Abstract
Bacteria of the genus Pseudomonas occupy diverse environments. The Pseudomonas fluorescens group is particularly well-known for its plant-beneficial properties including pathogen suppression. Recent observations that some strains of this group also cause lethal infections in insect larvae, however, point to a more versatile ecology of these bacteria. We show that 26 P. fluorescens group strains, isolated from three continents and covering three phylogenetically distinct sub-clades, exhibited different activities toward lepidopteran larvae, ranging from lethal to avirulent. All strains of sub-clade 1, which includes Pseudomonas chlororaphis and Pseudomonas protegens, were highly insecticidal regardless of their origin (animals, plants). Comparative genomics revealed that strains in this sub-clade possess specific traits allowing a switch between plant- and insect-associated lifestyles. We identified 90 genes unique to all highly insecticidal strains (sub-clade 1) and 117 genes common to all strains of sub-clade 1 and present in some moderately insecticidal strains of sub-clade 3. Mutational analysis of selected genes revealed the importance of chitinase C and phospholipase C in insect pathogenicity. The study provides insight into the genetic basis and phylogenetic distribution of traits defining insecticidal activity in plant-beneficial pseudomonads. Strains with potent dual activity against plant pathogens and herbivorous insects have great potential for use in integrated pest management for crops.
Introduction
Bacteria of the genus Pseudomonas occupy diverse terrestrial, aquatic and atmospheric environments, exhibiting a wide variety of ecological behaviors. Some are feared as human or plant pathogens such as Pseudomonas aeruginosa or Pseudomonas syringae; others are welcome agents for bioremediation of pollutants such as Pseudomonas putida. Members of the Pseudomonas fluorescens group are well-known for plant-beneficial effects that improve crop health and agricultural production. Many strains of fluorescent pseudomonads isolated from the rhizosphere have been studied for their ability to suppress root diseases, to promote plant growth and to induce systemic resistance (Haas and Défago, 2005; Bakker et al., 2007). They harbor strain-specific arsenals of antifungal metabolites, which enable them to inhibit pathogen growth through direct antibiosis (Haas and Keel, 2003; Raaijmakers et al., 2010). All these features make fluorescent pseudomonads interesting organisms for use as biofertilizers and biopesticides in sustainable agriculture and several products have been commercialized (Kupferschmied et al., 2013). On top of plant-beneficial activity, genomics has revealed unexpected and broader ecological versatility for these bacteria (Paulsen et al., 2005; Loper et al., 2012). Three of the best-characterized biocontrol strains, Pseudomonas protegens strains CHA0 and Pf-5 and Pseudomonas chlororaphis PCL1391, were shown to have potent insecticidal activity (Péchy-Tarr et al., 2008; Ruffner et al., 2013). When injected into the hemocoel of Galleria mellonella or Manduca sexta larvae, they rapidly multiply and cause larval death within a few hours (Péchy-Tarr et al., 2008). Ecologically more relevant, these strains are also able to infect and kill insect larvae, such as Drosophila melanogaster and the agricultural pests Spodoptera littoralis or Plutella xylostella, after oral uptake (Olcott et al., 2010; Ruffner et al., 2013). Oral insecticidal activity is considered a rare trait amongst bacteria and requires specific mechanisms to cope with host immune responses and to breach the gut epithelium in order to access the hemocoel (Vallet-Gely et al., 2008; Opota et al., 2011; Herren and Lemaitre, 2012). How P. protegens and P. chlororaphis overcome these barriers remains unclear. However, an association with insecticidal activity has been demonstrated for a set of genes termed the fit genes (P. fluorescens insecticidal toxin) (Péchy-Tarr et al., 2008; Péchy-Tarr et al., 2013; Ruffner et al., 2013). The unique virulence cassette harbors the fitD gene encoding the proteinaceous Fit toxin as well as regulatory genes and a type I secretion system (Péchy-Tarr et al., 2008; Péchy-Tarr et al., 2013; Kupferschmied et al., 2014). Nevertheless, fitD deletion mutants retain substantial toxicity, indicating the existence of additional virulence factors (Péchy-Tarr et al., 2008; Ruffner et al., 2013). Mutational analyses provide evidence that some of them are regulated by the global regulator GacA (Olcott et al., 2010; Ruffner et al., 2013).
Insecticidal activity is not universal to the P. fluorescens group. A survey by Ruffner et al. (2015) revealed that sub-clade 2 strains (Loper et al., 2012) neither harbor fit genes nor have ability to kill G. mellonella larvae. In contrast, all tested P. protegens and P. chlororaphis strains, which represent the sub-clade 1 (Loper et al., 2012), have both the toxin and injectable activity. Accordingly, Pseudomonas sp. strains Pf-01 and Q2-87 (formerly called P. fluorescens Pf-01 and Q2-87), both belonging to sub-clade 2, have no oral activity against larvae of D. melanogaster and several lepidopteran species, respectively (Olcott et al., 2010; Ruffner et al., 2013). Interestingly, Pseudomonas sp. SBW25 (formerly called P. fluorescens SBW25) of sub-clade 3, which does not harbor the fit genes, was shown to cause mortality and developmental delay in D. melanogaster larvae, but to a much lower extent than P. protegens Pf-5 (Olcott et al., 2010).
The discovery of insecticidal activity in fluorescent pseudomonads raises diverse ecological and agronomic questions. What ecological advantage may be gained by this ability to switch from a plant to an insect environment? Can we use these pseudomonads as double agents to fight both plant disease and insect pests? To date, our understanding of the interaction of plant-associated pseudomonads with insects is still very poor. Although large differences in their ability to infect insects were found between the strains investigated so far, no extensive data on frequency and distribution of insecticidal activity throughout the whole P. fluorescens group is available and individual strains with different phylogenetic background have never been compared directly. Moreover, the precise factors beyond the Fit toxin, which enable certain fluorescent pseudomonads to kill insects, and thereby to occupy a habitat alternative to plant roots, are still elusive. As a first step toward understanding the genomic features enabling insect pathogenicity we have taken an approach that combines bioassays with comparative genomics. We investigated 26 strains of fluorescent pseudomonads for their insecticidal activity and their biocontrol activity against root diseases. The strains included in our study are representative of the three phylogenetic sub-clades within the P. fluorescens group that harbor most plant-beneficial pseudomonads and were isolated from root but also from non-root habitats. Strong oral activity was found for all strains belonging to the phylogenetic sub-clade 1, which showed potent dual activity against insects and plant pathogens. However, we identified also a second phylogenetic group, sub-clade 3, containing strains with lower insecticidal activity. The strains were sequenced and comparative genomics revealed around 200 genes that are common and unique to the insecticidal strains and we hypothesize that this specific set of genes may represent major evolutionary events toward insect pathogenicity of Pseudomonas spp. Finally, we present first results from testing the involvement of some of the newly identified putative virulence factors in insecticidal activity using a mutational approach.
Materials and methods
Bacterial strains
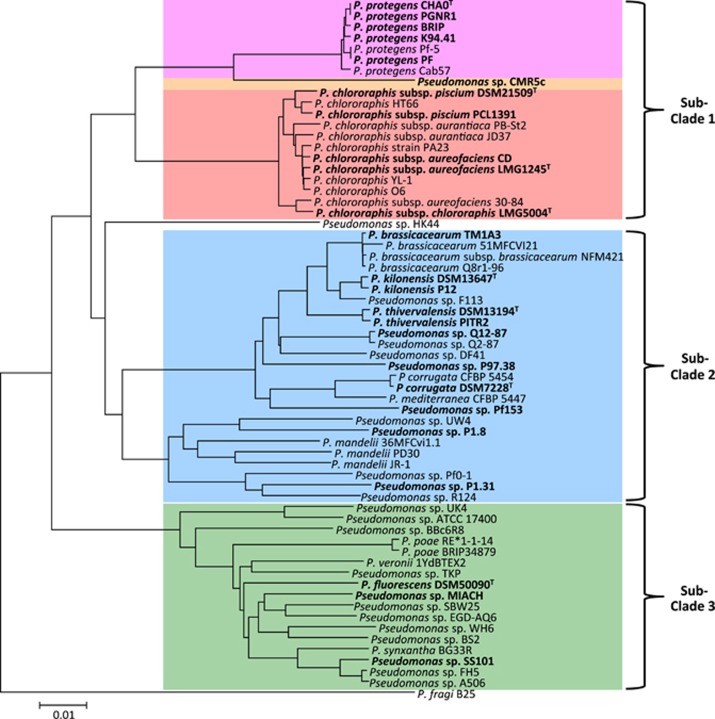
Strain names and origins are listed in Table 1. We use species names only for strains that cluster closely to a species-type strain in the phylogenetic tree we created based on core genomes (Figure 1) and thus can clearly be assigned to a certain species. All other strains are referred to as Pseudomonas sp. For sequencing, strains were taken from our long-term strain storage kept at −80 °C, or were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). If not otherwise stated bacterial cultures for bioassays and sequencing were grown in LB medium (Bertani, 1951) overnight on a rotary shaker (180 r.p.m.) at 24 °C. For bioassays cells were washed in sterile 0.9% NaCl. OD600 was measured and cells diluted to the desired concentration, while assuming that a cell suspension with an OD600 of 0.125 contains ~108 colony forming units (c.f.u.) per ml.
Table 1. Strain information.
| Strain | Former name | Geographic origin | Habitat/Hosta | Biocontrol ability | Genome sequenced | References |
|---|---|---|---|---|---|---|
| P. protegens CHA0T | P. fluorescens CHA0T | Switzerland | Tobacco | Cucumber-Pu, Tobacco-Tb, Wheat-Ggt, Tomato-Forl | Jousset et al. (2014) | Stutz et al. (1986); Keel et al. (1996); Haas and Défago (2005); Ramette et al. (2011) |
| P. protegens PGNR1 | P. fluorescens PGNR1 | Ghana | Tobacco | Cucumber-Pu, Tomato-Forl | This study | Keel et al. (1996) |
| P. protegens BRIP | Switzerland | Cyclops | ND | This study | Ruffner et al. (2015) | |
| P. protegens K94.41 | P. fluorescens K94.41 | Slovakia | Cucumber | Cucumber-Pu, Tomato-Forl | This study | Wang et al. (2001) |
| P. protegens PF | P. fluorescens PF | Oklahoma, USA | Wheat leaves | Wheat-St | This study | Levy et al. (1992); Keel et al. (1996) |
| Pseudomonas sp. CMR5c | Cameroon | Red cocoyam | Cocoyam-Pm | This study | Perneel et al. (2007) | |
| Pseudomonas sp. CMR12a | Cameroon | Red cocoyam | Cocoyam-Pm, Bean-Rs | This study | Perneel et al. (2007); D'Aes et al. (2011) | |
| P. chlororaphis subsp. piscium DSM 21509T | Lake of Neuchâtel, Switzerland | Intestine of European perch | ND | This study | Burr et al. (2010) | |
| P. chlororaphis subsp. piscium PCL1391 | Spain | Tomato | Tomato-Forl | This study | Chin-A-Woeng et al. (1998) | |
| P. chlororaphis subsp. aureofaciens LMG 1245T | Netherlands | River Clay | ND | This study | Kluyver (1956); Peix et al. (2007) | |
| P. chlororaphis subsp. aureofaciens CD | Switzerland | Cyclops (water) | ND | This study | Ruffner et al. (2015) | |
| P. chlororaphis subsp. chlororaphis LMG 5004T | — | Contaminated plate | ND | This study | Peix et al. (2007) | |
| P. brassicacearum TM1A3 | P. fluorescens TM1A3 | Switzerland | Tomato | Cucumber-Pu, Cotton-Rs | This study | Fuchs and Defago (1991); Keel et al. (1996) |
| P. thivervalensis DSM 13194T | France | Rapeseed | ND | This study | Achouak et al. (2000) | |
| P. thivervalensis PITR2 | P. fluorescens PITR2 | Albenga, Italy | Wheat | Cucumber-Pu, Tomato-Forl | This study | Keel et al. (1996) |
| P. kilonensis P12 | P. fluorescens P12 | Switzerland | Tobacco | Tobacco-Tb | This study | Keel et al. (1996) |
| P. kilonensis DSM 13647T | Germany | Agricultural soil | ND | This study | Sikorski et al. (2001) | |
| Pseudomonas sp. Q12-87 | P. fluorescens Q12-87 | Washington, USA | Wheat | Wheat-Ggt | This study | Keel et al. (1996) |
| Pseudomonas sp. P97.38 | P. fluorescens P97.38 | Switzerland | Cucumber | Cucumber-Pu, Tomato-Forl | This study | Wang et al. (2001) |
| P. corrugata DSM 7228T | United Kingdom | Tomato stem | ND | This study | Scarlett et al. (1978) | |
| Pseudomonas sp. Pf153 | P. fluorescens Pf153 | Switzerland | Tobacco | Cucumber-Pu | This study | Fuchs et al. (2000) |
| Pseudomonas sp. P1.8 | Switzerland | Earthworm | ND | This study | Ruffner et al. (2015) | |
| Pseudomonas sp. P1.31 | Switzerland | Woodlouse (dead) | ND | This study | Ruffner et al. (2015) | |
| P. fluorescens DSM 50090T | United Kingdom | pre-filter tanks | ND | This study | Rhodes (1959) | |
| Pseudomonas sp. MIACH | P. fluorescens MIACH | Switzerland | Wheat | ND | This study | Meyer et al. (2011) |
| Pseudomonas sp. SS101 | P. fluorescens SS101 | The Netherlands | Wheat | Cucumber-Pc, Tomato-Pi | Loper et al. (2012) | de Souza et al. (2003); Mazzola et al. (2007); Tran et al. (2007); Kruijt et al. (2009) |
Abbreviations: Forl, Fusarium oxysporum f. sp. radicis-lycopersici; Ggt, Gaeumannomyces graminis var. tritici; ND, not documented; Pc, Phytophthora capsici; Pi, Phytophthora infestans; Pm, Pythium myriotylum; Ps, Phomopsis sclerotioides; Pu, Pythium ultimum; Rs, Rhizoctonia solani; St, Septoria tritici; Tb, Thielaviopsis basicola.
Plant hosts: If not otherwise stated strains were isolated from roots or rhizosphere.
Figure 1.
Phylogeny of the P. fluorescens group based on the core genome. Genomes sequenced in this study and high-quality genomes that are publicly available by February 2015 were used to generate a core genome tree in EDGAR. Strains investigated in this study are depicted in bold. Sub-clades were defined after Loper et al. (2012). Sub-clade 1 corresponds to the P. chlororaphis subgroup, sub-clade 3 to the P. fluorescens subgroup and sub-clade 2 comprises strains belonging to three different subgroups within the P. fluorescens group according to Mulet et al. (2012), see also Supplementary Figure 1.
Genome sequencing, assembly and comparative genomics
For sequencing the genomes, DNA was extracted from overnight cultures in LB using the Wizard Genomic DNA Purification Kit (Promega AG, Dübendorf, Switzerland). All genomes apart from Pseudomonas sp. CMR5c (Supplementary Methods) were sequenced on an Illumina MiSeq (2 × 300 bp shotgun sequencing) at the Quantitative Genomics Facility (QGF) of the BioSystems Science and Engineering department (BSSE) of ETH Zürich located in Basel, Switzerland. Subsequently, the reads were de novo assembled using SeqMan NGen12 (DNASTAR, Madison, WI, USA) and further manually improved in silico using different subroutines of the Genomics Package of LASERGENE 12 (DNASTAR, Madison, WI, USA).
All genome sequences generated in this study and several database sequences that do not contain an annotation were automatically annotated in GenDB (Meyer et al., 2003). The annotation of the genome of P. chlororaphis subsp. piscium PCL1391 was manually improved and the whole Genome Shotgun project was deposited at DDBJ/EMBL/GenBank. Genomes of all other sequenced strains were deposited without annotations at DDBJ/EMBL/GenBank. Accession numbers are indicated in Supplementary Table S1.
Comparative genomics was done using EDGAR (Blom et al., 2009). Gene sets common to certain strains, but absent in other strains were calculated with a cut-off of 70% amino-acid identity over 70% of the gene length (Smits et al., 2010). For the phylogenetic tree of the core genomes, annotated assemblies and genomes from the public GenBank database (NCBI) were used. However, for quality reasons only genomes that consisted of <200 contigs and are thus classified as ‘high-quality draft genome sequences' by the NCBI, were included.
The phylogeny based on the core genome of all included strains was generated in EDGAR. The phylogenetic tree was created with the neighbor joining algorithm on a Kimura distance matrix as implemented in the PHYLIP package (Blom et al., 2009). Due to the huge size of the core alignment and the long resulting calculation time for a tree, bootstrapping was not performed.
Insect assays
Injection assays with G. mellonella were performed with small adaptations as described by Péchy-Tarr et al. (2008). More information is placed in Supplementary Methods.
Feeding assays: Eggs of P. xylostella were obtained from Syngenta Crop Protection AG (Stein, Switzerland). General growth conditions for larvae before and during the experiments were 26 °C, 60% humidity and a 16-h day, 8-h night cycle. Before experiments, boxes with larvae were placed at 18 °C in the dark for 48 h. For virulence assays, 1-week-old larvae were exposed to 10 μl washed bacterial cells adjusted to the desired concentration or 0.9% NaCl (controls) on a pellet of modified insect diet (Gupta et al., 2005; Ruffner et al., 2013). To prevent injuries each larva was kept separately in 128-cell bioassay trays (Frontier Agricultural Sciences, Delaware, USA). Each treatment was tested on four replicates of eight larvae. Mortality was defined as the inability to react to poking.
Construction of deletion mutants of P. protegens CHA0
The chiC, aprX and plcN genes and the rebB1-3 cluster of P. protegens CHA0 were deleted by an allelic replacement technique using the I-SceI system with the suicide vector pEMG (Martinez-Garcia and de Lorenzo, 2011) as detailed in previous work (Kupferschmied et al., 2014). To construct the pEMG-based plasmids, the 600–700-bp upstream and downstream regions flanking the genomic region to be deleted were amplified by PCR using the primer pairs specified in Supplementary Table S2. The obtained fragments were digested with the relevant restriction enzymes (Supplementary Table S2) and cloned into pEMG via triple ligation. Constructs were verified by sequencing. The obtained suicide plasmids served then to generate the deletion mutants CHA5099 (ΔchiC), CHA5222 (ΔaprX), CHA5223 (ΔplcN) and CHA5221 (ΔrebB1-3) (Supplementary Table S2), using the I-SceI system with the expression plasmid pSW-2.
Chitinase activity assay
Chitinase activity was measured in supernatants of cultures grown for 48 h in LB shaking with a methylumbelliferone-based chitinase assay kit (Sigma, St Louis, MO, USA) according to the manufacturer's instructions.
Statistics
Data analysis was performed in R version 3.1.1. (http://www.r-project.org). Mortality rates of the insect toxicity tests with wild-type strains were analyzed by multiple comparisons using Kruskal–Wallis adjusted by Bonferroni–Holm. Lethal time 50 (LT50) values were estimated based on the generalized linear model using the MASS package in R (Venables and Ripley, 2002). To test for significant differences between P. protegens CHA0 and its mutant strains the Log-Rank test of the Survival package of R and the Student's t-test were used in insect toxicity test and chitinase activity assays, respectively.
Results and discussion
Strain selection
To obtain an extensive overview of the occurrence of insecticidal activity within the P. fluorescens group, we selected 26 strains (Table 1). Many strains were isolated from roots and are well-known for their activity against plant pathogens, others were recently isolated from completely different habitats such as perch intestine and cyclops, for example, strains P. chlororaphis subsp. piscium DSM 21509T and P. protegens BRIP, respectively. Type strains were included in the study when considerable indications were present for close relationships of non-assigned strains to existing species.
The included strains, isolated on three different continents, belong to five subgroups within the P. fluorescens group (Supplementary Figure S1; Mulet et al., 2012; Gomila et al., 2015), that are covered in the three sub-clades defined by Loper et al. (2012) (Figure 1): 12 strains representing sub-clade 1, 11 strains representing 4 known and 5 new species in sub-clade 2 (including the P. corrugata, P. koreensis and P. jessenii subgroups) and 3 strains in sub-clade 3 (Figure 1 and Supplementary Figure S1). A detailed overview of the phylogeny of the included strains is given in Supplementary Results, Supplementary Figure S1 and Supplementary Table S3.
Insecticidal activity and presence of the Fit toxin
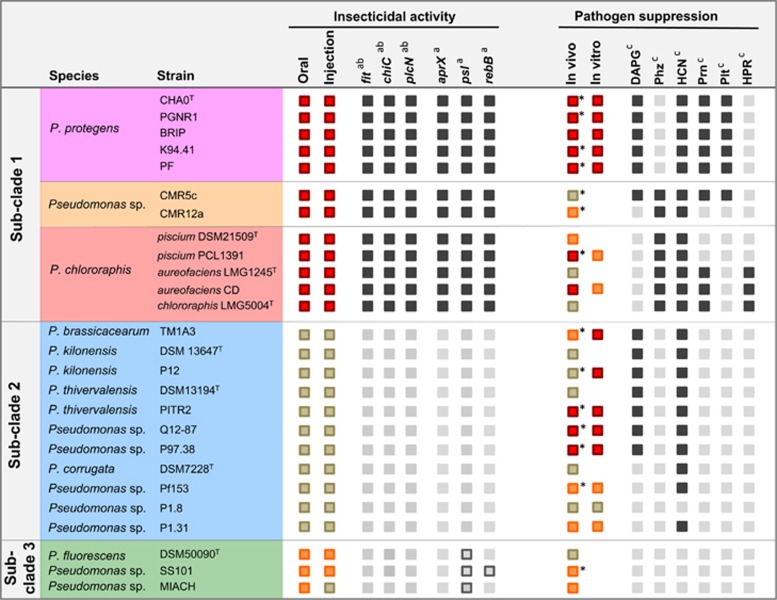
Functions encoded by the Fit gene cluster were demonstrated to contribute to insecticidal activity of the strains P. protegens strains CHA0 and Pf-5 and P. chlororaphis PCL1391 (Péchy-Tarr et al., 2008; Ruffner et al., 2013; Kupferschmied et al., 2014). Searching the genomes of the selected strains revealed that the gene cluster is present in all strains of sub-clade 1, but neither in sub-clade 2 nor sub-clade 3 (Figure 2), which is in line with results obtained by Ruffner et al. (2015).
Figure 2.
Overview on insecticidal activity, pathogen suppression and presence of associated gene clusters in 26 strains of the P. fluorescens group. Colored boxes represent activity against insects and plant pathogens as assessed within this study:  high activity,
high activity,  medium activity,
medium activity,  no activity. Insecticidal activity was assessed in injection assays against G. mellonella larvae and feeding assays against P. xylostella and S. littoralis larvae, and depicted activities are based on the results presented in Figure 3,Table 2, Supplementary Figure S2 and Supplementary Table S4. Disease suppression was assessed in a cucumber-Pythium ultimum assay and activities are based on the data depicted in Supplementary Table S5. Strains indicated by an asterisk were reported to have biocontrol activity against plant diseases in earlier studies (Table 1). In vitro inhibition of mycelial growth was assessed on two media against P. ultimum and Fusarium oxysporum f. sp. radicis-lycopersici and activities are based on the results shown in Supplementary Figure S3. Gray boxes represent presence of selected genes/gene clusters that were found to be associated with insecticidal strains (this study) or that are required for the production of the indicated antifungal metabolites.
no activity. Insecticidal activity was assessed in injection assays against G. mellonella larvae and feeding assays against P. xylostella and S. littoralis larvae, and depicted activities are based on the results presented in Figure 3,Table 2, Supplementary Figure S2 and Supplementary Table S4. Disease suppression was assessed in a cucumber-Pythium ultimum assay and activities are based on the data depicted in Supplementary Table S5. Strains indicated by an asterisk were reported to have biocontrol activity against plant diseases in earlier studies (Table 1). In vitro inhibition of mycelial growth was assessed on two media against P. ultimum and Fusarium oxysporum f. sp. radicis-lycopersici and activities are based on the results shown in Supplementary Figure S3. Gray boxes represent presence of selected genes/gene clusters that were found to be associated with insecticidal strains (this study) or that are required for the production of the indicated antifungal metabolites.  present,
present,  partially present,
partially present,  absent. Exact loci, which were checked for presence/absence, are indicated in Supplementary Table S1. There, additional genes as well as all additional strains are presented. aSelected genes that were identified by comparative genomics to be specific for strains that show insecticidal activity. A complete list is presented in Supplementary Table S6. P. fluorescens insecticidal toxin-cluster (fit), chitinase C (chiC), phospholipase C (plcN), metallopeptidase AprX (aprX), rebB-cluster (rebB), psl-cluster (psl). bGenes that were shown to contribute to insecticidal activity in this study (chiC and plcN) or elsewhere (fit) (Péchy-Tarr et al., 2008; Ruffner et al., 2013). cPresence/absence of gene clusters required for the production of the indicated antifungal metabolites. DAPG, 2,4-diacetylphloroglucinol; Phz, phenazine; HCN, hydrogen cyanide; Prn, pyrrolnitrin; Plt, pyoluteorin; HPR, 2-hexyl-5-propyl-alkylresorcinol.
absent. Exact loci, which were checked for presence/absence, are indicated in Supplementary Table S1. There, additional genes as well as all additional strains are presented. aSelected genes that were identified by comparative genomics to be specific for strains that show insecticidal activity. A complete list is presented in Supplementary Table S6. P. fluorescens insecticidal toxin-cluster (fit), chitinase C (chiC), phospholipase C (plcN), metallopeptidase AprX (aprX), rebB-cluster (rebB), psl-cluster (psl). bGenes that were shown to contribute to insecticidal activity in this study (chiC and plcN) or elsewhere (fit) (Péchy-Tarr et al., 2008; Ruffner et al., 2013). cPresence/absence of gene clusters required for the production of the indicated antifungal metabolites. DAPG, 2,4-diacetylphloroglucinol; Phz, phenazine; HCN, hydrogen cyanide; Prn, pyrrolnitrin; Plt, pyoluteorin; HPR, 2-hexyl-5-propyl-alkylresorcinol.
All 26 strains were tested for their injectable and oral activity against insect larvae. A summary of the results is given in Figure 2. All strains of sub-clade 1 exhibited strong injectable and oral insecticidal activity whereas no strain of sub-clade 2 had an effect on larval survival in any of the test systems. However, the presence of the fit cluster, although indicative of strong insecticidal activity, does not seem to be the sole factor associated with the ability to kill insects, as also the tested strains of sub-clade 3, which do not contain the fit cluster caused some mortality, but to a much lower extent than strains of sub-clade 1. Insecticidal activity was associated with specific phylogenetic subgroups, but did not correlate with the origin of the isolate (that is, root or non-root habitat).
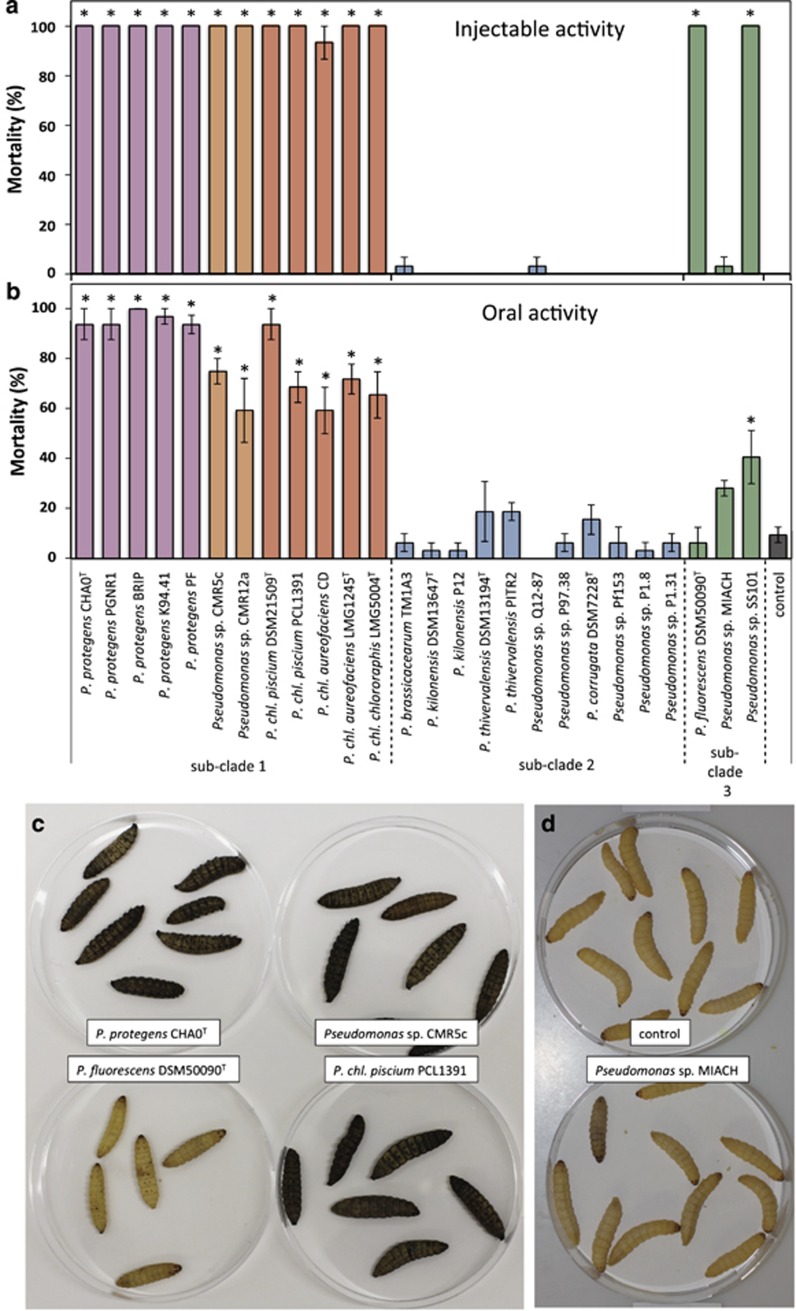
To mimic a systemic infection, bacteria were injected into the hemocoel of G. mellonella larvae (Figure 3a). All Fit-producing strains, that is, the entire sub-clade 1, were able to cause 100% mortality within the first 48 h, confirming and extending results of an earlier study demonstrating sub-clade 1 strains to cause 100% mortality when injected into G. mellonella (Ruffner et al., 2015). Although all strains in sub-clade 1 are highly insecticidal, strain-specific differences for killing rate were observed. The two strains Pseudomonas sp. CMR5c and CMR12a, that probably represent a new species within sub-clade 1, were killing more rapidly than P. protegens and P. chlororaphis as indicated by significantly shorter times to reach 50% larval mortality (LT50; Table 2). P. chlororaphis subsp. aureofaciens strains CD and LMG 1245T have higher LT50 values compared with strains of the P. chlororaphis subspecies piscium and chlororaphis (Table 2). Thus, the kill-time reflects phylogenetic relationships, which may be explained by the presumably multifactorial nature of insecticidal activity of fluorescent pseudomonads. Beside a common arsenal of contributing factors harbored by all insecticidal strains, some specific factors may exist, which enable certain strains or closely related groups of strains to kill more efficiently than others. No injectable activity was found for strains of sub-clade 2 (Figure 3a). In contrast, two strains of sub-clade 3, namely P. fluorescens DSM 50090T and Pseudomonas sp. SS101, both lacking the fit genes, caused lethal infections in G. mellonella. However, mortality caused by these strains was delayed compared with infections with most strains of sub-clade 1 (Table 2) and larvae lack the strong melanization response and the floppy phenotype observed after infection by sub-clade 1 (Figure 3c). These symptoms might be attributed to the Fit toxin as larvae injected with a fitD deletion mutant of P. protegens CHA0 lack these phenotypes (Péchy-Tarr et al., 2008) similarly to larvae injected with SS101 or DSM 50090T. Although injectable insecticidal activity seems to be universal to strains of sub-clade 1 this is not the case for sub-clade 3. The third tested strain of this sub-clade, Pseudomonas sp. MIACH, was not able to kill insect larvae upon injection (Figure 3a), although larvae started to slightly melanize at 1-day post infection (Figure 3d).
Figure 3.
Oral and systemic insecticidal activity is restricted to strains of specific phylogenetic subgroups within the P. fluorescens group. (a) Systemic activity against G. mellonella. Larvae were injected with 4 × 104 bacterial cells. Bars show average mortality of three replicates with 10 larvae after 48 h. The experiment was repeated twice and highly similar results were obtained. (b) Oral activity against P. xylostella. Larvae were exposed to artificial diet covered with 8 × 107 bacterial cells. Bars show average mortality of four replicates with eight larvae after 3 days. The experiment was repeated and similar results were obtained (Supplementary Table S4). Error bars show s.e.m. Asterisks indicate strains that were significantly different from control larvae treated with 0.9% NaCl based on multiple comparisons by Kruskal–Wallis adjusted by Bonferroni–Holm (P⩽0.05). (c) Typical melanization symptoms observed after 32 h in infections with P. protegens CHA0T, P. chlororaphis subsp. piscium PCL1391, Pseudomonas sp. CMR5c, but not with P. fluorescens DSM 50090T. (d) Although larvae injected with Pseudomonas sp. MIACH do not die, they become slightly melanized compared with control larvae. P. chl., Pseudomonas chlororaphis.
Table 2. Lethal time 50 (LT50) values for G. mellonella and P. xylostella larvae upon injection or oral uptake, respectively, of Pseudomonas strains.
| Sub-clade | Strain | Injection, LT50 (h) | Oral, LT50 (d) |
|---|---|---|---|
| Sub-clade 1 | P. protegens CHA0T | 26.3 (25.9; 26.6)b | 1.65 (1.44; 1.86)a |
| P. protegens CHA0T | 26.9 (26.5; 27.4)†γ | ||
| P. protegens PGNR1 | 29.3 (28.9; 29.8)de | 1.74 (1.56; 1.92)ab | |
| P. protegens BRIP | 29.0 (28.6; 29.4)d | 1.58 (1.40; 1.76)a | |
| P. protegens K94.41 | 26.3 (25.9; 26.7)b | 1.58 (1.40; 1.76)a | |
| P. protegens PF | 34.4 (33.3; 35.6)g | 1.63 (1.43; 1.83)a | |
| Pseudomonas sp. CMR5c | 24.5 (24.2; 24.9)†β | 2.24 (1.98; 2.49)c | |
| Pseudomonas sp. CMR12a | 22.0 (21.5; 22.6)†α | 2.63 (2.31; 2.95)c | |
| P. chl. piscium DSM 21509T | 27.2 (26.7; 27.6)c | 1.66 (1.46; 1.86)a | |
| P. chl. piscium PCL1391 | 24.9 (24.5; 25.3)a | 2.19 (1.87; 2.51)bc | |
| P. chl. aureofaciens LMG 1245T | 30.1 (29.6; 30.6)e | 2.56 (2.24; 2.89)c | |
| P. chl. aureofaciens CD | 33.7 (32.8; 34.7)g | 2.08 (1.76; 2.39)abc | |
| P. chl. chlororaphis LMG 5004T | 26.7 (26.2; 27.1)bc | 2.33 (1.97; 2.68)c | |
| Sub-clade 2 | P. brassicacearum TM1A3 | NA | NA |
| P. kilonensis DSM 13647T | NA | NA | |
| P. kilonensis P12 | NA | NA | |
| P. thivervalensis DSM 13194T | NA | NA | |
| P. thivervalensis PITR2 | NA | NA | |
| Pseudomonas sp. Q12-87 | NA | NA | |
| Pseudomonas sp. P97.38 | NA | NA | |
| P. corrugata DSM 7228T | NA | NA | |
| Pseudomonas sp. Pf153 | NA | NA | |
| Pseudomonas sp. P1.8 | NA | NA | |
| Pseudomonas sp. P1.31 | NA | NA | |
| Sub-clade 3 | P. fluorescens DSM 50090T | 32.0 (31.4; 32.7)f | NA |
| Pseudomonas sp. MIACH | NA | NA | |
| Pseudomonas sp. SS101 | 37.9 (36.9; 38.8)†δ | NA | |
| Control | 0.9% NaCl | NA | NA |
Abbreviations: NA, no LT50 value was calculated, because end mortality was <50% P. chl., Pseudomonas chlororaphis.
G. mellonella larvae were injected with 4 × 104 washed bacterial cells of the indicated strains. P. xylostella larvae were exposed to food pellets inoculated with 8 × 107 bacterial cells. LT50 values are estimates based on the generalized linear model using the MASS package in R (Venables and Ripley, 2002). LT50 estimates were calculated from three replicates with 10 larvae per replicate for G. mellonella and from four replicates with eight larvae per replicate for P. xylostella. Numbers in brackets depict 95% confidence intervals for LT50 and significantly different values (P⩽0.05) within the same column are followed by different letters.
†The strains for which significance is indicated by greek letters were tested in a separate experiment.
In natural infections, a bacterium first has to breach several barriers to reach the hemocoel. Therefore, the selected strains were further tested for oral activity against P. xylostella larvae. All strains that carry the fit genes were able to cause high mortality within 3 days (Figure 3b; Supplementary Table S4). In contrast to the injection assays, here P. protegens were the most efficient insect killers in terms of extent and pace (Table 2). None of the strains of sub-clade 2 caused higher mortality than observed for control larvae (Figure 3b). This result was confirmed in a second oral test system where a selection of 15 strains of sub-clades 1 and 2 was fed to S. littoralis larvae (Supplementary Methods). No sub-clade 2 strain was able to kill the larvae, whereas all Fit-producing strains showed strong insecticidal activity (Supplementary Figure S2A). Thus, sub-clade 2 strains lack crucial traits enabling them to kill lepidopteran larvae. However, the lack of killing potential does not necessarily mean that these strains might not be able to persist in the insect gut. Persistence without killing could be a clever strategy to use the insect as a means of dispersal as a living insect will transport the bacteria further than a dead one. Monitoring bacterial cells revealed that all strains of sub-clade 1 were able to multiply within the S. littoralis larvae (data only shown for CHA0, Supplementary Figure S2B) and to reach about 108 c.f.u. per larva whereas large differences were observed for strains in sub-clade 2. Several strains, namely Pseudomonas sp. P97.38, Q12-87 and P1.31, were indeed able to persist at levels of 106 to 107 c.f.u. per larva, whereas others such as P. thivervalensis PITR2, P. kilonensis P12 or Pseudomonas sp. P1.8 underwent a 1000-fold population decline within a few days (Supplementary Figure S2B) indicating that they were cleared from the gut. Thus, although not having the ability to kill insect larvae, some strains of sub-clade 2 seem to possess features allowing certain persistence in insects.
In contrast to sub-clade 2 strains, sub-clade 3 strains were found to cause lethal oral infections in P. xylostella, which is to our knowledge the first report for strains of this sub-clade to orally kill lepidopteran insect larvae. However, similar to the results of the injection assay, strains of sub-clade 3 appeared to have strongly reduced oral activity compared with strains of sub-clade 1. This is in line with observations of Olcott et al. (2010), who described that oral infections of D. melanogaster with Pseudomonas sp. SBW25 (sub-clade 3) were less detrimental than infections with P. protegens Pf-5 (sub-clade 1). Strain Pseudomonas sp. SS101 had significant oral insecticidal activity in all repetitions of the experiment (Figure 3b; Supplementary Table S4). However, killing occurred slower and to a lower extent than it was the case for infections with strains of sub-clade 1. P. fluorescens DSM 50090T which had injectable activity against G. mellonella showed either no or weak insecticidal activity when fed to P. xylostella. We hypothesize that this strain faces difficulties to breach the gut barrier on its own, but can act as an opportunistic pathogen taking the chance when an insect gets injured or weakened by other factors. More puzzling, is the outcome for Pseudomonas sp. MIACH, which, in spite of not killing larvae in injection experiments, seems to have slight oral activity. Strain MIACH caused mortality rates of 30–53% though the effect was significant only in one of the two experiments (Figure 3b; Supplementary Table S4). We hypothesize that this strain is able to do some damage to the insect gut, without killing the insect itself, thereby promoting a secondary infection by other microbes that invade the hemocoel and lead to larval death. Another explanation would be that this strain is less of a generalist and causes lethal infections only in certain insect species. As we tested injectable and oral activity in different insect species, we cannot exclude this possibility. Although some strain-specific differences exist, we conclude that strains in sub-clade 3 mostly possess some insecticidal activity but that it is by far less distinct than in Fit-producing strains of sub-clade 1. To date no factor contributing to pathogenicity of sub-clade 3 strains has been identified, but it was suggested that so-called toxin complexes (Tc), first discovered in the entomopathogen Photorhabdus luminescens, could have a role (Loper et al., 2012). In accordance to the study of Loper et al. (2012) different Tc-related genes could be identified in the genomes of the strains included in this study, but they were not restricted to the strains with insecticidal activity (data not shown). Hence, they might have a rather subtle role in Pseudomonas insect associations.
Plant-beneficial effects are phylogenetically less predictable than insecticidal activity
Although biocontrol activity against root pathogens has been demonstrated for many strains of the P. fluorescens group, most of the species-type strains have never been investigated. The lack of knowledge for these strains and for the new strains from non-root habitats led us to test all 26 strains investigated for insecticidal activity also for their biocontrol activity against the oomycete pathogen P. ultimum on cucumber roots and a subset of strains also for their in vitro inhibition of P. ultimum and F. oxysporum f. sp. radicis-lycopersici. Biocontrol activity appeared to be phylogenetically less predictable than insecticidal activity, as effective as well as poor biocontrol strains were found throughout all the three sub-clades (Supplementary Results; Figure 2; Supplementary Table S5; Supplementary Figure S3). Similar to the results on insecticidal activity, no connection between the original habitat and the degree of plant protection was observed. Together, the bioassays with insects and pathogens identified several strains of sub-clade 1, which exhibit potent dual activity against plant pests and diseases and therefore could be of interest for implementation in integrated crop protection strategies.
Comparative genomics to identify potential factors associated to insecticidal activity
Draft genomes of all selected strains were generated with exception of the strains P. protegens CHA0 and Pseudomonas sp. SS101, for which the genomes were already available (Loper et al., 2012; Jousset et al., 2014), and Pseudomonas sp. CMR12a, for which the genome description will be released elsewhere and which was therefore not included in the comparative genomics analysis. The average number of contigs per genome was 32 (Supplementary Table S6). The obtained genome sizes range between 6.06 and 7.07 Mbp, which is in accordance to genome sizes obtained for other fluorescent pseudomonads (Loper et al., 2012).
The next step was to search for genes that are common and unique to insecticidal strains, encoding candidate factors potentially involved during the infection of insect larvae. Using EDGAR (Blom et al., 2009), we identified 90 genes that are present in all highly insecticidal strains (sub-clade 1), but neither in moderately insecticidal strains (sub-clade 3) nor in non-insecticidal strains (sub-clade 2; Table 3). We further identified 117 genes that are present in all strains of sub-clade 1 as well as in one or several of the strains in sub-clade 3, but again in none of the strains of sub-clade 2 (Table 3). A full list of all identified genes can be found in Supplementary Table S7. It comprises about 28 putative transporters, 21 putative regulatory genes and over 100 enzymes and hypothetical proteins that are unique to insecticidal strains (Supplementary Table S7). Amongst the identified transporters, there are several putative amino-acid transporters. Insects are very rich in amino acids (Rumpold and Schluter, 2013) and thus these transporters might help to exploit the insect as a source of nutrients. The Fit toxin is specifically expressed in insects but not on plant roots (Péchy-Tarr et al., 2013). This could also be the case for other virulence factors and might involve some of the many regulatory genes that were found to be specific to insecticidal strains. However, besides the Fit toxin (Péchy-Tarr et al., 2008; Péchy-Tarr et al., 2013; Ruffner et al., 2013), no other insecticidal toxin was identified. For most of the 207 genes unique to insecticidal strains, a prediction on the biological function of the encoded product as well as on a possible role during the infection of insects would be very speculative at the present stage. Nevertheless, the comparative genomics also revealed several genes encoding proteins with homology to known virulence factors of other bacteria and that are of interest in terms of a possible association with insecticidal activity. Presence of those genes, which are discussed below, is indicated for our selection of strains in Figure 2 and for all strains included in the phylogeny of Figure 1 in Supplementary Table S1.
Table 3. Genes associated with insecticidal activity.
| Sub-clade 1 | Sub-clade 2 |
Sub-clade 3 |
Number of Genes | ||
|---|---|---|---|---|---|
| SS101 | DSM 50090T | MIACH | |||
| + | − | − | − | − | 90 |
| + | − | + | + | + | 57 |
| + | − | + | + | − | 20 |
| + | − | + | − | + | 7 |
| + | − | + | − | − | 20 |
| + | − | − | + | + | 13 |
| + | − | − | + | − | 0 |
| + | − | − | − | + | 0 |
Numbers of genes that are specific to insecticidal strains.
Presence of genes was defined as 70% amino-acid pairwise identity over at least 70% of gene length for the pairwise comparisons.
Only genes that were common to all strains of sub-clade 1 (highly insecticidal strains), but not found in any strain of sub-clade 2 (non-insecticidal strains) were considered.
+ Indicates genes present in all these strains.
− Indicates genes absent in all these strains.
Upon ingestion of pathogenic bacteria, insects produce reactive oxygen species, antimicrobial peptides and lysozymes to rapidly eliminate infesting bacteria (Lemaitre and Hoffmann, 2007). One mechanism to counter this first line of insect immunity is to produce enzymes degrading antimicrobial peptides. Exoproteases such as the Zn-dependent metallopeptidase AprX, also called serralysin and the AprA alkaline protease were suggested to have a role during the early phase of bacterial infections (Liehl et al., 2006). The gene aprA is present in all 25 genomes, except that of strain P1.8, whereas aprX was only detected in the genomes of strains belonging to sub-clade 1 (Supplementary Table S1). AprA and AprX belong to the M10 family that includes serralysin, aeruginolysin and other related exopeptidases that cause tissue damage and anaphylactic responses (Park and Ming, 2002). In Pseudomonas entomophila, an aprA mutant was shown to be slightly less virulent and to have a reduced persistence in D. melanogaster (Liehl et al., 2006). Serralysin of Serratia marcescens was shown to promote hemolymph bleeding in the silkworm (Bombyx mori) (Ishii et al., 2014).
If bacteria persist within the insect gut, living cells or their toxins must breach the peritrophic membrane, a gut-delimiting chitinous matrix, to access the hemocoel (Vallet-Gely et al., 2008). Chitinases affecting the peritrophic matrix are therefore potential virulence factors of entomopathogenic bacteria. For instance chitinases of B. thuringiensis subsp. israelensis IPS68 and B. thuringiensis subsp. aizawai HD133 were shown to contribute to insecticidal activity toward Culicuoides nubeculosus and S. littoralis, respectively (Sampson and Gooday, 1998). In insecticidal strains of the P. fluorescens group, we identified two chitinase genes. The chitinase gene chiC encoded next to a chitin-binding protein is present exclusively in genomes of sub-clade 1 strains, whereas the second chitinase is present in nearly all P. chlororaphis strains and some sub-clade 3 strains (Supplementary Table S1).
PCL1391_2966 encodes for a phosphocholine-specific phospholipase C. This gene, plcN, was detected only in sub-clade 1 strains. Phospholipases are recognized as major virulence determinants in a number of bacterial species, including human, animal and several invertebrate pathogens (Songer, 1997; Farn et al., 2001; Yang et al., 2012). The ymt gene encoding for a phospholipase D in Yersinia pestis, for example, is needed for persistence in the flea midgut (Hinnebusch et al., 2002). Phospholipase C produced by Mycobacterium abscessus is crucial for survival in amoeba and is suggested to cause damage to mouse macrophages presumably by hydrolysis of membrane phospholipids (N'goma et al., 2015).
Three small genes with homology to reb genes were found to be present in all strains of sub-clade 1 and in Pseudomonas sp. SS101, the strain with the highest insecticidal activity of sub-clade 3. Such reb genes have been mainly studied in Caedibacter taenospiralis, a Paramecium endosymbiont. They encode R-bodies, highly insoluble protein ribbons that are typically coiled into cylindrical structures but can unroll under certain conditions (Pond et al., 1989) and are associated with the killing trait toward sensitive Paramecia (Dilts and Quackenbush, 1986). Orthologs of reb were found to be present in many free-living bacteria, but their function remains unclear to date (Raymann et al., 2013).
A whole cluster of genes specific to insecticidal strains (loci PCL1391_4983 to PCL1391_4994) has high percentage of sequence identity to the psl gene cluster of P. aeruginosa which specifies the production of the extracellular polysaccharide Psl (Franklin et al., 2011). Psl was shown contributing to biofilm production, tolerance to oxidizing agents and host defensive processes (Friedman and Kolter, 2004; Jackson et al., 2004; Mishra et al., 2012), that is, traits likely useful in insect interactions.
Other factors, which still have to be kept in mind, are the antimicrobial compounds, such as 2,4-diacetylphloroglucinol, phenazine, pyoluteorin, pyrrolnitrin and hydrogen cyanide that are crucial for biocontrol activity against fungal diseases, although none is shared by all or unique to insecticidal strains (Figure 2; Supplementary Table S1; Haas and Défago, 2005; Mercado-Blanco and Bakker, 2007; Lugtenberg and Kamilova, 2009). However, some have activity against a broad spectrum of organisms including plants, nematodes, arthropods and even mammalian cells (Maurhofer et al., 1992; Devi and Kothamasi, 2009; Kwak et al., 2011; Neidig et al., 2011; Nisr et al., 2011; Jang et al., 2013) and thus could contribute to Pseudomonas-derived insecticidal activity.
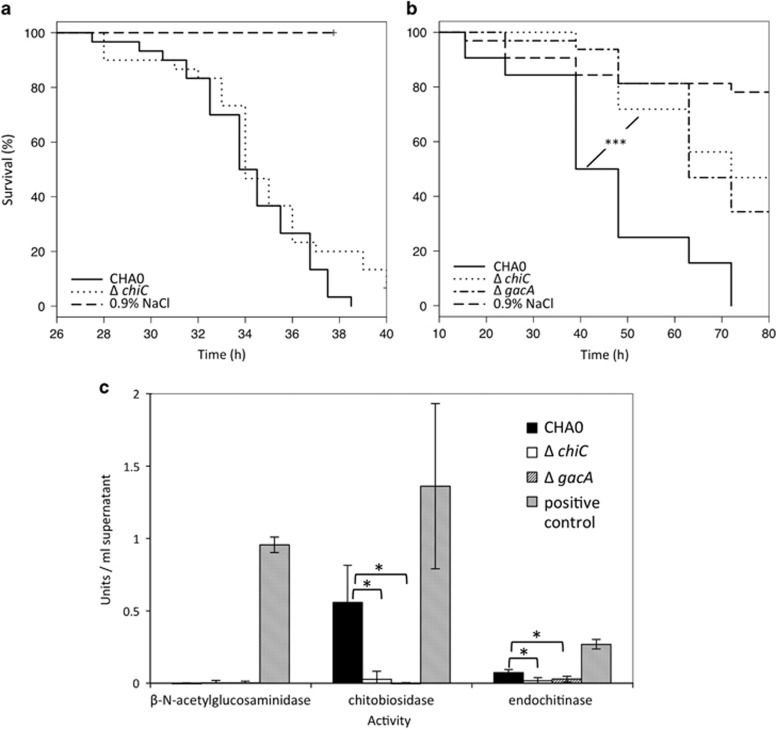
Chitinase ChiC and phospholipase PlcN contribute to oral insecticidal activity
In order to verify that our combination of bioassays and comparative genomics indeed led to the identification of valuable candidate genes associated with bacterial virulence toward insects, we generated, in model strain P. protegens CHA0, in-frame deletion mutants for selected genes: plcN, chiC, aprX and the cluster encoding homologs of rebB. None of the mutants differed in activity from the wild type when injected directly into the hemocoel of G. mellonella (Figure 4a; Supplementary Figures S4A and C). However, the chiC mutant was always significantly delayed in killing P. xylostella larvae upon ingestion (Figure 4b; Supplementary Figure S5A). We therefore conclude that the chitinase C, encoded by chiC, a gene common and unique to highly insecticidal strains, contributes to oral insect pathogenicity of P. protegens CHA0. Chitinase C was found to be responsible for chitobiosidase as well as endochitinase activity of P. protegens CHA0 as the chiC mutant exhibited no residual chitinase activity (Figure 4c). As also a gacA mutant completely lost chitinase activity (Figure 4c) we believe that we identified with the chiC one of the hitherto unknown Gac-regulated virulence factors involved in oral insecticidal activity. Furthermore, the mutant deficient for phopholipase C (plcN) also showed reduced oral activity against P. xylostella. Although less distinct than for the chiC mutant, a delay in killing was always observed for the plcN mutant but the effect was not significant in all experiments (Supplementary Figures S4B and S5B).
Figure 4.
A derivative of P. protegens CHA0 deficient for a specific chitinase is reduced in oral, but not in injectable activity against insect larvae. (a) Systemic activity against G. mellonella. Thirty larvae per treatment were injected with 2 × 103 bacterial cells and survival was recorded hourly. (b) Oral activity against P. xylostella. Larvae were exposed to artificial diet inoculated with 4 × 106 bacterial cells. Significant differences according to a Log-Rank test (Survival Package in R) between treatments with the wild-type CHA0 and the chitinase C-negative mutant (ΔchiC) are indicated with ***P<0.0001. Each mutant was tested at least three times with similar results. A repetition of the feeding assay is depicted in Supplementary Figure S5. (c) Chitinase activity of wild-type CHA0 and its chiC mutant was assessed using a chitinase assay kit (Sigma, St Louis, MO, USA). Three different substrates were used to test for exo- (β-N-acetylglucosaminidase and chitobiosidase) and endochitinase activity. Treatments indicated by an asterisk are significantly different based on a t-test (P⩽0.05). CHA0, wild type; ΔchiC, chitinase C-negative mutant; ΔgacA, GacA-negative mutant; 0.9% NaCl served as negative control in the virulence assay; a positive control for chitinase activity was provided by the chitinase assay kit.
In contrast, no difference to the wild-type strain CHA0 was found for the rebB1-3 and aprX mutants (Supplementary Figure S4D). However, these results do not exclude a role of factors encoded by these genes under different conditions or in an interaction with other insect species. Accordingly, the impact of the well-characterized Fit toxin on virulence toward insects also varies between insect species. Thus, a fitD mutant compared to the wild-type P. protegens CHA0 is more strongly reduced in virulence toward S. littoralis than toward P. xylostella (data not shown).
The mutational analysis performed in this study gives only a first insight into the possible contribution of interesting candidate genes identified in the comparative genomics approach to insecticidal activity. An in-depth analysis of the role of chitinase C and phospholipase C would include the complementation of these mutants and will be subject to further studies.
Conclusions
We provide the first extensive overview on insecticidal activity in the P. fluorescens group. Whilst biocontrol activity against fungal pathogens occurs throughout all studied sub-clades, insecticidal activity is unique to sub-clades 1 and 3. Only strains of sub-clade 1 display strong oral insecticidal activity and only they produce the Fit toxin. Intriguingly, Fit seems to contribute to the floppy and melanized phenotype associated with infections by highly pathogenic strains, however, the toxin is clearly not the major killing factor upon oral ingestion. Mutants of strains CHA0 and PCL1391 lacking the fit genes cause delayed, but still substantial mortality in S. littoralis (Ruffner et al., 2013) when acquired via the oral infection route. By comparative genomics we now identified several candidate genes that might contribute to insecticidal activity and we demonstrated that the absence of two of these genes, encoding a specific chitinase and a phospholipase, negatively affects insecticidal activity. We hypothesize that especially the chitinase C might be involved during the gut stage of the infection process, causing damage to the peritrophic membrane. However, to understand the exact mode of action of these pathogenicity factors during the infection process, further investigations are needed. Nevertheless, the presented data highly increases the knowledge on the genetic basis of insecticidal activity of fluorescent pseudomonads and points to a multifactorial nature of this trait.
Although we provide evidence that many strains of the P. fluorescens group can be insect pathogenic and others might persist in insects as commensals, the ecological relevance of insects as a host for these bacteria is still elusive and an intriguing field for future research. The fact that certain pseudomonads, to date considered to be plant-associated, perform very well in a completely different habitat such as an insect raises the question whether these bacteria are indeed mainly plant-associated. Insecticidal as well as biocontrol activity against plant diseases was found to be independent of the original habitat of a strain. For example, closely related strains can be isolated from fish or cyclops and behave similarly well on roots as root isolates. This observation is in line with other studies that found the isolation source of a bacterial strain not to be predictive for its performance in another habitat (Alonso et al., 1999; Grosso-Becerra et al., 2014). For instance, Hilker and colleagues found no correlation between original habitat and virulence in different test systems for clinical and environmental clones of P. aeruginosa (Hilker et al., 2015). In general, fluorescent pseudomonads might be quite ubiquitous and probably possess an arsenal of traits allowing them to easily switch niches and to conquer the habitat they encounter. Insects could be especially useful as a means of dispersal, a phenomenon documented for diverse plant-pathogenic bacteria (Nadarasah and Stavrinides, 2011). Some Pseudomonas syringae strains for instance can use the pea aphid as alternative primary host where they replicate to high numbers and can be deposited onto a new plant host via excreted honeydew (Stavrinides et al., 2009). Similarly, the rhizobacterium P. chlororaphis strain L11 was found to be transmittable from one plant to another by Diabrotica undecimpunctata subsp. howardi feeding on colonized plants (Snyder et al., 1998). In contrast to considering the insect as an alternative host, one could even speculate that the plant is not the primary host for species like P. protegens and P. chlororaphis, but rather a transient host on which they endure until they encounter the next insect host. As research to date is very much biased toward plant-association, future studies especially on strains actually isolated from insects will be required to gain a better understanding of the importance insects have as hosts for strains of the P. fluorescens group.
Besides its ecological relevance, insecticidal activity might be of great agronomical interest. Our bioassays revealed several strains, especially of the species P. protegens, that display potent dual activity, killing insect larvae and protecting plants against pathogens. Fluorescent pseudomonads are already commercially used for the biological control of plant diseases (Stockwell and Stack, 2007; Berg, 2009). Our discovery of strains with the capacity to control insect pests on top of fungal pathogens renders these bacteria highly interesting for a new field of application and an additional market.
Acknowledgments
We thank Peter Kupferschmied for his help with the development of mutational strategies and the annotation of the genome of P. chlororaphis PCL1391. We acknowledge Sandra Siegfried, Nicolas Hofer, Tania Meylan, Dominique Fuchs, Michael Schläfli, Benjamin Laubi, Simona Crivelli and Christelle Chanez for technical assistance. We acknowledge technical assistance by the Bioinformatics Core Facility/Professorship of Systems Biology at JLU Giessen and access to resources financially supported by the BMBF grant FKZ 031A533 within the de.NBI network. This study was financed by grants obtained from the Swiss National Foundation for Scientific Research SNSF (Projects 31003A-138248, 31003A-120121A0 and 31003A-159520). Funding for BD and THMS was provided by the SNSF NRP 68 (Project 406840-143144). Syngenta Crop Protection (Stein, Switzerland) generously provided Spodoptera littoralis and Plutella xylostella larvae.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Achouak W, Sutra L, Heulin T, Meyer JM, Fromin N, Degraeve S et al. (2000). Pseudomonas brassicacearum sp. nov. and Pseudomonas thivervalensis sp. nov., two root-associated bacteria isolated from Brassica napus and Arabidopsis thaliana. Int J Syst Evol Microbiol 50: 9–18. [DOI] [PubMed] [Google Scholar]
- Alonso A, Rojo F, Martinez JL. (1999). Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ Microbiol 1: 421–430. [DOI] [PubMed] [Google Scholar]
- Bakker PAHM, Pieterse CMJ, van Loon LC. (2007). Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97: 239–243. [DOI] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84: 11–18. [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951). Studies on lysogenesis.1. the mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter F-J, Zakrzewski M et al. (2009). EDGAR: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr SE, Gobeli S, Kuhnert P, Goldschmidt-Clermont E, Frey J. (2010). Pseudomonas chlororaphis subsp. piscium subsp. nov., isolated from freshwater fish. Int J Syst Evol Microbiol 60: 2753–2757. [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng TFC, Bloemberg GV, van der Bij AJ, van der Drift K, Schripsema J, Kroon B et al. (1998). Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact 11: 1069–1077. [Google Scholar]
- D'Aes J, Hua GK, De Maeyer K, Pannecoucque J, Forrez I, Ongena M et al. (2011). Biological control of Rhizoctonia root rot on bean by phenazine- and cyclic lipopeptide-producing Pseudomonas CMR12a. Phytopathology 101: 996–1004. [DOI] [PubMed] [Google Scholar]
- de Souza JT, de Boer M, de Waard P, van Beek TA, Raaijmakers JM. (2003). Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl Environ Microbiol 69: 7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi KK, Kothamasi D. (2009). Pseudomonas fluorescens CHA0 can kill subterranean termite Odontotermes obesus by inhibiting cytochromecoxidase of the termite respiratory chain. FEMS Microbiol Lett 300: 195–200. [DOI] [PubMed] [Google Scholar]
- Dilts JA, Quackenbush RL. (1986). A mutation in the R-body coding sequence destroys expression of the killer trait in P. tetraurelia. Science 232: 641–643. [DOI] [PubMed] [Google Scholar]
- Farn JL, Strugnell RA, Hoyne PA, Michalski WP, Tennent JM. (2001). Molecular characterization of a secreted enzyme with phospholipase B activity from Moraxella bovis. J Bacteriol 183: 6717–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Nivens DE, Weadge JT, Howell PL. (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. (2004). Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186: 4457–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, Defago G. (1991). Protection of cucumber plants against black root rot caused by Phomopsis sclerotioides with rhizobacteria. Bulletin SROP 14: 57–62. [Google Scholar]
- Fuchs JG, Moenne-Loccoz Y, Defago G. (2000). The laboratory medium used to grow biocontrol Pseudomonas sp. Pf153 influences its subsequent ability to protect cucumber from black root rot. Soil Biol Biochem 32: 421–424. [Google Scholar]
- Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E. (2015). Phylogenomics and systematics in Pseudomonas. Front Microbiol 6: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso-Becerra MV, Santos-Medellin C, Gonzalez-Valdez A, Mendez JL, Delgado G, Morales-Espinosa R et al. (2014). Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genomics 15: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Rani S, Birah A, Raghuraman M. (2005). Improved artificial diet for mass rearing of the tobacco caterpillar, Spodoptera litura (Lepidoptera: Noctuidae). Int J Trop Insect Sci 25: 55–58. [Google Scholar]
- Haas D, Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. [DOI] [PubMed] [Google Scholar]
- Haas D, Keel C. (2003). Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41: 117–153. [DOI] [PubMed] [Google Scholar]
- Herren J, Lemaitre B. (2012). Insect-microbe interactions: the good, the bad and the others. Curr Opin Microbiol 15: 217–219. [DOI] [PubMed] [Google Scholar]
- Hilker R, Munder A, Klockgether J, Losada PM, Chouvarine P, Cramer N et al. (2015). Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol 17: 29–46. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. (2002). Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296: 733–735. [DOI] [PubMed] [Google Scholar]
- Ishii K, Adachi T, Hara T, Hamamoto H, Sekimizu K. (2014). Identification of a Serratia marcescens virulence factor that promotes hemolymph bleeding in the silkworm, Bombyx mori. J Invertebr Pathol 117: 61–67. [DOI] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. (2004). Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol 186: 4466–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Yang SY, Kim YC, Lee CW, Park MS, Kim JC et al. (2013). Identification of orfamide A as an insecticidal metabolite produced by Pseudomonas protegens F6. J Agric Food Chem 61: 6786–6791. [DOI] [PubMed] [Google Scholar]
- Jousset A, Schuldes J, Keel C, Maurhofer M, Daniel R, Scheu S et al. (2014). Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc 2: e00322–00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel C, Weller DM, Natsch A, Defago G, Cook RJ, Thomashow LS. (1996). Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol 62: 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver AJ. (1956). Pseudomonas aureofaciens nov. spec. and its pigments. J Bacteriol 72: 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijt M, Tran H, Raaijmakers JM. (2009). Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J Appl Microbiol 107: 546–556. [DOI] [PubMed] [Google Scholar]
- Kupferschmied P, Maurhofer M, Keel C. (2013). Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci 4: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmied P, Pechy-Tarr M, Imperiali N, Maurhofer M, Keel C. (2014). Domain shuffling in a sensor protein contributed to the evolution of insect pathogenicity in plant-beneficial Pseudomonas protegens. PLoS Pathog 10: e1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y-S, Han S, Thomashow LS, Rice JT, Paulitz TC, Kim D et al. (2011). Saccharomyces cerevisiae genome-wide mutant screen for sensitivity to 2,4-diacetylphloroglucinol, an antibiotic produced by Pseudomonas fluorescens. Appl Environ Microbiol 77: 1770–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu Rev Immunol 25: 697–743. [DOI] [PubMed] [Google Scholar]
- Levy E, Gough FJ, Berlin KD, Guiana PW, Smith JT. (1992). Inhibition of Septoria tritici and other phytopathogenic fungi and bacteria by Pseudomonas fluorescens and its antibiotics. Plant Pathol 41: 335–341. [Google Scholar]
- Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B. (2006). Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog 2: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Hassan KA, Mavrodi DV, Davis 2nd EW, Lim CK, Shaffer BT et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8: e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia E, de Lorenzo V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol 13: 2702–2716. [DOI] [PubMed] [Google Scholar]
- Maurhofer M, Keel C, Schnider U, Voisard C, Haas D, Defago G. (1992). Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology 82: 190–195. [Google Scholar]
- Mazzola M, Zhao X, Cohen MF, Raaijmakers JM. (2007). Cyclic lipopeptide surfactant production by Pseudomonas fluorescens SS101 is not required for suppression of complex Pythium spp. populations. Phytopathology 97: 1348–1355. [DOI] [PubMed] [Google Scholar]
- Mercado-Blanco J, Bakker PA. (2007). Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92: 367–389. [DOI] [PubMed] [Google Scholar]
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J et al. (2003). GenDB - an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JB, Frapolli M, Keel C, Maurhofer M. (2011). Pyrroloquinoline quinone biosynthesis gene pqqC, a novel molecular marker for studying the phylogeny and diversity of phosphate-solubilizing pseudomonads. Appl Environ Microbiol 77: 7345–7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L et al. (2012). Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol 14: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet M, Gomila M, Scotta C, Sanchez D, Lalucat J, Garcia-Valdes E. (2012). Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol 35: 455–464. [DOI] [PubMed] [Google Scholar]
- N'goma JCB, Le Moigne V, Soismier N, Laencina L, Le Chevalier F, Roux AL et al. (2015). Mycobacterium abscessus phospholipase C expression is induced during coculture within Amoebae and enhances M. abscessus virulence in mice. Infect Immun 83: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarasah G, Stavrinides J. (2011). Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev 35: 555–575. [DOI] [PubMed] [Google Scholar]
- Neidig N, Paul RJ, Scheu S, Jousset A. (2011). Secondary metabolites of Pseudomonas fluorescens CHA0 drive complex non-trophic interactions with bacterivorous nematodes. Microb Ecol 61: 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisr RB, Russell MA, Chrachri A, Moody AJ, Gilpin ML. (2011). Effects of the microbial secondary metabolites pyrrolnitrin, phenazine and patulin on INS-1 rat pancreatic beta-cells. FEMS Immunol Med Microbiol 63: 217–227. [DOI] [PubMed] [Google Scholar]
- Olcott MH, Henkels MD, Rosen KL, Walker FL, Sneh B, Loper JE et al. (2010). Lethality and developmental delay in Drosophila melanogaster larvae after ingestion of selected Pseudomonas fluorescens strains. PloS One 5: e12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opota O, Vallet-Gely I, Vincentelli R, Kellenberger C, Iacovache I, Gonzalez MR et al. (2011). Monalysin, a novel ss-pore-forming toxin from the Drosophila pathogen Pseudomonas entomophila, contributes to host intestinal damage and lethality. PLoS Pathog 7: e1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HI, Ming LJ. (2002). Mechanistic studies of the astacin-like Serratia metalloendopeptidase serralysin: highly active (>2000%) Co(II) and Cu(II) derivatives for further corroboration of a "metallotriad" mechanism. J Biol Inorg Chem 7: 600–610. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV et al. (2005). Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23: 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix A, Valverde A, Rivas R, Igual JM, Ramirez-Bahena MH, Mateos PF et al. (2007). Reclassification of Pseudomonas aurantiaca as a synonym of Pseudomonas chlororaphis and proposal of three subspecies, P. chlororaphis subsp. chlororaphis subsp. nov., P. chlororaphis subsp. aureofaciens subsp. nov., comb. nov. and P. chlororaphis subsp. aurantiaca subsp. nov., comb. nov. Int J Syst Evol Microbiol 57: 1286–1290. [DOI] [PubMed] [Google Scholar]
- Perneel M, Heyrman J, Adiobo A, De Maeyer K, Raaijmakers JM, De Vos P et al. (2007). Characterization of CMR5c and CMR12a, novel fluorescent Pseudomonas strains from the cocoyam rhizosphere with biocontrol activity. J Appl Microbiol 103: 1007–1020. [DOI] [PubMed] [Google Scholar]
- Pond FR, Gibson I, Lalucat J, Quackenbush RL. (1989). R-body-producing bacteria. Microbiol Rev 53: 25–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péchy-Tarr M, Borel N, Kupferschmied P, Turner V, Binggeli O, Radovanovic D et al. (2013). Control and host-dependent activation of insect toxin expression in a root-associated biocontrol pseudomonad. Environ Microbiol 15: 736–750. [DOI] [PubMed] [Google Scholar]
- Péchy-Tarr M, Bruck DJ, Maurhofer M, Fischer E, Vogne C, Henkels MD et al. (2008). Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. Environ Microbiol 10: 2368–2386. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34: 1037–1062. [DOI] [PubMed] [Google Scholar]
- Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer J-M, Défago G et al. (2011). Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol 34: 180–188. [DOI] [PubMed] [Google Scholar]
- Raymann K, Bobay L-M, Doak TG, Lynch M, Gribaldo S. (2013). A genomic survey of Reb homologs suggests widespread occurrence of R-bodies in proteobacteria. G3 (Bethesda) 3: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes ME. (1959). The characterization of Pseudomonas fluorescens. J Gen Microbiol 21: 221–263. [DOI] [PubMed] [Google Scholar]
- Ruffner B, Péchy-Tarr M, Höfte M, Bloemberg G, Grunder J, Keel C et al. (2015). Evolutionary patchwork of an insecticidal toxin shared between plant-associated pseudomonads and the insect pathogens Photorhabdus and Xenorhabdus. BMC Genomics 16: 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner B, Péchy-Tarr M, Ryffel F, Hoegger P, Obrist C, Rindlisbacher A et al. (2013). Oral insecticidal activity of plant-associated pseudomonads. Environ Microbiol 15: 751–763. [DOI] [PubMed] [Google Scholar]
- Rumpold BA, Schluter OK. (2013). Nutritional composition and safety aspects of edible insects. Mol Nutr Food Res 57: 802–823. [DOI] [PubMed] [Google Scholar]
- Sampson MN, Gooday GW. (1998). Involvement of chitinases of Bacillus thuringiensis during pathogenesis in insects. Microbiology 144: 2189–2194. [DOI] [PubMed] [Google Scholar]
- Scarlett CM, Fletcher JT, Roberts P, Lelliott RA. (1978). Tomato pith necrosis caused by Pseudomonas corrugata n. sp. Ann Appl Biol 88: 105–114. [Google Scholar]
- Sikorski J, Stackebrandt E, Wackernagel W. (2001). Pseudomonas kilonensis sp. nov., a bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 51: 1549–1555. [DOI] [PubMed] [Google Scholar]
- Smits THM, Rezzonico F, Kamber T, Blom J, Goesmann A, Frey JE et al. (2010). Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol Plant Microbe Interact 23: 384–393. [DOI] [PubMed] [Google Scholar]
- Snyder WE, Tonkyn DW, Kluepfel DA. (1998). Insect-mediated dispersal of the rhizobacterium Pseudomonas chlororaphis. Phytopathology 88: 1248–1254. [DOI] [PubMed] [Google Scholar]
- Songer JG. (1997). Bacterial phospholipases and their role in virulence. Trends Microbiol 5: 156–161. [DOI] [PubMed] [Google Scholar]
- Stavrinides J, McCloskey JK, Ochman H. (2009). Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl Environ Microbiol 75: 2230–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell VO, Stack JP. (2007). Using Pseudomonas spp. for integrated biological control. Phytopathology 97: 244–249. [DOI] [PubMed] [Google Scholar]
- Stutz EW, Defago G, Kern H. (1986). Naturally-occuring fluorescent pseudomonads involved in suppression of black root-rot of tobacco. Phytopathology 76: 181–185. [Google Scholar]
- Tran H, Ficke A, Asiimwe T, Hofte M, Raaijmakers JM. (2007). Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol 175: 731–742. [DOI] [PubMed] [Google Scholar]
- Vallet-Gely I, Lemaitre B, Boccard F. (2008). Bacterial strategies to overcome insect defences. Nat Rev Microbiol 6: 302–313. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. (2002) Modern Applied Statistics with S, 4th edn. Springer: New York, NY, USA. [Google Scholar]
- Wang CX, Ramette A, Punjasamarnwong P, Zala M, Natsch A, Moenne-Loccoz Y et al. (2001). Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiol Ecol 37: 105–116. [Google Scholar]
- Yang GW, Hernandez-Rodriguez CS, Beeton ML, Wilkinson P, Ffrench-Constant RH, Waterfield NR. (2012). Pdl1 is a putative lipase that enhances Photorhabdus toxin complex secretion. PLoS Pathog 8: e1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.