Abstract
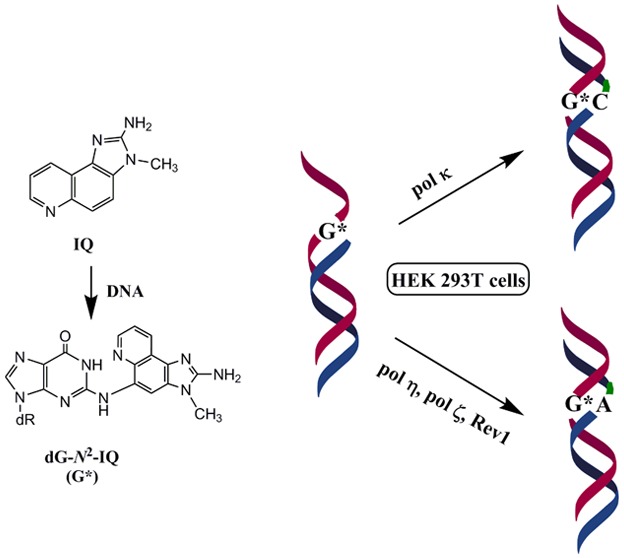
Translesion synthesis (TLS) of the N2-2′-deoxyguanosine (dG-N2-IQ) adduct of the carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) was investigated in human embryonic kidney 293T cells by replicating plasmid constructs in which the adduct was individually placed at each guanine (G1, G2, or G3) of the NarI sequence (5′-CG1G2CG3CC-3′). TLS efficiency was 38%, 29%, and 25% for the dG-N2-IQ located at G1, G2, and G3, respectively, which suggests that dG-N2-IQ is bypassed more efficiently by one or more DNA polymerases at G1 than at either G2 or G3. TLS efficiency was decreased 8–35% in cells with knockdown of pol η, pol κ, pol ι, pol ζ, or Rev1. Up to 75% reduction in TLS occurred when pol η, pol ζ, and Rev1 were simultaneously knocked down, suggesting that these three polymerases play important roles in dG-N2-IQ bypass. Mutation frequencies (MFs) of dG-N2-IQ at G1, G2, and G3 were 23%, 17%, and 11%, respectively, exhibiting a completely reverse trend of the previously reported MF of the C8-dG adduct of IQ (dG-C8-IQ), which is most mutagenic at G3 ((2015) Nucleic Acids Res. 43, 8340−8351). The major type of mutation induced by dG-N2-IQ was targeted G → T, as was reported for dG-C8-IQ. In each site, knockdown of pol κ resulted in an increase in MF, whereas MF was reduced when pol η, pol ι, pol ζ, or Rev1 was knocked down. The reduction in MF was most pronounced when pol η, pol ζ, and Rev1 were simultaneously knocked down and especially when the adduct was located at G3, where MF was reduced by 90%. We conclude that pol κ predominantly performs error-free TLS of the dG-N2-IQ adduct, whereas pols η, pol ζ, and Rev1 cooperatively carry out the error-prone TLS. However, in vitro experiments using yeast pol ζ and κ showed that the former was inefficient in full-length primer extension on dG-N2-IQ templates, whereas the latter was efficient in both error-free and error-prone extensions. We believe that the observed differences between the in vitro experiments using purified DNA polymerases, and the cellular results may arise from several factors including the crucial roles played by the accessory proteins in TLS.
Introduction
2-Amino-3-methylimidazo[4,5-f]quinoline (IQ), a heterocyclic amine found in cooked meat,1 is generated by the Maillard reaction upon pyrolysis of reducing sugars and amino acids. Although the exposure level of IQ to humans is low (∼60 ng/day),2 it is believed to be involved in the development of human cancer.3 In addition to cooked meats,4,5 it is present in tobacco smoke.6 IQ is a potent mutagen in Ames’ Salmonella typhimurium assay and is “reasonably anticipated to be a human carcinogen” according to the National Toxicology Program.3,7,8 While IQ induces tumors in various organs in rats, mice, and primates, it is principally a liver carcinogen.9−11
In cells, IQ must undergo bioactivation prior to DNA adduction.12 IQ is converted to its N-hydroxylamine by cytochrome P450 1A2, which is acetylated by N-acetyl transferase, predominantly NAT2 (Figure 1).13,14 The N-acetoxy-IQ (or the nitrenium ion formed from it) is the ultimate carcinogen, that forms DNA adducts (Figure 1).15 Metabolic activation of IQ also causes oxidative DNA damage, but this process is unlikely to play a role in IQ carcinogenesis.16 Metabolically activated IQ generates a main DNA adduct (dG-C8-IQ) at the C8-position of 2′-deoxyguanosine (dG) as well as a less abundant N2-dG adduct (dG-N2-IQ) (Figure 1).17−19 However, the latter is slowly repaired and persists for a long time in rodents.20 In Ames’ test, IQ induces high levels of dinucleotide deletions in the CpG repeat sequences of the HisD3052 target sequence (5′-CGCGCGCG-3′), which is common for many aromatic amines and nitroaromatic compounds.8,21 Single adduct mutagenesis studies showed that the C8-dG adducts derived from metabolically activated 1-nitropyrene, and 1,6- and 1,8-dinitropyrenes cause two-base deletions in repetitive CpG sequences in bacteria, whereas mostly base substitutions occur in simian kidney cells.22−24 Like the CpG repeat sequence, the NarI restriction site 5′-CG1G2CG3CC-3′ is a hot-spot for frameshift mutagenesis for the C8-dG adduct of N-acetyl-2-aminofluorene, primarily when the adduct is positioned at G3, but only base substitutions occur in simian kidney cells.25−27 The diversity in the types and frequencies of mutations caused by the DNA lesions in different cells and organisms may stem from the differences in DNA polymerases (pols) that bypass them.28 Bulky DNA adducts such as the ones formed by IQ are usually strong blocks of replicative DNA pols, but a group of specialized translesion synthesis (TLS) DNA pols can bypass them.29−32 In eukaryotic cells, TLS is carried out by the Y-family pols, pol η, pol κ, pol ι, and Rev1 and the B-family enzyme pol ζ.32,33 The TLS pols are characterized by high error rates on undamaged templates, although some of them are equipped to bypass specific DNA damages efficiently and with high fidelity.34,35 In eukaryotic cells, often efficient TLS involves two sequential steps.36,37 In the first step, the stalled replicative pol is replaced by a TLS pol, which inserts a nucleotide opposite the DNA lesion. In the subsequent step, this inserter pol may continue with the lesion bypass or be substituted by another TLS pol for extension of the primer a few nucleotides past the lesion site. This TLS pol is eventually replaced by the replicative pol to restart and continue DNA synthesis. In a cell, accessory proteins, such as proliferating cell nuclear antigen (PCNA) that is activated by monoubiquitination after DNA damage, are also required for efficient TLS.38,39
Figure 1.
Metabolic activation and the DNA adduct formation by IQ.
In vitro studies established that pol δ, a replicative B-family pol, cannot bypass the IQ adducts.40 In the NarI restriction site, human pol η (hpol η) is able to extend primers beyond both dG-C8-IQ and dG-N2-IQ more efficiently than pol κ and much more efficiently than pol ι and pol δ.40 TLS by pol η is largely error-free for dG-C8-IQ. A two-base deletion occurred with the dG-N2-IQ adduct at the G3 site, while the product was error-free when the adduct was located at G1 but pol η failed to carry out further extension. In a cell, however, TLS is far more complex, owing to the involvement of multiple proteins, and the mode of selection and recruitment of the TLS pol that bypasses the DNA lesion has not yet been established.41 In a recent work, we found that most of the error-prone TLS of dG-C8-IQ is performed by pol κ and pol ζ, whereas pol η carries out error-free bypass in human cells.42 We also determined that both TLS and the mutation frequency (MF) of dG-C8-IQ situated at the three guanines of the NarI site are significantly different.42 In the current investigation, using an approach similar to the prior study of dG-C8-IQ, we have explored the roles of different TLS pols in bypassing dG-N2-IQ located in the three different guanine sites of the NarI restriction sequence. We report herein that dG-N2-IQ is mutagenic in human embryonic kidney (HEK) 293T cells and induces mainly G → T transversions but the MF is different at the three guanine sites. We also show that pol κ is more efficient in its TLS than the other pols and that it performs TLS principally in an error-free manner. In contrast, pol η, pol ζ, and Rev1 cooperatively perform the majority of the mutagenic bypass.
Experimental Procedures
Materials
Yeast pol ζ, human pol κ, and Rev1 were obtained from Enzymax (Lexington, KY). The dNTP solutions (100 mM) were from New England Biolabs (Ipswich, MA) or GE Healthcare (Piscataway, NJ). [γ-32P]ATP was obtained from PerkinElmer (Waltham, MA). dG-N2-IQ modified oligonucleotides were synthesized according to a literature procedure.40 Unmodified oligonucleotides were from Midland Certified Reagents (Midland, TX).
siRNAs
Synthetic siRNA duplexes against POLH, POLK, POLI, REV1 and negative control (NC) siRNA were from Qiagen (Valencia, CA). siRNA for REV3 was acquired from Integrated DNA Technologies (Coralville, IA). Sequences of all the siRNAs are as reported in ref (43).
Methods
Construction and Characterization of dG-N2-IQ-Containing Plasmids and Their Replication in HEK293T Cells
Site-specific adduct-containing pMS2 vectors with neomycin and ampicillin resistance genes were constructed in a manner similar to the construction of the dG-C8-IQ-containing vectors.42 Transfection of 50 ng of each construct was performed in HEK293T cells, after they were grown to ∼90% confluency, using 6 μL of Lipofectamine cationic lipid reagent (Invitrogen, Carlsbad, CA). Subsequently, the cells were grown at 37 °C in 5% CO2 for 48 h, and the plasmid DNA was isolated and purified.44 It was used to transform E. coli DH10B, and the progeny were examined as described.24,45
Determination of TLS Efficiency
Single-stranded pMS2 DNA construct containing a different DNA sequence where the 12-mer oligonucleotide was inserted (i.e., 5′-GTGCGTGTTTGT-3′ in place of 5′-CTCG1G2CG3CCATC-3′) into the gapped plasmid was mixed with the adducted or control constructs (1:1). The DNA mixture was used to transfect HEK293T cells and processed as described above. Oligonucleotide probes for both the wild type and the mutant plasmid were used to analyze the progeny. The unmodified DNA was used an internal control so that TLS efficiency could be determined from the percentages of the colonies originating from the adduct-containing plasmid relative to that from the unmodified plasmid.
Mutational Analyses of TLS Products from Human Cells with Pol Knockdowns
HEK293T cells were transfected with synthetic siRNA duplexes and plated in 6-well plates at 50% confluence. Following a 24-h incubation, they were seeded in 24-well plates at 70% confluence a day before transfection of the plasmid. Subsequently, cells were cotransfected with another aliquot of siRNA and the adducted or control plasmid. Following a 24-h incubation, plasmid DNA was isolated and used to transform E. coli as described above. RT-PCR and Western blotting were performed as reported.43 Mutations were determined by oligonucleotide hybridization followed by DNA sequence analyses around the NarI site. Two 15-mer probes, complementary to part of the inserted oligonucleotide sequence on the left or right side of the plasmid where ligation took place and a part of the plasmid, were used to select plasmids containing the correct insert, and transformants that did not hybridize with both the left and right probes were omitted. A third probe containing the complementary 14-mer wild type sequence encompassing the entire inserted DNA sequence was used to analyze the progeny plasmids. Any clone that failed to hybridize with this probe was considered a putative mutant and subjected to DNA sequencing.
Single-Nucleotide Incorporation Assays by hRev1
32P-Labeling, annealing, and extension reactions of the primers on the unmodified or the dG-N2-IQ modified template by Rev1 were carried out in the presence of dCTPs in a manner similar to that in ref (42). Details are provided in the SI.
Full-Length Extension Assay by yPol ζ or Pol κ with All Four dNTPs
Annealing the 32P-labeled 0-primers (with a 3′-C or A) to the unmodified or dG-N2-IQ modified template and extension in the presence of all four dNTPs (100 μM each) at 37 °C for 5 h followed the method reported earlier.42 Details are provided in the SI.
Results
Roles of Pol η, κ, and ζ in TLS of dG-N2-IQ
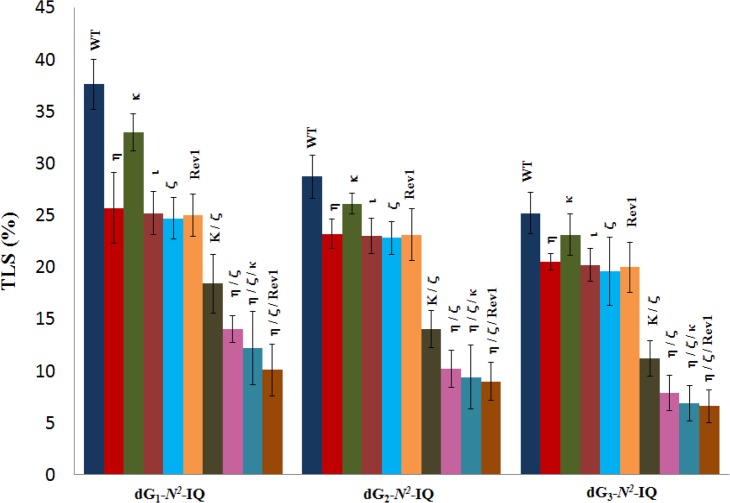
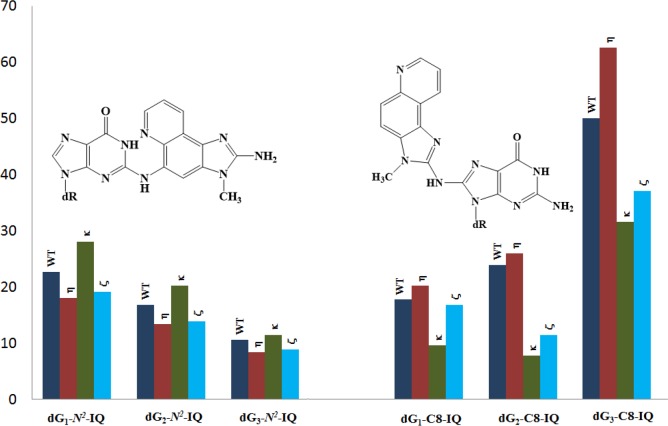
The construction of the adduct containing vector, its replication in human cells, and progeny analysis followed an approach described earlier.24,42,46 In order to identify the pols involved in TLS of dG-N2-IQ in human cells, the siRNA knockdown approach was used to limit the expression of specific TLS pol(s) in HEK293T cells.43 An internal control plasmid, containing an undamaged 12-mer of different sequence in place of the 12-mer adducted oligonucleotide, was mixed with the adduct-containing plasmid before transfection. TLS efficiency was calculated by the percentages of the colonies originating from each dG-N2-IQ-containing plasmid relative to that from the unmodified internal control. In HEK293T cells, the frequency of TLS was 38%, 29%, and 25% for the dG-N2-IQ placed at G1, G2, and G3, respectively, relative to 100% progeny derived from the undamaged plasmid construct (Figure 2). This suggests that dG-N2-IQ is bypassed considerably more efficiently by one or more pols at G1 than at either G2 or G3. TLS efficiency was decreased in cells with knockdown of each TLS pol. Only 8–12% reduction in TLS efficiency was observed in cells with knockdown of pol κ, whereas knockdown of pol η, pol ι, pol ζ, or Rev1 resulted in 20–35% reduction in TLS. In the pol knockdown experiments, the location of the adduct did not significantly influence the TLS efficiency. A more prominent effect on TLS was observed upon simultaneous knockdown of pol η/pol ζ and pol κ/pol ζ (Figure 2). Simultaneous knockdown of pol κ and pol ζ resulted in approximately 50% reduction in TLS, but simultaneous knockdown of pol η/pol ζ showed a more noticeable outcome, resulting in up to 70% reduction in TLS when dG-N2-IQ was located at G3. The most significant reduction (∼75%) in TLS occurred when pol η, pol ζ, and Rev1 were simultaneously knocked down. We conclude that each TLS pol takes part in TLS of dG-N2-IQ and that pol η, pol ζ, and Rev1 play complementary roles in TLS of dG- N2-IQ.
Figure 2.
Effects of siRNA knockdowns of TLS pols on the extent of replicative bypass of dG-N2-IQ. Percent TLS in various pol knockdowns was measured relative to an internal control in which a different 12-mer oligonucleotide was inserted (i.e., 5′-GTGCGTGTTTGT-3′ in place of 5′-CTCG1G2CG3CCATC-3′) into the gapped plasmid in a manner similar to the construction of the dG- N2-IQ (or control) construct. The data represents the mean of results from two independent experiments. HEK293T cells are treated with negative control (NC) siRNA, whereas the other single, double, and triple pol(s) knockdowns are indicated above the bar.
Error-Free and Error-Prone Bypass of dG-N2-IQ in HEK293T Cells
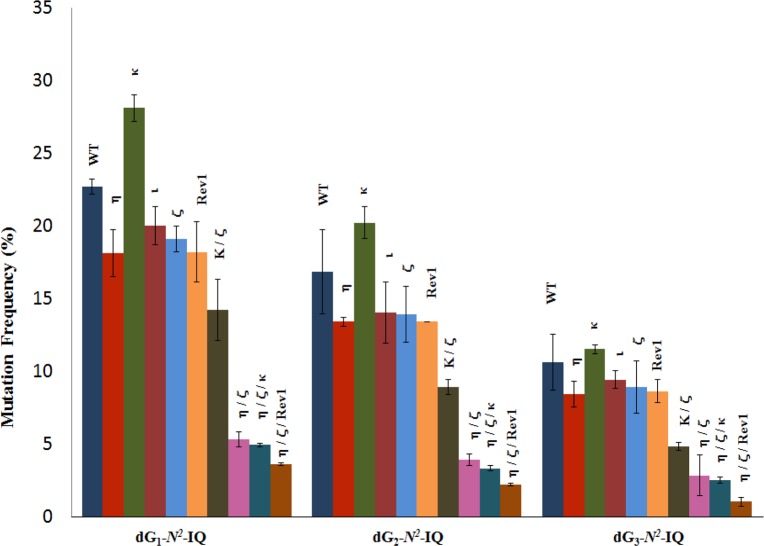
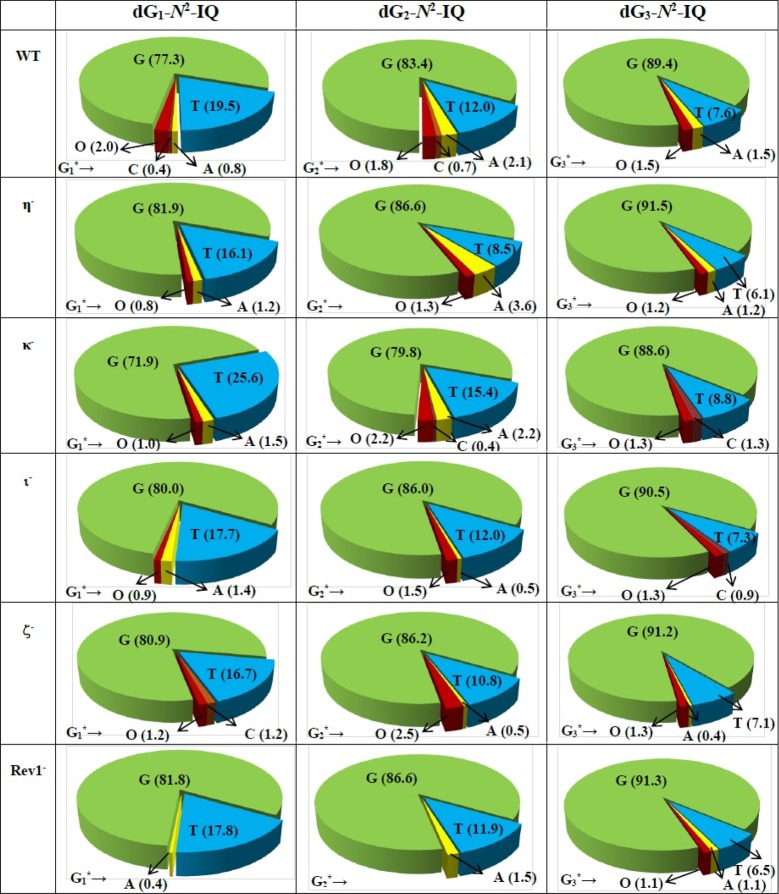
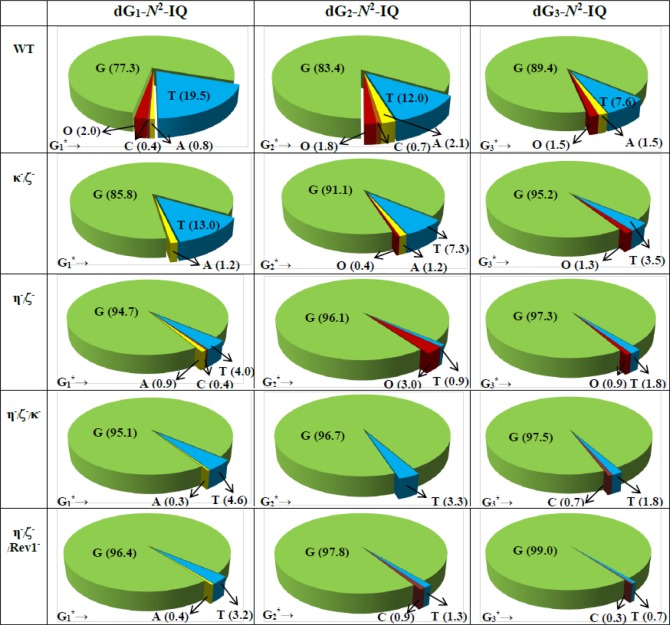
DNA sequence analysis showed that dG-N2-IQ is mutagenic in HEK293T cells in each of the three sites (Figure 3). As in the case of dG-C8-IQ, MFs were different at the three guanines, but they exhibited a completely reverse trend here. Unlike the dG-C8-IQ, in which the adduct at G3 is most mutagenic, MF of dG-N2-IQ at G1 (23%) was higher than that at G2 (17%), which, in turn, was higher than the adduct located at G3 (11%). However, the major types of mutations for dG-C8-IQ and dG-N2-IQ were the same in that they both induced targeted G → T transversions (Figures 4 and 5). In each site, knockdown of pol κ resulted in an increase in MF, whereas MF was reduced when pol η, pol ι, pol ζ, or Rev1 was knocked down (Figure 3). This suggests that pol κ is involved in a greater fraction of error-free bypass of dG-N2-IQ, whereas pol η, pol ι, pol ζ, and Rev1 each participate in the error-prone TLS of this adduct. Simultaneous knockdown of two and three pols resulted in further reduction in MF. The diminution in MF was most pronounced when pol η, pol ζ, and Rev1 were simultaneously knocked down and especially when the adduct was located at G3 (Figures 3 and 5), where 90% reduction in MF was observed. We conclude that the error-prone TLS of the dG-N2-IQ adduct occurs cooperatively by pols η, pol ζ, and Rev1.
Figure 3.
Mutational frequency of dG-N2-IQ derived from the progeny from G1, G2, and G3 constructs in HEK293T cells, also transfected with NC siRNA (WT) or siRNA for single, double, or triple pol(s) knockdowns, is shown. The data represent the average of two independent experiments (presented in Table S1A–J in SI).
Figure 4.
Types and frequencies of mutations induced by dG-N2-IQ in the progeny from the G1, G2, and G3 constructs in HEK293T cells also transfected with NC siRNA (293T) or siRNA for single pol knockdowns are shown in a pie chart. O represents other (i.e. semi-targeted) mutations. The data represent the average of two independent experiments (presented in Table S1A–F in the SI).
Figure 5.
Types and frequencies of mutations induced by dG-N2-IQ in the progeny from the G1, G2, and G3 constructs in HEK293T cells also transfected with NC siRNA (293T) or siRNA for double or triple pol(s) knockdowns are shown in a pie chart. O represents other (i.e. semi-targeted) mutations. The data represent the average of two independent experiments (presented in Table S1G–J in the SI).
In Vitro TLS of dG-N2-IQ by Eukaryotic Pols
The in vitro replication of dG-N2-IQ at the iterated G3- and noniterated G1-positions of the NarI recognition sequence has been previously examined with purified human TLS pols η, κ, and ι, which showed that hpol η is the most efficient in bypassing both dG-C8-IQ and dG-N2-IQ.40 Now, we also examined hRev1, which was reasonably efficient at inserting dCTP opposite the lesions (Figure S1 of the SI). In contrast, ypol ζ was unable to insert a nucleotide opposite the dG-N2-IQ in either sequence context. Interestingly, the efficiency of hRev1 was higher when the modification was at the G1-position than for G3 (Figure S1 of the SI). These results suggest that Rev1 could participate in nonmutagenic TLS. Further, the higher efficiency for insertion at G1 suggested a more prominent role in nonmutagenic bypass at this position.
In previous studies, steady-state kinetic analysis showed a significant sequence context effect for the misinsertion of dATP opposite the dG-N2-IQ adduct by hpol η.40 The misinsertion frequency for dATP opposite dG-N2-IQ was 0.042 and 0.71 when the lesion was situated at G3-, and G1-positions, respectively. Cellular data showing that more G → T mutations occurred at G1 relative to G3 is consistent with the in vitro kinetic result. However, only replication products from initial insertion of dCTP opposite the lesion were observed by mass spectrometry, suggesting that hpol η is inefficient in extending from an A opposite the dG-N2-IQ adduct.40 Pol ζ is typically implicated as an extender from the adduct:N template primer terminus. We, therefore, examined the ability of ypol ζ to extend the dG-N2-IQ adduct when paired with C or mispaired with A. We observed that ypol ζ was able to extend the primer by only one nucleotide in both sequence contexts (Figure 6). The one nucleotide extension appeared to be more efficient for the dG-N2-IQ:A mispair, particularly for the G3-modified template. In contrast, hpol κ was efficient at extending from the A opposite the dG-N2-IQ adduct in both sequence contexts (Figure 7). This suggests that pol κ may play a role in mutagenic bypass of the dG-N2-IQ adduct by extending from a mispaired A opposite the lesion. This would be in addition to its primary role in error-free replication of the lesion.
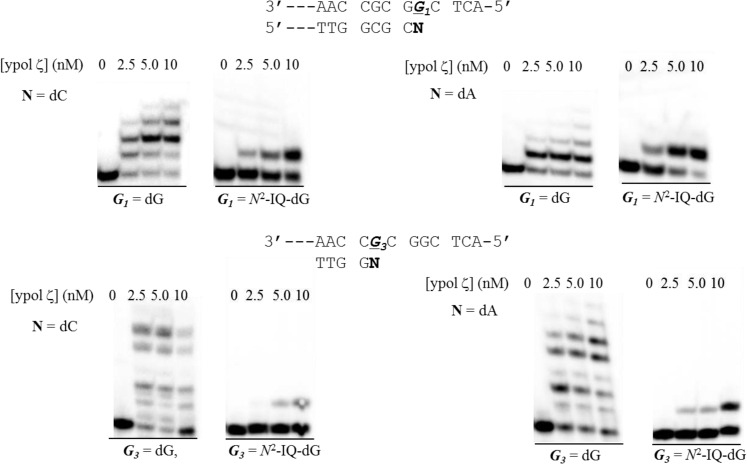
Figure 6.
Yeast pol ζ extension of a control and dG-N2-IQ modified template at the G1 (top) or G3 (bottom) positions when paired by dC (left) or mispaired with dA (right). Reactions were run with 10 nM DNA, 100 μM dNTPs, and 0, 2.5, 5.0, and 10 nM of ypol ζ.
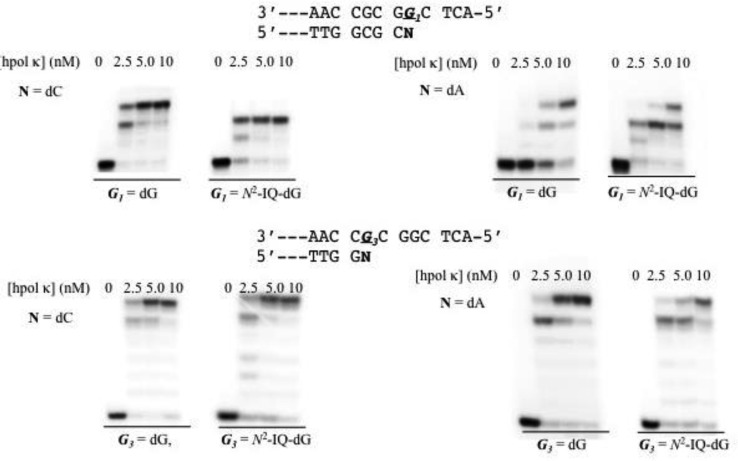
Figure 7.
Human pol κ extension of a control and dG-N2-IQ modified template at the G1 (top) or G3 (bottom) positions when paired by dC (left) or mispaired with dA (right). Reactions were run with 10 nM DNA, 100 μM dNTPs, and 0, 2.5, 5.0, and 10 nM of hpol κ.
Discussion
TLS Pols Active in Bypassing dG-N2-IQ
Like many other bulky DNA adducts, both dG-C8-IQ and dG-N2-IQ are strong blocks of replication in vitro.47 Pol δ was completely inhibited by these adducts, whereas hpol η could extend primers beyond the adduct site more proficiently than hpol κ and significantly more efficiently than hpol ι for each adducted G in the NarI sequence.40
As reported earlier with the dG-C8-IQ, our current study suggests that each TLS pol we examined, including pol η, pol κ, pol ι, and Rev1 of the Y-family and pol ζ of the B-family, has a role in TLS of dG-N2-IQ (Figure 2). Nevertheless, as with other bulky DNA adducts,43 none of the TLS pols was indispensable for TLS of dG-N2-IQ. Knockdown of multiple pols resulted in a more marked effect on TLS. For example, concurrent knockdown of pol η, pol ζ, and Rev1 reduced the TLS by ∼75%, suggesting that they work cooperatively.
The results also establish that the magnitudes of the TLS of dG-N2-IQ differ at different sequence contexts (Figure 2). The major conformations of dG-N2-IQ in the three different sequences were found to be similar, adopting an intercalated conformation as determined when positioned at G1 and G3 of the NarI sequence.48,49 However, these structures were determined on a fully duplex DNA, which may not be reflective of the structure at a replication fork bound to a pol. TLS pols have spacious active sites that can accommodate various conformation of the DNA adduct, and subtle conformational differences of the DNA adduct may influence their ability to bypass. Thus, the context effect of TLS likely relates to a small difference in the conformation of the DNA adduct. It is noteworthy that for dG-N2-IQ, the efficiency of TLS when the adduct was positioned at G1 was higher than that at G2, which, in turn, was higher than that when it was located at G3. This pattern was exactly opposite of what was observed with dG-C8-IQ.42
Error-Free versus Error-Prone TLS
Similar to the TLS result, MFs of dG-N2-IQ were different in the three sites, and it was most mutagenic when positioned at G1. Of the several single pol knockdown experiments, an increase in MF was seen in pol κ-knockdown cells, whereas the reduction in MF was approximately the same upon knockdown of pol η, pol ι, pol ζ, or Rev1 (Figures 3 and 4). Error-free TLS, therefore, is largely carried out by pol κ, as reported for many other dG-N2 adducts.40,50−52 However, pol η and pol ζ were found to be the most critical pols involved in erroneous TLS, as demonstrated by the greater synergy in lowering MF upon simultaneous knockdown of these two pols (Figures 3 and 5). A previous in vitro study using a single pol, the steady-state kinetic insertion frequency of A relative to C opposite dG-N2-IQ by hpol η at G1 was 71%, whereas the same at G3 was only 4%, suggesting that hpol η is more error-prone at G1,40 as observed in our current work. However, the extension of the dG-N2-IQ:A pair was inefficient by hpol η. Since pol ζ was reported to extend efficiently after a nucleotide has been inserted opposite a DNA lesion,53−55 based on this in vitro report and our current result, we postulate that pol η misincorporates dATP opposite dG-N2-IQ and the mispair is extended by pol ζ. Rev1 also has a role since further decrease in MF was noted when pol η, pol ζ, and Rev1 were simultaneously knocked down. Unfortunately, we were unable to validate the cellular results by in vitro experiments using ypol ζ, which was only able to carry out one base extension of the dG-N2-IQ:C and dG-N2-IQ:A pair at both G1 and G3 sites (Figure 6). In fact, hpol κ was more efficient in extension than ypol ζ. However, the ypol ζ we employed contained the catalytic subunit Rev3 and the accessory subunit Rev7 only, whereas Pol31 and Pol32 were reported to be indispensable for pol ζ function in TLS in yeast cells.56 Furthermore, despite sequence homologies, the mammalian Rev3 is twice as large as its yeast homologue. Therefore, one can anticipate substantial differences in TLS of dG-N2-IQ by the four subunit (Rev3, Rev7, PolD2, and PolD3) human pol ζ relative to the yeast enzyme.57
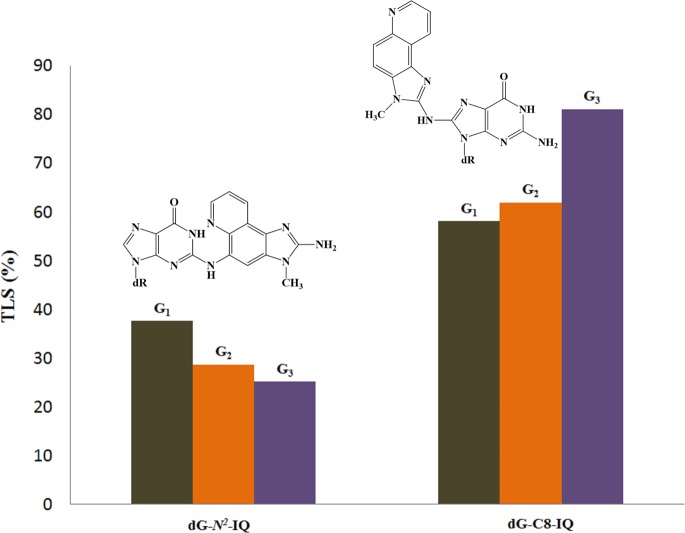
A comparison of the TLS of dG-N2-IQ with dG-C8-IQ is shown in Figure 8, demonstrating an opposite trend at the three sites by these two DNA adducts formed by the same carcinogen. Likewise, Figure 9 compares the MF of these two adducts located at G1, G2, and G3, which also exhibits a reverse trend. While an explanation for these trends must await future structural experiments, one particular noteworthy aspect in Figure 9 is that the role of pol η and pol κ were switched in the TLS of these two adducts. Pol η carried out predominantly error-prone and error-free TLS of dG-N2-IQ and dG-C8-IQ, respectively, whereas pol κ did exactly the opposite. It is also notable in Figure 9 that MF of dG-N2-IQ was only 28% higher than that of dG-C8-IQ at G1. In stark contrast, MF of dG-C8-IQ was 450% higher than that of dG-N2-IQ at G3. Since the decrease in TLS accompanied a decrease in MF from G1 to G2 to G3, one can normalize the MF by dividing it with TLS. The MF/TLS assessment provided values of 0.58, 0.52, and 0.36 for G1, G2, and G3, respectively, which may be considered a mutation per the TLS event. However, they still followed the same trend as MF alone. Taken together, these results underscore the importance of DNA sequence context on mutagenesis by carcinogen-DNA adducts. For both IQ-DNA adducts, MF was higher at the site where more facile TLS occurred.
Figure 8.
Comparison of TLS of dG-N2-IQ and dG-C8-IQ.
Figure 9.
Comparison of MF of dG-N2-IQ and dG-C8-IQ derived from the progeny from G1, G2, and G3 constructs in HEK293T cells, also transfected with NC siRNA (WT) or siRNA for single knockdowns of pol η, pol κ, or pol ζ.
High-resolution NMR studies revealed that dG-N2-IQ adopts a base-displaced intercalated conformation at both G1 and G3 but that in each case the modified guanine remains in the anti conformation about the glyosidic bond.48,49 In contrast, the torsion angle is in syn conformation for the dG-C8-IQ adduct at each of the three guanine sites of NarI with the modified guanine displaced into the major groove, but base-displaced intercalated IQ is favored only at G3, in which it is flanked by guanines on both sides in the complementary strand,58,59 and where it is most error-prone. When dG-C8-IQ was positioned at G1 or G2, the IQ moiety was characterized as a minor groove bound.59 DNA adducts localized in the solvent-exposed major groove, as in the case of dG-C8-IQ at G3, have been suggested to be better tolerated than adducts located in the minor groove as in the case of dG-N2-IQ that interact with the polymerase surface,60 although this observation was made with a replicative pol. A more pertinent study is the one in which the major benzo[a]pyrene adduct at the dG-N2 position in the minor groove is accommodated in anti conformation in the active site of pol κ.61 The hydrophobic polycyclic ring system of the adduct is stabilized by the protein residues along the minor groove side to allow Watson–Crick base pairing with an incoming dCTP for accurate replication. The N-clasp domain of pol κ supports an open conformation, and similar interactions can be anticipated for the error-free bypass of dG-N2-IQ. When the adduct maintains its syn conformation, efficient destacking of the polycyclic aromatic moiety on top of the primer–template junction was shown to be a necessary step for efficient TLS by pol η.62 The bypass, in such a case, is anticipated to be slow and error-prone. We also cannot rule out the possibility that of the various conformers of dG-N2-IQ at the three guanines of NarI sequence, the TLS pol can selectively bypass a minor conformer, which may escape detection in adduct structure investigations. For the erroneous TLS of dG-N2-IQ, the base-displaced intercalated anti orientation of the adduct may comprise a two or more polymerase bypass model,36,37,63,64 in which pol η incorporates dATP and pol ζ carries out extension of the mispair, whereas Rev1 is involved in a noncatalytic role.
In conclusion, dG-N2-IQ is mutagenic in HEK293T cells inducing mainly G → T transversions; no frameshift deletions were observed. In the NarI sequence, the lesion is most mutagenic when located at G1. Of the bypass pols, pol κ performs TLS of dG-N2-IQ in an error-free manner. In contrast, pol η, pol ζ, and Rev1 cooperatively carry out the mutagenic TLS.
Glossary
Abbreviations
- TLS
translesion synthesis
- IQ
2-amino-3-methylimidazo[4,5-f]quinoline
- HEK
human embryonic kidney
- MF
mutation frequency
- pol
polymerase
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.6b00221.
In vitro experimental details, autoradiogram of the gel electrophoresis of the insertion of dCTP by human Rev1 opposite a control dG or dG-N2-IQ, and tables detailing mutation data (PDF)
Author Present Address
§ A.B.: Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213.
Author Present Address
∥ A.D.: Nitto Denko Avecia Inc., Cincinnati, OH 45215.
This work was supported by the NIH grants ES09127 and ES021762 to A.K.B. and ES016561, P30 ES000267, and P30 CA068485 to C.J.R.
The authors declare no competing financial interest.
Supplementary Material
References
- Sugimura T.; Wakabayashi K.; Nakagama H.; Nagao M. (2004) Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 95, 290–299. 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M.; Hanaoka T.; Nishioka S.; Kataoka H.; Tsugane S. (2002) Estimation of dietary HCA intakes in a large-scale population-based prospective study in Japan. Mutat. Res., Fundam. Mol. Mech. Mutagen. 506–507, 233–241. 10.1016/S0027-5107(02)00170-7. [DOI] [PubMed] [Google Scholar]
- Program N. T. (2005) National Toxicology Program, Report on Carcinogenesis, 11th ed., U.S. Department of Health and Human Services, Public Health Service., Research Triangle Park, NC. [Google Scholar]
- Kataoka H.; Nishioka S.; Kobayashi M.; Hanaoka T.; Tsugane S. (2002) Analysis of mutagenic heterocyclic amines in cooked food samples by gas chromatography with nitrogen-phosphorus detector. Bull. Environ. Contam. Toxicol. 69, 682–689. 10.1007/s00128-002-0115-5. [DOI] [PubMed] [Google Scholar]
- Felton J. S.; Knize M. G.; Salmon C. P.; Malfatti M. A.; Kulp K. S. (2002) Human exposure to heterocyclic amine food mutagens/carcinogens: relevance to breast cancer. Environ. Mol. Mutagen. 39, 112–118. 10.1002/em.10070. [DOI] [PubMed] [Google Scholar]
- Yamashita M.; Wakabayashi K.; Nagao M.; Sato S.; Yamaizumi Z.; Takahashi M.; Kinae N.; Tomita I.; Sugimura T. (1986) Detection of 2-amino-3-methylimidazo[4,5-f]quinoline in cigarette smoke condensate. Jpn. J. Cancer Res. 77, 419–422. [PubMed] [Google Scholar]
- Sugimura T.; Sato S. (1983) Mutagens-carcinogens in foods. Cancer Res. 43, 2415s–2421s. [PubMed] [Google Scholar]
- Sugimura T. (1997) Overview of carcinogenic heterocyclic amines. Mutat. Res., Fundam. Mol. Mech. Mutagen. 376, 211–219. 10.1016/S0027-5107(97)00045-6. [DOI] [PubMed] [Google Scholar]
- Ohgaki H.; Kusama K.; Matsukura N.; Morino K.; Hasegawa H.; Sato S.; Takayama S.; Sugimura T. (1984) Carcinogenicity in mice of a mutagenic compound, 2-amino-3-methylimidazo[4,5-f]quinoline, from broiled sardine, cooked beef and beef extract. Carcinogenesis 5, 921–924. 10.1093/carcin/5.7.921. [DOI] [PubMed] [Google Scholar]
- Takayama S.; Nakatsuru Y.; Masuda M.; Ohgaki H.; Sato S.; Sugimura T. (1984) Demonstration of carcinogenicity in F344 rats of 2-amino-3-methyl-imidazo[4,5-f]quinoline from broiled sardine, fried beef and beef extract. Gann 75, 467–470. [PubMed] [Google Scholar]
- Adamson R. H.; Thorgeirsson U. P.; Snyderwine E. G.; Thorgeirsson S. S.; Reeves J.; Dalgard D. W.; Takayama S.; Sugimura T. (1990) Carcinogenicity of 2-amino-3-methylimidazo[4,5-f]quinoline in nonhuman primates: induction of tumors in three macaques. Jpn. J. Cancer Res. 81, 10–14. 10.1111/j.1349-7006.1990.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe Y.; Shimada M.; Kamataki T.; Kato R. (1983) Microsomal activation of 2-amino-3-methylimidazo[4,5-f]quinoline, a pyrolysate of sardine and beef extracts, to a mutagenic intermediate. Cancer Res. 43, 5768–5774. [PubMed] [Google Scholar]
- Boobis A. R.; Lynch A. M.; Murray S.; de la Torre R.; Solans A.; Farre M.; Segura J.; Gooderham N. J.; Davies D. S. (1994) CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 54, 89–94. [PubMed] [Google Scholar]
- Hein D. W.; Doll M. A.; Rustan T. D.; Gray K.; Feng Y.; Ferguson R. J.; Grant D. M. (1993) Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis 14, 1633–1638. 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. (2002) N-hydroxyarylamines. Drug Metab. Rev. 34, 607–623. 10.1081/DMR-120005663. [DOI] [PubMed] [Google Scholar]
- Wei M.; Wanibuchi H.; Nakae D.; Tsuda H.; Takahashi S.; Hirose M.; Totsuka Y.; Tatematsu M.; Fukushima S. (2011) Low-dose carcinogenicity of 2-amino-3-methylimidazo[4,5-f ]quinoline in rats: Evidence for the existence of no-effect levels and a mechanism involving p21(Cip/WAF1). Cancer Sci. 102, 88–94. 10.1111/j.1349-7006.2010.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyderwine E. G.; Roller P. P.; Adamson R. H.; Sato S.; Thorgeirsson S. S. (1988) Reaction of N-hydroxylamine and N-acetoxy derivatives of 2-amino-3-methylimidazolo[4,5-f]quinoline with DNA. Synthesis and identification of N-(deoxyguanosin-8-yl)-IQ. Carcinogenesis 9, 1061–1065. 10.1093/carcin/9.6.1061. [DOI] [PubMed] [Google Scholar]
- Snyderwine E. G.; Yamashita K.; Adamson R. H.; Sato S.; Nagao M.; Sugimura T.; Thorgeirsson S. S. (1988) Use of the 32P-postlabeling method to detect DNA adducts of 2-amino-3-methylimidazolo[4,5-f]quinoline (IQ) in monkeys fed IQ: identification of the N-(deoxyguanosin-8-yl)-IQ adduct. Carcinogenesis 9, 1739–1743. 10.1093/carcin/9.10.1739. [DOI] [PubMed] [Google Scholar]
- Turesky R. J.; Rossi S. C.; Welti D. H.; Lay J. O. Jr.; Kadlubar F. F. (1992) Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3-methylimidazo[4,5-f]quinoline and N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol. 5, 479–490. 10.1021/tx00028a005. [DOI] [PubMed] [Google Scholar]
- Turesky R. J.; Markovic J.; Aeschlimann J. M. (1996) Formation and differential removal of C-8 and N2-guanine adducts of the food carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in the liver, kidney, and colorectum of the rat. Chem. Res. Toxicol. 9, 397–402. 10.1021/tx950131r. [DOI] [PubMed] [Google Scholar]
- Purohit V.; Basu A. K. (2000) Mutagenicity of nitroaromatic compounds. Chem. Res. Toxicol. 13, 673–692. 10.1021/tx000002x. [DOI] [PubMed] [Google Scholar]
- Malia S. A.; Vyas R. R.; Basu A. K. (1996) Site-specific frame-shift mutagenesis by the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-y1)-1-aminopyrene located in the (CG)3 sequence: effects of SOS, proofreading, and mismatch repair. Biochemistry 35, 4568–4577. 10.1021/bi9525132. [DOI] [PubMed] [Google Scholar]
- Hilario P.; Yan S.; Hingerty B. E.; Broyde S.; Basu A. K. (2002) Comparative mutagenesis of the C8-guanine adducts of 1-nitropyrene and 1,6- and 1,8-dinitropyrene in a CpG repeat sequence. A slipped frameshift intermediate model for dinucleotide deletion. J. Biol. Chem. 277, 45068–45074. 10.1074/jbc.M208103200. [DOI] [PubMed] [Google Scholar]
- Watt D. L.; Utzat C. D.; Hilario P.; Basu A. K. (2007) Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem. Res. Toxicol. 20, 1658–1664. 10.1021/tx700131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert I. B.; Napolitano R. L.; Fuchs R. P. (1992) Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl. Acad. Sci. U. S. A. 89, 1310–1314. 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffel-Schwartz N.; Fuchs R. P. (1995) Sequence determinants for −2 frameshift mutagenesis at NarI-derived hot spots. J. Mol. Biol. 252, 507–513. 10.1006/jmbi.1995.0515. [DOI] [PubMed] [Google Scholar]
- Tan X.; Suzuki N.; Grollman A. P.; Shibutani S. (2002) Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry 41, 14255–14262. 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev. 13, 2191–2195. 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- Broyde S.; Wang L.; Rechkoblit O.; Geacintov N. E.; Patel D. J. (2008) Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 33, 209–219. 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C.; Wagner R.; Radman M. (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296, 1627–1630. 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- Fuchs R. P.; Fujii S. (2013) Translesion DNA Synthesis and Mutagenesis in Prokaryotes. Cold Spring Harbor Perspect. Biol. 5, a012682. 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J. E. (2013) Translesion DNA Synthesis and Mutagenesis in Eukaryotes. Cold Spring Harbor Perspect. Biol. 5, a012708. 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H.; Friedberg E. C.; Fuchs R. P.; Goodman M. F.; Hanaoka F.; Hinkle D.; Kunkel T. A.; Lawrence C. W.; Livneh Z.; Nohmi T.; Prakash L.; Prakash S.; Todo T.; Walker G. C.; Wang Z.; Woodgate R. (2001) The Y-family of DNA polymerases. Mol. Cell 8, 7–8. 10.1016/S1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Yang W.; Woodgate R. (2007) What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104, 15591–15598. 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. F.; Woodgate R. (2013) Translesion DNA Polymerases. Cold Spring Harbor Perspect. Biol. 5, a010363. 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S.; Prakash L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16, 1872–1883. 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Livneh Z.; Ziv O.; Shachar S. (2010) Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle 9, 729–735. 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- Ghosal G.; Leung J. W. C.; Nair B. C.; Fong K. W.; Chen J. J. (2012) Proliferating Cell Nuclear Antigen (PCNA)-binding Protein C1orf124 Is a Regulator of Translesion Synthesis. J. Biol. Chem. 287, 34225–34233. 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. S.; Ghosal G.; Chen J. J. (2012) The HARP-like Domain-Containing Protein AH2/ZRANB3 Binds to PCNA and Participates in Cellular Response to Replication Stress. Mol. Cell 47, 410–421. 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y.; Stover J. S.; Angel K. C.; Chowdhury G.; Rizzo C. J.; Guengerich F. P. (2006) Biochemical basis of genotoxicity of heterocyclic arylamine food mutagens: Human DNA polymerase eta selectively produces a two-base deletion in copying the N2-guanyl adduct of 2-amino-3-methylimidazo[4,5-f]quinoline but not the C8 adduct at the NarI G3 site. J. Biol. Chem. 281, 25297–25306. 10.1074/jbc.M605699200. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C.; Lehmann A. R.; Fuchs R. P. (2005) Trading places: how do DNA polymerases switch during translesion DNA synthesis?. Mol. Cell 18, 499–505. 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Bose A.; Pande P.; Jasti V. P.; Millsap A. D.; Hawkins E. K.; Rizzo C. J.; Basu A. K. (2015) DNA polymerases κ and ζ cooperatively perform mutagenic translesion synthesis of the C8–2′-deoxyguanosine adduct of the dietary mutagen IQ in human cells. Nucleic Acids Res. 43, 8340–8351. 10.1093/nar/gkv750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande P.; Malik C. K.; Bose A.; Jasti V. P.; Basu A. K. (2014) Mutational analysis of the C8-guanine adduct of the environmental carcinogen 3-nitrobenzanthrone in human cells: critical roles of DNA polymerases η and κ and Rev1 in error-prone translesion synthesis. Biochemistry 53, 5323–5331. 10.1021/bi5007805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26, 365–369. 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kalam M. A.; Basu A. K. (2005) Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells: tandem mutations and enhancement of G→T transversions. Chem. Res. Toxicol. 18, 1187–1192. 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- Colis L. C.; Raychaudhury P.; Basu A. K. (2008) Mutational specificity of gamma-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase eta. Biochemistry 47, 8070–8079. 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov P. P.; Chowdhury G.; Garmendia C. A.; Wang F.; Stover J. S.; Elmquist C. E.; Kozekova A.; Angel K. C.; Turesky R. J.; Stone M. P.; Guengerich F. P.; Rizzo C. J. (2010) The C8–2′-deoxyguanosine adduct of 2-amino-3-methylimidazo[1,2-d]naphthalene, a carbocyclic analogue of the potent mutagen 2-amino-3-methylimidazo[4,5-f]quinoline, is a block to replication in vitro. Chem. Res. Toxicol. 23, 1076–1088. 10.1021/tx100053n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavros K. M.; Hawkins E. K.; Rizzo C. J.; Stone M. P. (2015) Base-Displaced Intercalated Conformation of the 2-Amino-3-methylimidazo[4,5-f]quinoline N2-dG DNA Adduct Positioned at the Nonreiterated G1 in the NarI Restriction Site. Chem. Res. Toxicol. 28, 1455–1468. 10.1021/acs.chemrestox.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavros K. M.; Hawkins E. K.; Rizzo C. J.; Stone M. P. (2014) Base-displaced intercalation of the 2-amino-3-methylimidazo[4,5-f]quinolone N2-dG adduct in the NarI DNA recognition sequence. Nucleic Acids Res. 42, 3450–3463. 10.1093/nar/gkt1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avkin S.; Goldsmith M.; Velasco-Miguel S.; Geacintov N.; Friedberg E. C.; Livneh Z. (2004) Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J. Biol. Chem. 279, 53298–53305. 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Yang Y.; Tang T. S.; Zhang H.; Wang Z.; Friedberg E.; Yang W.; Guo C. (2014) Variants of mouse DNA polymerase kappa reveal a mechanism of efficient and accurate translesion synthesis past a benzo[a]pyrene dG adduct. Proc. Natl. Acad. Sci. U. S. A. 111, 1789–1794. 10.1073/pnas.1324168111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A.; Surugihalli C.; Pande P.; Champeil E.; Basu A. K. (2016) Comparative Error-Free and Error-Prone Translesion Synthesis of N2-2′-Deoxyguanosine Adducts Formed by Mitomycin C and Its Metabolite, 2,7-Diaminomitosene, in Human Cells. Chem. Res. Toxicol. 29, 933–939. 10.1021/acs.chemrestox.6b00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L.; Prakash S.; Prakash L. (2003) Yeast DNA polymerase ζ is an efficient extender of primer ends opposite from 7,8-dihydro-8-oxoguanine and O6-methylguanine. Mol. Cell. Biol. 23, 1453–1459. 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D. Y.; Wu X. H.; Rajpal D. K.; Taylor J. S.; Wang Z. G. (2001) Translesion synthesis by yeast DNA polymerase zeta from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 29, 2875–2883. 10.1093/nar/29.13.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. W.; Braithwaite E.; Guo D. Y.; Zhao B.; Geacintov N. E.; Wang Z. A. (2003) Mutagenesis of benzo[a]pyrene diol epoxide in yeast: Requirement for DNA polymerase ζ and involvement of DNA polymerase η. Biochemistry 42, 11253–11262. 10.1021/bi0346704. [DOI] [PubMed] [Google Scholar]
- Johnson R. E.; Prakash L.; Prakash S. (2012) Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. U. S. A. 109, 12455–12460. 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S.; Gregory M. T.; Yang W. (2014) Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. U. S. A. 111, 2954–2959. 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; DeMuro N. E.; Elmquist C. E.; Stover J. S.; Rizzo C. J.; Stone M. P. (2006) Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for −2 bp deletions. J. Am. Chem. Soc. 128, 10085–10095. 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Elmquist C. E.; Stover J. S.; Rizzo C. J.; Stone M. P. (2007) DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme. Biochemistry 46, 8498–8516. 10.1021/bi700361u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu G. W.; Huang X.; Luneva N. P.; Geacintov N. E.; Beese L. S. (2005) Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 280, 3764–3770. 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- Jha V.; Bian C.; Xing G.; Ling H. (2016) Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase κ. Nucleic Acids Res. 44, 4957–4967. 10.1093/nar/gkw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr S.; Schneider S.; Lammens K.; Hopfner K. P.; Carell T. (2010) Mechanism of replication blocking and bypass of Y-family polymerase η by bulky acetylaminofluorene DNA adducts. Proc. Natl. Acad. Sci. U. S. A. 107, 20720–20725. 10.1073/pnas.1008894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S.; Ziv O.; Avkin S.; Adar S.; Wittschieben J.; Reissner T.; Chaney S.; Friedberg E. C.; Wang Z.; Carell T.; Geacintov N.; Livneh Z. (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 28, 383–393. 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington M. T.; Minko I. G.; Johnson R. E.; Haracska L.; Harris T. M.; Lloyd R. S.; Prakash S.; Prakash L. (2004) Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase ζ. Mol. Cell. Biol. 24, 6900–6906. 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.