor suppressor associated with DNA repair and genome integrity. The DNA repair pathway was established as a crucial pathway for identifying the glioblastoma cases-control status, and the SNP, rs799917 of BRCA1 was shown to be involved in this pathway (36). Additionally, the methylation PRKCDBP was implicated in GBM and identified as a BRCA1-interacting protein (37), suggesting that BRCA1 exerts its functions in GBM via its interactions with other proteins. EGFR abnormalities occur frequently in GBM patients. The aP2 protein is an essential component of growth factor receptor endocytosis (38), implying that aP2 may also play important roles in the progression of GBM. Notably, in a previous study, aP2-α was shown to be one of the putative TFs of BRCA1 through the combination of microarray and ChIP-chip analysis (39). Considering that BRCA1 and aP2 were both specific in subgroup 2 in our study, it can be speculated that aP2 may be the TF regulating the expression of BRCA1 in GBM patients in this subgroup. With regard to the miRNAs in subgroup 2, miR-770-5P was identified as the specific one, which was also predominant in the miRNA co-expression network of GBM (40). Moreover, miR-770-5P has been shown to be upregulated in neuronal stem cells which progress into GBM (41). All these data suggest the potential use of miR-770-5P as a target for GBM subgroup 2.
The disruption of cell cycle program is often involved in the development of GBM. For instance, JSI-124 functions as an inhibitor of GBM cell proliferation by inducing cell cycle arrest at G2/M phase (42). The protein Sp1 is a zinc finger TF that is involved in multiple cellular processes related to the cell cycle. p53 acts as a tumor suppressor in the development of various types of cancer, such as lung cancer (43) and GBM (44). The loss of p53 is often the initiation of oncogenesis. A previous study demonstrated that the p53-mediated repression of cyclin B1 (CCNB1), a cell cycle-related transcript, required the binding of Sp1 to the CCNB1 prompter (45). Emerging evidence has verified the interactions of p53 and Sp1 in the regulation of other cell cycle related genes, such as CDC25B (46). E2F1 is another vital TF that promotes cell progression. Δ9-tetrahydrocannabinol functions as an inhibitor of cell growth via the downregulation of E2F1 in GBM (47). Furthermore, E2F1 was suggested to be involved in the mediation of telomerase activity in malignant glioma cells and the overexpression of E2F1 and telomerase were proposed as markers for the prognosis of glioblastoma (48). Notably, the cyclin-dependent kinase inhibitor, p18INK4c, which plays significant roles in cell cycle progression and cellular differentiation, is regulated by the synergistic action of E2F1 and Sp1 (49). In the present study, Sp1 was the hub node and E2F1 was the crucial TF in subgroup 3, suggesting that they may play significant roles in the progression of GBM in this subgroup, both via the involvement of cell cycle regulation. miR-220 was predicted as a remarkable miRNA in subgroup 3. In Chinese patients with prostate cancer, miR-220 was also suggested to be a crucial therapeutic target (50). Additionally, the elevated level of miR-220 was also predicted based on an miRNA array analysis in colorectal cancer (51). Therefore, dysregulated miR-220 may also be a therapeutic marker of GBM in subgroup 3.
The CHD3 encoded protein is related to chromatin remodeling. The GO functional terms of this gene are helicase activity and ATP-dependent DNA helicase activity (52). It was discovered that CHD3 was one of the 60 genes significantly associated with the overall survival (OS) of GBM (53). The TF encoded by CREB1 was reported to be involved in a large number of cellular processes, such as survival and proliferation. CREB was implicated in the development of myeloid leukemia for it positively increased the expression of cell cycle-related genes and thus promoted the cellular transformation and survival of myeloid cells (54). Companied with other synergetic factors, such as forkhead box protein A1 (FOXA1) and MYB proto-oncogene like 2 (MYBL2), CREB1 induced histone H3 acetylation, which contributed to the transition of G1 to S phase cell cycle stage in castration-resistant prostate cancer (55). Our study revealed the dysregulated CHD3 and CREB1 in GBM subgroup 3, suggesting their roles as potential biomarkers of GBM in this subgroup.
miR-1247 has been shown to be markedly dysregulated in the tumor tissues in GBM (56,57), implying its role as a therapeutic target in GBM in subgroup 4. However, only two validated targets of this miRNA have been reported, such as the cartilage transcription factor, SRY-box 9 (SOX9) (58) and Myc-binding protein 2 (MYCBP2) (59).
As one of the approaches that target a specific integrin-ligand interaction, natalizumab has been applied for the treatment of GBM by targeting α4 integrin, which results in the blocking of α4β1 or α4β7 binding to vascular cell adhesion molecule 1 (VCAM-1) (60). The integrin, integrin subunit alpha 4 (ITGA4), was revealed as a target of natalizumab in GBM of subgroup 2 in the present study (data not shown), suggesting that ITGA4 may be a novel target of the small molecule of natalizumab in this subgroup.
The administration of the monoclonal antibodies of natalizumab has been shown to be responsible for the suppression of T-lymphocyte trafficking into the central nervous system (CNS); however it is also related to the risk of progressive multifocal leukoencephalopathy (61). Considering the adverse side-effects, caution should excised as regards the application of this agent for the management of GBM in subgroup 2.
The overall survival of GBM is often distinct among different subgroups, due to the factors of population, age, recursive partitioning analysis (RPA) class and other genetic factors (62). Our findings indicated that the survival rate of m-group 1 (subgroups 1 and 4) was significantly higher than that of m-group 2 (subgroups 2 and 3), suggesting that the same agent of GBM may be more beneficial to individuals in m-group 1 than that of m-group 2. This also supports the necessity of the use of specific agents in different subgroups. In conclusion, the molecular regulatory mechanisms responsible for the pathogenesis GBM were distinct in the different subgroups. Several crucial genes, TFs, miRNAs and small molecules in the different subgroups of GBM were identified, which may be used as potential markers in the 4 subgroups, respectively. Among these, the LCP2- and PAX5-mediated immune pathway, the transcriptional regulation of BRCA1 by aP2 in subgroup 2, the synergistic action of Sp1 and E2F1 and CREB1-modulated cell cycle process in subgroups 1–4, respectively, may play significant roles in the progression of GBM. Moreover, natalizumab may be used as a potent therapeutic agent for GBM in subgroup 2. However, all these predicted results need to be further verified by extensive experiments.
References
- 1.Gollapalli K, Ray S, Srivastava R, Renu D, Singh P, Dhali S, Bajpai Dikshit J, Srikanth R, Moiyadi A, Srivastava S. Investigation of serum proteome alterations in human glioblastoma multiforme. Proteomics. 2012;12:2378–2390. doi: 10.1002/pmic.201200002. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Zhang W, Cao WD, Cheng G, Zhang YQ. Glioblastoma multiforme: molecular characterization and current treatment strategy (Review) Exp Ther Med. 2012;3:9–14. doi: 10.3892/etm.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahbar A, Orrego A, Peredo I, Dzabic M, Wolmer-Solberg N, Strååt K, Stragliotto G, Söderberg-Nauclér C. Human cytomegalovirus infection levels in glioblastoma multiforme are of prognostic value for survival. J Clin Virol. 2013;57:36–42. doi: 10.1016/j.jcv.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Carlsson A, Persson O, Ingvarsson J, Widegren B, Salford L, Borrebaeck CA, Wingren C. Plasma proteome profiling reveals biomarker patterns associated with prognosis and therapy selection in glioblastoma multiforme patients. Proteomics Clin Appl. 2010;4:591–602. doi: 10.1002/prca.200900173. [DOI] [PubMed] [Google Scholar]
- 5.Jung CS, Foerch C, Schänzer A, Heck A, Plate KH, Seifert V, Steinmetz H, Raabe A, Sitzer M. Serum GFAP is a diagnostic marker for glioblastoma multiforme. Brain. 2007;130:3336–3341. doi: 10.1093/brain/awm263. [DOI] [PubMed] [Google Scholar]
- 6.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, Schroeder MA, James CD. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncol. 2005;7:164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo F, Barzon L, Franchin E, Pacenti M, Pinna V, Danieli D, Zanusso M, Palù G. Combined HSV-TK/IL-2 gene therapy in patients with recurrent glioblastoma multiforme: biological and clinical results. Cancer Gene Ther. 2005;12:835–848. doi: 10.1038/sj.cgt.7700851. [DOI] [PubMed] [Google Scholar]
- 8.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 9.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galanis E, Buckner JC, Maurer MJ, Kreisberg JI, Ballman K, Boni J, Peralba JM, Jenkins RB, Dakhil SR, Morton RF, et al. Phase II trial of temsirolimus (CCI-779) in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2005;23:5294–5304. doi: 10.1200/JCO.2005.23.622. [DOI] [PubMed] [Google Scholar]
- 11.Galanis E, Jaeckle KA, Maurer MJ, Reid JM, Ames MM, Hardwick JS, Reilly JF, Loboda A, Nebozhyn M, Fantin VR, et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2009;27:2052–2058. doi: 10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang TW, Choi SW, Yang SR, Shin TH, Kim HS, Yu KR, Hong IS, Ro S, Cho JM, Kang KS. Growth arrest and forced differentiation of human primary glioblastoma multiforme by a novel small molecule. Sci Rep. 2014;4:5546. doi: 10.1038/srep05546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Lee Y, Cho HJ, Lee YE, An J, Cho GH, Ko YH, Joo KM, Nam DH. Recurrent fusion of NTRK1 in glioblastoma multiforme. PLoS One. 2014:e91940. doi: 10.1371/journal.pone.0091940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Campos CP, Rancoita PM, Kwee I, Zucca E, Zaffalon M, Bertoni F. Discovering subgroups of patients from DNA copy number data using NMF on compacted matrices. PLoS One. 2013;8:e79720. doi: 10.1371/journal.pone.0079720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal S, Bi Y, Macyszyn L, Showe LC, O'Rourke DM, Davuluri RV. Isoform-level gene signature improves prognostic stratification and accurately classifies glioblastoma subtypes. Nucleic Acids Res. 2014;42:e64. doi: 10.1093/nar/gku121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Gu Q, Hou JP, Han J, Ma J. A network-assisted co-clustering algorithm to discover cancer subtypes based on gene expression. BMC Bioinformatics. 2014;15:37. doi: 10.1186/1471-2105-15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 25.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database - 2009 update. Nucleic Acids Res. 2009;37(Database):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from argonaute CLIP-Seq and degradome-Seq data. Nucleic Acids Res. 2011;39(Database):D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao YY, Lee TS, Lin YM. A fisher exact test will be more proper. Radiology. 2006;239:300–301. doi: 10.1148/radiol.2391051114. author reply 301. [DOI] [PubMed] [Google Scholar]
- 29.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Cancer Genome Atlas Research Network: integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yano S, Miwa S, Kishimoto H, Toneri M, Hiroshima Y, Yamamoto M, Bouvet M, Urata Y, Tazawa H, Kagawa S, et al. Experimental curative fluorescence-guided surgery of highly invasive glioblastoma multiforme selectively labeled with a killer-reporter adenovirus. Mol Ther. 2015;23:1182–1188. doi: 10.1038/mt.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fadul CE, Fisher JL, Hampton TH, Lallana EC, Li Z, Gui J, Szczepiorkowski ZM, Tosteson TD, Rhodes CH, Wishart HA, et al. Immune response in patients with newly diagnosed glioblastoma multiforme treated with intranodal autologous tumor lysate-dendritic cell vaccination after radiation chemotherapy. J Immunother. 2011;34:382. doi: 10.1097/CJI.0b013e318215e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siggs OM, Miosge LA, Daley SR, Asquith K, Foster PS, Liston A, Goodnow CC. Quantitative reduction of the TCR adapter protein SLP-76 unbalances immunity and immune regulation. J Immunol. 2015;194:2587–2595. doi: 10.4049/jimmunol.1400326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan M, Himoudi N, Pule M, Sebire N, Poon E, Blair A, Williams O, Anderson J. Development of cellular immune responses against PAX5, a novel target for cancer immunotherapy. Cancer Res. 2008;68:8058–8065. doi: 10.1158/0008-5472.CAN-08-0153. [DOI] [PubMed] [Google Scholar]
- 34.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Seashols S. unpublished PhD thesis. Virginia Commonwealth University; 2013. Variation and modulation of microRNAs in prostate cancer and biological fluids. [Google Scholar]
- 36.Chang JS, Yeh RF, Wiencke JK, Wiemels JL, Smirnov I, Pico AR, Tihan T, Patoka J, Miike R, Sison JD, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–264. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- 38.Bradley SV, Holland EC, Liu GY, Thomas D, Hyun TS, Ross TS. Huntingtin interacting protein 1 is a novel brain tumor marker that associates with epidermal growth factor receptor. Cancer Res. 2007;67:3609–3615. doi: 10.1158/0008-5472.CAN-06-4803. [DOI] [PubMed] [Google Scholar]
- 39.Gorski JJ, Savage KI, Mulligan JM, McDade SS, Blayney JK, Ge Z, Harkin DP. Profiling of the BRCA1 transcriptome through microarray and ChIP-chip analysis. Nucleic Acids Res. 2011;39:9536–9548. doi: 10.1093/nar/gkr679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong H, Luo L, Hong S, Siu H, Xiao Y, Jin L, Chen R, Xiong M. Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst Biol. 2010;4:163. doi: 10.1186/1752-0509-4-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mokhtari T. Role of microRNAs in the development of cancer stem cells into glioblastomas. 2008. [Google Scholar]
- 42.Su Y, Li G, Zhang X, Gu J, Zhang C, Tian Z, Zhang J. JSI-124 inhibits glioblastoma multiforme cell proliferation through G(2)/M cell cycle arrest and apoptosis augment. Cancer Biol Ther. 2008;7:1243–1249. doi: 10.4161/cbt.7.8.6263. [DOI] [PubMed] [Google Scholar]
- 43.Antonia SJ, Mirza N, Fricke I, Chiappori A, Thompson P, Williams N, Bepler G, Simon G, Janssen W, Lee JH, et al. Combination of p53 cancer vaccine with chemotherapy in patients with extensive stage small cell lung cancer. Clin Cancer Res. 2006;12:878–887. doi: 10.1158/1078-0432.CCR-05-2013. [DOI] [PubMed] [Google Scholar]
- 44.Villalonga-Planells R, Coll-Mulet L, Martínez-Soler F, Castaño E, Acebes JJ, Giménez-Bonafé P, Gil J, Tortosa A. Activation of p53 by nutlin-3a induces apoptosis and cellular senescence in human glioblastoma multiforme. PLoS One. 2011;6:e18588. doi: 10.1371/journal.pone.0018588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Innocente SA, Lee JM. p53 is a NF-Y- and p21-independent, Sp1-dependent repressor of cyclin B1 transcription. FEBS Lett. 2005;579:1001–1007. doi: 10.1016/j.febslet.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 46.Dalvai M, Mondesert O, Bourdon JC, Ducommun B, Dozier C. Cdc25B is negatively regulated by p53 through Sp1 and NF-Y transcription factors. Oncogene. 2011;30:2282–2288. doi: 10.1038/onc.2010.588. [DOI] [PubMed] [Google Scholar]
- 47.Galanti G, Fisher T, Kventsel I, Shoham J, Gallily R, Mechoulam R, Lavie G, Amariglio N, Rechavi G, Toren A. Δ9-Tetrahydrocannabinol inhibits cell cycle progression by downregulation of E2F1 in human glioblastoma multiforme cells. Acta Oncol. 2008;47:1062–1070. doi: 10.1080/02841860701678787. [DOI] [PubMed] [Google Scholar]
- 48.Alonso MM, Fueyo J, Shay JW, Aldape KD, Jiang H, Lee OH, Johnson DG, Xu J, Kondo Y, Kanzawa T, et al. Expression of transcription factor E2F1 and telomerase in glioblastomas: mechanistic linkage and prognostic significance. J Natl Cancer Inst. 2005;97:1589–1600. doi: 10.1093/jnci/dji340. [DOI] [PubMed] [Google Scholar]
- 49.Blais A, Monté D, Pouliot F, Labrie C. Regulation of the human cyclin-dependent kinase inhibitor p18INK4c by the transcription factors E2F1 and Sp1. J Biol Chem. 2002;277:31679–31693. doi: 10.1074/jbc.M204554200. [DOI] [PubMed] [Google Scholar]
- 50.He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757. doi: 10.1186/1471-2164-14-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 52.Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS. The chromatin remodeling factor Mi-2α acts as a novel co-activator for human c-Myb. J Biol Chem. 2007;282:13994–14005. doi: 10.1074/jbc.M700755200. [DOI] [PubMed] [Google Scholar]
- 53.Bao ZS, Zhang CB, Wang HJ, Yan W, Liu YW, Li MY, Zhang W. Whole-genome mRNA expression profiling identifies functional and prognostic signatures in patients with mesenchymal glioblastoma multiforme. CNS Neurosci Ther. 2013;19:714–720. doi: 10.1111/cns.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 2005;15:457–459. doi: 10.1016/j.tcb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Zhang C, Wang L, Wu D, Chen H, Chen Z, Thomas-Ahner JM, Zynger DL, Eeckhoute J, Yu J, Luo J, et al. Definition of a FoxA1 Cistrome that is crucial for G1 to S-phase cell-cycle transit in castration-resistant prostate cancer. Cancer Res. 2011;71:6738–6748. doi: 10.1158/0008-5472.CAN-11-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sana J, Radova L, Lakomy R, Kren L, Fadrus P, Smrcka M, Besse A, Nekvindova J, Hermanova M, Jancalek R, et al. Risk score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis. 2014;35:2756–2762. doi: 10.1093/carcin/bgu212. [DOI] [PubMed] [Google Scholar]
- 57.Liu S, Yin F, Zhang J, Wicha MS, Chang AE, Fan W, Chen L, Fan M, Li Q. Regulatory roles of miRNA in the human neural stem cell transformation to glioma stem cells. J Cell Biochem. 2014;115:1368–1380. doi: 10.1002/jcb.24786. [DOI] [PubMed] [Google Scholar]
- 58.Martinez-Sanchez A, Murphy CL. miR-1247 functions by targeting cartilage transcription factor SOX9. J Biol Chem. 2013;288:30802–30814. doi: 10.1074/jbc.M113.496729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scaravilli M, Porkka KP, Brofeldt A, Annala M, Tammela TL, Jenster GW, Nykter M, Visakorpi T. MiR-1247-5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate. 2015;75:798–805. doi: 10.1002/pros.22961. [DOI] [PubMed] [Google Scholar]
- 60.Carbonell WS, DeLay M, Jahangiri A, Park CC, Aghi MK. β1 Integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res. 2013;73:3145–3154. doi: 10.1158/0008-5472.CAN-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carson KR, Focosi D, Major EO, Petrini M, Richey EA, West DP, Bennett CL. Monoclonal antibody-associated progressive multifocal leucoencephalopathy in patients treated with rituximab, natalizumab, and efalizumab: a review from the Research on Adverse Drug Events and Reports (RADAR) Project. Lancet Oncol. 2009;10:816–824. doi: 10.1016/S1470-2045(09)70161-5. [DOI] [PubMed] [Google Scholar]
- 62.Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M, Filka E, Yong WH, Mischel PS, Liau LM, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29:142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]