Abstract
Influenza viruses isolated from ducks are rarely able to infect chickens; it is therefore postulated that these viruses need to adapt in some way to be able to transmit to chickens in nature. Previous studies revealed that sialyl Lewis X (3′SLeX), which is fucosylated α2,3 sialoside was predominantly detected on the epithelial cells of the chicken trachea, whereas this glycan structure is not found in the duck intestinal tract. To clarify the mechanisms of the interspecies transmission of influenza viruses between ducks and chickens, we compared the receptor specificity of low pathogenic avian influenza viruses isolated from these two species. Glycan-binding analysis of the recombinant hemagglutinin (HA) of a chicken influenza virus, A/chicken/Ibaraki/1/2005 (H5N2), revealed a binding preference to α1,3 fucosylated sialosides. On the other hand, the HA of a duck influenza virus, A/duck/Mongolia/54/2001 (H5N2) (Dk/MNG), particularly bound to non-fucosylated α2,3 sialosides such as 3-sialyllactosamine (3′SLacNAc). Computational analyses along with binding analyses of the mutant HAs revealed that this glycan-binding specificity of the HA was determined by amino acid residues at positions 222 and 227. Inconsistent with the glycan-binding specificity of the recombinant HA protein, virions of Dk/MNG bound to both 3′SLacNAc and 3′SLeX. Glycan-binding analysis in the presence of a neuraminidase (NA) inhibitor revealed that the NA conferred binding to 3′SLeX to virions of Dk/MNG. The present results reveal the molecular basis of the interaction between fucosylated α2,3 sialosides and influenza viruses.
Keywords: influenza A virus, hemagglutinin, receptor specificity, fucosylation, sialyl Lewis X
Introduction
Influenza A viruses are zoonotic pathogens that are widely distributed among mammalian hosts such as humans, pigs and horses, as well as avian species such as chickens, ducks, many other poultry and wild birds [1]. The mechanism of the interspecies transmission of influenza viruses has been intensively analyzed for the control of human and animal influenza [2–6]. Influenza A virus is an enveloped virus that contains two envelope glycoproteins: hemagglutinin (HA) and neuraminidase (NA), both of which recognize sialoglycans [7–9]. HA is a viral lectin, which specifically binds to terminal sialic acid (N-acetylneuraminic acid or N-glycolylneuraminic acid: Sia) linked to penultimate galactose (Gal) with an α2,3 or 2,6 linkage and is responsible for the attachment of virions to host cell surface receptors. The glycan-binding specificity of HA varies depending on the host species from which the virus is isolated; avian influenza viruses preferentially bind α2,3 sialosides, whereas human influenza viruses prefer α2,6 sialosides [10–11]. This specificity is consistent with receptor distribution in host tissues; α2,3 sialosides are predominantly detected on epithelial cells of the duck intestine [12–13], whereas α2,6 sialosides are predominantly detected on epithelial cells of the human trachea [14].
Influenza A viruses of each of the known subtypes (H1 to H16 and N1 to N9) have been isolated from water birds, particularly migratory ducks [15–17]. Therefore, migratory ducks are the natural hosts for influenza A viruses. Chickens are rarely infected directly with viruses isolated from ducks, although most influenza viruses isolated from chickens prefer α2,3 sialosides, similar to the viruses of duck origin [10, 18–19]. It was previously reported that influenza viruses isolated from terrestrial poultry prefer fucosylated α2,3 sialosides such as sialyl Lewis X antigen (3′SLeX) or 6-O-sulfo-3′SLeX for the receptor [20]. In our previous study, we revealed that 3′SLeX was predominantly detected on epithelial cells of the upper respiratory tract of chickens, which is the primary replication site of chicken-adapted influenza viruses [21]. On the contrary, 3′SLeX was not detected on epithelial cells of duck colon, suggesting that α1,3 fucosylation of antepenultimate N-acetylglucosamine (GlcNAc) in α2,3 sialosides is a species barrier between ducks and chickens. Modifications on the antepenultimate GlcNAc of α2,3 sialosides is considered a key factor explaining differential susceptibility of ducks and chickens to influenza virus infection. A previous study on the structural basis of recognition of this particular glycan motif suggested that the fucose moiety of 3′SLeX is positioned close to the 220-loop of the HA [22]. Role of the 220-loop, particularly an amino acid residue at position 222, in recognition of α1,3 fucosylated sialosides have been previously discussed in H3, H5, and H7 HA [23–25]. However, effects of the particular amino acid motif in the 220-loop on the glycan-binding property of HA are totally unknown in H5 HA; thus, the detailed molecular basis of this interaction between H5 HA and α1,3 fucosylated sialosides should be analyzed.
The other envelope glycoprotein of influenza A virus, NA, cleaves terminal Sia from penultimate Gal, which is essential for virus release from host cells and also facilitates cell attachment by destroying “decoy” receptors [26–27]. Previous studies suggested that the functional balance between HA (receptor binding) and NA (receptor destroying) contributes to pathogenicity and host range of influenza A viruses [28]. Nevertheless, the mechanism by which NA contributes to virus–glycan interaction has not been extensively studied because analyses on interaction of NA with modified α2,3 sialosides are still limited [29].
Here we identified that two amino acid residues located at positions 222 and 227 in the HA of H5 influenza A viruses have a crucial role in recognition of 3′SLeX. In addition, the present results reveal a contribution of NA to the binding of virions of an influenza A virus to 3′SLeX.
Materials and methods
Viruses and cells
A/chicken/Ibaraki/1/2005 (H5N2) (Ck/IBR) [30], was kindly provided by National Institute of Animal Health, National Agriculture and Food Research Organization, Tsukuba, Ibaraki, Japan. A/duck/Mongolia/54/2001 (H5N2) (Dk/MNG), was isolated from fecal samples of migratory ducks in Mongolia in 2001 [31]. The viruses were propagated in 10-day-old embryonated chicken eggs at 35°C for 48 h, and the infectious allantoic fluids were used as virus stocks. Madin-Darby canine kidney (MDCK) cells were maintained in Minimum Essential Medium (MEM; Nissui Pharmaceutical, Tokyo, Japan) supplemented with 0.3 mg/ml L-glutamine, 100 U/ml penicillin G, 0.1 mg/ml streptomycin, 8 μg/ml gentamicin and 10% calf serum. Human embryonic kidney (HEK) 293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (D-MEM; Life Technologies, Carlsbad, CA, USA) supplemented with 0.3 mg/ml L-glutamine, 100 U/ml penicillin G, 0.1 mg/ml streptomycin, 8 μg/ml gentamicin and 10% fetal calf serum. HEK 293S GnTI−/− cells were maintained in pyruvate free D-MEM (Life Technologies) supplemented with 0.3 mg/ml L-glutamine, 10 U/ml penicillin G, 0.01 mg/ml streptomycin and 10% fetal calf serum.
Reverse genetics
Eight genes from Ck/IBR and Dk/MNG each were cloned to produce viruses by reverse genetics as described previously [21, 32–33]. Amino acid substitutions R222K and R227S in the HA of Ck/IBR, as well as K222R and S227R in the HA of Dk/MNG, were generated by site-directed mutagenesis using a QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s instructions (primer sequences are available upon request). The mutant viruses were rescued by reverse genetics. All eight segments of the genome were sequenced to confirm the existence of the introduced mutations and the absence of undesired mutations.
Expression of the recombinant HAs (rHAs)
The cDNAs of the HA genes of Ck/IBR, Dk/MNG and their mutants were cloned into the pCD5 expression vector as described previously [34]. The rHA proteins were expressed in HEK293S GnTI−/− cells [35] and purified from the cell culture supernatants as described previously [36].
Glycan microarray
The glycan microarray was carried out with 53 glycans focused on α2,3 sialosides which are the preferred receptors of avian influenza viruses. On the array slide, 10 non-sialylated glycans (glycans #1–10), 36 non-fucosylated α2,3 sialylated glycans (glycans #11–46) and 7 fucosylated and α2,3 sialylated glycans (glycans #47–53) were printed. Most glycans used were reported previously [34, 37], except for a few with extended glycan chains that were synthesized using methods reported previously (#31–35, 39–42, 51) [38–39]. The rHA was used at 50 μg/ml precomplexed with horseradish peroxidase (HRP)-linked anti-Strep-tag mouse antibody and with Alexa 488-linked anti-mouse IgG (4:2:1 molar ratio) prior to incubation for 30 min on ice in 100 μl of PBS containing 0.05% Tween 20 (PBST) and incubated on the array surface in a humidified chamber for 90 min. Slides were subsequently washed by successive rinses with PBST, PBS and de-ionized H2O. Washed arrays were dried by centrifugation and immediately scanned for fluorescence signals on a Perkin-Elmer ProScanArray Express confocal microarray scanner (Waltham, MA, USA). Fluorescence signal intensity was measured using Imagene (Biodiscovery, Hawthorne, CA, USA), and the mean intensity minus mean background was calculated and graphed using MS Excel. For each glycan, the mean signal intensity was calculated from six replicates. The highest and lowest signals of the six replicates were removed, and the remaining four replicates were used to calculate the mean signal, and standard error (SE).
In silico prediction of the binding of the HA of Ck/IBR and 3′SLeX
The three-dimensional (3D) structure of the H5 HA of Ck/IBR was constructed on the basis of the HA crystal structure of A/Vietnam/1194/2004 (H5N1) (PDB code 2IBX) [40] as described previously [41]. A predicted 3D structure of 3′SLeX was downloaded from GLYCAM-Web (http://glycam.org/). To obtain the structure of HA of Ck/IBR bound to 3′SLeX, each structure was first superimposed on the crystal structure of the H2 HA in complex with 3-sialyllactosamine (3′SLacNAc) (PDB code 2WR3) [42], and coordinates of 2WR3 were then removed using Discovery Studio version 4.1 (Dassault Systemes Biovia, San Diego, CA, USA). This model structure was refined by energy minimization followed by a 5 ns of molecular dynamics with the AMBER 14 software (Conflex USA, San Diego, CA, USA). The ff99SB force field and GLYCAM06 force field were used for the HA and 3′SLeX, respectively. The energy minimization and molecular dynamics calculations were conducted in a similar way, as described previously [43]. The definition of hydrogen bonds is the following: distance between donor and acceptor is less than 3.5 Å, and angle between donor, hydrogen and acceptor is greater than 120°.
Solid-phase direct binding assay
The receptor binding specificity of viruses was assessed using a solid-phase direct binding assay with sialylglycopolymers 3′SLacNAc-PAA and 3′SLeX-PAA (Cosmo Bio Co., Ltd., Tokyo, Japan) as described previously [6, 21]. Briefly, each sialylglycopolymer was serially diluted and added to each well of a Universal-BIND™, 96 well polystyrene strip well microplate (Corning, Corning, NY, USA). Each well was blocked with 1% bovine serum albumin (BSA) at room temperature for 1 h. After washing with PBST, a solution containing influenza viruses (16 HAU in PBS) was added to each well and the plates were incubated at 4°C for 12 h. After washing, mouse anti-HA monoclonal antibodies were added to each well and the plates were incubated at 4°C for 2 h. The wells were then washed and incubated with goat anti-mouse IgG-HRP conjugate (Bio-Rad, Hercules, CA, USA) at 4°C for 2 h. After washing, 100 μl of the substrates including 0.5 mM 3,3′-tetramethylbenzidine (TMB) and 0.04% H2O2, were added to each well. After incubation at room temperature for 10 min, the reactions were stopped using 50 μl of 2N H2SO4, and absorbance at 450/630 nm was measured using a MULTISCAN JX (Thermo Fisher Scientific, Waltham, MA, USA). The solid-phase direct binding assay was also performed in the presence of the NA inhibitor peramivir, which was a kind gift from Dr. Masanori Kobayashi of Shionogi & Co. Ltd. In this case, viruses were pre-incubated with either 2.5, 10 or 40 nM peramivir in PBS for 1 h on ice. The solution was used as a virus containing solution.
A solid-phase direct binding assay was also conducted using rHA. Each sialylglycopolymer was serially diluted and added to each well of a Nunc Immobilizer Amino C8, 96 well strip well microplate (Thermo Fisher Scientific) and the plates were incubated at 37°C for 1 h. Each well was then blocked with PBST containing 2% BSA at room temperature for 3 h. The rHA was used at 5 μg/ml precomplexed with HRP-linked anti-Strep-tag mouse antibody (1:2000 dilution) and with goat anti-mouse IgG-HRP conjugate (1:1000 dilution) prior to incubation for 30 min on ice in PBST containing 0.5% BSA with or without 40 nM peramivir. After washing the plates, the complexes were added and incubated at room temperature for 1 h. After washing, 100 μl of the TMB substrates were added to each well. The reactions were stopped using 50 μl of 2N H2SO4, and absorbance at 450/630 nm was measured using a MULTISCAN JX.
Amino acid sequence comparison of the H5 HA
A total of 2901 amino acid sequences of H5 HA were obtained from GenBank. Sequence data were aligned with mafft version 7.215 (http://mafft.cbrc.jp/alignment/software/). The amino acid sequence data of highly pathogenic avian influenza viruses were removed according to the sequence of HA cleavage site. The remaining 631 sequences were further divided into viruses isolated from Anseriformes, non-chicken Galliformes or chickens according to the original host of each virus determined by the strain name.
Results
Computational analysis of the binding of the HA of a chicken influenza A virus to 3′SLeX
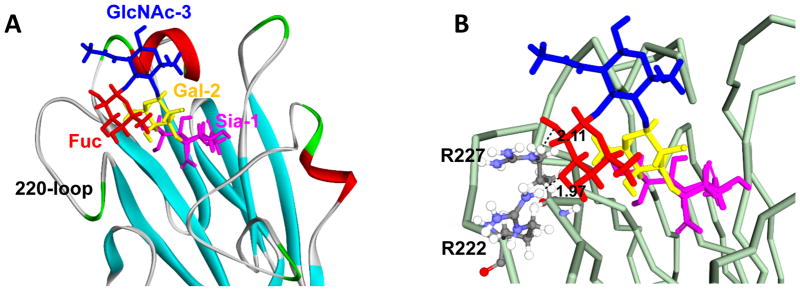
The binding model of an HA derived from a chicken influenza A virus, Ck/IBR to 3′SLeX was predicted using in silico analysis (Fig. 1A and B). The fucose moiety of 3′SLeX is positioned close to two arginine (R) residues at positions 222 and 227 (H3 numbering is used throughout) of the HA. The R residues at positions 222 and 227 are located within 3.5 Å of the C-2 and C-3 hydroxyl groups of the fucose, indicating potential hydrogen bonding. By comparing the amino acid sequence of Ck/IBR with that of a duck influenza virus, Dk/MNG, we observe a lysine (K) residue at position 222 and a serine (S) residue at position 227 instead of arginines. We hypothesized that these amino acid substitutions contribute to differences in receptor binding specificity.
Fig. 1.
Structure model of sialyl Lewis X (3′SLeX) bound to the hemagglutinin (HA) of Ck/IBR, which was taken from snapshot at 5 ns. The receptor binding site of the HA of Ck/IBR is shown in either a ribbon (A) or line (B) cartoon representation. In the glycan structure, N-acetylneuraminic acid (Sia) is shown in purple, galactose (Gal) is shown in yellow, N-acetylglucosamine (GlcNAc) is shown in blue and fucose (Fuc) is shown in red. In (B), dash lines indicate hydrogen bonds and the numbers indicate the predicted length of each hydrogen bond.
Glycan-binding specificity of the HA of Ck/IBR, Dk/MNG and their mutants
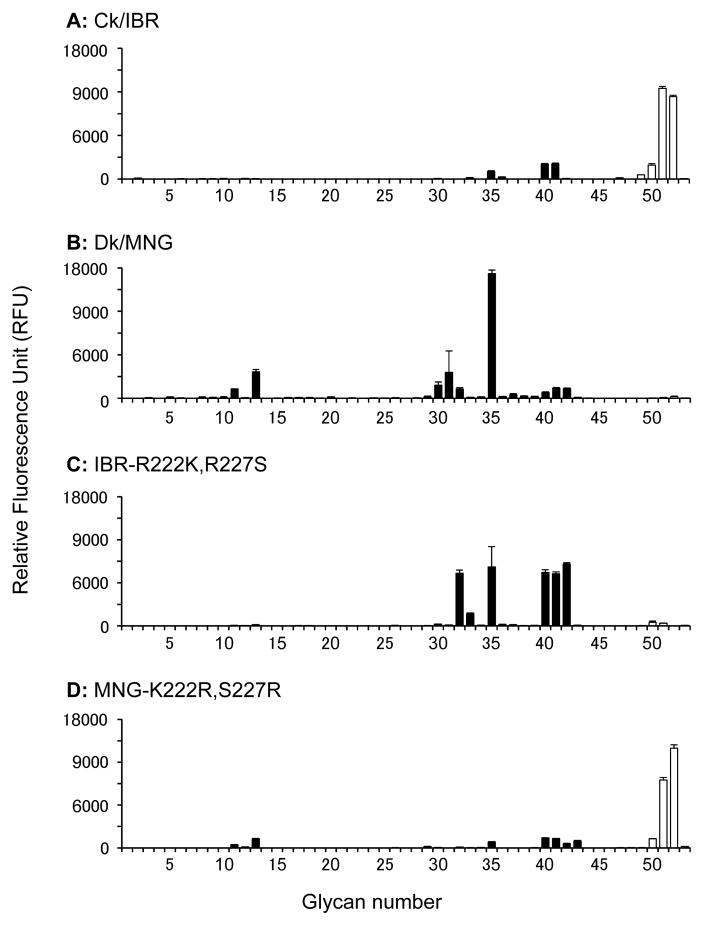
Soluble trimeric recombinant HAs (rHA) of Ck/IBR and Dk/MNG were generated and subjected to glycan microarray analysis to evaluate their glycan-binding specificity (Fig. 2A and B). The rHA of Ck/IBR interacted with glycans #51 (3′SLeXTriLN-Core4) and #52 [Siaα2,3Galβ1,4(Fucα1,3)(6O-sulfo)GlcNAcβ-propyl-NH2] with high relative avidity. Although the rHA of Ck/IBR slightly interacted with non-fucosylated α2,3 sialosides, these signals were much weaker than those of glycans #51 and #52. No interaction was observed between the rHA of Dk/MNG and fucosylated α2,3 sialosides. Interestingly, most of the glycans, which were bound by the rHA of Dk/MNG, are biantennary glycans with multiple lactosamine (LacNAc) repeats (e.g. #30–32 and #35). The results indicate that the rHA of Ck/IBR selectively binds α1,3 fucosylated sialosides, whereas that of Dk/MNG selectively binds non-fucosylated α2,3 sialosides. To evaluate the contribution of amino acid residues at positions 222 and 227 of the HA to this binding specificity of rHAs, we generated mutant rHAs of Ck/IBR and Dk/MNG, in which amino acid residues at positions 222 and 227 were altered (IBR-R222K,R227S and MNG-K222R,S227R respectively) and subjected these rHAs to glycan microarray analysis (Fig. 2C and D). The rHA of IBR-R222K,R227S interacted with non-fucosylated α2,3 sialosides, whereas no binding was observed with fucosylated α2,3 sialosides. The rHA of MNG-K222R,S227R interacted with glycans #51 and #52 with high specificity, whereas almost no binding was observed with non-fucosylated α2,3 sialosides. These results indicate that the glycan-binding specificity of rHA to fucosylated and non-fucosylated α2,3 sialosides is determined by the amino acid motifs at positions 222 and 227 of the HA.
Fig. 2.
Glycan-binding specificity of the soluble trimeric recombinant hemagglutinins (rHAs). The glycan-binding specificity of rHA of Ck/IBR (A), Dk/MNG (B), IBR-R222K,R227S (C) and MNG-K222R,S227R (D) was analyzed by glycan microarray. Non-sialylated controls are shown as grey bars (#1–10), non-fucosylated α2,3 sialylated glycans are presented as black bars (#11–46) and fucosylated and α2,3 sialylated glycans are shown in white bars (#47–53). Each bar represents the mean signal minus background for each glycan sample and error bars are the SE value. Glycans imprinted on the array are listed in Table S1.
Glycan-binding specificity of the virions of Ck/IBR, Dk/MNG and their mutants
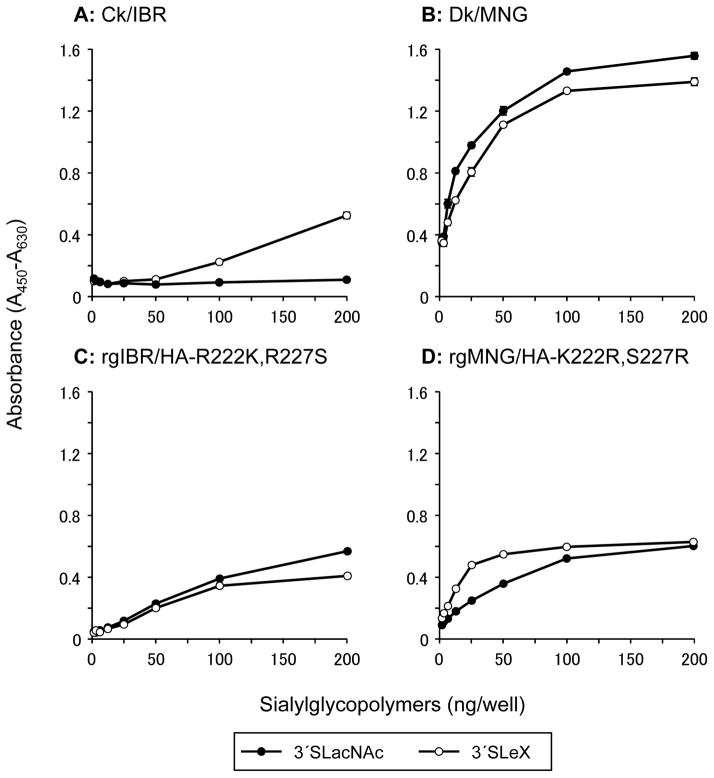
In our previous study, virions of Ck/IBR specifically bound to α1,3 fucosylated sialosides, 3′SLeX in a solid-phase direct binding assay; whereas those of Dk/MNG bound to both non-fucosylated α2,3 sialosides, 3 -sialyllactosamine (3′SLacNAc) and 3′SLeX [21]. To further elucidate the importance of the amino acid motifs at positions 222 and 227 of HA, mutant viruses, namely rgIBR/HA-R222K,R227S and rgMNG/HA-K222R,S227R were generated, and their receptor binding preferences were characterized by a solid-phase direct binding assay (Fig. 3). Consistent with our previous study, virions of Ck/IBR specifically bound to 3′SLeX, whereas those of Dk/MNG bound to both 3′SLacNAc and 3′SLeX. rgIBR/HA-222K,227S bound to both 3′SLacNAc and 3′SLeX, indicating that R222K and R227S mutations facilitate binding of non-fucosylated glycans. rgMNG/HA-222R,227R also bound to both 3′SLacNAc and 3′SLeX; however, binding avidity to 3′SLacNAc was significantly lower than that to 3′SLeX.
Fig. 3.
Glycan-binding specificity of virions. The glycan-binding specificity of Ck/IBR (A), Dk/MNG (B), rgIBR/HA-R222K,R227S (C) and rgMNG/HA-K222R,S227R (D) to sialylglycopolymers containing 3′sialyllactosamine (3′SLacNAc, black circles) and sialyl Lewis X (3′SLeX, white circles) was investigated using a solid-phase direct binding assays. The data are presented as the mean ± SE of triplicate experiments.
Dk/MNG virions bind to fucosylated α2,3 sialosides through the neuraminidase
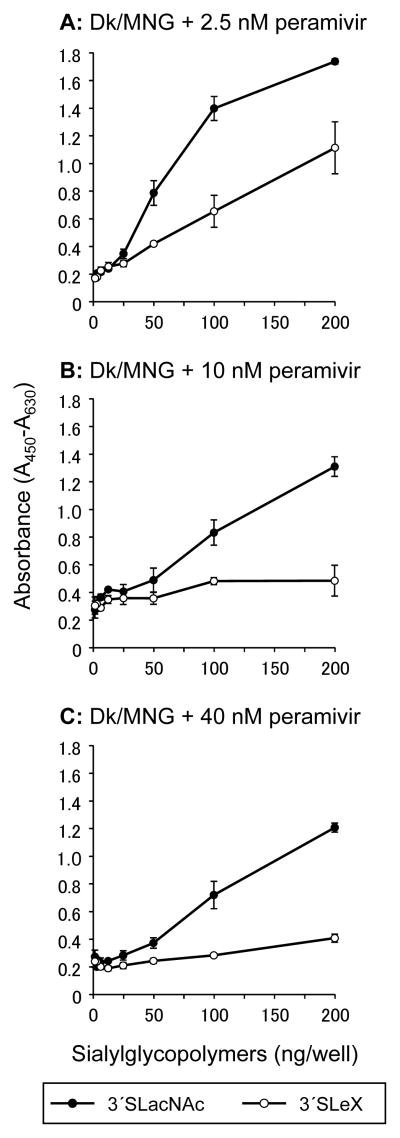
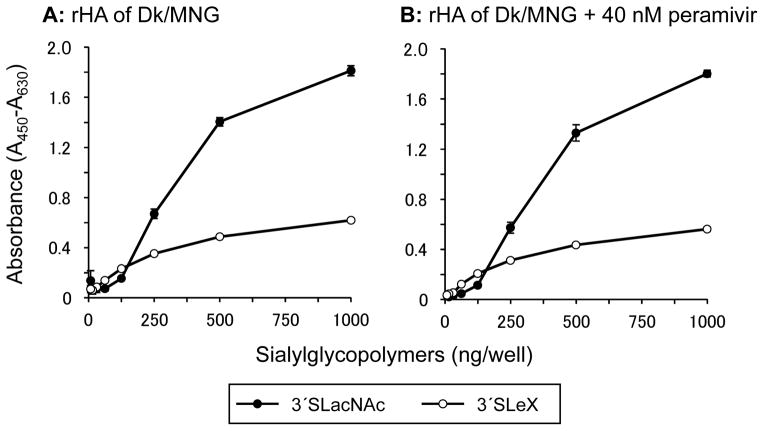
The rHA of Dk/MNG specifically bound to non-fucosylated α2,3 sialosides; whereas virions of Dk/MNG bound to both fucosylated and non-fucosylated sialosides (Fig. 2B and 3B). To investigate contribution of NA, which is the other Sia-interacting viral protein of influenza A viruses, to glycan-binding by influenza virus virions, a solid-phase direct binding assay was conducted in the presence of the NA inhibitor, peramivir (Fig. 4). Binding of Dk/MNG to 3′SLeX was diminished in the presence of 2.5 nM peramivir. Although binding to 3′SLacNAc was slightly affected in the presence of higher concentration of peramivir, almost no binding to 3′SLeX was observed in the presence of 10 or 40 nM peramivir. To confirm that peramivir specifically inhibit the glycan-binding by the NA and has no effect on the glycan-binding mediated by the HA of the HA in the assay, a solid-phase direct binding assay was conducted with the rHA of Dk/MNG in the presence of peramivir (Fig. 5). Consistent with the result of glycan microarray (Fig. 2B), the rHA of Dk/MNG preferentially bound 3′SLacNAc in the absence of peramivir (Fig. 5A). The binding of the rHA to 3′SLacNAc and 3′SLeX was similar in the presence of 40 nM peramivir, and no inhibition was observed (Fig. 5B). These results confirmed that peramivir has no effect on the interaction between sialosides and the rHA of Dk/MNG. Accordingly, the present result indicates that the NA of Dk/MNG binds to fucosylated α2,3 sialosides.
Fig. 4.
Glycan-binding specificity of virions of Dk/MNG in the presence of peramivir. The glycan-binding specificity of virions of Dk/MNG to sialylglycopolymers containing 3′sialyllactosamine (3′SLacNAc, black circles) and sialyl Lewis X (3′SLeX, white circles) was investigated in the presence of either 2.5 (A), 10 (B) or 40 (C) nM of a neuraminidase inhibitor peramivir. The data are presented as the mean ± SE of triplicate experiments.
Fig. 5.
Glycan-binding specificity of the rHA of Dk/MNG in the presence of peramivir. The glycan-binding specificity of the rHA of Dk/MNG to sialylglycopolymers containing 3′sialyllactosamine (3′SLacNAc, black circles) and sialyl Lewis X (3′SLeX, white circles) was investigated in the absence (A) or presence (B) of a neuraminidase inhibitor peramivir. The data are presented as the mean ± SE of triplicate experiments.
Amino acid sequence comparison of HA of H5 avian influenza viruses
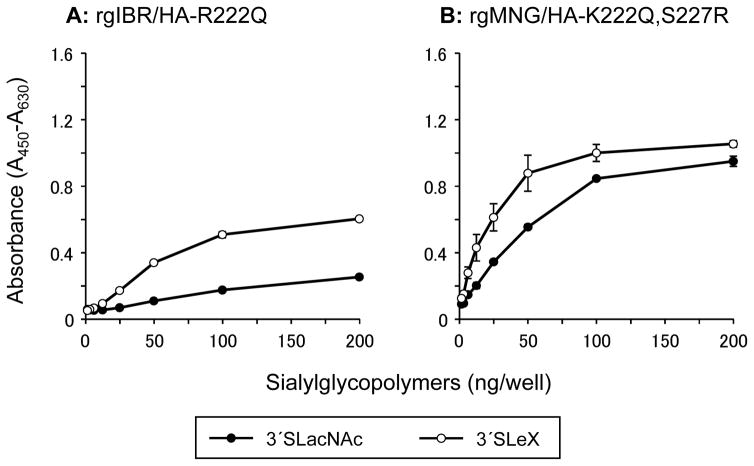
The amino acid motifs at positions 222 and 227 in HA of H5 avian influenza viruses isolated from Galliformes and Anseriformes were analyzed (Table 1). The majority of low pathogenic avian influenza viruses (LPAIVs) isolated from chickens have a glutamine (Q) residue at position 222 and an arginine (R) residue at position 227. On the other hand, the majority of viruses isolated from Anseriformes have a lysine (K) residue at position 222 and a serine (S) residue at position 227. Interestingly, most of the viruses isolated from non-chicken Galliformes have a K residue at position 222 and an S residue at position 227, which is the same amino acid sequence present in isolates from Anseriformes. An R residue at position 222 as observed in Ck/IBR is a relatively rare motif in H5 avian influenza viruses. We also generated mutant viruses, which possess same amino acid residues as consensus motif of chicken LPAIVs at positions 222 and 227 of the HA, namely rgIBR/HA-R222Q and rgMNG/HA-K222Q,S227R. Glycan-binding analysis by solid-phase direct binding assay revealed that rgIBR/HA-R222Q specifically bound to 3′SLeX (Fig. 6A). Also, glycan-binding preference of rgMNG/HA-K222Q,S227R was similar to that of rgMNG/HA-222R,227R (Fig. 6B). These results indicate that Q and R residue at position 222 of the HA have similar effects on the glycan-binding property of HA in combination with R residue at position 227.
Table 1.
Amino acid sequence comparison of 222/227 motifs in HA of H5 LPAIVs.
| Host | Amino acid residue
|
Number of strains | |
|---|---|---|---|
| 222 | 227 | ||
| Chicken (98 strains) | Q | R | 70 |
| K | S | 26 | |
| R | S | 1 | |
| R | R | 1 | |
|
| |||
| Anseriformes (506 strains) | K | S | 471 |
| R | S | 18 | |
| Q | S | 13 | |
| E | R | 2 | |
| N | S | 1 | |
| R | R | 1 | |
|
| |||
| Non-chicken Galliformes (27 strains) | K | S | 25 |
| R | S | 1 | |
| Q | R | 1 | |
Amino acid sequences of H5 HA were obtained form GenBank.
Amino acid motifs of Ck/IBR and Dk/MNG were shown in bold.
Fig. 6.
Glycan-binding specificity of rgIBR/HA-R222Q and rgMNG/HA-K222Q,S227R. Glycan-binding specificity of virions of rgIBR/HA-R222Q (A) and rgMNG/HA-K222Q,S227R (B) to sialylglycopolymers containing 3′sialyllactosamine (3′SLacNAc, black circles) and sialyl Lewis X (3′SLeX, white circles) was investigated using a solid-phase direct binding assays. The data are presented as the mean ± SE of triplicate experiments.
Discussion
We previously revealed that α2,3 sialosides expressed on epithelial cells of the chicken trachea are fucosylated [21]. Although modification of α2,3 sialosides has a critical impact on the glycan-binding specificity of influenza viruses [20, 21, 23], the interaction of these modified α2,3 sialosides with HA is not fully understood. In the present study, we analyzed binding specificity of two H5 LPAIVs, Ck/IBR and Dk/MNG, to fucosylated or non-fucosylated α2,3 sialosides. The present results reveal that R222 and R227 in the HA of Ck/IBR are located close to the α1,3 fucose linked to antepenultimate GlcNAc (Fig. 1). A previous structural analysis indicated that this fucose moiety is positioned close to K222 in the HA of a highly pathogenic avian influenza virus and that it de-stabilized the interaction of the HA with glycan [22]. Glycan microarray analysis of rHAs revealed that K222R substitution in combination with S227R in the HA of Dk/MNG altered its glycan-binding specificity from non-fucosylated to fucosylated α2,3 sialosides (Fig. 2B and D). This suggests that these substitutions in the HA alter the interaction with fucose and stabilizes the binding.
Sequence comparison of HA of H5 influenza A viruses revealed that the amino acid motif K222 and S227 were highly conserved within LPAIVs circulating among migratory ducks (Table 1). Receptor binding analyses indicate that viruses with K222 and S227 specifically bound non-fucosylated α2,3 sialosides, whereas viruses with R222 and R227 as well as Q222 and R227 specifically bound fucosylated α2,3 sialosides (Fig. 3 and 6). These facts suggest that H5 LPAIVs isolated from chickens prefer fucosylated α2,3 sialosides and those isolated from ducks prefer non-fucosylated α2,3 sialosides. Interestingly, viruses isolated from non-chicken Galliformes species, such as quails or turkeys have the same amino acid motif as LPAIVs isolated from Anseriformes. Influenza viruses circulating among migratory ducks are transmitted to chickens via terrestrial poultry such as quails or turkeys [1, 44]. These facts suggest that the acquisition of “chicken type receptor specificity” may not occur during adaptation in ducks or later in non-chicken terrestrial poultry, but occurs during the multiple replication and transmission events in chickens.
Glycan-binding analysis of virions and rHAs revealed different glycan-binding preferences (Fig. 2 and 3). Interestingly, virions of Dk/MNG bound to both fucosylated α2,3 sialosides, 3′SLeX and non-fucosylated α2,3 sialosides, 3′SLacNAc, whereas the rHA of Dk/MNG bound only non-fucosylated α2,3 sialosides. A solid-phase direct binding assay in the presence of peramivir suggests that the NA contributes to receptor binding specificity of Dk/MNG virions to 3′SLeX (Fig. 4). A solid-phase direct binding assay using rHA confirmed that the rHA of Dk/MNG preferentially bound non-fucosylated α2,3 sialosides (Fig. 5A). The presence of peramivir had no effect on the glycan-binding of the rHA of Dk/MNG (Fig. 5B), indicating that peramivir specifically inhibited glycan-binding of the NA in the solid-phase direct binding assay using virions of Dk/MNG (Fig. 4). Accordingly, we concluded that the glycan-binding specificity of virions of Dk/MNG results from additive effects of glycan-binding to 3′SLacNAc mediated by the HA and to 3′SLeX mediated by the NA, and thus, the binding to 3′SLeX was diminished by the NA inhibitor. NA of influenza viruses has sialidase activity, which means that NA also binds sialylated glycans [7, 9]. In the case of H7N9 influenza viruses recently isolated in China, it was proposed that the preferential cleavage of α2,3 sialosides by the NA along with intrinsic weak binding to α2,6 sialosides by the HA exaggerated receptor binding preference of these viruses to α2,6 sialosides [34]. Thus, the NA modulates the receptor binding preference of influenza virus by cancelling certain receptor binding with its sialidase activity. The NA of Dk/MNG modulates receptor specificity via a mechanism which is distinct from the previous model, as the NA obviously promote binding to 3′SLeX (Fig. 2B and 3B). However, we could not elucidate whether glycan binding by the NA is functional in that it would lead to infection. Thus, determining the receptor binding preferences of influenza viruses requires complementary assays using rHA and complete virions for the elucidation of fine receptor binding preferences of influenza viruses.
It should also be pointed out that the rHA of Dk/MNG did not exhibit binding to glycan #15 (Fig. 2B), which has exactly same glycan structure with the non-fucosylated α2,3 sialoside used in solid-phase direct binding assay. Virions of Dk/MNG strongly bound this glycan structure in the solid-phase direct binding assay (Fig. 3B). In the case of solid-phase direct binding assay, glycans are polymerized with polyacrylamide. In addition, we used whole virus particle for the solid-phase direct binding assay; thereby interaction of HA and glycans are multivalent in this assay. On the other hand, monomeric glycans were printed in the case of glycan microarray. These difference in the settings of each assay may lead the difference of the results in these two assays.
The present results reveal the molecular basis of the interaction between α1,3 fucosylated sialosides and HA of influenza viruses. α2,3 sialosides are structurally divergent; penultimate Gal and the antepenultimate GlcNAc can be sulfated, the linkage between Gal and GlcNAc can be a β1,3 or β1,4 linkage and the core structure of glycans can be N-linked, O-linked or glycolipid. It was proposed reported that presentation and internal complexity of glycan structure influenced their interaction with HA (See [39] and references therein). Indeed, the internal structure of glycans affected binding specificity of HA; the rHA of Dk/MNG preferred non-fucosylated α2,3 sialosides with multiple LacNAc repeats (Fig. 2B). Similarly, rHA of Ck/IBR bound to glycan #51 with high avidity, whereas binding to glycan #47, 49 and 50, which contain the same glycan sequence with glycan #51 on the non-redacting end, was much weaker (Fig. 2A). The importance of the internal glycan structure for influenza virus infection is not at all understood, even though glycan microarray data on the receptor binding specificity of influenza viruses are widely available. The present results demonstrated the significance of the structural diversity of sialosides on the interaction of influenza A viruses with host cell surface receptors.
Supplementary Material
Acknowledgments
We thank Ms. Yuki Maki and Ms. Ana L. Tran-Crie for their kind help in organizing this collaborative effort. We thank Drs. Masanori Kobayashi and Keiichi Taniguchi of Shionogi & Co. Ltd., for providing peramivir and their technical advice. The present work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI (grant number 26850178 to M.O.) and by the Program for Leading Graduate Schools from JSPS (grant number F01). This work was funded in part by the National Institutes of Health Grant R56 (grant number AI099274 to J.C.P). Several glycans used for the HA binding assays were partially provided by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/) funded by National Institute of General Medical Sciences (NIGMS) (grant number GM62116 to J.C.P.). R.P.d.V. is a recipient of Rubicon and VENI grants from the Netherlands Organization for Scientific Research (NWO). T.H. is supported by JSPS Research Fellowships for young scientists.
References
- 1.Kida H. Ecology of influenza viruses in nature, birds, and humans. Global Environmental Research. 2008;12:9–14. [Google Scholar]
- 2.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada S, Shinya K, Takada A, Ito T, Suzuki T, Suzuki Y, Le QM, Ebina M, Kasai N, Kida H, Horimoto T, Rivailler P, Chen LM, Donis RO, Kawaoka Y. Adaptation of a duck influenza A virus in quail. J Virol. 2012;86:1411–1420. doi: 10.1128/JVI.06100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulson JC, de Vries RP. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 2013;178:99–113. doi: 10.1016/j.virusres.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shichinohe S, Okamatsu M, Sakoda Y, Kida H. Selection of H3 avian influenza viruses with SAα2,6Gal receptor specificity in pigs. Virology. 2013;444:404–408. doi: 10.1016/j.virol.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Lamb RA, Choppin PW. The gene structure and replication of influenza virus. Annu Rev Biochem. 1983;52:467–506. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- 8.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 9.Gong J, Xu W, Zhang J. Structure and functions of influenza virus neuraminidase. Curr Med Chem. 2007;14:113–122. doi: 10.2174/092986707779313444. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 11.Rogers GN, D’Souza BL. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito T, Suzuki Y, Suzuki T, Takada A, Horimoto T, Wells K, Kida H, Otsuki K, Kiso M, Ishida H, Kawaoka Y. Recognition of N-glycolylneuraminic acid linked to galactose by the alpha2,3 linkage is associated with intestinal replication of influenza A virus in ducks. J Virol. 2000;74:9300–9305. doi: 10.1128/jvi.74.19.9300-9305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 15.Kida H, Yanagawa R. Isolation and characterization of influenza a viruses from wild free-flying ducks in Hokkaido, Japan. Zentralbl Bakteriol Orig A. 1979;244:135–143. [PubMed] [Google Scholar]
- 16.Kida H, Yanagawa R, Matsuoka Y. Duck influenza lacking evidence of disease signs and immune response. Infect Immun. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, Guan Y, Peiris M, He S, Webby RJ, Perez D, Webster RG. The quest of influenza A viruses for new hosts. Avian Dis. 2003;47:849–856. doi: 10.1637/0005-2086-47.s3.849. [DOI] [PubMed] [Google Scholar]
- 20.Gambaryan AS, Tuzikov AB, Pazynina GV, Desheva JA, Bovin NV, Matrosovich MN, Klimov AI. 6-sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J. 2008;5:85. doi: 10.1186/1743-422X-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiono T, Okamatsu M, Nishihara S, Takase-Yoden S, Sakoda Y, Kida H. A chicken influenza virus recognizes fucosylated α2,3 sialoglycan receptors on the epithelial cells lining upper respiratory tracts of chickens. Virology. 2014;456–457:131–138. doi: 10.1016/j.virol.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Xiong X, Tuzikov A, Coombs PJ, Martin SR, Walker PA, Gamblin SJ, Bovin N, Skehel JJ. Recognition of sulphated and fucosylated receptor sialosides by A/Vietnam/1194/2004 (H5N1) influenza virus. Virus Res. 2013;178:12–14. doi: 10.1016/j.virusres.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Gambaryan A, Yamnikova S, Lvov D, Tuzikov A, Chinarev A, Pazynina G, Webster R, Matrosovich M, Bovin N. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology. 2005;334:276–283. doi: 10.1016/j.virol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, Bovin NV, Klenk HD, Matrosovich MN. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang G, Li S, Blackmon S, Ye J, Bradley KC, Cooley J, Smith D, Hanson L, Cardona C, Steinhauer DA, Webby R, Liao M, Wan XF. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol. 2013;94:2599–2608. doi: 10.1099/vir.0.054692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohuchi M, Asaoka N, Sakai T, Ohuchi R. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 2006;8:1287–1293. doi: 10.1016/j.micinf.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 28.de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heider A, Mochalova L, Harder T, Tuzikov A, Bovin N, Wolff T, Matrosovich M, Schweiger B. Alterations in hemagglutinin receptor-binding specificity accompany the emergence of highly pathogenic avian influenza viruses. J Virol. 2015;89:5395–5405. doi: 10.1128/JVI.03304-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamatsu M, Saito T, Yamamoto Y, Mase M, Tsuduku S, Nakamura K, Tsukamoto K, Yamaguchi S. Low pathogenicity H5N2 avian influenza outbreak in Japan during the 2005–2006. Vet Microbiol. 2007;124:35–46. doi: 10.1016/j.vetmic.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Sakoda Y, Sugar S, Batchluun D, Erdene-Ochir TO, Okamatsu M, Isoda N, Soda K, Takakuwa H, Tsuda Y, Yamamoto N, Kishida N, Matsuno K, Nakayama E, Kajihara M, Yokoyama A, Takada A, Sodnomdarjaa R, Kida H. Characterization of H5N1 highly pathogenic avian influenza virus strains isolated from migratory waterfowl in Mongolia on the way back from the southern Asia to their northern territory. Virology. 2010;406:88–94. doi: 10.1016/j.virol.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 34.Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science. 2013;342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Vries RP, de Vries E, Bosch BJ, de Groot RJ, Rottier PJ, de Haan CA. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology. 2010;403:17–25. doi: 10.1016/j.virol.2010.03.047. [DOI] [PubMed] [Google Scholar]
- 37.de Vries RP, Zhu X, McBride R, Rigter A, Hanson A, Zhong G, Hatta M, Xu R, Yu W, Kawaoka Y, de Haan CA, Wilson IA, Paulson JC. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J Virol. 2014;88:768–773. doi: 10.1128/JVI.02690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng W, Pranskevich J, Nycholat C, Gilbert M, Wakarchuk W, Paulson JC, Razi N. Helicobacter pylori β1,3-N-acetylglucosaminyltransferase for versatile synthesis of type 1 and type 2 poly-LacNAcs on N-linked, O-linked and I-antigen glycans. Glycobiology. 2012;22:1453–1464. doi: 10.1093/glycob/cws101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA, Paulson JC. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem Int Ed Engl. 2012;51:4860–4863. doi: 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, Shinya K, Sawada T, Usui T, Murata T, Lin Y, Hay A, Haire LF, Stevens DJ, Russell RJ, Gamblin SJ, Skehel JJ, Kawaoka Y. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 41.Motohashi Y, Igarashi M, Okamatsu M, Noshi T, Sakoda Y, Yamamoto N, Ito K, Yoshida R, Kida H. Antiviral activity of stachyflin on influenza A viruses of different hemagglutinin subtypes. Virol J. 2013;10:118. doi: 10.1186/1743-422X-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009;106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takano R, Kiso M, Igarashi M, Le QM, Sekijima M, Ito K, Takada A, Kawaoka Y. Molecular mechanisms underlying oseltamivir resistance mediated by an I117V substitution in the neuraminidase of subtype H5N1 avian influenza A viruses. J Infect Dis. 2013;207:89–97. doi: 10.1093/infdis/jis633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertran K, Dolz R, Majó N. Pathobiology of avian influenza virus infection in minor gallinaceous species: a review. Avian Pathol. 2014;43:9–25. doi: 10.1080/03079457.2013.876529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.