Activation of DP1 by PGD2 in macrophages induces the binding of PRKAR2A to the IFN-γR2 transmembrane region, inhibits JAK2/STAT1 signaling, and triggers the expression of antiinflammatory and reparative genes in myocardial infarction and sepsis.
Abstract
The kinetic participation of macrophages is critical for inflammatory resolution and recovery from myocardial infarction (MI), particularly with respect to the transition from the M1 to the M2 phenotype; however, the underlying mechanisms are poorly understood. In this study, we found that the deletion of prostaglandin (PG) D2 receptor subtype 1 (DP1) in macrophages retarded M2 polarization, antiinflammatory cytokine production, and resolution in different inflammatory models, including the MI model. DP1 deletion up-regulated proinflammatory genes expression via JAK2/STAT1 signaling in macrophages, whereas its activation facilitated binding of the separated PKA regulatory IIα subunit (PRKAR2A) to the transmembrane domain of IFN-γ receptor, suppressed JAK2–STAT1 axis–mediated M1 polarization, and promoted resolution. PRKAR2A deficiency attenuated DP1 activation–mediated M2 polarization and resolution of inflammation. Collectively, PGD2–DP1 axis–induced M2 polarization facilitates resolution of inflammation through the PRKAR2A-mediated suppression of JAK2/STAT1 signaling. These observations indicate that macrophage DP1 activation represents a promising strategy in the management of inflammation-associated diseases, including post-MI healing.
INTRODUCTION
Prostaglandin (PG) D2 is a bioactive lipid mediator derived from arachidonic acid through the sequential reaction of cyclooxygenases (COXs) and PGD2 synthases (PGDSs; Ricciotti and FitzGerald, 2011). PGD2 exerts its functions through activation of D prostanoid receptors (DP1 and DP2; Joo and Sadikot, 2012). Recently, increasing evidence demonstrates that PGD2 is involved in resolution of inflammation (Buckley et al., 2014). For instance, a second increase in both COX-2 expression and PGD2 production is observed at later stages (48 h) of carrageenan-induced inflammation, and treatment with COX-2 inhibitor exacerbates inflammation during resolution, which can be reversed by addition of exogenous PGD2 (Gilroy et al., 1999). Both PGD2 and PGE2 regulate and coordinate with lipoxins (LXs) to promote resolution of inflammation (Levy et al., 2001).Consistently, hematopoietic PGDS (H-PGDS) deficiency results in impaired inflammatory resolution in mice (Rajakariar et al., 2007). However, the detailed molecular mechanisms of PGD2-mediated resolution remain unclear.
Macrophages play an important role in innate and adaptive immunity in response to exogenous pathogens, mediate inflammatory processes, and participate in the active resolution of inflammation by secreting antiinflammatory cytokines and eliminating dead cells and matrix debris (Headland and Norling, 2015). Plasticity and heterogeneity are the characteristic features of macrophages, which allow cells to undergo phenotypic shifts in response to different environmental signals. Two distinct subpopulations of macrophages are involved in recovery after myocardial infarction (MI), a sterile inflammatory reaction (Nahrendorf et al., 2007; Yan et al., 2013). During this process, early responding macrophages (Ly6ChighCD11b+CD206−) predominate 1–3 d after MI and express high levels of M1 signature genes, such as TNF-α, IL-6, chemokine C-C motif ligand 2 (CCL2), and nitric oxide synthase 2 (NOS2), whereas late responders (Ly6ClowCD11b+CD206+) appear 4–7 d after MI and stimulate inflammatory resolution via the increased expression of M2-like reparative genes, including IL-10, arginase 1 (arg1), and TGFβ1.
DP1 is highly expressed in monocytes/macrophages (Rajakariar et al., 2007; Sandig et al., 2007). Yet, the role of the PGD2–DP1 axis in the shaping of macrophage polarization and inflammatory resolution remains to be determined. In this study, we found that targeting DP1, but not DP2, resulted in a striking bias toward M1-like macrophage polarization in the presence of inducing stimuli. Moreover, the macrophage-specific genetic deletion of DP1 (DP1flox/floxLysMCre; termed macrophage-DP1 [Mac-DP1] KO) led to delayed resolution in multiple mouse models of inflammation. Notably, macrophage DP1 activation attenuated JAK2/STAT1 signaling and accelerated resolution in a PKA regulatory IIα subunit (PRKAR2A)–dependent manner. Collectively, all the data suggest that DP1 activation–induced M2 polarization facilitates inflammatory resolution through the PRKAR2A-mediated suppression of JAK2/STAT1 signaling.
RESULTS
Deletion of DP1 receptor, but not DP2, promotes M1 macrophage polarization
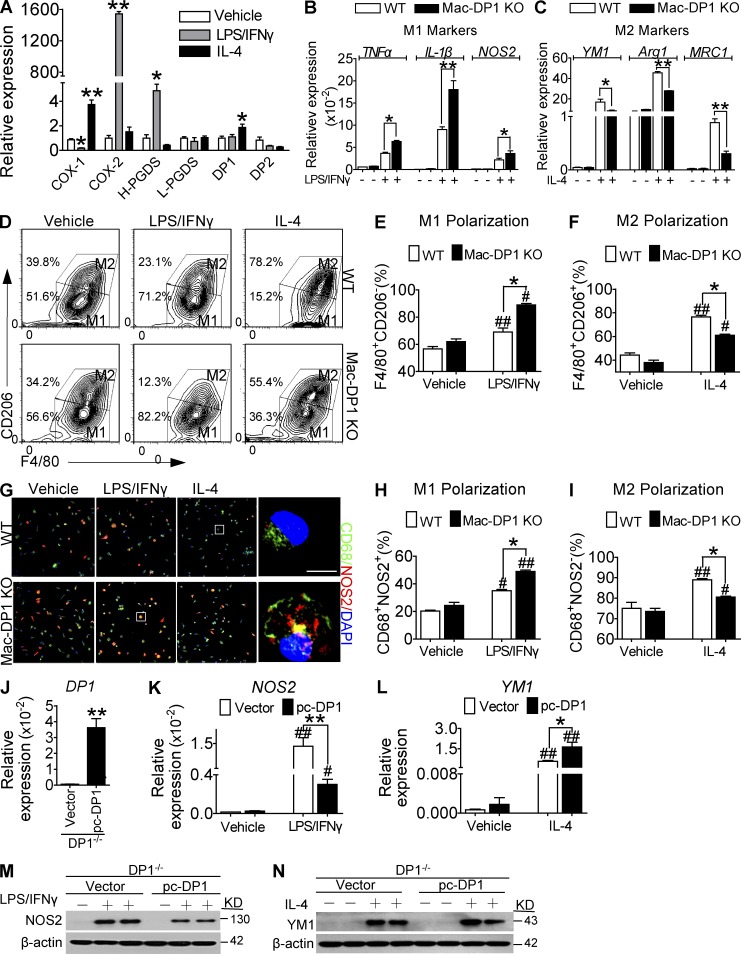
Macrophage polarization is rapid and capable of dynamic interconversion in response to environmental stimuli, as exemplified by induction of the M1 phenotype by LPS/IFN-γ and of the M2 phenotype by IL-4 (Sica et al., 2014). To assess the role of PGD2 in macrophage polarization, we first examined the expression of COXs, PGDSs, and DP receptors during macrophage polarization in vitro. Expression of COX-2 and H-PGDS (Fig. 1 A) and PGD2 production (not depicted) were markedly increased in response to LPS/IFN-γ, whereas IL-4 increased expression of DP1. Expression of DP2, in contrast, was down-regulated during both M1 and M2 polarization (Fig. 1 A). Interestingly, deletion of DP1 in macrophages attained with LysMCre mice (DP1flox/floxLysMCre; Mac-DP1 KO; Fig. S1, A–C) and pharmacological inhibition of DP1 (not depicted) markedly augmented M1 marker expression (Tnfa, Il1b, and Nos2) in response to LPS/IFN-γ stimulation (Fig. 1 B) but suppressed M2 marker expression (Ym1, Arg1, and Mrc1) in response to IL-4 stimulation (DP1flox/flox littermate; WT; Fig. 1 C). Similarly, we also observed that macrophage DP1 deficiency suppressed IL-4–induced M2 polarization and promoted LPS/IFN-γ–mediated M1 polarization by flow cytometry and immunostaining (Fig. 1, D–I). In contrast, DP1 reconstitution in DP1-deficient macrophages (Fig. 1 J) restrained M1 marker (Fig. 1, K and M) and enhanced M2 marker (Fig. 1, L and N) expression induced by LPS/IFN-γ and IL-4, respectively. However, we failed to detect any significant effects of DP2 deficiency on macrophage polarization (not depicted). Collectively, these results indicate that the PGD2–DP1 axis directs macrophage polarization toward an M2-like phenotype.
Figure 1.
DP1 targeting promotes M1 macrophage polarization. (A) Expression of COXs, PGDSs, and PGD2 receptors in macrophages in response to LPS/IFN-γ and IL-4 treatment. L-PGDS, lipocalin PGDS. (B) Effect of DP1 deficiency on macrophage proinflammatory M1 marker expression in response to LPS/IFN-γ treatment. (C) Effect of DP1 deficiency on macrophage antiinflammatory M2 marker expression in response to IL-4 treatment. (A–C) *, P < 0.05; **, P < 0.01 versus vehicle (A) or WT (B and C). n = 6. (D–F) Effect of DP1 deficiency on M1 (F4/80+CD206−) and M2 (F4/80+CD206+) macrophage distributions analyzed by flow cytometry. (D) Representative flow cytometry profiles. (E and F) Quantification of F4/80+CD206− (M1) and F4/80+CD206+ (M2) macrophages. (G–I) Effect of DP1 deficiency on M1 (CD68+NOS2+) and M2 (CD68+NOS2−) macrophage distributions analyzed by immunofluorescence. (G) Representative immunofluorescent images. Bar, 5 µm. (H and I) Quantification of M1 (CD68+NOS2+) and M2 (CD68+NOS2−) macrophages. (J–N) Effect of DP1 reconstitution on macrophage polarization. pc, pc-DNA3.1 vector. DP1 mRNA levels (J), NOS2 (K) and YM1 (L) mRNA levels, and protein levels (M and N) were examined in DP1-reexpressed DP1−/− macrophages. *, P < 0.05; **, P < 0.01 versus WT (D–F, H, and I) or vector (J–N). #, P < 0.05; ##, P < 0.01 versus vehicle. n = 4. All graphs are shown as mean ± SEM. Data are representative of at least two independent experiments. Statistical significance was determined using unpaired Student's t tests.
Mac-DP1 deletion delays resolution of inflammation by promoting M1 macrophage polarization in mice
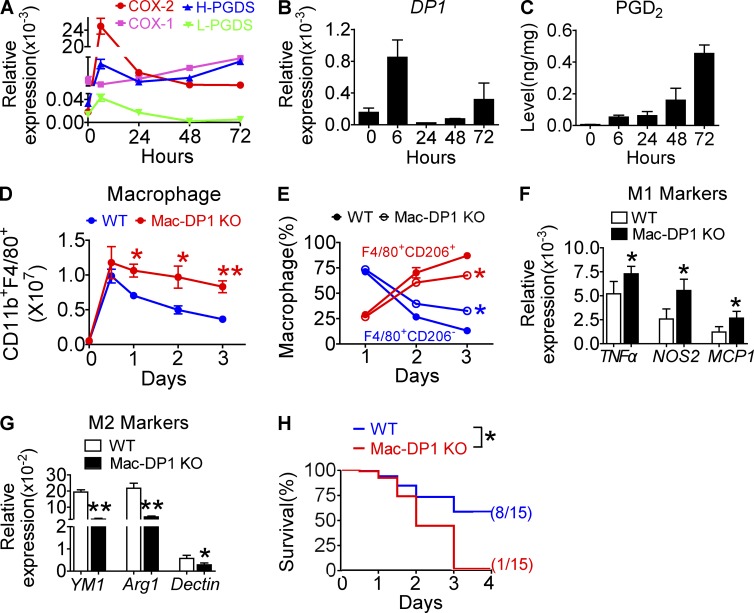
An increase in PGD2 production in the later stages of a model of acute inflammation favors resolution of inflammation in mice (Gilroy et al., 1999). To address the role of DP1, we examined the dynamic alterations of macrophage proinflammatory and reparative genes expression with respect to the COX–PGDS–DP axis during the resolution of zymosan-induced peritonitis. Consistent with a previous study (Gilroy et al., 1999), the second peak of COX-2 expression in peritoneal macrophages coincided with inflammatory resolution 24 h after zymosan treatment (Fig. 2 A). COX-1 and lipocalin-type PGDS expression were also up-regulated, albeit at relatively low levels (Fig. 2 A). Similarly, macrophage PGD2 production was increased at both 3 h and 24 h (Fig. 2 B), whereas DP1 expression inversely correlated with PGD2 production (Fig. 2 C). Then, we examined the resolution indices (Bannenberg et al., 2005; Schwab et al., 2007) of PGD2 and DP1 agonist BW245C (BW) using a zymosan-induced peritonitis model. As shown in Fig. 2 D, the resolution interval Ri of PGD2 and BW was shortened ∼2–3 h, and moreover, macrophage resolution was also marked accelerated (not depicted). Moreover, DP1 disruption increased accumulation of peritoneal leukocytes and macrophages at later stages of inflammation (Fig. 2, E and F). Flow cytometry analysis revealed that M1-like macrophages (F4/80+CD206−) were significantly increased, whereas M2-like macrophages (F4/80+CD206+) were decreased (Fig. 2, G and H) during resolution (24–72 h) in zymosan-treated Mac-DP1 KO mice as compared with DP1flox/flox littermates (WT). Consistent with these observations, all the tested proinflammatory genes (Tnfa, Mcp1, and Nos2 [Fig 2. I] and Il-6, Il-12β, Tlr2, Tlr4, Tlr9, and Ccr2 [not depicted]) were elevated, whereas the reparative genes (i.e., Ym1, Arg1, and Mrc1; Fig. 2 J) were depressed in DP1-deficient peritoneal macrophages. Immunofluorescence assays also revealed more M1-like residential macrophages (CD68+NOS2+) during resolution in Mac-DP1 KO mice than that in WT controls (not depicted). Furthermore, DP1 blockade by MK0524 24 h after zymosan challenge prolonged the resolution of inflammation (Fig. 2 K), promoted proinflammatory macrophage accumulation (Fig. 2 L), elevated proinflammatory gene expression, and suppressed reparative gene expression (Fig. 2, M and N).
Figure 2.
DP1 activation promotes timely resolution by triggering M2 polarization in zymosan-induced peritonitis in mice. (A–N) Peritoneal macrophages were collected at the indicated time points after zymosan challenge. (A–C) COX and PGDS mRNA levels in peritoneal macrophages (A), PGD2 production in peritoneal lavages (B), and DP1 expression in peritoneal macrophages (C) are shown. L-PGDS, lipocalin PGDS. (D) Resolution indices including Tmax (time point when PMNs reach maximal level),T50 (time point corresponding to ∼50% PMN reduction), and Ri (resolution interval, the interval between Tmax and T50) were calculated when 100 µg/kg PGD2 or 1 mg/kg BW was given at peak (12 h) of inflammation. For zymosan-induced inflammation: Tmax, ∼12 h; T50, ∼24h; Ri, ∼12 h. For plus PGD2: Tmax, ∼12 h; T50, ∼21 h; Ri, ∼9 h. For plus BW: Tmax, ∼12 h; T50, ∼22 h; Ri, ∼10 h. n = 4–6. (E–J) Total infiltrated leukocytes (E) and total macrophages (F), F4/80+CD206+ and F4/80+CD206− cells (G) and ratio (H), and mRNA levels of proinflammatory (I) and antiinflammatory (J) gene expression were analyzed from the initiation to resolution of zymosan-induced peritonitis in mice. (K–N) 4 mg/kg/day of DP1 antagonist MK0524 was administrated 24 h after zymosan challenge in mice, and peritoneal macrophages were collected at the indicated time points. Total macrophages (K) and F4/80+CD206+ and F4/80+CD206− cells (L) were calculated at different time points. Proinflammatory (M) and antiinflammatory (N) gene expression were examined in macrophages collected at 72 h after zymosan challenge. (E–J and K–N) *, P < 0.05; **, P < 0.01 versus WT (E–J) or vehicle (K–N). n = 4–6. All data are expressed as mean ± SEM. Statistical analysis was performed using unpaired Student’s t test. Data are representative of two independent experiments.
We also examined Escherichia coli infection–induced peritonitis in a cecal ligation and puncture (CLP) mouse model. During the resolution process, COX-1, H-PGDS, and DP1 mRNA expression in peritoneal macrophages and PGD2 production were elevated (Fig. 3, A–C). However, macrophage-specific DP1 deletion exaggerated CLP-induced peritonitis (Fig. 3 D), stimulated M1-like bias (Fig. 3 E), increased proinflammatory cytokine expression (Fig. 3, F and G), and, thus, reduced the overall survival rate of CLP (Fig. 3 H). Collectively, these data suggest that the PGD2–DP1 axis directs macrophage polarization toward a reparative phenotype to promote inflammatory resolution.
Figure 3.
DP1 deficiency aggravates inflammation and mortality in a CLP mouse model. (A–E) Peritoneal macrophages were sorted at different post-CLP time points and subjected to mRNA expression analysis of COXs and PGDSs (A) and DP1 (B). L-PGDS, lipocalin PGDS. (C) PGD2 production in peritoneal lavage fluid was also measured. (D and E) Total macrophages (D) and ratio of F4/80+CD206+ and F4/80+CD206− cells (E) were calculated. n = 4–6. (F and G) mRNA levels of proinflammatory genes and antiinflammatory genes were examined in peritoneal macrophages sorted 72 h after CLP in mice. n = 6. (H) Effect of macrophage DP1 deficiency on survival rates of mice with CLP-induced sepsis. n = 15. *, P < 0.05; **, P < 0.01 versus WT. Graphs are shown as mean ± SEM. Results are representative of two independent experiments. (D–G) Statistical significance was determined using unpaired Student's t tests. (H) Survival rate was compared using the log-rank test.
Mac-DP1 deficiency impairs recovery from experimental MI by delaying resolution of inflammation
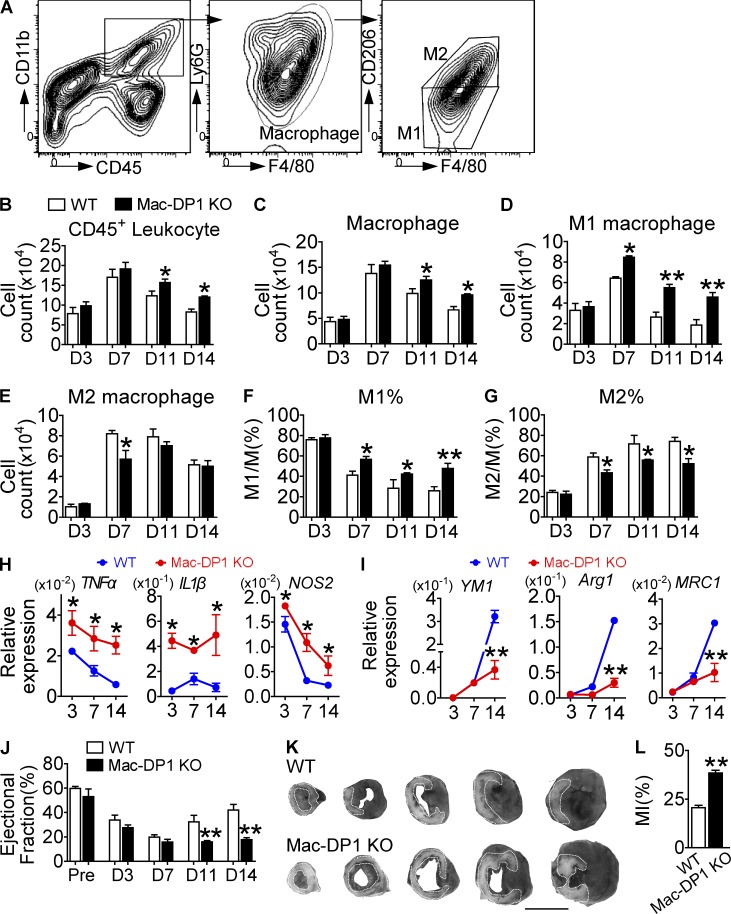
Monocytes/macrophages are involved in the healing process after MI, which involves proteolysis, phagocytosis, angiogenesis, and myocardial remodeling (Ben-Mordechai et al., 2015). In mice, reparative M2-like monocytes/macrophages (Ly-6Clow) predominate in the infarct zone during the resolution of inflammation (day 4–7 after MI) and propagate after MI recovery (Nahrendorf et al., 2007). Flow cytometry analysis (Fig. 4 A) revealed an accumulation of inflammatory cells (leukocytes and macrophages) in the hearts of Mac-DP1 KO mice 11 and 14 d after MI, when compared with control mice (Fig. 4, B and C). Interestingly, infiltrating M1-like macrophages (CD68+D206−) appeared more frequently, whereas M2-like macrophages (CD68+D206+) were reduced at days 11 and 14 after MI in Mac-DP1 KO hearts compared with those from WTs (Fig. 4, D–G), consistent with marker gene expression (Fig. 4, H and I). Both functional recovery (Fig. 4 J) and the extent of tissue damage after MI (Fig. 4, K and L) were markedly altered in Mac-DP1 KO mice consistent with delayed healing caused by disruption of M2-dependent resolution of inflammation.
Figure 4.
DP1 deletion in macrophages impairs MI healing by suppressing M2 polarization and resolution in ischemic hearts. (A–G) Effect of DP1 deficiency in macrophages on inflammatory resolution during post-MI recovery. (A) Inflammatory cell infiltrate in infarcted hearts was isolated from both WT and Mac-DP1 KO mice. (B–G) Total CD45+ leukocytes (B), CD45+CD11b+ F4/80+Ly6G− macrophages (C), and F4/80+CD206− and F4/80+ CD206+ cells (D and E) and their ratios (F and G) were calculated at different time points. n = 4–6. (H and I) Effect of DP1 deletion on mRNA expression of proinflammatory genes (H) and antiinflammatory genes (I) in macrophages infiltrated in hearts. n = 6. (J and K) Effect of DP1 deficiency in macrophages on post-MI recovery in mice. (J) The ejection fraction was evaluated by echocardiography. Pre, pre-ligation. n = 10–14 per group. (K) Infarct areas were analyzed at day 14 by Evans blue dye and tetrazolium chloride staining, followed by OPTIMAS software analysis for color enhancement. Dotted lines denote the infarct zone. The darkest area is normal heart tissue, whereas the gray area represents at-risk myocardial tissue. Bar,10 mm. (L) Quantitation of infarcted areas. n = 6. *, P < 0.05; **, P < 0.01 versus WT. Representative data are shown as mean ± SEM derived from two independent experiments. Statistical significance was determined using unpaired Student's t tests.
DP1-mediated inflammation resolution is dependent on JAK2/STAT1 activity in macrophages
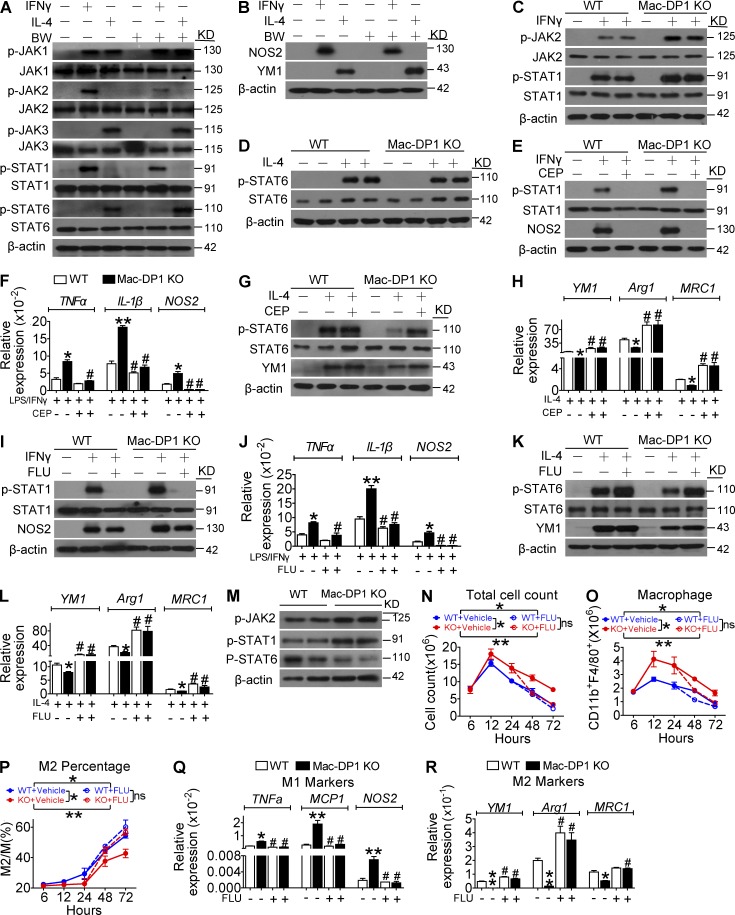
It has been proposed that STAT1 activation is indispensable for M1 macrophage polarization, whereas M2 macrophage differentiation requires STAT6 activity (Sica and Mantovani, 2012). The DP1 agonist BW inhibited JAK2 and STAT1 phosphorylation in response to IFN-γ treatment but had no influence on that of JAK1 and JAK3. In contrast, DP1 activation augmented IL-4–induced STAT6 phosphorylation (Fig. 5 A). Consistently, BW pretreatment suppressed IFN-γ–induced NOS2 expression but enhanced IL-4–induced YM1 expression (Fig. 5 B). Moreover, DP1 disruption amplified JAK2 and STAT1 phosphorylation (Fig. 5 C) and attenuated IL-4–mediated STAT6 activation in macrophages (Fig. 5 D) without overt effects on JAK1 and JAK3 phosphorylation. We then examined whether excessive JAK2–STAT1 pathway activation contributed to M1 bias in DP1-deficient macrophages. As shown in Fig. 5 (E–H), the JAK2 specific inhibitor CEP33791(CEP) completely inhibited STAT1 activation, attenuated M1 gene expression, and rescued STAT6 phosphorylation and M2 marker gene expression in DP1-deficient macrophages. Likewise, the STAT1 inhibitor Fludara (FLU) also attenuated excessive STAT1 activity and M1 gene expression and rescued M2 gene expression in DP1-deficient macrophages (Fig. 5, I–L). Moreover, increased JAK2 and STAT1 phosphorylation and decreased STAT6 phosphorylation were also observed in Mac-DP1 KO peritoneal macrophages harvested after zymosan treatment (Fig. 5 M). Collectively, these observations are consistent with PGD2–DP1 axis promotion of M2 polarization via modulation of JAK2/STAT1 signaling.
Figure 5.
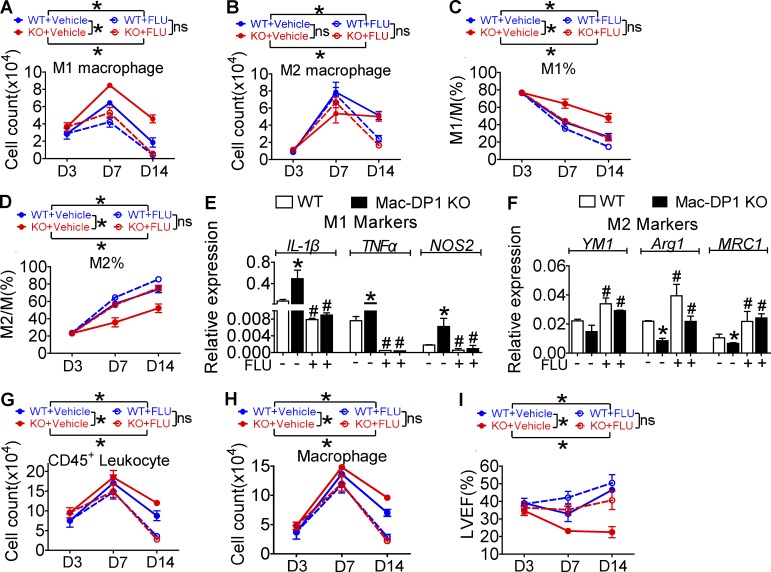
DP1 disruption suppresses M2 polarization and postpones resolution through JAK2/STAT1 pathway activation. (A and B) DP1 agonist (BW)–treated macrophages were analyzed for IFN-γ–induced JAK2/STAT1 phosphorylation (A) and NOS2 expression (B). (C and D) DP1 KO macrophages were examined for IFN-γ–induced JAK2/STAT1 phosphorylation (C) and IL-4–induced STAT6 phosphorylation (D). (E–H) DP1-deficient and WT macrophages were pretreated with JAK2 inhibitor CEP and monitored for JAK2/STAT1 phosphorylation (E), proinflammatory gene expression (F), STAT6 phosphorylation (G), and antiinflammatory gene expression (H). *, P < 0.05; **, P < 0.01 versus WT. #, P < 0.05 versus without CEP. n = 4. (I–L) DP1-deficient and WT macrophages were pretreated with the STAT1 inhibitor FLU and assessed for JAK2/STAT1 phosphorylation (I), proinflammatory gene expression (J), STAT6 phosphorylation (K), and antiinflammatory gene expression (L). *, P < 0.05; **, P < 0.01 versus WT. #, P < 0.05; ##, P < 0.01 vs. without FLU. n = 4. (M) Sorted peritoneal macrophages harvested after zymosan treatment in WT and DP1-deficient mice were assessed for JAK2/STAT1/STAT6 phosphorylation. (N–R) 40 mg/kg/12 h of FLU was administered 24 h after zymosan challenge in WT and Mac-DP1 KO mice. Peritoneal cells were then collected at the indicated time points. Total infiltrated cells (N), total CD11b+F4/80+ cells (O), M2 ratio (P), M1 genes expression (Q), and M2 genes expression (R) of macrophages from Mac-DP1 KO mice at resolution phase are shown. *, P < 0.05; **, P < 0.01 as indicated. #, P < 0.05 versus without FLU. n = 4. Data are expressed as mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by a Bonferroni posthoc test (N–P) or unpaired Student’s t test (F, H, J, L, Q, and R). All Western blots were repeated three times, and the others were also verified in two independent experiments.
Because loss of DP1 represses M2 macrophage polarization by activating JAK2/STAT1 signaling, retarding resolution of inflammation, we next examined whether STAT1 inhibitor treatment could rescue delayed resolution in vivo. Again, Mac-DP1 deletion postponed inflammatory resolution (increase of total inflammatory cells and macrophages at both 48 h and 72 h), and this was restored by STAT1 inhibition (Fig. 5, N and O). Likewise, the STAT1 inhibitor corrected the reduction in M2-like macrophages and elevated proinflammatory and attenuated antiinflammatory gene expression in Mac-DP1 KO mice at both 48 and 72 h (Fig. 5, P–R). Thus, the STAT1 inhibitor could rescue impaired resolution in Mac-DP1 KO mice by rectifying macrophage polarization in the zymosan-induced peritonitis model. Moreover, in experimental MI, an example of sterile inflammation, STAT1 inhibition suppressed infiltration of M1-like macrophages (Fig. 6, A–D), corrected the M1 polarization bias (Fig. 6, E and F), and blunted the prolonged retention of infiltrating leukocytes and macrophages in Mac-DP1 KO mice (Fig. 6, G and H), resulting in restoration of myocardial function (Fig. 6 I).
Figure 6.
FLU treatment attenuates exacerbated LAD ligation–induced inflammation and heart failure caused by macrophage-DP1 deletion in mice. (A–D) 40 mg/kg FLU twice daily was administrated to mice at day 4 after LAD ligation. Infiltrating macrophages isolated from infarcted hearts were sorted to calculate F4/80+CD206− and F4/80+CD206+ macrophages and ratios at the indicated time points. (E and F) mRNA levels of proinflammatory (E) and antiinflammatory (F) genes were examined in macrophages harvested at day 14. *, P < 0.05 versus WT. #, P < 0.05 versus without FLU. n = 6. (G and H) Total CD45+ leukocytes (G) and macrophages (H) per heart were also compared. *, P < 0.05 as indicated. n = 5–6. (I) Effect of FLU on heart function in Mac-DP1 KO and WT mice after LAD ligation. *, P < 0.05. n = 10–12. LVEF, left ventricular ejection fraction. All data are expressed as mean ± SEM. P-values were calculated using two-way ANOVA followed by a Bonferroni posthoc test (A–D and G–I) or unpaired Student’s t test (E and F).
DP1 activation facilitates PRKAR2A–IFN-γR2 binding to suppress JAK2/STAT1 signaling in macrophages
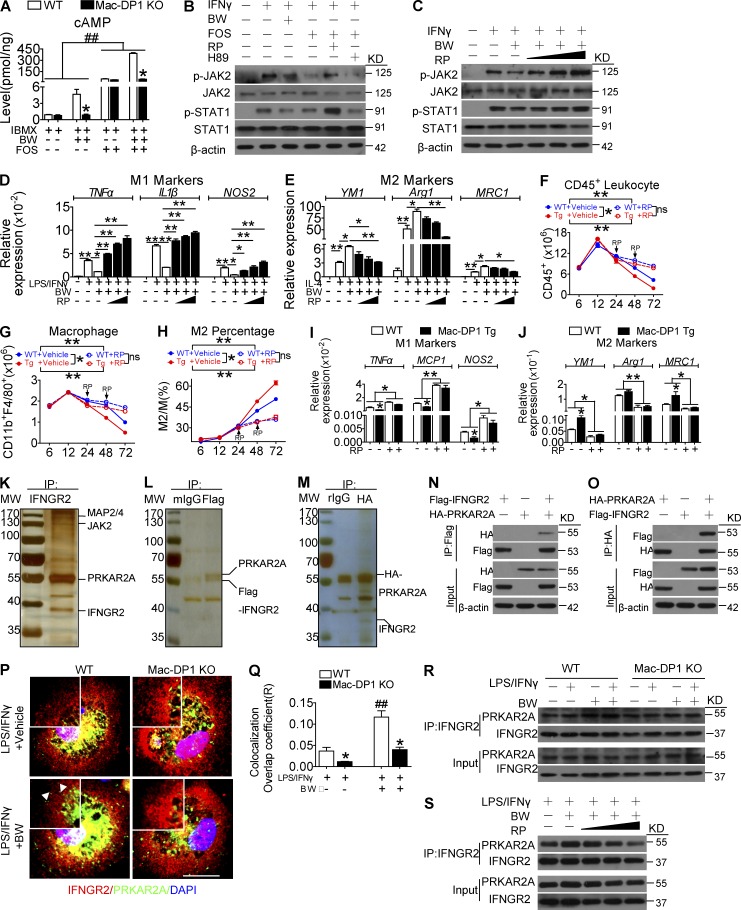
The DP1 receptor couples to Gs to trigger cyclic adenosine monophosphate (cAMP)/PKA-dependent cellular functions (Breyer et al., 2001). As anticipated, DP1 activation increased intracellular cAMP levels, whereas its ablation markedly reduced cAMP generation (Fig. 7 A). To evaluate whether DP1-mediated M2 macrophage polarization is cAMP/PKA dependent, Rp-cAMP and H89, which inhibit PKA regulatory and catalytic subunit dissociation and its kinase activity, respectively, were used to treat macrophages cultured in the presence of M1-polarizing stimuli. Notably, the DP1 agonist BW and adenylyl cyclase activator Forskolin both inhibited JAK2/STAT1 activation, which was attenuated by Rp-cAMP but not H89 (Fig. 7 B). Moreover, Rp-cAMPs rescued JAK2/STAT1 activity, reduced M1 marker expression, and augmented M2 marker expression in BW-treated macrophages in a dose-dependent manner (Fig. 7, C–E), whereas H89 had no significant effect on DP1-mediated M2 polarization (not depicted). Conversely macrophage-specific DP1 overexpression (Mac-DP1 Tg; two lines with >20-fold increase of DP1 expression in macrophages; Fig. S2, A–C) promoted the resolution of zymosan-induced peritonitis by inhibiting leukocyte and macrophage infiltration, suppressing proinflammatory genes expression, and enhancing reparative marker expression (Fig. 7, F–J). These phenomena were specifically attenuated by Rp-cAMP treatment (Fig. 7, F-J). Thus, DP1-mediated M2 polarization and JAK2/STAT1 depression relies on the dissociation of the PKA regulatory and catalytic subunits, rather than PKA catalytic activity.
Figure 7.
DP1 activation facilitates PRKAR2A–IFN-γR2 binding to suppress macrophage JAK2/STAT1 signaling. (A) WT and DP1-deficient macrophages were treated with DP1 agonist and monitored for changes in intracellular cAMP levels. *, P < 0.05 versus WT. ##, P < 0.01. n = 6. FOS, Forskolin. (B) Effect of Rp-cAMP (RP) and H89 on Forskolin-mediated suppression of JAK2/STAT1 phosphorylation in macrophages. (C–E) Effect of Rp-cAMP (10 µM, 50 µM, and 100 µM) on BW-mediated JAK2/STAT1 phosphorylation (C), M1 gene expression (D), and M2 gene expression (E) in macrophages. *, P < 0.05; **, P < 0.01 as indicated. n = 4. (F–J) Rp-cAMP was administrated 24 h after zymosan challenge in Mac-DP1 transgenic (Tg) mice. Peritoneal cells were collected to monitor CD45+ cells (F), total CD11b+F4/80+ cells (G), M2 percentage (H), and M1 (I) and M2 (J) gene expression. *, P < 0.05; **, P < 0.01 as indicated. n = 4–6. (K) Representative silver staining of anti–IFN-γR2 (IFNGR2) immunoprecipitate (IP). The marked proteins were resulted from LC-MS/MS. (L–O) The IFN-γR2–PRKAR2A interaction was analyzed by anti-FLAG (L and N) and anti-HA (M and O) immunoprecipitation. (P and Q) Effect of BW treatment on PRKAR2A and IFN-γR2 colocalization in macrophages (P) and quantification (Q). *, P < 0.05 versus WT. ##, P < 0.01 versus without BW. n = 4. Bar, 5 µm. Arrowheads indicate colocalization of PRKAR2A and IFN-γR2 in the membrane. (R) Analysis of the IFN-γR2–PRKAR2A interaction in BW-treated WT and Mac-DP1 KO macrophages. (S) Effect of Rp-cAMP (10 µM, 50 µM, and 100 µM) on the BW-induced IFN-γR2–PRKAR2A interaction in macrophages. Data are presented as mean ± SEM and are representative of at least two independent experiments. Statistical analysis was performed using a two-way ANOVA followed by a Bonferroni posthoc test (F–H) or unpaired Student’s t test (A, D, E, I, J, and Q).
JAK activity can be regulated either by intrinsic regulatory elements, such as its transphosphorylation and inhibitory pseudokinase domain, or by direct binding of extrinsic negative modulators, including suppressor of cytokine signaling (SOCS) family members and SH2B3 (Babon et al., 2014). We then examined whether PKA subunits may modulate JAK2 activity through direct interactions with JAK2 or IFN-γR2. Interestingly, we did not find any PKA subunits in the anti-JAK2 immunoprecipitate, whereas PRKAR2A protein was discovered in that of anti–IFN-γR2 (Fig. 7 K). Subsequently, we confirmed this interaction in an exogenous overexpression system with FLAG-tagged IFN-γR2 and hemagglutinin (HA)-tagged PRKAR2A in RAW264.7 macrophages (Fig. 7, L and M), as well as by Western blotting in 293T cells (Fig. 7, N and O). DP1 activation increased the membrane mobilization of free PRKAR2A in macrophages, as well as its colocalization with IFN-γR2, whereas this was markedly decreased in Mac-DP1 KO macrophages (Fig. 7, P and Q). Consistent with these observations, BW augmented the PRKAR2A–IFN-γR2 interaction (Fig. 7 R), which was blunted by Rp-cAMP in a dose-dependent manner (Fig. 7 S). These results indicate that PRKAR2A dissociation and the subsequent binding of IFN-γR2 may contribute to DP1-mediated M2 polarization. Thus, DP1-facilitated M2 polarization appears to be contingent on the PRKAR2A–IFN-γR2 interaction–mediated suppression of JAK2/STAT1 signaling.
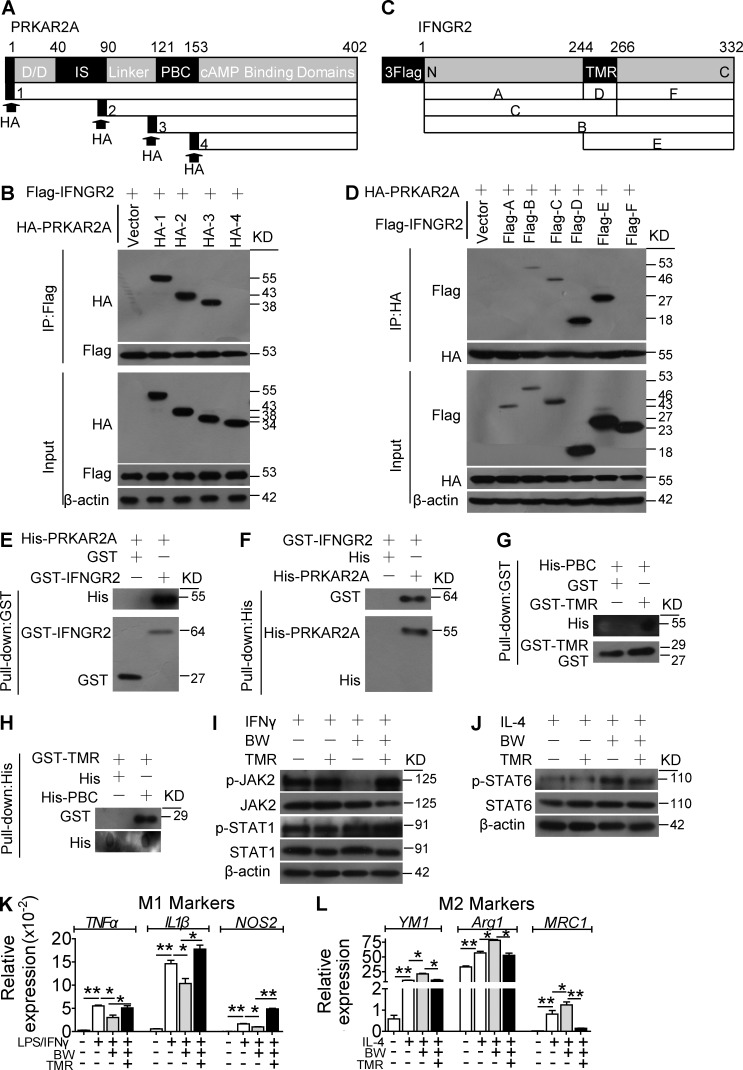
Truncated protein pull-down assays in 293T cells indicated that PRKAR2A binds the IFN-γR2 transmembrane region (TMR) through its phosphate-binding cassette (PBC) domain (Fig. 8, A–D), which was confirmed by in vitro binding assays with recombinant purified protein (Fig. 8, E–H). Consistent with this observation, overexpression of the TMR fragment in macrophages blocked DP1-mediated JAK2/STAT1 suppression, STAT6 activation, and M2 polarization (Fig. 8, I–L). Thus, this evidence supports the contention that DP1 activation facilitates binding between the PRKAR2A PBC and the IFN-γR2 TMR domains to suppress JAK2/STAT1 signaling–mediated M1 macrophage polarization.
Figure 8.
PRKAR2A binding to the IFN-γR2 TMR is required for the DP1-mediated suppression of JAK2/STAT1 signaling and M2 polarization in macrophages. (A) FLAG–IFN-γR2 was cotransfected with HA-PRKAR2A–truncated mutants into 293T cell. D/D, dimerization/docking domain; IS, inhibitor site domain. (B) Immunoprecipitation (IP) assays were performed with FLAG antibody, and Western blot analysis was conducted with HA antibody. IFNGR2, IFN-γGR2. (C) HA-PRKAR2A was cotransfected with FLAG−IFN-γR2–truncated mutants into 293T cells. (D) Immunoprecipitation assays were performed with HA antibody, and Western blot analysis was conducted using FLAG antibody. (E–H) GST and His pulldown assays were used to examine direct interactions of purified IFN-γR2 with PRKAR2A, including full-length proteins (E and F) and the TMR fragment of IFN-γR2 with PBC fragment of PRKAR2A (G and H). (I–L) Effect of IFN-γR2 TMR domain overexpression on BW-induced suppression of JAK2/STAT1 activity (I), STAT6 phosphorylation (J), proinflammatory gene expression (K), and antiinflammatory gene expression (L) in macrophages. *, P < 0.05; **, P < 0.01 as indicated. n = 4. Data are presented as mean ± SEM. (A–J) Data are representative of two independent experiments. (K and L) Data are repeated three times, and statistical significance was determined using unpaired Student's t tests.
PRKAR2A disruption abolishes PGD2–DP1 axis–induced M2 macrophage polarization and resolution of inflammation
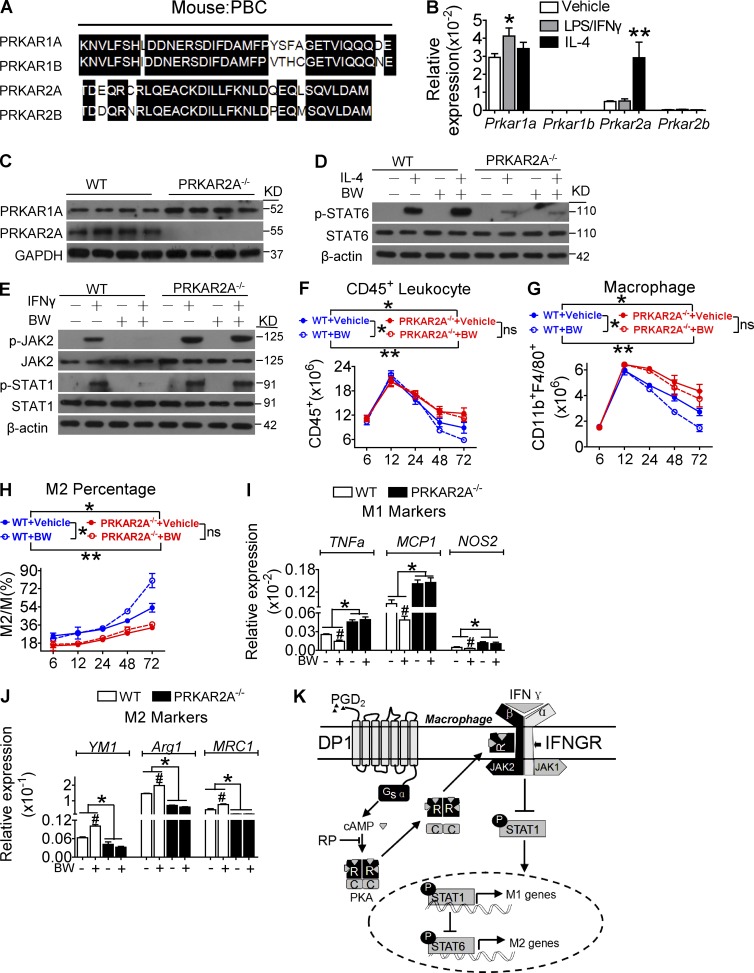
Both PRKAR2A and PRKAR1A were the most abundant of the four PKA regulatory isoforms in macrophages, although they have little homology in their PBC domains (Fig. 9 A). Interestingly, Prkar2a was up-regulated upon M2 polarization (Fig. 9 B). PRKAR2A deletion (Fig. 9 C; Saloustros et al., 2015) resulted in JAK2/STAT1 activation and M1 polarization in macrophages (Fig. 9, D and E). BW suppressed JAK2/STAT1 activation and M1 polarization, which was entirely abolished by deletion of PRKAR2A (Fig. 9, D and E). Interestingly, Prkar1a was up-regulated in PRKAR2A-deleted macrophages (Fig. 9 C). Then, we explored the role of PRKAR2A deficiency on DP1 activation–mediated resolution. As expected, DP1 agonist BW significantly accelerated the resolution of zymosan-induced peritonitis by inhibiting leukocyte and macrophage infiltration, suppressing proinflammatory genes expression, and enhancing reparative markers expression (Fig. 9, F–J). Strikingly, PRKAR2A deficiency attenuated BW-induced M2 polarization and resolution of inflammation (Fig. 9, G–K), whereas knockdown of Prkar1a had no effect on macrophage polarization (Fig. 10, A–G). Collectively, PRKAR2A–IFN-γR2 interaction in macrophages is required for PGD2–DP1 axis–mediated resolution of inflammation.
Figure 9.
PRKAR2A deficiency attenuates BW245C-induced M2 polarization and resolution of inflammation in mice. (A) PBC domain of four PKA R subunits (RIα, RIβ, RIIα, and RIIβ). (B) Prkar1a and Prkar2a expression in macrophages in response to LPS/IFN-γ and IL-4 treatment. **, P < 0.01. n = 6. (C) Prkar1a and Prkar2a expression in Prkar2a−/− macrophages. (D and E) Effect of Prkar2a deletion on STAT6 (D) and JAK2/STAT1 activity (E) in BW-pretreated macrophages. (F–J) BW was administrated 24 h after zymosan challenge in PRKAR2A−/− mice. Peritoneal inflammatory cells were collected to monitor CD45+ cells (F), total CD11b+F4/80+ cells (G), M2 ratio (H), and M1 (I) and M2 (J) gene expression. *, P < 0.05; **, P < 0.01 as indicated. #, P < 0.05 versus vehicle. n = 4–6. (K) Schematic of PGD2–DP1 axis–mediated JAK2/STAT1 inhibition and M2 polarization. IFNGR, IFN-γR; RP, Rp-cAMP. Data are presented as mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by a Bonferroni posthoc test (F–H) or unpaired Student’s t test (B, I, and J). All Western blots were repeated for three times, and the others were also verified in two independent experiments.
Figure 10.
Effect of Prkar1a knockdown on BW254C-induced M2 macrophage polarization. (A) Efficiency of Prkar1a knockdown in macrophages. **, P < 0.01 versus scramble. n = 4. (B–G) Effect of Prkar1a knockdown on proinflammatory (B–D) and antiinflammatory (E–G) gene expression in BW-pretreated macrophages. *, P < 0.05; **, P < 0.01 versus vehicle. n = 4–6. All graphs are shown as mean ± SEM. Data are representative of two independent experiments. Statistical significance was determined using unpaired Student's t tests.
DISCUSSION
Monocyte/macrophage infiltration is critical for inflammatory resolution (Serhan et al., 2007) and MI recovery (Ben-Mordechai et al., 2015). Here, we observed enhanced COX-2 and DP1 expression as well as PGD2 biosynthesis in macrophages during inflammatory resolution. Disruption of the DP1 receptor in macrophages postponed inflammatory resolution in multiple peritonitis models, promoting M1 polarization, and thus increased CLP-induced peritonitis mortality and impaired MI healing. DP1 deficiency also enhanced JAK2/STAT1 transcriptional activity in macrophages by suppressing PKA complex disaggregation and subsequent binding of the PRKAR2A to the IFN-γ receptor. These observations indicate that macrophage DP1 activation represents a promising strategy in the management of inflammation-associated diseases, including post-MI healing.
In a rodent model of carrageenin-induced pleurisy, PGD2 is reported to be produced both in the initiation phase and in the resolution phase (biphasic; Gilroy et al., 1999). COX-2 inhibitors aggravate the carrageenin-induced inflammation at the later stage. This aggravation is associated with reduced PGD2 and 15-d PGJ2 and can be relieved by replacement of these PGs (Gilroy et al., 1999). Consistently, we observed both PGD2 and DP1 receptor agonist BW accelerate resolution in a zymosan-induced peritonitis mouse model. In addition, COX-2–derived LXA4 and 15-epi-LXA4 (in the presence of aspirin) contribute to the resolution of acute lung injury (Fukunaga et al., 2005). Along with LXA4, COX-2–derived PGE2 appears to promote the resolution of allergic pleuritis in rats (Bandeira-Melo et al., 2000).
Plasticity and heterogeneity are key features of macrophages. In tissues, macrophages embrace diverse functional phenotypes in response to environmental cues (Sica and Mantovani, 2012). IFNs and TLR ligands activate traditional IFN regulatory factor/STAT1 signaling pathways to drive macrophages toward a classic M1 phenotype, whereas IL-4 and IL-13 induce STAT6 activation to skew macrophages to an alternative M2 phenotype. Moreover, IL-10 can markedly suppress LPS-induced transcription and enhance antiinflammatory gene expression to direct macrophages to an M2 phenotype via STAT3 signaling (Lang et al., 2002). STAT1 (via IFNs) and STAT6 activation in macrophages also facilitates expression of SOCS1 (Whyte et al., 2011) and SOCS3 (Liu et al., 2008), which in turn restrain STAT6 and STAT1 activity, respectively. Furthermore, the DP1 agonist suppressed JAK2 and STAT1 phosphorylation, also increasing that of STAT6 in macrophages. In contrast, Mac-DP1 disruption led to enhanced JAK2/STAT1 activity and repressed STAT6 activity without any significant influence on STAT3 signaling. Therefore, DP1 deficiency promoted macrophage M1 polarization by disturbing the balance between STAT1 and STAT6/STAT3 pathways.
DP1 activation–induced JAK2/STAT1 suppression was interrupted by Rp-cAMP, but not H89, suggesting that JAK2/STAT1 signaling was modulated by PKA subunit dissociation rather than its own kinase activity. PKA is a holoenzyme comprised of two regulatory (R) and two catalytic (C) subunits. Multiple isoforms have been identified for R (RIα, RIβ, RIIα, and RIIβ) and C (Cα, Cβ, and Cγ) subunits (Taylor et al., 2013). Through various approaches, only RIIα was identified to bind directly the IFN-γR2 TMR domain through its PBC domain, and blocking this binding attenuated PGD2–DP1 axis–mediated JAK2/STAT1 inhibition and M2 polarization. Consistently, only RIIα expression was up-regulated in M2 macrophages. With respect to the cAMP docking site, the RIIα PBC domain does not share structural homology with other R subunits (Su et al., 1995). Generally, R units mediate PKA compartmentalization through their physical binding to A-kinase anchoring proteins (AKAPs) in different subcellular compartments (Skroblin et al., 2010). Indeed, several AKAPs, such as AKAP-2, AKAP-4, and MEKK (also known as MAP3K1, short for mitogen-activated protein kinase kinase kinase 1), also coimmunoprecipitated with FLAG-PRKAR2A in macrophages. Besides regulating PKA activity, PKA R units also directly participate in many biological processes (Filteau et al., 2015), including transcriptional co-repression (Elliott et al., 2003) and calcium signal modulation (Manni et al., 2008). Normally, the hydrophobic cluster in the PBC domain of PKA R units binds to an extensive hydrophobic surface that surrounds Tyr247 in the G helix of PKA C units (Wu et al., 2007). Upon stresses, cAMP incorporates the PBC domain of PKA R units, which leads to stretch away of PKA C units and facilitates PBC domain binding to other hydrophobic proteins, such as the IFN-γR2 receptor. In an M1-prone state, JAK2 phosphorylation is triggered by IFN-γ binding to its cognate receptor dimer (IFN-γR2:IFN-γR1), which elicits conformational change of IFN-γ receptor dimer and repositioning of the associated JAK2 and ultimately transphosphorylation of a tyrosine residue in the conserved activation loop of JAK2 (Babon et al., 2014). Thus, we deduce that the enhanced binding of PRKAR2A to IFN-γR2 by DP1 activation may stabilize IFN-γ receptor dimerization within the macrophage membrane, thereby inhibiting IFN-γ–induced conformation changes of IFN-γR2:IFN-γR1 dimer and JAK2/STAT1 activation during M2 polarization. Likewise, this PKA-dependent suppression of IFN-induced JAK/STAT activity was also observed in Ehrlichia chaffeensis–infected THP-1 human monocytes and U266 myeloma cells (David et al., 1996; Lee and Rikihisa, 1998). Consistent with these observations, PGD2 induces 15-lypoxygenase expression and generation of pro-resolution LXA4 in polymorphonuclear neutrophils dependent on intracellular cAMP accumulation (Levy et al., 2001), perhaps through activation of a Gs-coupled DP1 receptor (Breyer et al., 2001).
Cardiac healing after MI requires a balanced inflammatory response. Macrophages are the dominant cells orchestrating the initiation, amplification, and resolution of the inflammation in the infracted myocardium (Ben-Mordechai et al., 2015). Proinflammatory M1 macrophages are recruited shortly after neutrophils, peaking around day 3 after MI, and display phagocytic and proteolytic functions, whereas antiinflammatory M2 macrophages accumulate at later points and facilitate tissue repair by secreting angiogenic, trophic, and profibrotic cytokines (Nahrendorf et al., 2007; Ben-Mordechai et al., 2013). We observed that DP1 targeting promoted proinflammatory monocyte recruitment and suppressed that of reparative monocytes, resulting in prolonged resolution during MI recovery in mice. Monocytes/macrophages reside within the infarcted heart for relatively short periods (∼20 h), with most (<95%) undergoing apoptosis (Leuschner et al., 2012). Interestingly, DP1 ablation did not alter the apoptosis of infiltrated macrophages but inhibited M2 polarization and antiinflammatory cytokine expression both in vitro and in vivo. Thus, DP1-mediated M2 polarization contributes to MI recovery by promoting timely resolution of inflammation.
In summary, DP1 activation in macrophages promotes M2 polarization and timely inflammatory resolution through the PKA regulatory unit–mediated suppression of JAK2/STAT1 signaling. These observations indicate that targeting the PGD2–DP1 pathway may represent a promising therapeutic strategy for inflammatory disease.
MATERIALS AND METHODS
Animals
DP1flox/flox mice, possessing two loxP sites flanking exon 1 of the DP1 gene (Fig. S1 A; provided by R. Breyer, Vanderbilt University Medical Center, Nashville, TN), were maintained on C57BL/6 genetic background and crossed with C57BL/6 LysMCre mice to generate DP1flox/floxLysMCre (Mac-DP1 KO) mice. DP1flox/flox littermates (WT) served as experimental controls. Macrophage-specific DP1 transgenic (Mac-DP1 Tg) mice were generated by Caygen Biosciences on the C57BL/6 genetic background using macrophage-specific synthetic promoter 146 (SP146; Kang et al., 2014). PRKAR2A−/− mice were maintained on C57BL/6 genetic background. All WT littermates were used as experimental controls. All animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, University of Chinese Academy of Sciences.
Reagents
IFN-γ and IL-4 were from PeproTech. LPS, zymosan, thioglycolate, IBMX, and the ProteoSilver Plus Stain kit were from Sigma-Aldrich. PGD2, H89, and BW245C were from Cayman Chemical. Forskolin was obtained from Enzo Life Sciences. FLU and CEP33791 were from Selleck Chemical. MK0524 was from Santa Cruz Biotechnology, Inc.
Peritoneal macrophage isolation and treatment
Peritoneal macrophages were induced and prepared by an i.p. injection of 3% Brewer’s thioglycolate as described previously (Yang et al., 2010). Macrophages were allowed to adhere overnight at 37°C and 5% CO2 and were washed with fresh medium to remove unattached cells before use. Adherent macrophages were pretreated with 1 µM BW245C, 0.1 mM CEP33791, 100 µM FLU, or 10 µM H89 2 h before stimulation. Macrophage polarization was subsequently induced with 1 µg/ml LPS plus 20 ng/ml IFN-γ or IL-4 and analyzed with real-time PCR (RT-PCR; all primers used in this study are described in Table S1).
Zymosan-induced peritonitis model
1 mg type A zymosan in 0.5 ml PBS (Sigma-Aldrich) was injected to the peritoneal cavity to induce peritonitis in mice (Rajakariar et al., 2007). 40 mg/kg FLU was then administrated as indicated. Peritoneal inflammatory cells were obtained by using 8 ml of sterile PBS to wash out the inflamed peritoneal cavity at progressive time points. The cells were then collected by centrifugation at 800 g for 5 min, and supernatants were stored at −80°C until PG analysis. Pelleted cells were counted with a hemocytometer (02270113; QIUJING) and stained with antibodies for flow cytometry. Macrophages were also sorted at the different time points to examine intracellular cAMP levels and M1/M2 gene expression, as well as JAK2, STAT1, and STAT6 phosphorylation.
Resolution indices of peritonitis model
Resolution indices were calculated as previously described (Bannenberg et al., 2005; Schwab et al., 2007). 100 µg/kg PGD2 or 1mg/kg BW was given at peak (12 h) of zymosan-induced inflammation. After 12 h, peritoneal inflammatory cells were obtained by using 8 ml of sterile PBS to wash out the inflamed peritoneal cavity. The cells were then collected by centrifugation at 800 g for 5 min. Pelleted cells were counted with a hemocytometer and stained with antibodies (PE-conjugated anti–mouse CD45 antibody and APC-conjugated anti–mouse Ly6G antibody) for flow cytometry.
CLP mouse model
CLP surgeries were performed on 2-mo-old female mice as previously described (Hubbard et al., 2005; Ishii et al., 2012). Peritoneal lavage fluid was collected at different time points after CLP surgery and centrifuged at 800 g for 5 min at 4°C, and the supernatants were stored at −80°C until PG analysis. Pelleted peritoneal inflammatory cells were counted with a hemocytometer and stained with specific antibodies for flow cytometry. Sorted macrophages were subjected to quantitative RT-PCR (qRT-PCR) analysis to examine the mRNA expression of different genes. For survival studies, cecal were punctured five times with a 21-gauge needle after ligation.
MI mouse model
Female, 6–8-wk-old mice were subjected to a permanent ligation of the left anterior descending (LAD) coronary (Gao et al., 2010). In brief, mice were fully anesthetized with 1–1.5% isoflurane gas before being mechanically ventilated with a rodent respirator. The chest cavity was opened via left thoracotomy to expose the heart so that LAD could be visualized and permanently ligated with a 6–0 silk suture at the site of its emergence from the left atrium. Complete vessel occlusion was confirmed by the presence of myocardial blanching in the perfusion bed. Mice that died within 24 h after surgery were excluded from the experiment. Sham-operated animals underwent the same procedure without coronary artery ligation. Mice were later euthanized at different time points to analyze immune cell infiltration.
Cell sorting from post-MI hearts
Female, 6–8-wk-old mice were anesthetized and intracardially perfused with 40 ml of ice-cold PBS to exclude blood cells. The heart was dissected, minced with fine scissors, and enzymatically digested with a cocktail of 450 U/ml collagenase I, 125 U/ml collagenase XI, 60 U/ml DNase I, and 60 U/ml hyaluronidase (Sigma-Aldrich) for 1.5 h at 37°C with gentle agitation. After digestion, the tissue was triturated and passed through a 70-µm cell strainer. Leukocyte-enriched fractions were isolated by 37–70% Percoll (GE Healthcare) density gradient centrifugation as described previously (Yan et al., 2013). Cells were removed from the interface and washed with RPMI 1640 cell culture medium before staining with CD45 and CD11b and sorting into three populations: CD45+CD11b+ (myeloid cells), CD45+CD11blow (lymphocytes), and CD45−CD11blow (nonleukocytes). The CD45+CD11b+ population could be further divided into CD11b+F4/80+ macrophages and CD11b+Ly-6G+ neutrophils.
Infarct size evaluation
To evaluate infarct size after MI, 0.2 ml of 2% Evans blue dye was injected into the tail vain, and then, hearts were removed and frozen at −80°C. The frozen heart was then cut transversely into 1-mm thick slices using a Mouse Heart Slicer Matrix (Leica Biosystems) and stained with 2% tetrazolium chloride in PBS (pH 7.4) for 20 min in a 37°C water bath. After fixation for 4–6 h in 10% neutral buffered formaldehyde, each slice was weighed and photographed. Infarct areas were measured by automated planimetry using ImageJ software (National Institutes of Health) and calculated as the ratio of infarct weight to total weight.
Echocardiography
Transthoracic echocardiography was performed at different time points after surgery using an echocardiograph (Vevo2100). The investigator was blinded to group assignment. Mice were anesthetized by isoflurane inhalation. Two-dimensional parasternal long axis views of the left ventricle were obtained for guided M-mode measurements of the left ventricle internal diameter at the diastole end and the systole end, as well as the interventricular septal wall thickness and posterior wall thickness.
Flow cytometry and immunofluorescence
Flow cytometry and cell populations sorting were performed on a FACSAria flow cytometer (BD), using a range of antibodies. Flow cytometry data were analyzed using FlowJo (v.9 or v.10; Tree Star). Confocal microscopy was used to image immunofluorescence using a range of antibodies and quantified using ImageJ software.
Reconstitution of mouse DP1
The mouse DP1 cDNA was subcloned into pcDNA3.1, and a HA tag was added at the extracellular N terminus. Peritoneal macrophages from DP1−/− mice were transiently transfected with pcDNA3.1/DP1 plasmid or pcDNA3.1 empty vector using PT-103-01N jetPEI-Macrophage reagent (PolyPlus-transfection), according to the manufacturer’s protocol.
PG extraction and analysis
500 µl of cell supernatant or peritoneal exudates was used for PG extraction after protein quantification. 2 µl of an internal standard was added to the sample in 40 µl of 1 M citric acid and 5 µl of 10% butylated hydroxytoluene, and the sample was strenuously vibrated with 1 ml solvent (normal hexane/ethyl acetate, 1:1) for 1 min. The organic phase supernatant was collected after centrifugation at 6,000 g/min for 10 min The eluate was dried under nitrogen and analyzed by electrospray triple/quadruple liquid chromatography (LC) coupled with tandem mass spectrometry (MS/MS; 4000Q Trap AB; Sciex). Chromatographic separation was performed on ZORBAX SB-Aq HPLC columns (3 × 250 mm, 5 µm; Agilent Technologies), using 0.1% acetic acid in water (mobile phase A) and acetonitrile (mobile phase B) as the mobile phase for binary gradient elution. The column flow rate was 0.4 mL/min, the column temperature was 25°C, and the autosampler was kept at 4°C. The binary elution gradient was 30% B to 53% B in 15 min and then was to 90% B in 1 min, maintained at 90% B for 3 min. The column was equilibrated for 10 min with the initial solvent composition between injections. PGD2 was detected and quantified in negative ion mode, and the electrospray potential was maintained at −4.5 kV and heated to 500°C. For MS/MS analysis, PGD2/internal standards were subjected to collision-induced fragmentation. PG production was normalized to total protein or per mouse.
Immunofluorescence staining
Primary and sorted macrophages were plated on coverslips, washed twice with PBS, fixed in 4% formaldehyde for 30 min at room temperature, and then washed three times with PBS. Cells were blocked with 3% bovine serum albumin and permeabilized with 0.1% Triton X-100 in PBS for 30 min at room temperature before incubating with rat anti–mouse CD68 antibody (1:400) and rabbit anti–mouse NOS2 (1:500) antibody or rabbit anti–mouse IFN-γR2 (1:200) antibody and mouse anti–mouse PRKAR2A (1:200) antibody at 4°C for 18–20 h. Cells were washed and incubated with appropriate secondary antibody (1:1,000) for 2 h at room temperature and then rinsed in PBS, counter-stained with DAPI, sealed with antifade reagent, and visualized using a laser scanning confocal microscope (Olympus). Images were analyzed with ImagePro Plus software (Media Cybernetics).
Measurement of cellular cAMP Levels
Cells were washed with cold PBS and then harvested in ice-cold lysis buffer. The amount of intracellular cAMP was determined using a cAMP Parameter Assay kit (R&D Systems), according to the manufacturer’s instructions.
RNA extraction and qRT-PCR
Total RNA samples from sorted cells or adherent macrophages were prepared using an RNeasy Mini kit (QIAGEN) or TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. Total RNA (1 µg) was reverse-transcribed to cDNA with a Reverse Transcription Reagent kit (Takara Bio Inc.), according to the manufacturer’s instructions. The resulting cDNA was amplified for 40 cycles. L32 and GAPDH RNA were amplified as internal controls. Each sample was analyzed in triplicate and normalized to a reference RNA. PCR products were confirmed by a single band of expected size on a 2% agarose gel. The primer sequences for PCR are summarized in Table S1.
Western blotting
The protein concentrations of adherent or sorted macrophage lysates were determined using a bicinchoninic acid assay protein assay kit (Thermo Fisher Scientific). Equal quantities of proteins were denatured and resolved by 10% SDS-PAGE gels, transferred to nitrocellulose membranes, incubated with 5% skimmed milk for 1–1.5 h, and then incubated with primary antibodies overnight at 4°C. Primary antibodies were diluted as follows: COX-1 (1:1,000; Cayman Chemical), COX-2 (1:1,000; Cayman Chemical), DP1 (1:200; Cayman Chemical), HA-tag (1:1,000; Cell Signaling Technology), phospho-JAK1 (1:1,000; Cell Signaling Technology), JAK1 (1:1,000; Cell Signaling Technology), phospho-JAK2 (1:1,000; Cell Signaling Technology), JAK2 (1:1,000; ABclonal), phospho-JAK3 (1:1,000; Cell Signaling Technology), JAK3 (1:1,000; Cell Signaling Technology), phospho-STAT1 (1:1,000; Cell Signaling Technology), STAT1 (1:1,000; ABclonal), phospho-STAT6 (pY641) (1:1,000; BD), STAT6 (1:1,000; ABclonal), Ym1 (1:1,000; R&D Systems), NOS2 (1:500; Abcam), FLAG M2 (1:2,000; ABclonal), His-tag (1:2,000; Cell Signaling Technology), glutathione S-transferase (GST)–tag (1:2,000; ABclonal), IFN-γR2 (1:500; Santa Cruz Biotechnology, Inc.), and PRKAR2A (1:500; Santa Cruz Biotechnology, Inc.). Actin (1:2,000; Sigma-Aldrich) or GAPDH (1:2,000; Cell Signaling Technology) was used as the load control. The membranes were then incubated in HRP-labeled secondary antibody in blocking buffer for 2 h at room temperature. Blots were developed using an enhanced chemiluminescence reagent (Thermo Fisher Scientific).
Micro–LC-MS/MS protein preparation
Peritoneal macrophage, RAW264.7, or 293T cells transfected with mock or FLAG-tagged IFN-γR2 and HA-tagged PRKAR2A were cultured in the presence of LPS/IFN-γ and BW245C. Protein extracts were then incubated with anti–IFN-γR2, followed by anti-FLAG M2 beads or anti-HA beads. Samples were separated by 10% SDS-PAGE, the gels were subjected to silver staining, and then bands of interest were excised for further micro–LC-MS/MS analysis.
Immunoprecipitation
Cell lysates were incubated with the indicated antibodies for 12 h followed by protein A/G beads for another 3 h. Beads were washed three times and eluted with SDS loading buffer. Native PAGE analysis was performed as described in the Western blotting section..
Recombinant protein expression and purification
BL21 E. coli cells were transformed with the expression plasmids pGEX-4T-2–IFN-γR2, pGEX-4T-2-TMR, pET-28a-PRKAR2A, and pET-28a-PBC. Recombinant proteins were purified using GST-Bind agarose (ELPIS Biotech) or Ni–nitrilotriacetic acid agarose (QIAGEN) resin according to manufacturer’s protocol.
RNA interference
Peritoneal macrophages were grown to ∼80% confluence for siRNA transfection by RNAiFect Transfection reagent (QIAGEN) according to the manufacturer’s instructions with either a siRNA pool (Genepharma) containing three pairs of specific sequences for prkar1a (100 nmol/L), prkar1a splice variant targeting sequences, or a scrambled siRNA (as a negative control; 100 nmol/L). The mouse prkar1a siRNAs were designed to target 21-nucleotide sequences. Control siRNAs included scrambled derivatives of the prkar1a siRNA sequences, an unrelated siRNA labeled by FAM, and a positive control GAPDH siRNA. In brief, knockdown efficiency was assessed by qRT-PCR (si-prkar1a-FGCGAAAGAGGAAGAUGUAUTTsi-prkar1a-RAUACAUCUUCCUCUUUCGCTT). 1 µg siRNA and RNAiFect Transfection reagent were mixed and incubated for 10–15 min at room temperature to allow formation of transfection complexes before delivery on the cells. 48–72 h after interference, the cells were treated for further experiments.
Statistical analysis
All data are expressed as the mean ± SEM. Data were analyzed in Prism 5 (GraphPad Software). Two-tailed Student’s t testing and ANOVA were used for comparisons between different groups. Survival rates were compared using the log-rank test. P-values <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows the generation and characterization of myeloid cell–specific DP1-deficient mice. Fig. S2 shows the generation of Mac-DP1-Tg mice. Table S1 lists the primers used for quantitative PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20160459/DC1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Chinese Ministry of Science and Technology (2012CB945100), the National Natural Science Foundation of China (81525004, 91439204, 81030004, and 31200860), the Science and Technology Commission of Shanghai Municipality key program (14JC1407400 and 15140902000), the Science and Technology Service Network Initiative (KFJ-EW-STS-099), the postdoctoral fellowship program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (2012KIP514 and 2013KIP312), the National Institutes of Health (GM15431 and DK37097), and a merit award from the Department of Veterans Affairs (1BX000616 to R.M. Breyer). Y. Ji and Y. Yu are fellows at the Jiangsu Collaborative Innovation Center for Cardiovascular Disease Translational Medicine.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AKAP
- A-kinase anchoring protein
- cAMP
- cyclic adenosine monophosphate
- CLP
- cecal ligation and puncture
- COX
- cyclooxygenase
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- H-PGDS
- hematopoietic PGDS
- LAD
- left anterior descending
- LC
- liquid chromatography
- LX
- lipoxin
- MI
- myocardial infarction
- MS/MS
- tandem mass spectrometry
- NOS2
- nitric oxide synthase 2
- PBC
- phosphate-binding cassette
- PG
- prostaglandin
- PGDS
- PGD2 synthase
- qRT-PCR
- quantitative RT-PCR
- RT-PCR
- real-time PCR
- SOCS
- suppressor of cytokine signaling
- TMR
- transmembrane region
References
- Babon J.J., Lucet I.S., Murphy J.M., Nicola N.A., and Varghese L.N.. 2014. The molecular regulation of Janus kinase (JAK) activation. Biochem. J. 462:1–13. 10.1042/BJ20140712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira-Melo C., Serra M.F., Diaz B.L., Cordeiro R.S., Silva P.M., Lenzi H.L., Bakhle Y.S., Serhan C.N., and Martins M.A.. 2000. Cyclooxygenase-2-derived prostaglandin E2 and lipoxin A4 accelerate resolution of allergic edema in Angiostrongylus costaricensis-infected rats: relationship with concurrent eosinophilia. J. Immunol. 164:1029–1036. 10.4049/jimmunol.164.2.1029 [DOI] [PubMed] [Google Scholar]
- Bannenberg G.L., Chiang N., Ariel A., Arita M., Tjonahen E., Gotlinger K.H., Hong S., and Serhan C.N.. 2005. Molecular circuits of resolution: formation and actions of resolvins and protectins. J. Immunol. 174:4345–4355. 10.4049/jimmunol.174.7.4345 [DOI] [PubMed] [Google Scholar]
- Ben-Mordechai T., Holbova R., Landa-Rouben N., Harel-Adar T., Feinberg M.S., Abd Elrahman I., Blum G., Epstein F.H., Silman Z., Cohen S., and Leor J.. 2013. Macrophage subpopulations are essential for infarct repair with and without stem cell therapy. J. Am. Coll. Cardiol. 62:1890–1901. 10.1016/j.jacc.2013.07.057 [DOI] [PubMed] [Google Scholar]
- Ben-Mordechai T., Palevski D., Glucksam-Galnoy Y., Elron-Gross I., Margalit R., and Leor J.. 2015. Targeting macrophage subsets for infarct repair. J. Cardiovasc. Pharmacol. Ther. 20:36–51. 10.1177/1074248414534916 [DOI] [PubMed] [Google Scholar]
- Breyer R.M., Bagdassarian C.K., Myers S.A., and Breyer M.D.. 2001. Prostanoid receptors: subtypes and signaling. Annu. Rev. Pharmacol. Toxicol. 41:661–690. 10.1146/annurev.pharmtox.41.1.661 [DOI] [PubMed] [Google Scholar]
- Buckley C.D., Gilroy D.W., and Serhan C.N.. 2014. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity. 40:315–327. 10.1016/j.immuni.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Petricoin E. III, and Larner A.C.. 1996. Activation of protein kinase A inhibits interferon induction of the Jak/Stat pathway in U266 cells. J. Biol. Chem. 271:4585–4588. 10.1074/jbc.271.9.4585 [DOI] [PubMed] [Google Scholar]
- Elliott M.R., Tolnay M., Tsokos G.C., and Kammer G.M.. 2003. Protein kinase A regulatory subunit type IIβ directly interacts with and suppresses CREB transcriptional activity in activated T cells. J. Immunol. 171:3636–3644. 10.4049/jimmunol.171.7.3636 [DOI] [PubMed] [Google Scholar]
- Filteau M., Diss G., Torres-Quiroz F., Dubé A.K., Schraffl A., Bachmann V.A., Gagnon-Arsenault I., Chrétien A.E., Steunou A.L., Dionne U., et al. . 2015. Systematic identification of signal integration by protein kinase A. Proc. Natl. Acad. Sci. USA. 112:4501–4506. 10.1073/pnas.1409938112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K., Kohli P., Bonnans C., Fredenburgh L.E., and Levy B.D.. 2005. Cyclooxygenase 2 plays a pivotal role in the resolution of acute lung injury. J. Immunol. 174:5033–5039. 10.4049/jimmunol.174.8.5033 [DOI] [PubMed] [Google Scholar]
- Gao E., Lei Y.H., Shang X., Huang Z.M., Zuo L., Boucher M., Fan Q., Chuprun J.K., Ma X.L., and Koch W.J.. 2010. A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circ. Res. 107:1445–1453. 10.1161/CIRCRESAHA.110.223925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy D.W., Colville-Nash P.R., Willis D., Chivers J., Paul-Clark M.J., and Willoughby D.A.. 1999. Inducible cyclooxygenase may have anti-inflammatory properties. Nat. Med. 5:698–701. 10.1038/9550 [DOI] [PubMed] [Google Scholar]
- Headland S.E., and Norling L.V.. 2015. The resolution of inflammation: Principles and challenges. Semin. Immunol. 27:149–160. 10.1016/j.smim.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Hubbard W.J., Choudhry M., Schwacha M.G., Kerby J.D., Rue L.W. III, Bland K.I., and Chaudry I.H.. 2005. Cecal ligation and puncture. Shock. 24:52–57. 10.1097/01.shk.0000191414.94461.7e [DOI] [PubMed] [Google Scholar]
- Ishii M., Asano K., Namkoong H., Tasaka S., Mizoguchi K., Asami T., Kamata H., Kimizuka Y., Fujiwara H., Funatsu Y., et al. . 2012. CRTH2 is a critical regulator of neutrophil migration and resistance to polymicrobial sepsis. J. Immunol. 188:5655–5664. 10.4049/jimmunol.1102330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M., and Sadikot R.T.. 2012. PGD synthase and PGD2 in immune resposne. Mediators Inflamm. 2012 10.1155/2012/503128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W.S., Kwon J.S., Kim H.B., Jeong H.Y., Kang H.J., Jeong M.H., Cho J.G., Park J.C., Kim Y.S., and Ahn Y.. 2014. A macrophage-specific synthetic promoter for therapeutic application of adiponectin. Gene Ther. 21:353–362. 10.1038/gt.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R., Patel D., Morris J.J., Rutschman R.L., and Murray P.J.. 2002. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 169:2253–2263. 10.4049/jimmunol.169.5.2253 [DOI] [PubMed] [Google Scholar]
- Lee E.H., and Rikihisa Y.. 1998. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 66:2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner F., Rauch P.J., Ueno T., Gorbatov R., Marinelli B., Lee W.W., Dutta P., Wei Y., Robbins C., Iwamoto Y., et al. . 2012. Rapid monocyte kinetics in acute myocardial infarction are sustained by extramedullary monocytopoiesis. J. Exp. Med. 209:123–137. 10.1084/jem.20111009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B.D., Clish C.B., Schmidt B., Gronert K., and Serhan C.N.. 2001. Lipid mediator class switching during acute inflammation: signals in resolution. Nat. Immunol. 2:612–619. 10.1038/89759 [DOI] [PubMed] [Google Scholar]
- Liu Y., Stewart K.N., Bishop E., Marek C.J., Kluth D.C., Rees A.J., and Wilson H.M.. 2008. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J. Immunol. 180:6270–6278. 10.4049/jimmunol.180.9.6270 [DOI] [PubMed] [Google Scholar]
- Manni S., Mauban J.H., Ward C.W., and Bond M.. 2008. Phosphorylation of the cAMP-dependent protein kinase (PKA) regulatory subunit modulates PKA-AKAP interaction, substrate phosphorylation, and calcium signaling in cardiac cells. J. Biol. Chem. 283:24145–24154. 10.1074/jbc.M802278200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M., Swirski F.K., Aikawa E., Stangenberg L., Wurdinger T., Figueiredo J.L., Libby P., Weissleder R., and Pittet M.J.. 2007. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 204:3037–3047. 10.1084/jem.20070885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakariar R., Hilliard M., Lawrence T., Trivedi S., Colville-Nash P., Bellingan G., Fitzgerald D., Yaqoob M.M., and Gilroy D.W.. 2007. Hematopoietic prostaglandin D2 synthase controls the onset and resolution of acute inflammation through PGD2 and 15-deoxyΔ12–14 PGJ2. Proc. Natl. Acad. Sci. USA. 104:20979–20984. 10.1073/pnas.0707394104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciotti E., and FitzGerald G.A.. 2011. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31:986–1000. 10.1161/ATVBAHA.110.207449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloustros E., Salpea P., Qi C.F., Gugliotti L.A., Tsang K., Liu S., Starost M.F., Morse H.C. III, and Stratakis C.A.. 2015. Hematopoietic neoplasms in Prkar2a-deficient mice. J. Exp. Clin. Cancer Res. 34:143 10.1186/s13046-015-0257-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandig H., Pease J.E., and Sabroe I.. 2007. Contrary prostaglandins: the opposing roles of PGD2 and its metabolites in leukocyte function. J. Leukoc. Biol. 81:372–382. 10.1189/jlb.0706424 [DOI] [PubMed] [Google Scholar]
- Schwab J.M., Chiang N., Arita M., and Serhan C.N.. 2007. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 447:869–874. 10.1038/nature05877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., Brain S.D., Buckley C.D., Gilroy D.W., Haslett C., O’Neill L.A., Perretti M., Rossi A.G., and Wallace J.L.. 2007. Resolution of inflammation: state of the art, definitions and terms. FASEB J. 21:325–332. 10.1096/fj.06-7227rev [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A., and Mantovani A.. 2012. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122:787–795. 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A., Invernizzi P., and Mantovani A.. 2014. Macrophage plasticity and polarization in liver homeostasis and pathology. Hepatology. 59:2034–2042. 10.1002/hep.26754 [DOI] [PubMed] [Google Scholar]
- Skroblin P., Grossmann S., Schäfer G., Rosenthal W., and Klussmann E.. 2010. Mechanisms of protein kinase A anchoring. Int. Rev. Cell Mol. Biol. 283:235–330. 10.1016/S1937-6448(10)83005-9 [DOI] [PubMed] [Google Scholar]
- Su Y., Dostmann W.R., Herberg F.W., Durick K., Xuong N.H., Ten Eyck L., Taylor S.S., and Varughese K.I.. 1995. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science. 269:807–813. 10.1126/science.7638597 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Zhang P., Steichen J.M., Keshwani M.M., and Kornev A.P.. 2013. PKA: lessons learned after twenty years. Biochim. Biophys. Acta. 1834:1271–1278. 10.1016/j.bbapap.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte C.S., Bishop E.T., Rückerl D., Gaspar-Pereira S., Barker R.N., Allen J.E., Rees A.J., and Wilson H.M.. 2011. Suppressor of cytokine signaling (SOCS)1 is a key determinant of differential macrophage activation and function. J. Leukoc. Biol. 90:845–854. 10.1189/jlb.1110644 [DOI] [PubMed] [Google Scholar]
- Wu J., Brown S.H., von Daake S., and Taylor S.S.. 2007. PKA type IIα holoenzyme reveals a combinatorial strategy for isoform diversity. Science. 318:274–279. 10.1126/science.1146447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Anzai A., Katsumata Y., Matsuhashi T., Ito K., Endo J., Yamamoto T., Takeshima A., Shinmura K., Shen W., et al. . 2013. Temporal dynamics of cardiac immune cell accumulation following acute myocardial infarction. J. Mol. Cell. Cardiol. 62:24–35. 10.1016/j.yjmcc.2013.04.023 [DOI] [PubMed] [Google Scholar]
- Yang P., An H., Liu X., Wen M., Zheng Y., Rui Y., and Cao X.. 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nat. Immunol. 11:487–494. 10.1038/ni.1876 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.