Abstract
Objective
Delayed diagnosis of Kawasaki disease (KD) may lead to serious cardiac complications. We sought to create and test the performance of a natural language processing (NLP) tool, the KD-NLP, in the identification of emergency department (ED) patients for whom the diagnosis of KD should be considered.
Methods
We developed an NLP tool that recognizes the KD diagnostic criteria based on standard clinical terms and medical word usage using 22 pediatric ED notes augmented by Unified Medical Language System vocabulary. With high suspicion for KD defined as fever and three or more KD clinical signs, KD-NLP was applied to 253 ED notes from children ultimately diagnosed with either KD or another febrile illness. We evaluated KD-NLP performance against ED notes manually reviewed by clinicians and compared the results to a simple keyword search.
Results
KD-NLP identified high-suspicion patients with a sensitivity of 93.6% and specificity of 77.5% compared to notes manually reviewed by clinicians. The tool outperformed a simple keyword search (sensitivity = 41.0%; specificity = 76.3%).
Conclusions
KD-NLP showed comparable performance to clinician manual chart review for identification of pediatric ED patients with a high suspicion for KD. This tool could be incorporated into the ED electronic health record system to alert providers to consider the diagnosis of KD. KD-NLP could serve as a model for decision support for other conditions in the ED.
BACKGROUND AND IMPORTANCE
For the emergency department (ED) or urgent care provider, a major obstacle to timely diagnosis of Kawasaki disease (KD) is omission of KD from the differential diagnosis of a child with fever and mucocutaneous signs. Yet the stakes for a missed or delayed diagnosis are high as KD is the most common cause of acquired heart disease in children in developed countries.1 Although timely administration of intravenous immunoglobulin (IVIG) reduces the rate of coronary artery aneurysms, the major cardiovascular sequela of KD, from 25% to 5%,2,3 diagnosis beyond 10 days of fever increases the risk of coronary aneurysms by 2.8- to 7.1-fold.4,5
Although a definitive diagnostic laboratory test for KD has not yet been found, improved diagnostic algorithms have been created to aid in the diagnosis of KD. Ling et al.6,7 created a diagnostic algorithm using seven clinical variables and 12 laboratory variables that performed well in differentiating between KD cases and febrile controls. However, to utilize such diagnostic tools, clinicians must consider KD as part of the differential diagnosis in the febrile child. This is not always straightforward in the diagnosis of an illness that evolves over several days with the sequential appearance and disappearance of key clinical signs in the out-patient setting where different providers may evaluate the patient on different days.4 Among patients ultimately diagnosed with KD, 95% had seen a medical provider during the first 5 days of illness with a mean time to first visit of 2.5 days.5 However, only 4.7% had received the correct diagnosis on the first medical visit, with an average of three visits prior to diagnosis. Thus, delayed recognition of the constellation of signs may increase the likelihood of cardiovascular sequelae in infants and children with KD. In addition, the majority of children seen in an ED present to nonpediatric centers that may be less familiar with pediatric patients and the diagnostic evaluation of KD.8 Therefore, the diagnosis of KD in the urgent care or ED setting continues to be challenging.
Delayed diagnosis is particularly problematic among patients with incomplete and initially misleading presentations.5,9 An incomplete presentation (<4 of 5 clinical criteria) occurs in 16%–33% of KD patients and is a risk factor for delayed diagnosis.4,5,10 For many children, the cardiovascular damage following missed, untreated KD does not manifest until early adulthood. Increasing numbers of young adults in the United States and Japan present with myocardial infarction, congestive heart failure, arrhythmias, and sudden death as late sequelae of missed KD.11,12 Coronary angiography reveals aneurysms attributable to antecedent KD in 5%–7% of adults < 40 years of age evaluated for suspected myocardial ischemia.13,14 Unless clinicians consistently identify children at risk for KD from clinical features, children will experience delays in diagnosis and suffer potentially preventable morbidity and mortality.
Natural language processing (NLP) is a field of computer science and artificial intelligence utilizing linguistic analysis to gain “understanding” from unstructured free text. A familiar example of NLP is the ability of Internet search engines such as Google or Bing to correctly interpret complex queries and return the most relevant results. NLP algorithms are varied and include pattern recognition and machine learning (i.e., Google), such as decision trees with if–then branch points or probabilistic models that place a weight on a particular variable. The use of NLP in extracting phenotypic information from electronic health records is an increasing area of interest throughout medicine. The widespread adoption of electronic medical records (EMRs) has provided an immense amount of clinical data to be analyzed, both in discrete fields such as problem lists, billing codes, and laboratory values and in the narrative forms of clinician notes. However, leveraging these data for clinical research and improving patient care can be diffficult as hurdles include integration of NLP into and across multiple EMRs, patient privacy, and prospective validation of a tool.
Because the diagnosis of KD still rests primarily on the recognition of fever and the five classical signs described by Dr. Kawasaki nearly 50 years ago, it is an ideal opportunity to apply NLP techniques to screen patients and aid clinicians in considering KD in their differential diagnosis. Thus, we postulated that NLP using straight pattern recognition could be successfully utilized to evaluate narrative text from ED providers for the signs of KD.
Goals of This Investigation
The goal of this study was to create and test the performance of an NLP tool, KD-NLP, for early and rapid detection of subjects with a high suspicion for KD from text in clinical notes in the EMR. KD-NLP was developed as a screening tool to prompt the subsequent laboratory and/or echocardiographic testing required for the accurate diagnosis of KD. Our NLP screening tool is intended to identify patients with fever and three or more clinical signs of KD in whom the diagnosis of KD should be considered.
METHODS
Study Design
This retrospective study was conducted on notes collected from the EMR at Site 1 (Rady Children’s Hospital San Diego) and Site 2 (Children’s Healthcare of Atlanta) EDs from January 2010 to December 2014. The Site 1 ED evaluates 82,000 children per year and 80–90 children with acute KD are hospitalized at Site 1 annually and cared for by a specialized KD team led by two pediatric infectious disease physicians. The EDs of Site 2 evaluate over 220,000 children per year and 100–120 children with acute KD are cared for by the hospitalist service at these three hospitals. The study was reviewed and approved by the institutional review boards at both sites. Written informed consent was obtained from the parents or legal guardians of all subjects.
Study Setting and Population
Inclusion criteria for subjects with KD were based on the American Heart Association (AHA) guidelines as confirmed by KD expert clinicians (JCB, AHT, JK, and DL).1 We included subjects with KD diagnosed between illness day 3 and 10 (first day of fever = illness day 1) who had fever and four of five standard criteria (Table 1) or three or fewer criteria with coronary artery abnormalities documented by echocardiogram. Febrile children (FC) were children evaluated in the ED and enrolled in a KD study as febrile controls and met the following criteria: fever for at least 3 days and at least one of the KD clinical criteria. FC were found ultimately to not have KD. FC were specifically excluded if they had: 1) prominent respiratory or gastrointestinal symptoms, 2) treatment with IVIG within previous month, 3) treatment with steroids or other immunomodulatory agents within the previous week, or 4) any serious underlying medical condition. The criteria for FC were chosen to enrich the cohort for patients who might have KD considered in their differential diagnosis or who had been specifically referred into the ED for evaluation for KD. Because our goal was to create a screening tool that could be used in EDs or urgent care centers, a single clinical ED note for each subject was used for our analysis. For KD subjects, the ED note leading to hospital admission was used; for FC, the ED note at time of enrollment was used.
Table 1.
Diagnostic Criteria for KD1 and List of Semantic Tags for KD Tagger With Examples
| Clinical Criterion | Tag name | Description and examples |
|---|---|---|
| Fever | FEVER | Fever or temperature at least 100.4°F or 38°C Examples:
|
| Bilateral conjunctival injection | CONJUNCTIVAL_INJECTION | Bilateral bulbar conjunctival injection without exudate Examples:
|
| Changes of the oropharynx: injected pharynx, injected, fissured lips, strawberry tongue | ORAL_CHANGES | Changes in lips and oral cavity, including erythema, cracked lips, strawberry tongue, diffuse injection of oral and pharyngeal mucosae Examples:
|
| Changes of the peripheral extremities: peripheral edema, palm/sole erythema, periungual desquamation | EXTREMITY_CHANGES | Changes in extremities: palms, soles, hands, feet, or periungual peeling of fingers or toes Examples:
|
| Polymorphous rash | POLYMORPHOUS_EXANTHEMA | Polymorphous exanthema Examples:
|
| Cervical adenopathy > 1.5 cm | CERVICAL_LYMPHADENOPATHY | Cervical lymphadenopathy (≥1.5 cm diameter), usually unilateral Examples:
|
KD = Kawasaki disease.
In the absence of standards for the number of patients and documentation instances (number of times a sign is found in documentation) needed to develop the NLP tool, we selected a convenience sample over a 3-month period of 22 ED notes at Site 1 from children diagnosed with KD. The performance of KD-NLP was evaluated using a convenience sample of a consecutive series of notes from two additional cohorts of subjects (n = 166 from Site 1 and n = 87 from Site 2).
Study Protocol
Creation of a Criterion Standard
Two physicians (CKM, JDC), the latter with an expertise in pediatric infectious diseases, individually and manually reviewed each of the 253 ED notes and assessed them for the presence or absence of each of the five KD signs. Physician reviewers did not have access to patient information outside of the findings included in the finalized version of the note. After individual review, the two assessments were compared and any disagreements, such as ambiguous semantics, missing keywords, hypothetical statements, and negations, were discussed and resolved by consensus during an in-person meeting to create a criterion standard of manually reviewed notes.
Developing the KD Tagger
The main component of KD-NLP is a KD tagger, a software program that can identify a predefined KD sign within clinical text. To devise the KD tagger, we created annotation guidelines from 22 ED notes from children diagnosed with KD to identify KD signs from the clinical text. The annotation guidelines established a list of clinical signs that should be identified by the KD tagger. The purpose of the annotation guidelines was to translate medical language into linguistic signals that could be recognized by NLP software. We followed the AHA guidelines for KD and defined semantics tags for the five principal KD signs: EXTREMITY_CHANGES,POLYMORPHOUS_EXANTHE-MA, ORAL_ CHANGES, CONJUNCTIVAL_INJECTION, and CERVICAL_LYMPHADENOPATHY, in addition to FEVER.1 KD signs with examples of corresponding KD semantic tags are listed in Table 1. The annotation guidelines consisted of four parts for each tag: Definitions, Patterns, Examples, and Negations. Definitions were established from the AHA guidelines. Patterns indicate a word or phrase describing a KD sign in clinical text and were determined based on experience of the KD expert coauthors (AHT, JCB) as well as from the training notes. Negations in the annotation guidelines refer to which parts of the text should not be annotated. The annotation guidelines were revised and modified after several iterations of feedback from the KD experts. These guidelines served as the main reference for the development of the KD tagger. Two physicians among the authors (CKM, JDC) annotated all instances of relevant tags in the 22 ED notes. We then created a Web-based, HIPAA-compliant environment within the iDASH cloud15 and used Brat,16 a visual text annotation tool, available at http://brat.nlpla-b.org, to annotate the notes.
Overview of KD-NLP
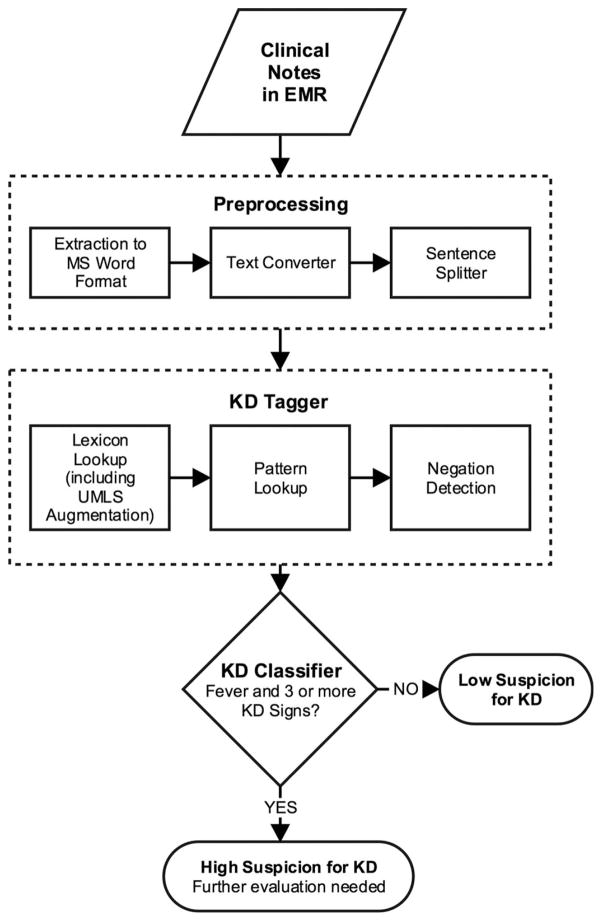
The pipeline of KD-NLP consists of three main modules (Figure 1):
Figure 1.
Algorithm followed by the KD-NLP tool. Preprocessing: This module includes converting text from MS Word format into plain-text files and a sentence splitter that breaks text into individual sentences. KD tagger: The KD tagger recognizes fever and KD signs from clinical text. KD classifier: This classifies a subject as high suspicion for KD if the number of KD signs detected by the KD tagger is at least three in addition to fever; otherwise, it assigns the subject as a low suspicion for KD. KD-NLP = Kawasaki disease natural language processing tool; MS = Microsoft; UMLS = Unified Medical Language System.
Preprocessing: ED provider notes were copied in their entirety from the EMR and saved in Microsoft (MS) Word format. We used the doc2txt package, written in Perl programming language, to convert text from MS Word format to plain-text format (available at http://docx2txt.sourceforge.net). We then used a Perl-based sentence splitter program developed by our group to divide text into individual sentences.
KD tagger: The KD tagger recognizes fever and KD signs from clinical text.
KD classifier: This module assigns a subject as high suspicion for KD if the KD tagger detects fever and three or more KD signs; otherwise, it assigns the subject as low suspicion for KD.
The KD tagger consists of three main components: lexicon look-up, pattern look-up, and negation detection (Figure 1).
Lexicon Look-up
We selected all keywords and key phrases of KD signs from 22 annotated training notes. There were a total of 313 keywords in the initial lexicon. We expanded this list by including synonyms according to the Unified Medical Language System (UMLS) dictionary.17 Each keyword/phrase in the initial lexicon was mapped into UMLS’s Concept Unified Identifier (CUI) using MetaMap.18 We extracted all synonyms from the UMLS’s CUI database. For example, “skin flaking” and “desquamation” are both synonyms of “peeling of skin” according to UMLS. We obtained a final lexicon of 28,580 keywords.
Pattern Look-up
We used regular expressions, sequences of characters that form search patterns, to represent patterns of KD signs according to the annotation guideline. For example, “Extremity Changes” with the semantic tag EXTREMITY_CHANGES in the sentence “hands and feet appeared red and swollen” matches the regular expression ^(hands and feet | hands or feet | hands | feet | palmar | plantar) [\w| ]*(red and swollen | red | swollen)$.
The source code for KD-NLP contained a total of 10 patterns for the semantic tag EXTREMITY_CHANGES, seven patterns for POLYMORPHOUS_EXANTHEMA, 17 patterns for CONJUNCTIVAL_INJECTION, 13 patterns for ORAL_CHANGES, and four patterns for CERVICAL_-LYMPHADENOPATHY. To recognize FEVER, we simply used a list of keywords including “fever” and “febrile.”
Negation Detection. The negation detection program finds negation linked to KD signs from clinical text such as “no red eyes” and does not annotate them as a KD semantic tag. To achieve this, we used NegEx19 (Python version implementation), available at https://code.-google.com/p/negex/. We added a negation pattern to identify a KD sign followed by “absent” as a negation, e.g., “Cervical adenopathy: absent.” We also included a negation pattern to identify KD signs in a sentence containing “if” as negations, e.g., “return if any new symptom develop (including new rashes, hand/foot swelling, lymphadenopathy).” The negation detection program follows the lexicon and pattern look-up.
KD Classifier
We sought to create a highly sensitive screening tool to detect patients with high suspicion for KD. In a separate analysis of all patients in the preexisting KD registry at Site 1, we found that among 455 patients with KD, 83 (18%) presented with five clinical signs, 257 (56%) with four signs, 80 (18%) with three signs, 30 (7%) with two signs, and five (1%) with one sign, with a cumulative 92% of patients presenting with three or more KD signs. Because minimal additional sensitivity was gained with the addition of patients with only one or two signs and inclusion of patients with so few signs would likely result in a significant loss of specificity, we classified as high suspicion those patients who had fever and three or more KD signs.
Outcomes
We evaluated KD-NLP’s ability to identify patients with fever and three or more KD signs, and thus a high suspicion for KD, compared to the manual review of ED notes. We also compared KD-NLP to a simple keyword search approach, which performs exact string matching using predefined keywords derived from the AHA guidelines for KD (Table 2).1
Table 2.
Keywords Used in the Simple Keyword Search Method
| Tag name | Keyword(s) |
|---|---|
| FEVER | fever, febrile |
| CONJUNCTIVAL_INJECTION | conjunctival injection, conj injection, red eyes, redness of eyes |
| ORAL_CHANGES | red lips, strawberry tongue |
| EXTREMITY_CHANGES | erythema of palms, erythema of soles, edema of hands, edema of feet, peeling of fingers, peeling of toes |
| POLYMORPHOUS_ EXANTHEMA | rash |
| CERVICAL_ LYMPHADENOPATHY | neck adenopathy, cervical adenopathy |
The list of terms was based on the American Heart Association guidelines for Kawasaki disease.
Data Analysis
We calculated sensitivity and specificity for the ability of KD-NLP and keyword search to classify suspicion for KD and identify individual signs.20 We calculated differences in proportions and the corresponding 95% confidence intervals (CIs). We used R package (version 2.15.1), available at https://www.r-project.org/, for statistical analysis.
RESULTS
Characteristics of Study Subjects
A total of 253 subjects were included in our evaluation cohort from the two clinical sites. There were no significant differences in sex between the sites (Table 3). The median age for the cohort at Site 1 was significantly higher than the median age for Site 2. While Hispanic patients predominated at Site 1, African Americans constituted the largest racial group at Site 2. A total of 93 unique authors created the 253 ED notes. We found that clinicians reliably documented the signs of KD in their text; 91.7% of the notes included documentation, whether positive or negative, for at least four of the five KD signs. The sign that was least frequently documented was extremity changes, found in only 65% of notes. The two physician reviewers found the mention of at least three KD signs in the notes of 173 subjects.
Table 3.
Demographic Characteristics of Study Subjects in the Evaluation Cohort
| Site 1 (n = 166) | Site 2 (n = 87) | 95% CI Difference Between Two Sites | |
|---|---|---|---|
| Sex, male, n (%) | 106 (63.9) | 46 (52.9) | −2.6 to 24.6 |
| Age (y), median (IQR) | 4.0 (2.1–5.9) | 3.0 (1.7–5.0) | −2.0 to −0.3 |
| Ethnicity, n (%) | |||
| Asian | 9 (5.4) | 3 (3.5) | −4.1 to 8.0 |
| African American | 8 (4.8) | 41 (47.1) | −54.2 to −30.4 |
| Caucasian | 47 (28.3) | 33 (37.9) | −3.5 to 22.7 |
| Hispanic | 57 (34.3) | 4 (4.6) | 20.4 to 39.1 |
| Mixed | 30 (18.1) | 2 (2.3) | 8.3 to 23.3 |
| Unknown | 15 (9.1) | 4 (4.6) | −2.6 to 11.5 |
IQR = interquartile range.
Classification of ED Notes for Suspicion of KD
Compared to manual review, the KD-NLP tool had a sensitivity of 93.6% (95% CI = 90.0% to 97.3%) and a specificity of 77.5% (95% CI = 68.4 to 86.7%) with no differences in performance between sites (Table 4). The simple keyword search had a lower sensitivity of 41.0% (95% CI = 33.7% to 48.4%) for identifying subjects with a high suspicion for KD (Data Supplement S1, available as supporting information in the online version of this paper).
Table 4.
Performance by Site of KD-NLP in Identifying Patients With at Least Three Clinical Signs of KD Compared to Manual Tagging by the Two Physician Reviewers of ED Clinical Notes
| Sites | Sensitivity | Specificity |
|---|---|---|
| Site 1 | 92.7 (87.8–97.6) | 79.0 (68.4–89.5) |
| Site 2 | 95.3 (90.1–100.0) | 73.9 (56.0–91.9) |
| Sites 1 and 2 combined | 93.6 (90.0–97.3) | 77.5 (68.4–86.7) |
Data are reported as % (CI).
KD-NLP = Kawasaki disease natural language processing tool.
Classification of Individual KD Signs
In the recognition of individual KD signs, sensitivity of the KD-NLP tool was highest for conjunctival injection, cervical lymphadenopathy, and rash (>95%; Data Supplement S2, available as supporting information in the online version of this paper). For FEVER, KD-NLP was able to correctly identify all subjects as having fever.
Sources of Error
An analysis of misclassification for high or low suspicion for KD revealed 11/173 (6.4%) false-negative and 18/180 (10%) false-positive errors. Among false-negative cases, nine (81.8%) were due to omission or misclassification of string patterns or keywords (e.g., “erythema of palms and soles” was classified as POLYMORPHOU-S_EXANTHEMA rather than EXTREMITY_CHANGES due to the word erythema being common to both tags), one (9.1%) due to misspelling (e.g., “midl swellign to hands”), and one (9.1%) due to preprocessing (e.g., line break in sentence splitting). Among the false-positive cases, 12 (66.6%) were due to assigning the wrong KD sign to a pattern (e.g., “erythema of pharynx” or “erythematous pharynx” as POLYMORPHOUS_EX-ANTHEMA), five (27.8%) were due to failing to recognize the negation (e.g., “neck without rigidity or adenopathy”), and one (5.6%) was due to a hypothetical sentence in the discharge instructions (e.g., “monitor at home for peeling of hands and feet”).
DISCUSSION
We developed KD-NLP as a screening tool to identify subjects with a high suspicion for KD in whom the diagnosis should be considered. It is important to recognize that this tool was evaluated not on the ability to diagnose KD based on a single clinical note but as a screening tool to reliably recognize signs of KD and to identify patients of higher suspicion for KD that may merit further laboratory or imaging evaluation. Although KD-NLP was trained on ED notes from a single institution, it performed equally well on clinical notes from a second, geographically and demographically distinct cohort. In fact when analyzed by final diagnosis, 83.3% of patients who were eventually diagnosed with KD and 47% of FC had three or more KD signs mentioned in their ED note.
NLP systems have previously successfully identified fever,21 diseases/treatments,22 medication information,23 and clinical phenotypes24–26 from clinical text. Keyword searches have identified pediatric cohorts for retrospective studies on complex febrile seizure.27,28 Predictive models using machine learning had an accuracy of 90% in distinguishing tractable and intractable epilepsy patients.29 An NLP system to categorize patients into low-, high-, and equivocal-risk categories for acute appendicitis had a sensitivity of 89.7% and positive predicted value of 95.2%.30 NLP systems have also determined disease severity in epilepsy31 and asthma status32 and have been applied to multiple sources of clinical notes including radiology reports, ED notes, discharge summaries, clinical visits, and progress notes.33
NLP has also been used for a variety of phenotypic identification tasks from analyzing chest radiograph reports to identify patients with possible tuberculosis to recognizing postoperative complications in surgical patients.34,35 More recently, a group at Cincinnati Children’s Hospital utilized a combination of clinical variables extracted from physician and nursing notes by NLP and discrete variables such as vital signs and laboratory results to automate the calculation of the previously validated Pediatric Appendicitis Score and assign risk categories for patients seen in the ED for abdominal pain.30 The automated scoring system was able to classify patients as well as physician experts.
The high sensitivity (>92%) observed at both sites suggests that the tool can be useful in the ED setting to identify subjects with a high suspicion for KD. The incorporation of UMLS lexicon into KD-NLP increased its sensitivity compared to the simple keyword search based on the AHA clinical criteria for KD. Although we have focused on fever plus the five major clinical criteria for KD, future iterations could include more subtle examination findings such as perilimbic sparing, perineal accentuation of rash, or arthritis. A difficulty with all NLP tools is the inherent reliance on clinicians to adequately document their history and physical findings. However, > 90% of notes documented the presence or absence of at least four of the five KD signs.
Incorporated into the electronic health record, KD-NLP could drive clinical decision support by identifying patients who might benefit from further evaluation for KD. The tool and annotation guidelines are publicly available at https://idash-data.ucsd.edu/community/68. Our ultimate intention is to implement this in a real-time or near-real-time manner in departments with initial patient contact (urgent care, ED, or primary pediatric offices) such that the clinical decision support occurs at the point of care. Possible implementations we are considering include the use of an internal program placed upon the Epic EMR architecture that can evaluate note files as they are intermittently autosaved to the shadow server such that there is not an excessive processing burden placed upon the production system.
LIMITATIONS
We recognize several strengths and limitations to our study. This is the first NLP tool designed to aid health care providers in the timely recognition of patients with KD. A limitation in the development of the KD-NLP tool was the limited variety of syntax we encountered within our training set. Missed signs and the resultant misclassification occurred because of spelling errors, hypothetical clauses, and syntax for which we had not trained the tool. As we continue to analyze additional clinical documentation and increase our syntactic repertoire, future iterations of the tool will be better able to identify these variations.
Our patient population was drawn from pediatric facilities and chosen to maximize the frequency of KD signs to test the tool’s ability to identify them. However, this enriched cohort may limit the applicability of the tool to the general ED population where prevalence of KD and similar febrile illnesses is lower. The clinical notes, authored by clinicians familiar with KD, may not represent the documentation styles of ED physicians working in nonpediatric facilities.
Lastly, our analysis was done retrospectively with finalized documentation from a single encounter and was not performed on a real-time basis. As physician documentation may not be completed prior to medical decision-making (and occasionally up to 24 hours after the encounter), we recognize that this tool may not provide support at the time of the encounter. However, we believe that identifying high suspicion patients even with a short delay can shorten time to diagnosis and ultimately improve outcomes. We plan to perform timestamp analysis in future studies to evaluate how the tool performs on a real-time basis to address this limitation. Because patients with KD often present multiple times for medical care prior to diagnosis, incorporation of data from several visits may improve the performance of our tool and will be part of our future plans. It is also important to note that the type of EMR documentation aids, free text, and templates as well as documentation workflow such as the use of scribes might impact the feasibility of the KD-NLP tool and will need to be considered in future versions.
CONCLUSIONS
The Kawasaki disease natural language processing tool had performance comparable to manual chart review in identifying pediatric ED patients with three or more Kawasaki disease clinical signs. The Kawasaki disease natural language processing tool could be incorporated into the electronic health record to screen for patients for whom KD should be considered in the differential diagnosis and for whom additional testing or referral may be appropriate. A prospective validation study of the Kawasaki disease natural language processing tool in nonpediatric EDs and urgent care settings is warranted as is further testing of integrating this natural language processing pattern recognition algorithm into the electronic medical record architecture.
Supplementary Material
Acknowledgments
This work was supported in part by the Patient-Centered Outcomes Research Institute (PCORI), contract CDRN-1306-04819 and NIH grant U54HL108460.
We thank Divya Chhabra, Amanda Ritchart, and Mark Myslín for their contributions to the project.
Footnotes
The following supporting information is available in the online version of this paper:
Data Supplement S1. Performance by site of the simple keyword search in classification of suspicion for KD compared to manual tagging by the two physician reviewers of Emergency Department clinical notes.
Data Supplement S2. Performance of KD-NLP and simple keyword search in identification of individual KD clinical criteria.
Part of this work was presented in poster format at the Pediatric Academic Societies Annual Meeting in San Diego, CA, on April 27, 2015.
The authors have no potential confiicts to disclose. Supervising Editor: Damon Kuehl, MD.
References
- 1.Newburger JW, Takahashi M, Gerber MA, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 2004;110:2747–71. doi: 10.1161/01.CIR.0000145143.19711.78. [DOI] [PubMed] [Google Scholar]
- 2.Newburger JW, Takahashi M, Burns JC, et al. The treatment of Kawasaki syndrome with intravenous gamma globulin. N Engl J Med. 1986;315:341–7. doi: 10.1056/NEJM198608073150601. [DOI] [PubMed] [Google Scholar]
- 3.Newburger JW, Takahashi M, Beiser AS, et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N Engl J Med. 1991;324:1633–9. doi: 10.1056/NEJM199106063242305. [DOI] [PubMed] [Google Scholar]
- 4.Wilder MS, Palinkas LA, Kao AS, Bastian JF, Turner CL, Burns JC. Delayed diagnosis by physicians contributes to the development of coronary artery aneurysms in children with Kawasaki syndrome. Pediatr Infect Dis J. 2007;26:256–60. doi: 10.1097/01.inf.0000256783.57041.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson MS, Todd JK, Glodé MP. Delayed diagnosis of Kawasaki syndrome: an analysis of the problem. Pediatrics. 2005;115:e428–33. doi: 10.1542/peds.2004-1824. [DOI] [PubMed] [Google Scholar]
- 6.Ling XB, Lau K, Kanegaye JT, et al. A diagnostic algorithm combining clinical and molecular data distinguishes Kawasaki disease from other febrile illnesses. BMC Med. 2011;9:130. doi: 10.1186/1741-7015-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling XB, Kanegaye JT, Ji J, et al. Point-of-care differentiation of Kawasaki disease from other febrile illnesses. J Pediatr. 2013;162:183–8. e3. doi: 10.1016/j.jpeds.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gausche-Hill M, Schmitz C, Lewis RJ. Pediatric pre-paredness of US emergency departments: a 2003 survey. Pediatrics. 2007;120:1229–37. doi: 10.1542/peds.2006-3780. [DOI] [PubMed] [Google Scholar]
- 9.Kanegaye JT, Van Cott E, Tremoulet AH, et al. Lymphnode-first presentation of Kawasaki disease compared with bacterial cervical adenitis and typical Kawasaki disease. J Pediatr. 2013;162:1259–63. 1263.e1–2. doi: 10.1016/j.jpeds.2012.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minich LL, Sleeper LA, Atz AM, et al. Delayed diagnosis of Kawasaki disease: what are the risk factors? Pediatrics. 2007;120:e1434–40. doi: 10.1542/peds.2007-0815. [DOI] [PubMed] [Google Scholar]
- 11.Senzaki H. Long-term outcome of Kawasaki disease. Circulation. 2008;118:2763–72. doi: 10.1161/CIRCULATIONAHA.107.749515. [DOI] [PubMed] [Google Scholar]
- 12.Gordon JB, Kahn AM, Burns JC. When children with Kawasaki disease grow up: myocardial and vascular complications in adulthood. J Am Coll Cardiol. 2009;54:1911–20. doi: 10.1016/j.jacc.2009.04.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels LB, Tjajadi MS, Walford HH, et al. Prevalence of Kawasaki disease in young adults with suspected myocardial ischemia. Circulation. 2012;125:2447–53. doi: 10.1161/CIRCULATIONAHA.111.082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizk SR, El Said G, Daniels LB, et al. Acute myocardial ischemia in adults secondary to missed Kawasaki disease in childhood. Am J Cardiol. 2015;115:423–7. doi: 10.1016/j.amjcard.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno-Machado L, Bafna V, Boxwala AA, et al. iDASH: integrating data for analysis, anonymization, and sharing. J Am Med Inform Assoc. 19:196–201. doi: 10.1136/amiajnl-2011-000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenetorp P, Pyysalo S, Topic G, Ohta T, Ananiadou S, Tsujii J. BRAT: a web-based tool for NLP-assisted text annotation. Proc Demonstr 13th Conf Eur Chapter Assoc Comput Linguist; 2012; pp. 102–7. [Google Scholar]
- 17.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the AMIA Symposium. American Medical Informatics Association; 2001; pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 20.Manning CD, Schutze H. Foundations of Statistical Natural Language Processing. Cambridge (MA): The MIT Press; 1999. [Google Scholar]
- 21.Chapman WW, Dowling JN, Wagner MM. Fever detection from free-text clinical records for biosurveillance. J Biomed Inform. 2004;37:120–7. doi: 10.1016/j.jbi.2004.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/ VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17:514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathak J, Wang J, Kashyap S, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc. 2011;18:376–86. doi: 10.1136/amiajnl-2010-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 27.Kimia AA, Capraro AJ, Hummel D, Johnston P, Harper MB. Utility of lumbar puncture for first simple febrile seizure among children 6 to 18 months of age. Pediatrics. 2009;123:6–12. doi: 10.1542/peds.2007-3424. [DOI] [PubMed] [Google Scholar]
- 28.Kimia A, Ben-Joseph EP, Rudloe T, et al. Yield of lumbar puncture among children who present with their first complex febrile seizure. Pediatrics. 2010;126:62–9. doi: 10.1542/peds.2009-2741. [DOI] [PubMed] [Google Scholar]
- 29.Savova G, Deleger L, Solti I, Pestian J, Dexheimer J. Pediatric Biomedical Informatics: Computer Applications in Pediatric Research (Translational Bioinformatics) New York: Springer Verlag; 2013. Natural Language Processing - Applications in pediatric research; pp. 173–92. [Google Scholar]
- 30.Deleger L, Brodzinski H, Zhai H, et al. Developing and evaluating an automated appendicitis risk stratification algorithm for pediatric patients in the emergency department. J Am Med Inform Assoc. 2013;20:e212–20. doi: 10.1136/amiajnl-2013-001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connolly B, Matykiewicz P, Cohen K, et al. Assessing the similarity of surface linguistic features related to epilepsy across pediatric hospitals. J Am Med Inform Assoc. 2014;21:866–70. doi: 10.1136/amiajnl-2013-002601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu ST, Juhn YJ, Sohn S, Liu H. Patient-level temporal aggregation for text-based asthma status ascertainment. J Am Med Inform Assoc. 2014;21:876–84. doi: 10.1136/amiajnl-2013-002463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doan S, Conway M, Phuong TM, Ohno-Machado L. Natural language processing in biomedicine: a unified system architecture overview. Methods Mol Biol. 2014;1168:275–94. doi: 10.1007/978-1-4939-0847-9_16. [DOI] [PubMed] [Google Scholar]
- 34.Jain NL, Knirsch CA, Friedman C, Hripcsak G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp. American Medical Informatics Association; 1996; pp. 542–6. [PMC free article] [PubMed] [Google Scholar]
- 35.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306:848–55. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.