Abstract
Colibactin represents an as-yet uncharacterized genotoxic secondary metabolite produced by human gut bacteria. Here we report the biosynthetic discovery of two new precolibactin molecules from Escherichia coli, including precolibactin-886 that uniquely incorporates the highly sought genotoxicity-associated aminomalonate building block in its unprecedented macrocyclic structure. This work provides new insights into the biosynthetic logic and mode of action of this colorectal cancer-linked microbial chemical.
The cryptic natural product colibactin is a human gut bacterial genotoxin that remains structurally undefined a decade after its discovery1–4. Based on the widespread distribution of its biosynthetic gene cluster in pathogenic and probiotic human enterobacteria, colibactin is suggested to perform diverse roles in human health1,3,5,6. The observation of these seemingly paradoxical effects implies that the biological activity of colibactin may result from a mixture of compounds that are differentially expressed under distinct growth conditions2,3,7,8. Intriguingly, gut symbionts from other animal hosts, including honeybees, also produce clb products, suggesting that colibactin might function more comprehensively in bacteria-host interactions9.
While traditional discovery efforts to directly isolate colibactin from Escherichia coli have been unsuccessful, recent investigations employing modern genome mining methods have facilitated the isolation of precolibactin biosynthetic intermediates. The assembly line construction of colibactin via the Clb hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) utilizes a recently recognized ‘prodrug’ maturation strategy in which the peptidase ClbP hydrolyzes offloaded pathway products containing an N-terminal fatty acyl-asparagine residue10,11. None of the previously reported precolibactin structures, however, are products of clb enzymes known to be essential for genotoxicity (e.g., ClbD–F)1,12. Recently, in vitro enzymatic studies revealed that ClbD–G enzymes are responsible for the synthesis and attachment of the rare PKS substrate aminomalonyl-ACP12,13. This precursor was shown to be indispensable for genotoxicity1,12 and suggested that the production of a larger clb pathway product incorporating an aminomalonyl moiety could be the long sought precolibactin genotoxin. Herein we present the discovery of the first member of the precolibactin family that incorporates an aminomalonyl unit, which, as a consequence, unexpectedly results in a dramatically rearranged chemical structure unique amongst the precolibactins (Fig. 1a–d).
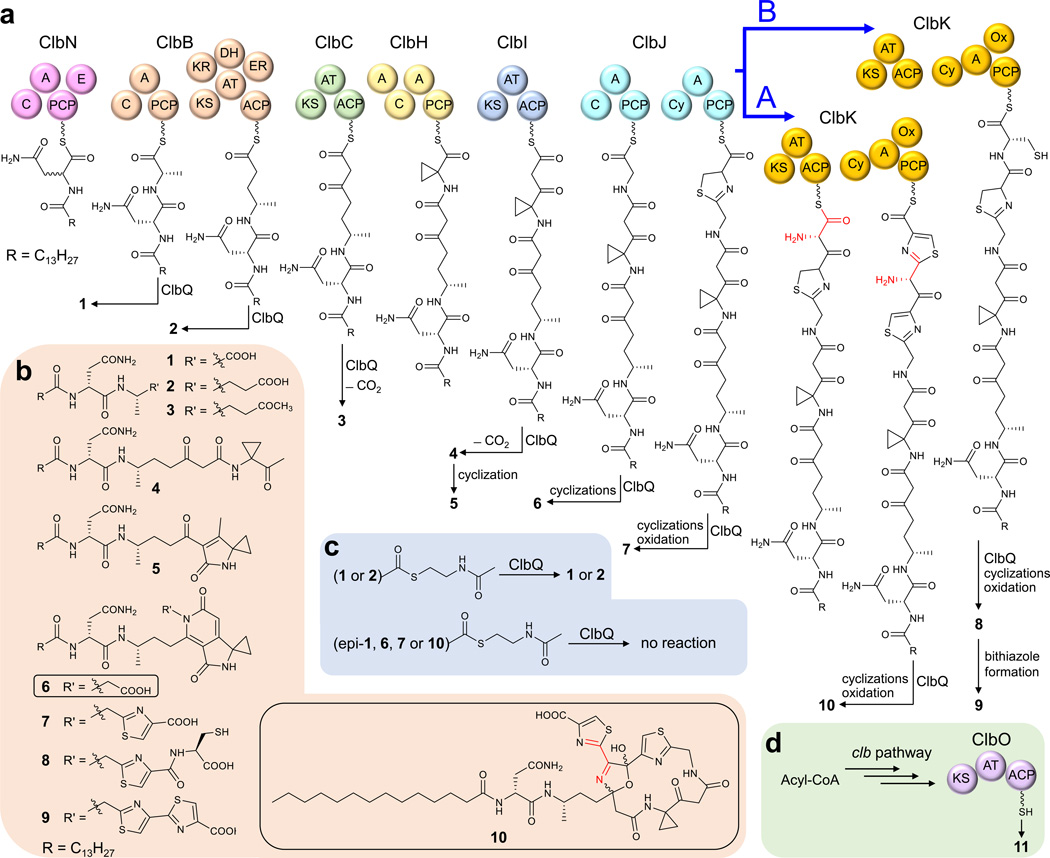
Figure 1. Structures and biosynthesis of precolibactins.
(a) Proposed biosynthesis and ClbQ-mediated offloading of precolibactins previously reported (1–5, 7–9) and newly characterized in this study (6, 10). The ClbK PKS module incorporates the extender unit aminomalonyl-ACP into 10 (path A, highlighted red) or is biochemically skipped to yield 8 and 9 (path B). Domain abbreviations: A, adenylation; ACP, acyl carrier protein; AT, acyltransferase; C, condensation; Cy, cyclization; DH, dehydratase; E, epimerase; ER, enoyl reductase; KR, ketoreductase; KS, ketosynthase; Ox, oxidase; PCP, peptidyl carrier protein. (b) Structures of candidate precolibactins with respect to the clb pathway. The new structures (6 and 10) reported here are boxed. We propose a new precolibactin naming nomenclature to reflect the growing number of clb pathway products: “precolibactin-molecular weight”, as exemplified in the new “precolibactin-886” (10), which has a mass of 886 Da. (c) Summary of in vitro biochemical reactions of precolibactin-SNAC derivatives with the type II thioesterase ClbQ. (d) Domain organization of ClbO and the detection of as-yet uncharacterized precolibactin-969 (11).
We profiled the native colibactin producer E. coli CFT073 strain and the clb+ heterologous expression host E. coli DH10B/pCAP01-clb8 in which clbP was inactivated to accumulate precolibactins. As expected7,8,14–16, numerous derivatives were detected by ultra-performance liquid chromatography–mass spectrometry, including a molecule with a m/z of 887 [M+H]+ (designated as precolibactin-886 hereinafter) that was found to be present in both E. coli CFT073 and DH10B ΔclbP mutants (Supplementary Results, Supplementary Fig. 1). Since this molecule is larger in mass than all other known precolibactins, we attempted to isolate it for structure elucidation but were unsuccessful due to low production levels.
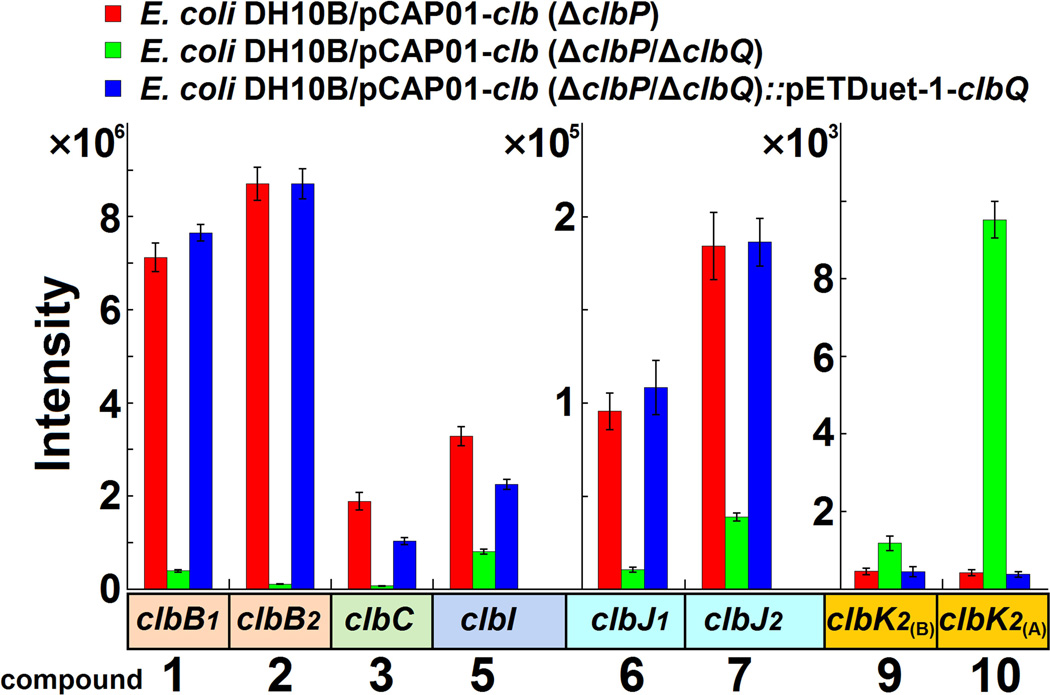
Given that substantially more upstream precolibactin intermediates were detected compared to downstream metabolites (Fig. 2), we speculated that limiting the release of biosynthetic intermediates might facilitate the increased production of late stage products. Although the pathway off-loading mechanism of the various precolibactin intermediates has not been reported, bioinformatics analyses suggested that the type II thioesterase (TE) ClbQ might be involved (Supplementary Fig. 2). Inactivation of clbQ notably resulted in the dramatic decrease of upstream intermediates (e.g., 1, 2 and 3) and the 22-fold average increase in precolibactin-886 (10) (Fig. 2 and Supplementary Figs. 3–6). To support the hypothesis that ClbQ mediates the off-loading of clb pathway intermediates, we genetically complemented the ΔclbP/ΔclbQ double mutant with clbQ and restored precolibactin production yields to their original levels (Fig. 2 and Supplementary Figs. 5 and 7).
Figure 2. Effect of ClbQ on the production of different precolibactins.
Relative abundances based on extracted ion chromatograms of individual precolibactin derivatives from extracts of clbQ-containing (pCAP01-clb (ΔclbP)), clbO-knockout (pCAP01-clb (ΔclbP/ΔclbQ)) and clbO-complemented (pCAP01-clb (ΔclbP/ΔclbQ)::pETDuet-1-clbQ) strains. Data are shown as mean ± SD (n = 5).
We next examined the in vitro function of ClbQ against N-acetylcysteamine (SNAC) precolibactin thioester derivatives (Supplementary Note) to explore its substrate scope (Fig. 1c). As anticipated, ClbQ readily hydrolyzed the SNAC derivatives 12a and 13 to the corresponding linear precolibactin-413 (1) and -441 (2), respectively (Supplementary Figs. 8 and 9). In contrast, we did not observe hydrolysis of 12b (C-19 epimer of 12a) or 14–16, the SNAC derivatives of the corresponding cyclized precolibactin-629 (6), -712 (7, also called precolibactin B), and -886 (10). As ClbQ is related to the amicoumacin thioesterase AmiD17, we also explored its catalytic properties, yet did not observe any hydrolytic activity with the precolibactin-SNAC derivatives (Supplementary Figs. 8 and 9). Together these results illuminated ClbQ’s specific function in mediating the off-loading of early stage clb pathway metabolites, and featured a unique function of the type II TE in releasing normal pathway intermediates rather than removal of aberrant byproducts18.
The pronounced production of 10 in the ΔclbP/ΔclbQ mutants allowed us to examine its biogenesis in relation to the aminomalonyl building block. Upon deletion of the aminomalonyl-ACP biosynthetic gene cassette clbD–F, the production of 10 was specifically abolished amongst the other characterized precolibactins (Supplementary Fig. 10). Moreover, l-[1-13C]- and l-[15N]serine were incorporated into 10 (Supplementary Figs. 11 and 12), consistent with aminomalonyl’s biogenesis from serine.
From 1000 L of fermentation broth we isolated 2.8 mg of 10 for structure elucidation. Precolibactin-886 was isolated as an approximately equal mixture of two isomers, 10a and 10b, as evidenced by a paired set of 13C NMR signals (Supplementary Note). The molecular formula of 10a was determined as C41H58N8O10S2 by high resolution mass spectrometry based on the protonated molecular ion peak at m/z 887.3787 (calculated 887.3796). The analysis of extensive NMR spectra (Supplementary Note) indicated that although 10a contained the same N-myristoyl-d-asparagine residue as in all other precolibactins previously characterized, neither the 7-methyl-4-azaspiro[2.4]hept-6-en-5-one moiety nor the 1H-pyrrolo[3,4-c]pyridine-3,6(2H,5H)-dione unit were present as in other cyclized precolibactins7,8,13,15,16. Instead, this region (C-24 to C-36) of 10a is assembled in a linear pattern as indicated by comprehensive HMBC correlations along with a characteristic ketone resonance at δC 205.4 (C-29). Key 1H-13C long range HMBC correlations of the H-34 and H-39 thiazole singlets helped establish the unique connectivity of the C-32 to C-41 terminus. Unlike precolibactin-795 (9, also called precolibactin C), neither of the thiazole moieties showed HMBC correlations to each other. Together, these data support the insertion of the aminomalonyl extender unit between the two thiazole rings where it is imbedded in a new ring associated with C-23 (δC 107.8) and C-36 (δC 107.6) that are suggestive of hemiacetal functionalities. We envision that the precursor carbonyl at C-23, the point of cyclization in other cyclic precolibactins, bonds to the aminomalonyl-derived nitrogen atom, which in turn further connects to the C-36 carbonyl to form an unusual heterocycle-fused macrocycle unprecedented in the natural products literature. Based on the chemical shift of C-37 (δC 160.4), the absence of 1H-15N HSQC correlation for N-37, and the additional degree of unsaturation indicated by the molecular formula of 10a, we deduced a double bond between C-37 and N-37. We speculate that this unsaturation is introduced by the oxidation domain of ClbK that has been proposed to install both the Δ34 and Δ39 double bonds of the two thiazoles13. This proposed structure, which is further supported by extensive MSn fragmentation data (Supplementary Note), is reminiscent of synthetic 2,5-dihydro-5-hydroxyoxazoles in terms of both construction and NMR chemical shifts (Supplementary Fig. 13)19,20.
Along with the discovery of precolibactin-886, we characterized precolibactin-629 (6) as another missing member of the clb assembly line (Supplementary Note 3). We envision that 6 is biosynthetically derived from the first NRPS module of ClbJ and fills the assembly gap between 5 and 7.
After establishing the chemical structure of 10, its biosynthetic logic was further investigated. In the clb locus, ClbKPKS and ClbO are the only two remaining PKS modules that had not been functionally connected to any previously reported precolibactin, although both were enzymatically established to accommodate aminomalonate as an extender unit13. We thus individually inactivated all clb PKS/NRPS encoding genes and established that disruption of clbO resulted in the only mutant that retained 10 (Supplementary Fig. 14). Inactivation of clbG, on the other hand, abolished the production of 10 while significantly increasing the yield of 9 by twenty-fold (Supplementary Fig. 10), suggesting that ClbKPKS accepts aminomalonyl-ACP from the trans-AT ClbG and that blocking this transfer redirects the overall pathway flux via path B to 9 (Fig. 1a and Supplementary Fig. 15).
Realizing that the mature precolibactin molecule is the product of the final modular PKS enzyme ClbO that also incorporates an aminomalonate unit13 (Fig. 1d), we examined the ΔclbP/ΔclbQ/ΔclbO and ΔclbP/ΔclbQ/ΔclbG mutants for the selective loss of a yet uncharacterized metabolite (Supplementary Fig. 16), and identified a precolibactin analog (11) with m/z 970 whose structure elucidation and biological evaluation are actively underway.
Precolibactin-886 is the largest and most complete colibactin derivative characterized to date, differing from the next-largest 9 and its precursor precolibactin-815 (8, also called precolibactin A) that derive from a skipped PKS module in ClbK (Fig. 1a). The addition of the aminomalonyl moiety transforms the structure of 10 to adopt a macrocyclic scaffold, dramatically altering the previously characterized aza-spirocyclopropane and bithiazole functionalities that were hypothesized as the DNA-targeting electrophilic warhead and intercalating elements, respectively7,8,13,21. Precolibactins 2, 5, 9, and 10 representing linear, aza-spirocyclopropane, bithiazole, and aminomalonate-containing structural derivatives, respectively, were evaluated for their biological activity against HCT-116 human colon cancer and HeLa human cervical cancer cells. We observed about 5-fold greater cytotoxic potency of 10 (IC50 22.3 and 34.0 µM against HCT-116 and HeLa cells, respectively) as compared to 2, 5, and 9 (Supplementary Fig. 17). Intriguingly, although 10 and 11 were detected in the ΔclbP mutant of uropathogenic E. coli CFT073, they were not observed in the ΔclbP mutant of probiotic E. coli Nissle 1917 despite an intensive molecular networking analysis7. Thus, the divergent biosynthesis of 10 and 11 may provide the missing link that correlates the clb pathway to both the beneficial and harmful effects of colibactin molecules on human health.
ONLINE METHODS
Bacterial strains, plasmids and general methods
Bacterial strains and plasmids used for colibactin production, gene inactivation and protein expression are listed in Supplementary Table 1. All PCRs were performed using Phusion polymerase (Thermo Scientific), 200 µM dNTPs (Thermo Scientific) and 0.5 µM of each oligonucleotide primer (ordered from GenScript, Supplementary Table 2). The sequences of the homology arms used for recombination are underlined in Supplementary Table 2. T4 DNA ligase (Thermo Scientific) was used for construction of the plasmids. E. coli strains were cultivated in Luria-Bertani (LB) medium (Affymetrix) while Bacillus subtilis 1779 strain was cultivated in sea salt-based SYT culture medium (10 g starch, 4 g yeast extract, 2 g tryptone, and 17 g sea salts per liter of deionized water). DNA isolation was performed as previously reported22. Gene inactivation was performed via the PCR targeting system23. Antibiotics required for plasmid maintenance were used at the concentrations listed: ampicillin (100 µg/mL), apramycin (50 µg/mL), chloramphenicol (25 µg/mL) and kanamycin (50 µg/mL).
Amino acid homology analysis of the thioesterase (TE) ClbQ
The accession numbers of the amino acid sequences used for alignment can be found in Supplementary Table 3, including the amino acid sequences of ClbQ and 33 other sequences of type I TE domains or type II TEs from various PKS, NRPS, and hybrid PKS/NRPS biosynthetic systems. Alignment was conducted using the CLUSTALW algorithm implemented in MEGA (version 7.0)24 based on the following parameters: gap opening penalty of 10 and gap extension penalty of 0.2 in pairwise alignment, gap opening penalty of 5 and gap extension penalty of 0.2 in multiple alignment, gap separation distance of 1, and delay divergent cutoff of 30%. Type I and type II TEs were aligned separately. The alignment results were visualized in UGENE (version 1.22)25.
Inactivation of chromosomal clbP and clbQ in the native colibactin producer E. coli CFT073
Using λ Red-mediated recombination26, the peptidase-encoding gene, clbP, in the clb locus of E. coli CFT073 was replaced by the aac(3)IV apramycin resistance gene (apraR) flanked with homologous arms. In brief, the primer pair clbP-knock out-F and clbP-knock out-R were used for PCR amplification of the apraR gene from a plasmid pIJ77323. The purified 1 kb PCR product was transformed by electroporation into l-arabinose induced E. coli CFT073 cells harboring λ Red recombinase expression plasmid pIJ790. The homologous recombination between the PCR product and the genomic DNA resulted in an in-frame deletion of clbP. The recombinants were selected on LB agar plates containing apramycin, followed by the elimination of the temperature-sensitive plasmid pIJ790 by incubation at 37 °C. The deletion mutant was examined by colony PCR (primers clbP-knock out check-F and clbP-knock out check-R). The resulting mutant was designated as E. coli CFT073 ΔclbP. Using a similar approach, clbP and the thioesterase-encoding gene clbQ, which are adjacent in the clb locus, were replaced together by an apraR cassette amplified by PCR using primers clbPQ-knock out-F and clbPQ-knock out-R. The resulting mutant was confirmed by colony PCR (primers clbPQ-knock out check-F and clbPQ-knock out check-R), and was assigned the name E. coli CFT073 ΔclbP/ΔclbQ.
Capture, expression and genetic manipulation of the clb gene cluster in E. coli heterologous expression system
As previously described, transformation-associated recombination (TAR) cloning technique27,28 was applied to capture the intact clb pathway from the NotI-digested genomic DNA of E. coli CFT073 (accession no. AE014075)5,8. The plasmid pCAP01-clb generated was introduced into E. coli DH10B cells by electroporation for heterologous expression. Benefiting from this heterologous expression system, the yields of clb pathway-related metabolites increased significantly (about five-fold), as reported in our previous study8.
Using the same PCR targeting system23, gene inactivation of clbP or clbPQ was separately performed in E. coli BW25113 cells carrying both pCAP01-clb and pIJ790 plasmids. The resulting constructs were confirmed by colony PCR and restriction analysis with KpnI and XhoI, generating pCAP01-clb (ΔclbP) and pCAP01-clb (ΔclbP/ΔclbQ), respectively. A further individual gene disruption of clbB, clbC, clbD-F, clbG, clbH, clbI, clbJ, clbK or clbO was performed in pCAP01-clb (ΔclbP/ΔclbQ). Each of these genes (or gene cassette in the case of clbD-F) was replaced by an ampicillin resistance marker to generate nine constructs named pCAP01-clb (ΔclbP/ΔclbQ/ΔclbB), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbC), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbDEF), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbG), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbH), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbI), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbJ), pCAP01-clb (ΔclbP/ΔclbQ/ΔclbK) and pCAP01-clb (ΔclbP/ΔclbQ/ΔclbO), respectively. All of these newly created plasmids BACs were isolated from E. coli BW25113 and introduced individually into E. coli DH10B cells by electroporation for heterologous expression.
Metabolite profile analysis by UPLC-MS
A 10 mL LB medium in a 50 mL sterile Falcon tube was inoculated with a single colony of the colibactin native producer E. coli CFT073, clb+ heterologous expression host E. coli DH10B, or various mutants mentioned above. Kanamycin was used to maintain the plasmids in the heterologous host E. coli DH10B and the mutants. After an incubation of 18 h at 37 °C and 250 rpm, each culture supernatant was extracted with an equal volume of EtOAc. The organic phase was separated by centrifugation (4200 rpm) for 10 min, evaporated, resuspended in MeOH (200 µL), and filtered through a 0.2 µm regenerated cellulose filter (Agilent Technologies, Inc.) prior to UPLC-MS analysis. For UPLC-MS analysis, a 5 µL aliquot was injected onto a Waters BEH C18 reversed-phase UPLC column (1.7 µm, 150 mm × 2.1 mm i.d.), and analyzed with a Bruker microTOF-q II mass spectrometer (Bruker Daltonics GmbH) coupled to a Waters ACQUITY UPLC system (Waters ACQUITY) by a gradient elution (A: CH3CN with 0.1% formic acid (FA); B: H2O with 0.1% FA: 5% A over 2 min, 5–100% A from 2 to 22 min, 100% A from 22 to 25 min, and 100–5% A from 25 to 27 min; flow rate 250 µL/min). TOF-MS settings during the UPLC gradient were as follows: Acquisition—mass range m/z 200–2000 Da, MS scan rate 1s−1; Source—gas temperature 200 °C, gas flow 8 L/min; nebulizer 4 Bar, ion polarity positive; Scan source parameters—capillary exit 140 V, skimmer 50 V. Hystar chromatography software (Bruker Daltonik) was used to control the system and the data was analyzed with Bruker Compass DataAnalysis 4.0 software (Bruker Daltonik).
Genetic complementation of the ΔclbQ mutant with clbQ and amiD
The genomic DNAs from E. coli CFT073 and B. subtilis 1779 were isolated as templates for the PCR amplifications of clbQ and amiD, respectively8,17, using the primers shown in Supplementary Table 2. The PCR products were cloned into the expression vector pETduet-1 to generate pETDuet-1-clbQ and pETDuet-1-amiD, respectively. These two expression plasmids were Sanger sequenced to confirm construction, and the expression of corresponding protein was confirmed by SDS-PAGE analysis.
Each construct described above was transferred into competent E. coli DH10B cells harboring pCAP01-clb (ΔclbP/ΔclbQ) by electroporation. Transformed cells were grown on 20 mL LB agar plates containing kanamycin and ampicillin at 37 °C overnight. Single colonies were picked and inoculated into 10 mL LB broth containing kanamycin and ampicillin, and incubated for 18 h at 37 °C and 250 rpm. Using the UPLC-MS analysis method mentioned above, the chemical profiles of the EtOAc extracts of the complementation strains were detected.
Construction, expression and purification of ClbQ and AmiD
The genes clbQ and amiD were amplified by PCR from the genomic DNAs of E. coli CFT073 and B. subtilis 1779, and cloned using appropriate restriction sites into pET28a(+) expression vector (Supplementary Table 2). The constructs were identified and confirmed by PCR screening, restriction analysis, and sequencing, followed by transformation into competent E. coli BL21 (DE3)/pLysE for recombinant protein expression.
The same protocol was used for expression and purification of N-His6-ClbQ and N-His6-AmiD. LB broth cultures were initially grown at 37 °C and 180 rpm in the presence of kanamycin and chloramphenicol until an optical density of 0.5 was reached. Then, the cultures were moved to 18 °C. After 1 h, protein expression was induced by the addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Further incubation was maintained at 16 °C and 120 rpm for 16 h. The purification of N-His6-ClbQ and N-His6-AmiD was performed at 4 °C. Cells were harvested by centrifugation, resuspended in Lysis buffer (50 mM sodium phosphate buffer, pH 8.0, with 300 mM NaCl and 10 mM imidazole), and lysed via ultrasound sonication for 30 min. The resulting lysate was centrifuged at 16,000 G for 45 min, and the supernatant was collected and applied to a 2-mL bed volume Ni-NTA column (Qiagen) equilibrated in the harvest buffer. The column was extensively washed with Wash buffer (50 mM sodium phosphate buffer, pH 8.0, with 300 mM NaCl and 20 mM imidazole), and eluted with a stepwise gradient of Elution buffer (50 mM sodium phosphate buffer, pH 8.0, with 300 mM NaCl and 50 to 250 mM imidazole). 1 mL fractions were collected and pure fractions detected by SDS-PAGE analysis were combined (Supplementary Fig. 18), dialyzed overnight in 20 mM Tris buffer (PH 8.0) with 100 mM NaCl and 10% glycerol, and stored at −80 °C.
Synthesis of N-acetylcysteamine thioesters of selected precolibactins
Compounds 1, 2, and 7 were isolated as previously reported8. Compounds 6 and 10 were isolated in the present study. Using 1, 2, 6, 7 or 10 as starting material, their corresponding N-acetylcysteamine (SNAC) thioesters (12–16, respectively) were synthesized based on a standard EDC and DMAP coupling procedure29,30. SNAC was added to a solution of individual precolibactin compound, EDC·HCl, and DMAP in CH2Cl2 (0.2–1 mL). The mixture was shaken at 100 r/min for 16 h at 23 °C, and the resultant reaction mixture was dissolved in 40% MeOH and loaded onto an open column packed with 40 g Sepra C18 (50 µm) sorbent. The SNAC thioester was eluted by a MeOH–H2O solvent system. The SNAC thioester-containing fractions eluted with 70–90% MeOH and were collected and dried under reduced pressure (approximately 75–90% yields). The resulting residue was further purified by HPLC (Waters 600 apparatus) using a semi-preparative C18 column (Phenomenex Luna 5 µm, 150 mm × 10 mm i.d.) eluted by CH3CN–H2O gradient mixtures (A: CH3CN with 0.1% formic acid (FA); B: H2O with 0.1% FA: 45% A over 5 min, and 45–100% A from 5 to 60 min) at a flow rate of 4.0 mL/min to yield the pure SNAC thioesters (12a, tR = 25.2 min; 12b, tR = 26.7 min; 13, tR = 28.0 min; 14, tR = 19.7 min; 15, tR = 24.2 min; 16, tR = 21.2 min;). These precolibactin-SNACs were analyzed by UPLC-MS (Supplementary Note 1), and the chemical structures of 13 and 14 were further determined by one- and two-dimensional NMR as representative examples (Supplementary Note 1).
Marfey’s analysis of precolibactin-413 SNAC isomers
Two isomers (approximately 3:1 in ratio) of precolibactin-413 SNAC thioester were observed after the synthesis which showed the same exact mass, MSn fragmentation pattern and 1H NMR spectra, but different UPLC retention times (Supplementary Note 1), likely derived from an α-epimerization during the synthesis.
Marfey’s method31 was used to determine the amino acid configurations of these two isomers, namely 12a and 12b. Both compounds (0.2 mg) were hydrolyzed in 6 M HCl at 110 °C overnight. Each hydrolysate was evaporated to dryness under a N2 stream to remove traces of HCl and redissolved in 50 µL of acetone. To each solution was added 50 µL of 1 % acetone solution of Nα-(2,4-dinitro-5-fluorophenyl)-l-alaninamide (l-FDAA, Marfey’s reagent, Sigma), followed by 50 µL of 1 M NaHCO3. The mixtures were incubated at 80 °C for 1 h with frequent mixing, then cooled to room temperature and quenched by addition of HCl (3 M, 20 µL). After being dried, each residue, i.e., 2,4-dinitrophenyl-5-l-alaninamide (DNPA)-amino acid diastereomer, was dissolved in 50 µL MeOH and analyzed by UPLC-MS. The retention times of the DNPA-12a Ala (tR = 9.1 min) and DNPA-12b Ala (tR = 9.9 min) diastereomers corresponded to those of the DNPA-standard l-Ala (tR = 9.1 min) and DNPA-standard D-Ala (tR = 9.9 min) (Supplementary Note 1).
Hydrolytic activity assay of ClbQ and AmiD
The thioesterase activity of ClbQ and AmiD towards each of the prepared precolibactin SNAC thioesters (12–16) was assayed according to a method previously reported29. A reaction mixture (100 µL) containing 50 mM phosphate buffer (pH 8.0), 2 µM protein (ClbQ or AmiD), and 40 µM substrate (dissolved in 1 µL DMSO) was incubated at 30 °C for 30 min and extracted with 1 mL EtOAc. The EtOAc extract was separated by centrifugation and evaporated to dryness using a vacuum concentrator. The dried residue was resuspended in 50 µL MeOH for analysis using the UPLC-MS detection method described above to directly detect the conversion of SNAC thioesters to the corresponding precolibactins.
Feeding experiments for biosynthetic pathway study
The heterologous expression host E. coli DH10B harboring pCAP01-clb (ΔclbP/ΔclbQ) was cultivated in 10 mL LB media supplemented with kanamycin and 0.5 mg/mL isotope-labeled l-[15N2]asparagine, [2,2-D2]glycine, l-[1-13C]serine or l-[15N]serine, and incubated at 37 °C on a shaker (250 rpm). After an incubation of 18 h, each culture supernatant was extracted with an equal volume of EtOAc. By centrifugation (4200 rpm) for 10 min, the organic layer was separated, collected and evaporated to dryness using a vacuum concentrator. Each extract prepared was redissolved in 100 µL MeOH for UPLC-ESI-MS analysis using the method described above.
Fermentation and isolation of precolibactin-629 (6) and -886 (10)
A single colony of E. coli DH10B harboring pCAP01-clb (ΔclbP/ΔclbQ) was picked from a freshly streaked kanamycin and apramycin selective plate and inoculated into 10 mL LB media containing kanamycin and apramycin in a sterile 50 mL sterile Falcon tube. The culture was incubated overnight at 37 °C and 250 rpm. One milliliter of the preculture was inoculated into 100 mL of LB media containing kanamycin and apramycin for another overnight incubation at 37 °C and 250 rpm. Each 10 mL of this preculture was added to l L LB media in a large Fernbach flask (2.8 L) containing only kanamycin. In total, 1000 L LB was cultured at an optimal condition of 28 °C and 160 rpm for 3 days, followed by extraction with EtOAc (2 × 1000 L) to afford a dried crude extract (130 g). This crude extract was subjected to chromatography over 2500 g Sepra C18 (50 µm) sorbent, and eluted with a MeOH–H2O gradient (10:90, 30:70, 50:50, 80:20 and 100:0) to afford five fractions (F01–F05). Fraction F04 (eluted with MeOH–H2O, 80:20; 36 g) was detected to contain 6 and 10 by UPLC-MS. It was then chromatographed over the same Sepra C18 column with isocratic elution (MeOH–H2O 65:35) to give 15 subfractions (F0401–F0415). F0408 (12 g) containing 6 and 10 was further subjected to three rounds of HPLC purification all using the same semi-preparative C18 Phenomenex Luna column (5 µm, 250 mm × 10 mm i.d.) along with a Waters 600 HPLC apparatus and a Waters 996 UV detector. The first round of HPLC separation was eluted by CH3CN–H2O gradient mixtures (A: CH3CN with 0.1% formic acid (FA); B: H2O with 0.1% FA: 40% A over 5 min, and 40–80% A from 5 to 100 min) at a flow rate of 4.0 mL/min, to afford subfraction F040809 (tR = 34–38 min; 150 mg) that was detected to contain 6 and 10 by UPLC-MS. This subfraction was purified by HPLC again with a MeOH–H2O gradient elution system (A: MeOH with 0.1% FA; B: H2O with 0.1% FA: 50% A over 5 min, and 50–100% A from 5 to 120 min, flow rate 2.5 mL/min), to further yield two subfractions, F04080901 (tR = 78 min; 8.0 mg) and F04080902 (tR = 80 min; 9.3 mg), which were detected to contain 10 and 6, respectively. F04080901 and F04080902 was individually subjected to a final round of HPLC purification using the same CH3CN–H2O elution condition (A: CH3CN with 0.1% FA; B: H2O with 0.1% FA: 50% A over 5 min, and 50–90% A from 5 to 100 min, flow rate 4.0 mL/min) to yield 10 (tR = 15.5 min; 2.8 mg) and 6 (tR = 16.5 min; 3.2 mg).
HRESIMS, MSn and NMR characterization of 6, 10 and precolibactin SNAC thioesters
HR-ESI-MS spectra were recorded on a Bruker microTOF II ESI-TOF-MS spectrometer. MSn analysis was carried out on a LTQ Velos dual-pressure ion trap mass spectrometer (Thermo Fisher Scientific)32 for compound 10. 1H, 13C, 1H-1H COSY, 1H-1H TOCSY, 1H-1H NOESY, 1H-13C HSQC, 1H-13C HMBC and 1H-15N HSQC NMR spectra for compound 10 were recorded on a Bruker Ascend® 850 NMR spectrometer (850 MHz for 1H and 212.5 MHz for 13C) or a 600 MHz Varian NMR spectrometer (Topspin 2.1.6 software, Bruker) with a 1.7 mm cryoprobe, respectively, using DMSO-d6 as solvent. 1H, 13C, 1H-1H COSY, 1H-1H NOESY, 1H-13C HSQC and 1H-13C HMBC spectra for compounds 6, 13, and 14 were all performed on a Bruker Ascend® 600 NMR spectrometer (600 MHz for 1H and 150 MHz for 13C) using DMSO-d6. Data were collected and reported as follows: chemical shift, integration multiplicity (s = singlet, d = doublet, t = triplet, m = multiplet), coupling constant. Chemical shifts were reported using the DMSO-d6 resonance as internal standard for 1H-NMR DMSO-d6: δ = 2.50 ppm and 13C-NMR DMSO-d6: δ = 39.6 ppm.
Cytotoxicity evaluation of selected precolibactins
The potential cytotoxic activity of precolibactins 2, 5, 9, and 10 was assayed according to a method described previously33. Briefly, authenticated and uncontaminated human HCT-116 colon carcinoma (ATCC CCL-247) and HeLa cervical carcinoma (ATCC CCL-2) cells were separately seeded onto 96-well plates at a density of 2.5 × 104 cells/mL in 190 µL of cell culture medium/well. After incubation for 24 h, the cells were then dosed with 10 µL of each test compound in 15% DMSO, or the positive control (etoposide), or the negative control (15% DMSO), and were incubated for 72 h. Using MTS/PMS as an indicator, the cytotoxic activity was measured at 490 nm with a Molecular Devices plate reader. Cytotoxicity was expressed as IC50, the concentration inhibiting cell growth by 50%. Data were processed using nonlinear regression analysis (TableCurve2DV4; AISN Software Inc.).
Data analysis
To compare the relative abundance of each colibactin pathway-related compound, the chemical profiles of all strains and their mutants were recorded in quintuplicate (N = 5), and standard deviations (SD) were calculated. All data analyses were performed using the Statistical Package for Social Sciences, SPSS Version 17.0 for Windows (SPSS Inc.).
Supplementary Material
Acknowledgments
This work was generously supported by grants from the China Ocean Mineral Resources Research and Development Association (COMRRDA12/SC.01 to P.Y.Q.) and the NIH (R01-GM85770 to B.S.M.). We thank P.R. Jensen. J. Busch, C.B. Naman, and Y.K. Tam for technical advice and access to equipment.
Footnotes
Author contributions
Z.R.L., J.L., J.P.G., J.Y.H.L., B.M.D., W.P.Z., Z.L.L., Y.X.L., R.B.T., Y.X. and D.H.L. performed experiments and collected data; all authors analyzed data; Z.R.L., J.L., B.S.M. and P.Y.Q. designed the study and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information and chemical compound information is available in the online version of the paper.
References
- 1.Nougayrède JP, et al. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 2.Bode HB. Angew. Chem. Int. Ed. 2015;54:10408–10411. doi: 10.1002/anie.201505341. [DOI] [PubMed] [Google Scholar]
- 3.Balskus EP. Nat. Prod. Rep. 2015;32:1534–1540. doi: 10.1039/c5np00091b. [DOI] [PubMed] [Google Scholar]
- 4.Arthur C, et al. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch RA, et al. Proc. Natl. Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olier M, et al. Gut Microb. 2012;3:501–509. doi: 10.4161/gmic.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vizcaino MI, Crawford JM. Nat. Chem. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li ZR, et al. ChemBioChem. 2015;16:1715–1719. doi: 10.1002/cbic.201500239. [DOI] [PubMed] [Google Scholar]
- 9.Engel P, Vizcaino MI, Crawford JM. Appl. Environ. Microbiol. 2015;81:1502–1512. doi: 10.1128/AEM.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brotherton CA, Balskus EP. J. Am. Chem. Soc. 2013;135:3359–3362. doi: 10.1021/ja312154m. [DOI] [PubMed] [Google Scholar]
- 11.Bian X, et al. ChemBioChem. 2013;14:1194–1197. doi: 10.1002/cbic.201300208. [DOI] [PubMed] [Google Scholar]
- 12.Brachmann AO, et al. Chem. Commun. 2015;51:13138–13141. doi: 10.1039/c5cc02718g. [DOI] [PubMed] [Google Scholar]
- 13.Zha L, Wilson MR, Brotherton CA, Balskus EP. ACS Chem. Biol. 2016;11:1287–1295. doi: 10.1021/acschembio.6b00014. [DOI] [PubMed] [Google Scholar]
- 14.Vizcaino MI, Engel P, Trautman E, Crawford JM. J. Am. Chem. Soc. 2014;136:9244–9247. doi: 10.1021/ja503450q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brotherton CA, Wilson M, Byrd G, Balskus EP. Org. Lett. 2015;17:1545–1548. doi: 10.1021/acs.orglett.5b00432. [DOI] [PubMed] [Google Scholar]
- 16.Bian X, Plaza A, Zhang Y, Müller R. Chem. Sci. 2015;6:3154–3160. doi: 10.1039/c5sc00101c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Sci. Rep. 2015;5:9383. doi: 10.1038/srep09383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du L, Lou L. Nat. Prod. Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 19.Patonay T, Hoffman RV. J. Org. Chem. 1995;60:2368–2377. [Google Scholar]
- 20.Pinho e Melo TMVD, et al. J. Org. Chem. 2002;67:66–71. doi: 10.1021/jo010504v. [DOI] [PubMed] [Google Scholar]
- 21.Healy AR, Vizcaino MI, Crawford JM, Herzon SB. J. Am. Chem. Soc. 2016;138:5426–5432. doi: 10.1021/jacs.6b02276. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 22.Aslan K, et al. Anal. Chem. 2008;80:4125–4132. doi: 10.1021/ac800519r. [DOI] [PubMed] [Google Scholar]
- 23.Gust B, et al. Proc. Natl. Acad. Sci. USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, Tamura K. Mol. Biol. Evol. 2016 Mar 22; doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okonechnikov K, Golosova O, Fursov M. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouprina N, Larionov V. Nat. Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 28.Yamanaka K, et al. Proc. Natl. Acad. Sci. USA. 2013;111:1957–1962. [Google Scholar]
- 29.Liu T, et al. Chem. Biol. 2006;13:945–955. doi: 10.1016/j.chembiol.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Rohm B, Scherlach K, Hertweck C. Org. Biomol. Chem. 2010;8:1520–1522. doi: 10.1039/b925483h. [DOI] [PubMed] [Google Scholar]
- 31.Fujii K, et al. Anal. Chem. 1997;69:5146–5151. [Google Scholar]
- 32.Ross AC, et al. J. Am. Chem. Soc. 2013;135:1155–1162. doi: 10.1021/ja3119674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lock RB, Stribinskiene L. Cancer Res. 1996;56:4006–4012. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.