Abstract
Mosquitoes are a diverse group of invertebrates, with members that are among the most important vectors of diseases. The correct identification of mosquitoes is paramount to the control of the diseases that they transmit. However, morphological techniques depend on the quality of the specimen and often unavailable taxonomic expertise, which may still not be able to distinguish mosquitoes among species complexes (sibling and cryptic species). High resolution melting (HRM) analyses, a closed-tube, post-polymerase chain reaction (PCR) method used to identify variations in nucleic acid sequences, has been used to differentiate species within the Anopheles gambiae and Culex pipiens complexes. We validated the use of PCR-HRM analyses to differentiate species within Anopheles and within each of six genera of culicine mosquitoes, comparing primers targeting cytochrome b ( cyt b), NADH dehydrogenase subunit 1 (ND1), intergenic spacer region (IGS) and cytochrome c oxidase subunit 1 ( COI) gene regions. HRM analyses of amplicons from all the six primer pairs successfully differentiated two or more mosquito species within one or more genera ( Aedes ( Ae. vittatus from Ae. metallicus), Culex ( Cx. tenagius from Cx. antennatus, Cx. neavei from Cx. duttoni, cryptic Cx. pipiens species), Anopheles ( An. gambiae s.s. from An. arabiensis) and Mansonia ( Ma. africana from Ma. uniformis)) based on their HRM profiles. However, PCR-HRM could not distinguish between species within Aedeomyia ( Ad. africana and Ad. furfurea), Mimomyia ( Mi. hispida and Mi. splendens) and Coquillettidia ( Cq. aurites, Cq. chrysosoma, Cq. fuscopennata, Cq. metallica, Cq. microannulatus, Cq. pseudoconopas and Cq. versicolor) genera using any of the primers. The IGS and COI barcode region primers gave the best and most definitive separation of mosquito species among anopheline and culicine mosquito genera, respectively, while the other markers may serve to confirm identifications of closely related sub-species. This approach can be employed for rapid identification of mosquitoes.
Keywords: High resolution melting analysis, molecular identification, mosquitoes, Aedes, Culex, Mansonia, Anopheles
Introduction
Mosquitoes are among the most important disease vectors, known to transmit and maintain the circulation of pathogens that cause both global and neglected tropical diseases in humans and animals 1. The correct identification of different field-collected mosquito species, endemic to distinct ecologies, with high parasite and arthropod-borne virus (arbovirus) diversities is crucial to the planning of targeted vector control strategies to mitigate disease transmission 2. The last and most comprehensive Afrotropical mosquito identification keys were published in 1941 for culicines 3 and in 1987 for anophelines 4. Molecular approaches that efficiently differentiate conspecific mosquitoes such as the barcode region 5 improve identification accuracy considerably 6, but are time consuming, expensive in terms of post-polymerase chain reaction (post-PCR) processing and depend heavily on DNA sequencing.
Recent approaches have taken advantage of the unique melting profiles generated by homologous PCR products with small sequence differences during high resolution melting (HRM) analysis 7, 8. Indeed, PCR-HRM has been used to differentiate mosquito transmitted arboviruses 9– 11 and malaria Plasmodium 12, 13, vertebrate blood meals of mosquitoes 10, between two members of the Anopheles gambiae complex 14 and amongst three members of the Culex pipiens complex 15. HRM analysis has proven to offer higher resolution of PCR product based species identification on sequence variants than electrophoretic methods by revealing even single nucleotide polymorphisms (SNPs) in the simple sequence repeats (SSRs) among products of similar sizes 16, 17. Conventional detection of specific PCR products sequence relies on costly molecular probes and/or product sequencing 18. For species identification 16, only representative samples with distinct HRM profiles need to be sequenced, thereby reducing reagent and sample consumption costs 10– 11. Combining HRM analysis of barcode region sequences (Bar-HRM) has been successfully used to rapidly and accurately distinguish between closely related antelope species 19 and medicinal plants 20, 21 and to authenticate the source of vegetable oils 22.
Although HRM has been successfully used to differentiate between specific Anopheles and Culex mosquitoes, the approach’s broader applicability and most suitable markers have not been evaluated. Previously, only the ribosomal DNA was targeted for An. gambiae sensu lato ( s.l.) 14 and only the acetylcholinesterase gene was used in distinguishing the Cx. pipiens complex 15. This study aimed at validating the use of HRM analysis for high throughput molecular culicine and anopheline mosquito identification and differentiation, comparing the utility of one ribosomal IGS (previously used to differentiate An. gambiae s.l.) 14 and three mitochondrial (COI, ND1, cyt b) gene markers.
Methods
Sample collection and identification
We used 109 mosquitoes ( Table 1 and Table 2) that were collected in 2012 during the rainy seasons near Lake Baringo from March 2–4, July 16–24 and October 12–21 and Lake Victoria from April 2–15, May 18–31 and November 12–29 during a mosquito diversity study around the islands and mainland shores of Lake Baringo in Baringo County ( Table 1) and Lake Victoria in Homa Bay County ( Table 2) in Kenya 6. Before sampling, we obtained ethical clearance for the study from the Kenya Medical Research Institute (KEMRI) ethics review committee (Approval Ref: Non-SSC Protocol #310). These mosquitoes were morphologically identified during a previous study 6. Baringo County is a known hotspot for arbovirus outbreaks 23, while Homa Bay County is endemic to malaria and is located in a region with a history of arbovirus activity 10. One sample each of Anopheles gambiae sensu stricto ( s.s.) and An. arabiensis, Aedes aegypti and Culex pipiens from laboratory colonies maintained in the Insectary of the International Centre of Insect Physiology and Ecology ( icipe), Nairobi, Kenya, were used as controls. Also, specimens with confirmed identity that have been previously sequenced and submitted to GenBank ( Table 1 and Table 2) were used as both controls and samples.
Table 1. Number (N) of mosquito species (GenBank accessions) used for HRM analyses from Baringo County, Kenya.
| Mosquito species | N | Logumgum
0.455 N, 36.078 E |
Sirata
0.462 N, 36.097 E |
Kampi ya Samaki
0.620 N, 36.028 E |
Nosuguro
0.605 N, 36.126 E |
|---|---|---|---|---|---|
| Ad. africana | 4 | 4 (KU186980, KU186981,
KU186982, KU186985) |
|||
| Ad. furfurea | 4 | 4 (KU186979, KU186983,
KU186984, KU186986) |
|||
| An. funestus | 3 | 3 (KU187102, KU187103,
KU187105) |
|||
| An. gambiae s.l. | 3 | 1 | 2 | ||
| Cq. aurites | 2 | 2 (KU187114, KU187117) | |||
| Cq. chrysosoma | 1 | 1 (KU187115) | |||
| Cq. fuscopennata | 1 | 1 (KU187116) | |||
| Cq. metallica | 2 | 2 (KU187112, KU187113) | |||
| Cx. antennatus | 2 | 1 | 1 (KU187050) | ||
| Cx. perexiguus | 4 | 4 (KU380423, KU380348,
KU380476, KU380382) |
|||
| Cx. tenagius | 1 | 1 (KU187054) | |||
| Ma. africana | 4 | 2 (KU187124, KU187130) | 2 (KU187127,
KU187128) |
||
| Ma. uniformis | 3 | 2 (KU187170, KU187171) | 1 (KU187164) | ||
| Total | 34 | 8 | 16 | 6 | 4 |
GenBank accessions are provided only for samples with confirmed identity and from which the COI DNA sequences were obtained during a previously published mosquito diversity study 6.
Table 2. Number (N) of mosquito species (GenBank accessions) used for HRM analyses from Homa Bay County, Kenya.
| Mosquito species | N | Mbita
0.432 S, 34.210 E |
Luanda
Nyamasare 0.478 S, 34.287 E |
Ngodhe
0.505 S, 34.363 E |
Ungoye
0.612 S, 34.098 E |
Mfangano Island
0.462 S, 33.999 E |
Rusinga Island
0.399 S, 34.193 E |
Chamaunga
Island 0.431 S, 34.228 E |
Takawiri
Island 0.472 S, 34.091 E |
|---|---|---|---|---|---|---|---|---|---|
| Ae. metallicus | 3 | 1 (KU187014) | 1
(KU187013) |
1 | |||||
| Ae. vittatus | 8 | 6 (KU187004, KU187006,
KU187008-KU187011) |
2 (KU187005,
KU187007) |
||||||
| An. coustani s.l. | 10 | 3
(KU187098, KU187100, KU187101) |
2 (KU187095,
KU187096) |
2 (KU187097, KU187099) | 2 | 1 | |||
| An. funestus | 2 | 1 (KU187104) | 1 | ||||||
| An. gambiae s.l. | 2 | 2 (KU187108, KU187109) | |||||||
| Cq. aurites | 1 | 1 (KU187121) | |||||||
| Cq. microannulatus | 2 | 1 | 1 (KU187118) | ||||||
| Cq. pseudoconopas | 1 | 1 | |||||||
| Cq. versicolor | 2 | 2 (KU187119,
KU187120) |
|||||||
| Cx. antennatus | 4 | 4 (KU187037,
KU187038, KU187048) |
|||||||
| Cx. duttoni | 6 | 1 (KU187075) | 5 (KU187067, KU187068,
KU187070-KU187072) |
1 | |||||
| Cx. neavei | 3 | 3 (KU187032, KU187040,
KU187046) |
|||||||
| Cx. perexiguus | 1 | 1 (KU380445) | |||||||
| Cx. pipiens | 6 | 1 (KU187083) | 5 (KU187077,
KU380366, KU380372, KU380431, KU380444) |
||||||
| Culex sp. GPA | 5 | 3 (KU380352,
KU380455, KU380394) |
2 (KU380377,
KU380413) |
||||||
| Cx. univittatus | 3 | 3 (KU187056,
KU187059, KU187060) |
|||||||
| Cx. watti | 5 | 2 (KU187063,
KU187064) |
2 | 1 | |||||
| Ma. africana | 2 | 1 | 1 (KU187153) | ||||||
| Ma. uniformis | 3 | 1
(KU380460) |
1 | 1 (KU187175) | |||||
| Mi. hispida | 2 | 2 | |||||||
| Mi. splendens | 3 | 3 (KU187093,
KU187094) |
|||||||
| Total | 75 | 21 | 5 | 5 | 1 | 22 | 12 | 6 | 3 |
GenBank accessions are provided only for samples with confirmed identity and from which the COI DNA sequences were obtained during a previously published mosquito diversity study 6.
DNA extraction
From each mosquito, we extracted DNA according to the hot sodium hydroxide and Tris (HotSHOT) DNA extraction protocol 24 from a single mosquito leg that was detached from the rest of the body using a pair of forceps and dissecting pin. Without crushing, the mosquito leg was put in a 0.2 ml microcentrifuge tube containing 30 µl of Alkaline Lysis buffer (25 mM NaOH (Thermo Fisher Scientific, Pittsburgh, USA), 0.2 mM disodium EDTA (Thermo Fisher Scientific), pH 8.0) and incubated in a thermocycler at 95°C for 30 minutes and cooled at 4°C for 5 minutes. Then, 30 µl neutralising solution (40 mM Tris-HCl (Thermo Fisher Scientific)) was added. The resulting DNA was stored at -20°C until required as templates for PCR assays.
Primer design, PCR and HRM analyses
Based on multiple alignments using Geneious software version 8.1.4 25 of mitochondrial genomes of mosquitoes (GenBank accessions NC_015079, NC_028616, NC_028223, KR068634, NC_010241, NC_014574, EU352212, NC_008070, KT358413, KT382816, KU494979, JX040513, AY729979, KU494979), we designed four sets of primers from two mitochondrial gene regions: COI (COI-AnophF/HCO2108R; Uni-Minibar-JVF/Uni-Minibar-JVR; Mos-CO1-JVF/Mos-CO1-JVR) and ND1 (Mos-ND1F/Mos-ND1R) genes ( Table 3). The COI AnophF primer was initially designed specifically for Anopheles mosquitoes to be used with the HCO2108R primer 26, but tested on other species as well. Using samples of morphologically and molecularly identified Culex, Aedeomyia, Mimomyia, Coquillettidia, Mansonia, Aedes, and Anopheles mosquito species ( Table 1 and Table 2), we amplified different gene regions of their genomes using six pairs of primers ( Table 3) in three replicate runs of single-plex PCRs in a Rotor-Gene Q HRM real time PCR thermocycler (QIAGEN, Hannover, Germany). PCR grade water was used as negative control while mosquito species from Ae. aegypti, An. gambiae s.s., An. arabiensis and Cx. Pipiens quinquefasciatus colonies maintained in the International Centre of Insect Physiology and Ecology ( icipe) Insectary Unit were used as positive controls. The PCR mix contained 5 µl of 5X Hot Firepol EvaGreen HRM Mix (Solis BioDyne, Tartu, Estonia), 0.5 µM of each primer, 1 µl of DNA template and distilled water in a final volume of 10 µl. The thermal cycling conditions involved an initial denaturation for 1 minute at 95°C, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 50°C for 20 seconds, and extension at 72°C for 30 seconds, and a final extension at 72°C for 7 minutes. Without stopping the reaction, the PCR amplicons were denatured at 95°C for 1 minute, held for another minute at 40°C and melted by gradually raising the temperature from 70°C to 95°C by 0.1°C in 2 second steps, waiting for 90 seconds of pre-melt conditioning on first step and 2 seconds in subsequent steps. The outcome was automatically plotted on a connected computer and visually observed and analysed using the Rotor-Gene Q Series software v2.1. Representative samples of differentiated mosquito species that had similar HRM curves were purified with ExoSAP-IT (USB Corporation, Cleveland, OH) and submitted for DNA sequencing at Macrogen (South Korea). To confirm the identity of PCR-HRM differentiated mosquitoes, DNA sequences were edited with Geneious version 8.1.4 25 and queried against the GenBank nr database ( http://www.ncbi.nlm.nih.gov/) using the Basic Local Alignment Search Tool (BLAST N) version 2.3.0 27.
Table 3. Primers used for the amplification of gene fragments.
| Target gene | Primer name | Primer Sequence (5’ to 3’) | Reference
genome |
Primer
coordinates |
Amplicon
size (bp) |
|---|---|---|---|---|---|
| Mitochondrial COI
(within barcode region) |
COI-AnophF | GCAGGAATTTCTTCTATTTTAGG | L20934 | 1,874–1,896 | 275 |
| HCO2198R 26 | TAAACTTCAGGGTGACCAAAAAATCA | L20934 | 2,148–2,123 | ||
| Mitochondrial COI | Uni-Minibar-JVF | ACAAATCATAARGATATTGGAAC | L20934 | 1,445–1,467 | 173 |
| Uni-Minibar-JVR | AAAATTATAATAAAWGCATGAGC | L20934 | 1,617–1,55 | ||
| Mitochondrial COI | Mos-Co1-JVF | ATAGTWATACCTATYATAATTGG | L20934 | 1,622–1,644 | 299 |
| Mos-Co1-JVR | ACWGTAGTAATAAAATTTACTGC | L20934 | 1,920–1,898 | ||
| Mitochondrial ND1 | Mos-ND1F | TATGTCTTGAAAACATAAGAAAG | L20934 | 11,569–11,591 | 173 |
| Mos-ND1R | CGDTATGATAAATTAATGTAATTAG | L20934 | 11,717–11,741 | ||
| Mitochondrial cyt b | CYT BF 35 | GGACAAATATCATTTTGAGGAGCAACAG | L20934 | 10,821–10,848 | 470 |
| CYT BR 35 | ATTACTCCTCCTAGCTTATTAGGAATTG | L20934 | 11,290–11,263 | ||
| Ribosomal DNA IGS | AgamUni F 2 | GTGAAGCTTGGTGCGTGCT | KT284724 | 126–174 | 169 |
| AgamUni R 2 | GCACGCCGACAAGCTCA | KT284724 | 319–303 |
F is forward primer direction; R is reverse primer direction.
Results
‘Contents.csv’ contains a description of the files.
Copyright: © 2016 Ajamma YU et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
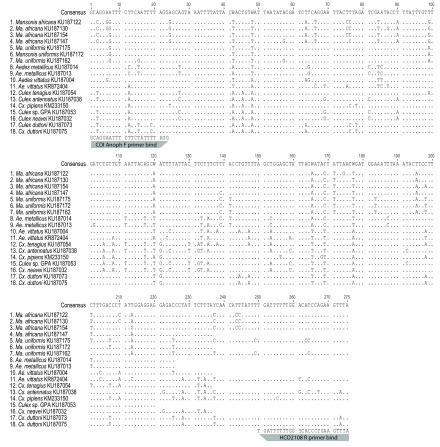
We differentiated 12 mosquito species in the Aedes (two) , Anopheles (two) , Culex (six), and Mansonia (two) genera by HRM analyses ( Table 4). The COI sequences of some of the mosquito samples analyzed and differentiated were obtained during a previously published mosquito diversity study 6 and their respective GenBank Accession numbers are listed in Table 1 and Table 2. Despite the fact that the COI-AnophF/HCO2198R primers were originally designed based on Anopheles mitochondria genome alignments, they were most efficient in differentiating among Mansonia ( Ma. africana and Ma. uniformis ( Figure 1A)), Culex ( Cx. neavei and Cx. duttoni, Cx. tenagius and Cx. antennatus, and two genetic variants of Cx. pipiens ( Figure 2A)), and Aedes ( Ae. vittatus and Ae. metallicus ( Figure 3)) mosquitoes ( Table 4). Indeed, the DNA sequences flanked by the COI-AnophF/HCO2198R primers included multiple polymorphic sites in species within these genera ( Figure 4). Although there are SNPs within species DNA that resulted to the slight changes observed in their HRM profiles, the SNPs across species were enough to distinguish between them.
Figure 1. HRM profiles of two Mansonia species.
Mansonia uniformis and Ma. africana mosquitoes were differentiated by PCR-HRM using the ( A) COI-AnophF/HCO2198R, ( B) MOS-CO1 and ( C) CYT B primer pairs.
Figure 2. HRM profiles of Culex species.
Culex species were differentiated by PCR-HRM using the ( A) COI-AnophF/HCO2198R, ( B) CYT B, ( C) Uni-Minibar-JV, and ( D) Mos-ND1 primer pairs.
Figure 3. HRM profiles of Aedes mosquitoes.
Aedes vittatus and Ae. metallicus were differentiated by PCR-HRM using the COI-AnophF/HCO2198R primer pair.
Figure 4. Single nucleotide polymorphisms (SNP) between mosquito species separated by the COI-AnophF/HCO2108R primer pair amplicons.
Polymorphic sites vary more between than within species.
Table 4. Differentiation of mosquito species using the six primer pairs amplifying four loci.
| Mosquito genera | COI | cyt b | ND1 | IGS | ||
|---|---|---|---|---|---|---|
| COI-AnophF/HCO2198R | Mos-CO1-JV | Uni-Minibar-JV | CYT B | Mos-ND1 | AgamUni | |
| Anopheles | DNS | DNS | DNS | DNS | DNS | Separated
An. gambiae from An. arabiensis |
| Mansonia | Separated
Ma. africana
from Ma. uniformis |
Separated
Ma. africana from Ma. uniformis |
DNS | Separated
Ma. africana from Ma. uniformis |
DNS | DNS |
| Aedes | Separated
Ae. vittatus from
Ae. metallicus |
DNS | DNS | DNS | DNS | DNS |
| Culex | Separated
Cx. tenagius
from Cx. antennatus, Separated Cx. pipiens from Culex sp. GPA, Separated Cx. neavei from Cx. duttoni |
DNS | Separated
Cx. pipiens from Culex sp. GPA |
Separated
Cx. tenagius from Cx. antennatus |
Separated
Cx. tenagius from Cx. antennatus, Separated Cx. pipiens from Culex sp. GPA |
DNS |
| Aedeomyia | DNS | DNS | DNS | DNS | DNS | DNS |
| Mimomyia | DNS | DNS | DNS | DNS | DNS | DNS |
| Coquillettidia | DNS | DNS | DNS | DNS | DNS | DNS |
DNS means did not separate. COI means cytochrome c oxidase subunit 1. cyt b means cytochrome B. ND1 means NADH dehydrogenase subunit 1. IGS means intergenic spacer region.
Mansonia africana and Ma. uniformis could also be differentiated by Mos-COI-JV ( Figure 1B) and CYT B ( Figure 1C) PCR-HRM analysis. Some Culex species were similarly differentiated by HRM based on their CYT B, Uni-Minibar-JV and Mos-ND1 ( Figure 2B–D) primer pair PCR products. The morphologically indistinguishable Cx. tenagius and Cx. antennatus were distinguished only by the COI-AnophF/HCO2198R, CYT B and ND1 primers ( Figure 2A, B and D). Similarly, HRM analysis of only two of the COI (COI-AnophF/HCO2198R and Uni-Minibar JV) and the ND1 primer pairs grouped morphologically identical and difficult to differentiate Cx. pipiens into two distinct clades: one with Cx. pipiens voucher sequences from GenBank (KF919189) and those with a sequence that we identified as Culex sp. GPA 6 (GenBank accessions KU380352, KU380455, KU380394) ( Figure 2A, C and D; Table 4). However, unlike the COI HRM profiles ( Figure 2A, B), the ND1 HRM profiles ( Figure 2D) of Cx. pipiens amplicons showed a melting temperature shift of to the right (higher temperature) compared to the Culex sp. GPA amplicons, possibly due to greater GC richness of Cx. pipiens at this locus 28. Similarly, the IGS primers (AgamUni) differentiated Anopheles gambiae s.s. from An. arabiensis ( Figure 5). In addition, the COI-AnophF/HCO2198R primers were also used to separate Cx. neavei from Cx. duttoni ( Figure 2A), which belong to the same subgenus of Culex mosquitoes.
Figure 5. HRM profiles of Anopheles mosquitoes.
Two sibling species of Anopheles gambiae s.l. were differentiated by PCR-HRM using the AgamUni primer pair.
HRM analysis of all the six primer pairs could not differentiate Aedeomyia ( Ad. africana and Ad. furfurea), Mimomyia ( Mi. hispida and Mi. splendens) and Coquillettidia ( Cq. aurites, Cq. chrysosoma, Cq. fuscopennata, Cq. metallica, Cq. microannulatus, Cq. pseudoconopas and Cq. versicolor) species ( Table 4) or among An. funestus and An. coustani species complexes.
Discussion
We compared six pairs of primers for their potential to differentiate at least two morphologically similar mosquito species within each of seven mosquito genera by PCR-HRM analysis and identified suitable markers for differentiating species within Anopheles, Aedes, Culex and Mansonia mosquitoes. However, none of the markers were suitable for HRM analysis to distinguish among species of Aedeomyia, Mimomyia or Coquillettidia genera mosquitoes. Also, Cx. watti, which can be misidentified morphologically as Cx. duttoni or Cx. pipiens, could not be differentiated by PCR-HRM analyses. Nonetheless, we were able to distinguish Ma. africana from Ma. uniformis, An. gambiae s.s. from An. arabiensis (sibling species of An. gambiae s.l.), Ae. vittatus from Ae. metallicus, as well as Cx. neavei from Cx. duttoni, Cx. tenagius from Cx. antennatus and two cryptic sympatric species of morphologically identical Cx. pipiens. Most notably, the two Cx. pipiens species with distinct COI barcode sequences 6 were indeed first identified by HRM analysis of numerous samples 6. Thus, the relative economy of HRM analysis compared to sequencing facilitates the rapid identification of cryptic species.
Surprisingly, HRM analysis of PCR products from the COI-AnophF/HCO2198R primers, which were designed for Anopheles, could not distinguish between these sibling species, yet were most effective in discriminating species within the Mansonia, Aedes and Culex genera, including between the cryptic Culex pipiens species. Anopheles gambiae and An. arabiensis were only distinguished using the IGS gene, which was also designed for An. gambiae 2 and is routinely used for distinguishing these sibling species by conventional PCR 29 and HRM analysis 14. In contrast, species complexes of An. coustani and An. funestus were not differentiated with any of the primers. The data suggest that COI 30, cyt b and ND1 loci may be unsuitable for distinguishing among Anopheles sibling species. Similarly, the Aedes species could only be differentiated by the COI-AnophF/HCO2198R primers. This could be as a result of more recent speciation, insufficient to allow for sibling species resolution at these markers. Such scenarios have been observed for recent or rapidly evolving groups, such as the Cichlid fishes of eastern Africa, where mitochondrial divergence is not concordant with morphological variations 31.
In contrast, Ma. africana and Ma. uniformis were separated by the COI and cyt b loci, but not by the ND1 and IGS gene primers and Culex species were variably differentiable by all markers, except IGS. For both Mansonia and Culex, as with Aedes, the COI-AnophF/HCO2198R primers were most sensitive in discriminating morphologically indistinct species. This highlights the power of the COI barcode region for identifying diverse cryptic species 32. The SNPs present in the COI genes of the ten mosquito species confirms that the COI gene is conserved and polymorphic enough to differentiate these species even in cases of morphological misidentification. The polymorphisms between species were enough to robustly separate them based on their HRM profiles, while sequence polymorphisms within species were too few to significantly alter their HRM profiles.
We, therefore, recommend the initial use of the COI-AnophF/HCO2198R primers Bar-HRM to differentiate Mansonia, Culex and Aedes mosquito species and the IGS primers for anopheline mosquito identification 2, 14, 33 by HRM. The inability of all the six primer pairs to differentiate many mosquito species among all seven genera tested is an indication that the genetic diversity of many mosquito species is complicated and still poorly understood. Also, the number (sample size) of many of the analyzed mosquito species was small (<3) because these species were scarcely present in the study areas. More samples (≥3) should be used and more study areas should be sampled in subsequent studies to test genetic differentiation of mosquito species 34. Additional polymorphic DNA loci should also be identified, tested and used in combination with existing ones for the identification of mosquito species, especially among species complexes and across genera.
Conclusions
This study shows that specific PCR markers can be used to distinguish closely related species of mosquitoes using HRM analysis. We distinguished two sibling species of An. gambiae s.l., two species each of Mansonia and Aedes, and six species, including cryptic species, of Culex using six pairs of primers targeting the mitochondrial and ribosomal genes. HRM is a low cost (<$1 per reaction), effective tool that enhances culicine and anopheline mosquito identification and may also reveal population differences in conserved mitochondrial sequences. This approach can improve vector surveillance associated with Plasmodium (malaria) or arbovirus transmission and inform targeted vector control strategies.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Ajamma YU et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
All sequence data associated with this manuscript are freely available in GenBank. All relevant accession numbers are listed in Table 1 and Table 2.
F1000Research: Dataset 1. Raw Rotor-Gene Q HRM data files (.rex), viewable using Rotor-Gene Q software (Qiagen), 10.5256/f1000research.9224.d130565 36
Acknowledgements
We thank Laban Njoroge of National Museums of Kenya for helping with morphological identifications of mosquitoes. We acknowledge John Tilion of Ruko Conservancy in Baringo County and Phillip Ojunju of Rusinga Island in Homa Bay County, for helping with mosquito sample collection in the two study areas respectively. We acknowledge the support of Milcah Gitau of icipe’s Insectary Unit in providing the Ae. aegypti, Cx. pipiens, An. gambiae s.s. and An. arabiensis controls. We also thank Esther Waweru of icipe’s Molecular Biology and Bioinformatics Unit (MBBU), Gerard Ronoh, Caroline Tigoi and Geoffrey Jagero of icipe’s ML-EID Laboratory, Lillian Igweta, Lisa Omondi and Margaret Ochanda icipe’s of Capacity Building & Institutional Development (CB&ID) Unit for assisting with logistics.
Funding Statement
We gratefully acknowledge the financial support for this research by the following organizations and agencies: Swedish International Development Cooperation Agency (SIDA), grant number 75000529 to YUA as an African Regional Postgraduate Programme in Insect Science (ARPPIS) student; Funds from Training Health Researchers into Vocational Excellence (THRiVE) in East Africa (grant number 087540) funded by Wellcome Trust to JV and DM supported part of the field sampling. We also acknowledge funding from UK’s Department for International Development (DFID); the Swiss Agency for Development and Cooperation (SDC); and the Kenyan Government.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Weaver SC, Reisen WK: Present and future arboviral threats. Antiviral Res. 2010;85(2):328–345. 10.1016/j.antiviral.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walker ED, Thibault AR, Thelen AP, et al. : Identification of field caught Anopheles gambiae s.s. and Anopheles arabiensis by TaqMan single nucleotide polymorphism genotyping. Malar J. 2007;6:23. 10.1186/1475-2875-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edwards FW: Mosquitoes of the Ethiopian Region. III.- Culicine adults and pupae. British Museum (Natural History), London;1941. Reference Source [Google Scholar]
- 4. Gillies MT, Coetzee M: A Supplement to the Anophelinae of Africa South of the Sahara. Publications of the South African Institute for Medical Research. 1987; (55):1–143. Reference Source [Google Scholar]
- 5. Hebert PD, Cywinska A, Ball SL, et al. : Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270(1512):313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ajamma YU, Villinger J, Salifu D, et al. : Abundance and species composition of mosquito vectors of arboviruses in the Lake Victoria and Lake Baringo regions of Kenya. J Med Entomol. 2016. In Press. [Google Scholar]
- 7. Ririe KM, Rasmussen RP, Wittwer CT: Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245(2):154–160. 10.1006/abio.1996.9916 [DOI] [PubMed] [Google Scholar]
- 8. Vossen RH, Aten E, Roos A, et al. : High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat. 2009;30(6):860–866. 10.1002/humu.21019 [DOI] [PubMed] [Google Scholar]
- 9. Naze F, Le Roux K, Schuffenecker I, et al. : Simultaneous detection and quantitation of Chikungunya, dengue and West Nile viruses by multiplex RT-PCR assays and dengue virus typing using high resolution melting. J Virol Methods. 2009;162(1–2):1–7. 10.1016/j.jviromet.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 10. Omondi D, Masiga DK, Ajamma YU, et al. : Unraveling Host-Vector-Arbovirus Interactions by Two-Gene High Resolution Melting Mosquito Bloodmeal Analysis in a Kenyan Wildlife-Livestock Interface. PLoS One. 2015;10(7):e0134375. 10.1371/journal.pone.0134375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villinger J, Mbaya MK, Ouso DO, et al. : Arbovirus and insect-specific virus discovery in Kenya by novel six genera multiplex high resolution melting analysis. Mol Ecol Resour. 2016. In Press. [DOI] [PubMed] [Google Scholar]
- 12. Kipanga PN, Omondi D, Mireji PO, et al. : High-resolution melting analysis reveals low Plasmodium parasitaemia infections among microscopically negative febrile patients in western Kenya. Malar J. 2014;13:429. 10.1186/1475-2875-13-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chua KH, Lim SC, Ng CC, et al. : Development of High Resolution Melting Analysis for the Diagnosis of Human Malaria. Sci Rep. 2015;5: 15671. 10.1038/srep15671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zianni MR, Nikbakhtzadeh MR, Jackson BT, et al. : Rapid discrimination between Anopheles gambiae s.s. and Anopheles arabiensis by high-resolution melt (HRM) analysis. J Biomol Tech. 2013;24(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang D, Sim C: Identification of Culex complex species using SNP markers based on high-resolution melting analysis. Mol Ecol Resour. 2013;13(3):369–376. 10.1111/1755-0998.12083 [DOI] [PubMed] [Google Scholar]
- 16. Winder LC, Phillips C, Richards N, et al. : Evaluation of DNA melting analysis as a tool for species identification. Methods Ecol Evol. 2011;2(3):312–320. 10.1111/j.2041-210X.2010.00079.x [DOI] [Google Scholar]
- 17. Distefano G, Caruso M, La Malfa S, et al. : High resolution melting analysis is a more sensitive and effective alternative to gel-based platforms in analysis of SSR--an example in citrus. PLoS One. 2012;7(8):e44202. 10.1371/journal.pone.0044202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wittwer CT: High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30(6):857–859. 10.1002/humu.20951 [DOI] [PubMed] [Google Scholar]
- 19. Tong Y, Jiang C, Yuan Y, et al. : Molecular identification of antelope horn by melting curve analysis. Mitochondrial DNA. 2014;26:1–7. 10.3109/19401736.2014.989500 [DOI] [PubMed] [Google Scholar]
- 20. Osathanunkul M, Suwannapoom C, Osathanunkul K, et al. : Evaluation of DNA barcoding coupled high resolution melting for discrimination of closely related species in phytopharmaceuticals. Phytomedicine. 2016;23(2):156–165. 10.1016/j.phymed.2015.11.018 [DOI] [PubMed] [Google Scholar]
- 21. Li J, Song M, Xiong C, et al. : Application of barcode high-resolution melting for rapid authentication of the medicinal plant Psammosilene tunicoides. Biotechnol Biotechnol Equip. 2016;30(4):790–796. 10.1080/13102818.2016.1181988 [DOI] [Google Scholar]
- 22. Ganopoulos I, Bazakos C, Madesis P, et al. : Barcode DNA high-resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J Sci Food Agric. 2013;93(9):2281–2286. 10.1002/jsfa.6040 [DOI] [PubMed] [Google Scholar]
- 23. Sang R, Kioko E, Lutomiah J, et al. : Rift Valley fever virus epidemic in Kenya, 2006/2007: the entomologic investigations. Am J Trop Med Hyg. 2010;83(2 Suppl):28–37. 10.4269/ajtmh.2010.09-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montero-Pau J, Gómez A, Muñoz J: Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnol Oceanogr Methods. 2008;6:218–222. 10.4319/lom.2008.6.218 [DOI] [Google Scholar]
- 25. Kearse M, Moir R, Wilson A, et al. : Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Folmer O, Black M, Hoeh W, et al. : DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- 27. Zhang Z, Schwartz S, Wagner L, et al. : A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7(1–2):203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- 28. Garritano S, Gemignani F, Voegele C, et al. : Determining the effectiveness of High Resolution Melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 2009;10:5. 10.1186/1471-2156-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–529. [DOI] [PubMed] [Google Scholar]
- 30. Tahir HM, Mehwish, Kanwal N, et al. : Genetic diversity in cytochrome c oxidase I gene of Anopheles mosquitoes. Mitochondrial DNA. 2015;12:1–4. 10.3109/19401736.2015.1082104 [DOI] [PubMed] [Google Scholar]
- 31. McGee MD, Neches RY, Seehausen O: Evaluating genomic divergence and parallelism in replicate ecomorphs from young and old cichlid adaptive radiations. Mol Ecol. 2016;25(1):260–268. 10.1111/mec.13463 [DOI] [PubMed] [Google Scholar]
- 32. Hebert PD, Penton EH, Burns JM, et al. : Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci U S A. 2004;101(41):14812–14817. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruiz F, Linton YM, Ponsonby DJ, et al. : Molecular comparison of topotypic specimens confirms Anopheles ( Nyssorhynchus) dunhami Causey (Diptera: Culicidae) in the Colombian Amazon. Mem Inst Oswaldo Cruz. 2010;105(7):899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murugan K, Vadivalagan C, Karthika P, et al. : DNA barcoding and molecular evolution of mosquito vectors of medical and veterinary importance. Parasitol Res. 2016;115(1):107–121. 10.1007/s00436-015-4726-2 [DOI] [PubMed] [Google Scholar]
- 35. Lyman DF, Monteiro FA, Escalante AA, et al. : Mitochondrial DNA sequence variation among triatomine vectors of Chagas' disease. Am J Trop Med Hyg. 1999;60(3):377–386. [DOI] [PubMed] [Google Scholar]
- 36. Ajamma YU, Mararo E, Omondi D, et al. : Dataset 1 in: Rapid and high throughput molecular identification of diverse mosquito species by high resolution melting analysis. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]