Abstract
Preclinical studies have suggested that the pancreatic tumor microenvironment both inhibits and promotes tumor development and growth. Here we establish the role of stromal fibroblasts during acinar-to-ductal metaplasia (ADM), an initiating event in pancreatic cancer formation. The transcription factor V-Ets avian erythroblastosis virus E26 oncogene homolog 2 (ETS2) was elevated in smooth muscle actin–positive fibroblasts in the stroma of pancreatic ductal adenocarcinoma (PDAC) patient tissue samples relative to normal pancreatic controls. LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre (KPC) mice showed that ETS2 expression initially increased in fibroblasts during ADM and remained elevated through progression to PDAC. Conditional ablation of Ets-2 in pancreatic fibroblasts in a KrasG12D-driven mouse ADM model decreased the amount of ADM events. ADMs from fibroblast Ets-2–deleted animals had reduced epithelial cell proliferation and increased apoptosis. Surprisingly, fibroblast Ets-2 deletion significantly altered immune cell infiltration into the stroma, with an increased CD8+ T-cell population, and decreased presence of regulatory T cells (Tregs), myeloid-derived suppressor cells, and mature macrophages. The mechanism involved ETS2-dependent chemokine ligand production in fibroblasts. ETS2 directly bound to regulatory sequences for Ccl3, Ccl4, Cxcl4, Cxcl5, and Cxcl10, a group of chemokines that act as potent mediators of immune cell recruitment. These results suggest an unappreciated role for ETS2 in fibroblasts in establishing an immune-suppressive microenvironment in response to oncogenic KrasG12D signaling during the initial stages of tumor development.
Introduction
Pancreatic cancer 5-year survival rates have remained around 5% for the last 40 years despite efforts to better understand the underpinnings of this disease [1], [2]. Research for the last 2 decades has concentrated on determining the major oncogenic signaling pathways within the evolving tumor cell [3], [4], [5]. These efforts have revealed that activating mutations in Kras are early events during malignant transformation and have suggested that acinar cells are a potential pancreatic ductal adenocarcinoma (PDAC) cell-of-origin [6], [7]. For example, genetically engineered mouse models demonstrated that mutant Kras targeted to the acinar lineage is sufficient to induce pancreatic intraepithelial neoplasia (PanIN) lesions with ductal morphology [8], [9], [10]. Additionally, second-hit mutations, such as activating Notch mutations, synergize with Kras in acinar cells to drive ductal reprogramming and PanIN lesion formation [11]. Lineage tracing experiments have shown that acinar cells readily transdifferentiate into ductal cells (commonly referred to as acinar-to-ductal metaplasia or ADM) and also demonstrated that oncogenic Kras mutations in mature acinar cells formed PanINs at a significantly higher frequency relative to the ductal or centroacinar cell lineages [12]. This paradigm-shifting study has sparked newfound interest into the signaling networks that collaborate with Kras to drive ADM in acini [13], [14], [15], [16], [17]. Other reports have solidified the notion that ADM progresses to PanIN and PDAC and show that ADM occurs in human PDAC patients [18], [19], [20], [21]. During the transformation from ADM to PanIN to PDAC, there is a concurrent increase in the desmoplastic reaction that is a cardinal feature of this disease [22], [23], [24].
The pancreatic stroma has been scrutinized as a “partner in crime” for PDAC development, but efforts to elucidate the role of the stroma in pancreatic cancer to date have relied primarily on in vitro or transplanted immunodeficient in vivo models of pancreatic cancer [25], [26], [27], [28]. More recent reports have revealed that stromal fibroblasts are comprised of a heterogeneous population that possesses both pro- and antitumor characteristics [29], [30], [31]. For example, the presence of smooth muscle actin (SMA)–positive fibroblasts in human PDAC samples has been correlated with reduced overall survival in one patient cohort and with increased patient survival in an independent patient cohort [31], [32]. Furthermore, stromal gene expression in 145 human PDAC patient samples revealed two distinct stroma-specific subtypes (termed normal and activated) with median survivals of 24 or 15 months, respectively [33]. Collectively, these observations have fueled interest in studying the molecular interactions between the epithelium and recruited stromal fibroblasts. Surprisingly, the contribution of fibroblasts to ADM has not garnered the same attention, and noncell autonomous signaling that drives ADM is not well described.
ETS2 is a member of the ETS transcription factor family that has been studied as a downstream regulator of RAS-mediated transformation [34], [35], [36], [37]. ETS2 also interacts specifically with mutant p53 to redirect promoter recruitment of p53, and further studies have shown that ETS2 is crucial for mutant tp53 oncogenic functions [38], [39], [40]. The ability of ETS2 to regulate both RAS- and p53-mediated signaling places it at a crossroads between these two powerful oncogenic signaling pathways that are misregulated in pancreatic cancer. Previous studies have revealed that ETS2 also regulates epithelial cell fate from the stromal compartment [41], [42]. Specifically, deletion of Ets2 in the epithelium of mouse mammary tumor models had no significant effect, whereas fibroblast-specific Ets2 deletion led to markedly decreased tumor growth [42]. These studies have established that fibroblast ETS2-mediated signaling is an important regulator of the complex cross talk between stromal cells and epithelial cells. ETS factors in general (especially ETS1 and ETS2) are enriched in stromal cell populations in many cancers through a variety of mechanisms [41], [43], [44], [45], [46].
In this report, we show that ETS2 signaling is activated in human and mouse pancreatic fibroblasts during ADM. We further use genetically engineered mouse models of ADM to demonstrate that deletion of Ets-2 in fibroblasts leads to decreased acinar cell transformation, decreased immune cell infiltration, and decreased cytokine and chemokine production by fibroblasts. These results define a fibroblast-specific ETS2-choreographed immune response that leads to an immune-suppressive microenvironment during the earliest stages of preneoplastic transformation of the pancreas.
Materials and Methods
Animal Strains, Husbandry, and Maintenance
The use of animals was in compliance with federal and Ohio State University Laboratory Animal Resources regulations. Mist1KrasG12D; Fsp-Cre; Ets2db/loxP and Mist1KrasG12D; Ets2db/loxP animals were generated by crossing the previously described Mist1KrasG12D, FspCre, Ets-2loxP, and Ets-2db strains [10], [47], [48], [49]. The experiments were performed using littermate mice from a mixed C57BL/6; 129/Sv and FVBN genetic background.
Multispectral Analysis of Dual-Color Immnohistrochemistry (IHC)
Dual-stained samples were imaged using the PerkinElmer's Vectra multispectral slide analysis system. For the mouse samples, at least three multispectral images per animal for at least three mice per genotype (unless otherwise noted) were manually taken. For the human PDAC Tissue Microarray, one field of interest per core was automatically acquired. The image acquisition workflow consisted of the following: 1) monochrome imaging of the entire slide, 2) RGB low-power imaging of the tumor tissue using an inForm tissue finding algorithm, and 3) multispectral high-power imaging of one field containing tumor epithelium and stroma by means of an inForm HPF finding algorithm.
For quantification of the DAB staining, the multispectral images were reviewed and analyzed using inForm Tissue Finder software. A pattern recognition algorithm was used for processing as follows: 1) trainable tissue segmentation to segment the SMA-positive regions from the tumor epithelium; 2) cell segmentation of the SMA-positive tissue category to locate the subcellular compartments; and 3) scoring to bin the spectrally unmixed DAB signal into four categories depending on the staining intensity (0+, 1+, 2+, and 3+), providing data in percent. The H-score, which ranges from 0 to 300, was calculated using following formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)]. Thus, H-score measures staining intensity as well as percentage of positive cells in a given cellular compartment.
Histology Analysis, Immunohistochemistry, and Immunofluorescent Staining
Dissected mouse pancreas tissues were fixed in 10% neutral-buffered formalin solution for 48 hours and transferred to 70% ethanol. Tissues were processed, embedded in paraffin, cut in 5 μm sections on positively charged slides, deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E).
For immunohistochemistry, all sections were stained using a Bond Rx autostainer (Leica) unless otherwise noted. Briefly, slides were baked at 65°C for 15 minutes, and automated software performed dewaxing, rehydration, antigen retrieval, blocking, primary antibody incubation, post primary antibody incubation, detection (DAB or RED), and counterstaining using Bond reagents (Leica). Samples were then removed from the machine, dehydrated through ethanols and xylenes, mounted, and coverslipped. Antibodies for the following markers were diluted in antibody diluent (Leica): rabbit anti-bodies αSMA (1:1500, Abcam), Ki67 (1:100, Abcam), and amylase (1:400, CST), ETS2 (SC-351) and rat antibodies cytokeratin 19 (TROMA-III) (1:150, DSHB, University of Iowa), and F4/80 (1:50 Invitrogen).
For fluorescent immunostaining, following deparaffinization, rehydration, and PBS washes, sections were blocked with serum-free Protein block (Dako) for 30 minutes at room temperature. Cells grown on coverslips were fixed with 4% paraformaldehyde and permeabilized with 100% ice-cold methanol prior to blocking. Tissue immunostaining required antigen retrieval, which was performed in a steamer (Black & Decker) using a 1× Target Retrieval solution (pH 6.0) (Dako). Antibodies for the following markers were diluted in antibody diluent (Dako) and applied overnight at 4°C: rabbit antibodies Ki67 (1:100, Abcam) and amylase (1:400, CST), rat antibodies cytokeratin 19 (TROMA-III) (1:150, DSHB, University of Iowa) and cytokeratin 8 (TROMA-I) (1:300, DSHB, University of Iowa), and mouse antibody α-SMA (1:400 Sigma). Secondary antibodies conjugated with Alexa Fluor 488 and Alexa Fluor 594 were placed on tissue sections for 1 hour at room temperature (1:300, Invitrogen). Nuclei were counterstained using SlowFade Gold Antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). Tissue sections for fluorescence microscopy images were mounted with SlowFade Gold Antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen) and coverslipped, and images were obtained using an Eclipse E800 microscope (Nikon) equipped with a CoolSNAP cf. monochrome digital camera Peltier cooled to 20°C (Photometrics). All images were captured with MetaVue version 6.2r6 software (Universal Imaging Corp.) and resized and formatted with Adobe Photoshop CS5 software (Adobe Systems Incorporated).
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed as previously described [43]. Briefly, cells were cross-linked for 10 minutes with 1% formaldehyde and lysed. Chromatin was collected and sonicated. Anti-Ets2 ChIP grade antibody (sc-351X) and protein G agarose slurry (Millipore 16-266) were used to pull down chromatin. After reverse cross-linking, DNA was recovered, purified, and analyzed by reverse transcriptase polymerase chain reaction (RT-PCR).
Pancreatic Fibroblast Isolation and Culture
Primary pancreatic fibroblasts were purified based on the following protocol. Briefly, pancreata were dissected from 6-month-old mice, minced, and digested with collagenase (0.5% Collagenase II, 120 U ml−1 DNase I in 1× PBS) shaking at 225 rpm for an hour at 37°C. Collagenase was neutralized with 10% FBS-Dulbecco’s modified Eagle’s medium. Digested tissue was resuspended in medium and gravity purified for 10 minutes. Supernatants were aspirated, and pellets were washed three times and subjected to two additional gravity sedimentations and then seeded on tissue-culture dishes.
RNA, Microarray, and Real-Time PCR
RNA was harvested with Trizol according to the manufacturer's instructions (Invitrogen). RNA quality and concentration were assessed with Bioanalyser and Nanodrop RNA 6000 nanoassays. RNA samples were hybridized to Affymetrix GeneChip Mouse genome 430 2.0 platform at the Microarray Shared Resource Facility, Ohio State University Comprehensive Cancer Center. The microarray data were deposited with Gene Expression Omnibus (GEO) and can be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=afqbqaeoxbgltor&acc=GSE61707.
The RNA-sequencing data were deposited with GEO under Series GSE76498 and can be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=mlwjyqiqplmfbyd&acc=GSE76498. Real-time quantification of mRNA was performed using Roche Universal Primer Library probes and probe-specific primers. Applied Biosystems StepOnePlus real time PCR machines were used. Normalization and target to reference ratios were calculated according to the manufacturer's instructions (Roche). Total RNA (1 μg) was converted to cDNA using the Super-script Reverse Transcriptase kit (Invitrogen). List of quantitative (q)RT-PCR and Roche Universal Probe library number is indicated in the table below:
| Gene Name | Forward Primer | Reverse Primer | Probe # |
|---|---|---|---|
| hRpl4 | AGCGTGGCTGTCTCCTCTC | GGACCCATCAAAGGTGTCAA | 40 |
| hEts2 | CCCCTGTGGCTAACAGTTACA | AAGGGAGCACAGCAAACAGA | 20 |
| hGli1 | CAGGGAGGAAAGCAGACTGA | ACTGCTGCAGGATGACTGG | 76 |
| hGli2 | ACTCCACACACGCGGAAC | CCACTGAAGTTTTCCAGGATG | 16 |
| hGli3 | TCAAACCCGATGAAGACCTC | TTGTTCCTTCGGGCTGTT | 26 |
| hStat3 | CCTCTGCCGGAGAAACAG | CTGTCACTGTAGAGCTGATGGAG | 1 |
| hJun | CCAAAGGATAGTGCGATGTTT | CTGTCCCTCTCCACTGCAAC | 19 |
| hMyc | CGGTTTTCGGGGCTTTAT | GGCTCTTCCACCCTAGCC | 13 |
| hSox11 | GAGCTGAGCGAGATGATCG | GAACACCAGGTCGGAGAAGT | 19 |
| hMed1 | AACACCCTCATTGGAAGCTG | GGACACACTTCAAATTGGAGAA | 2 |
| hp65 | CGGGATGGCTTCTATGAGG | GGATGCGCTGACTGATAGC | 1 |
| mRpl4 | GATGAGCTGTATGGCACTTGG | CTTGTGCATGGGCAGGTTA | 38 |
| mCcl3 | TGCCCTTGCTGTTCTTCTCT | GTGGAATCTTCCGGCTGTAG | 40 |
| mCcl4 | AGGAGGAGCCACTTCAGGA | GAGCAAGGACGCTTCTCAGT | 73 |
| mCxcl4 | AGCGGTGGTTGCTGTCAC | TGGTGATGTGCTTAAGATGGA | 82 |
| mCxcl5 | TAGAGCCCCAATCTCCACAC | GAGCTGGAGGCTCATTGTG | 67 |
| mCxcl10 | GCTGCCGTCATTTTCTGC | TCTCACTGGCCCGTCATC | 3 |
| chipCcl3 | GTCCTACCTCCTCCTGCTCA | TCAGCTCTCAACTCGTGACC | 60 |
| chipCcl4 | CCACTCCATCTCCCTCCTTT | GAAAGTGAGGAGTTCCTGACAGA | 3 |
| chipCxcl4 | CACATCTCCAACCCCACAG | CACGTAACTCCAAAGCCACTG | 4 |
| chipCxcl5 | GGCAAAGGGTGCAAAGATT | CAACTTCACAGATGACTCAGCA | 69 |
| chipCxcl10 | CTCAGCGGTGGATGAAGC | GGACTCAGGGAGGGAAACTC | 73 |
| hCcl3 | TCTTCCTAACCAAGCGAAGC | GAAGCTTCTGGACCCCTCA | 1 |
| hCcl4 | CTCTCCAGCGCTCTCAGC | ACCACAAAGTTGCGAGGAAG | 40 |
| hCxcl4 | ACTGAGATCCTGCTGGAAGC | AAGTGGCAGGAGCAGCAA | 66 |
| hCxcl5 | TGACACTTGTGAAAAGGCTTGTA | AGCAAAAATAGAAATTCACAACCA | 52 |
| hCxcl10 | GATGCAGTGCTTCCAAGGAT | TGACATATACTCCATGTAGGGAAGTG | 34 |
Statistics
Wilcoxon rank sum test and Student's t test were calculated using R 3.0.1. The P values from Student's t tests are listed unless otherwise specified. In all graphs, median, means (bar), and standard deviations (lines) are denoted. Microarray data were processed by Robust Multiarray Average method and analyzed using the moderated t test approach [50]. P value of .05 was considered significant.
Results
Ets-2 Signaling Is Activated in Pancreatic Fibroblasts
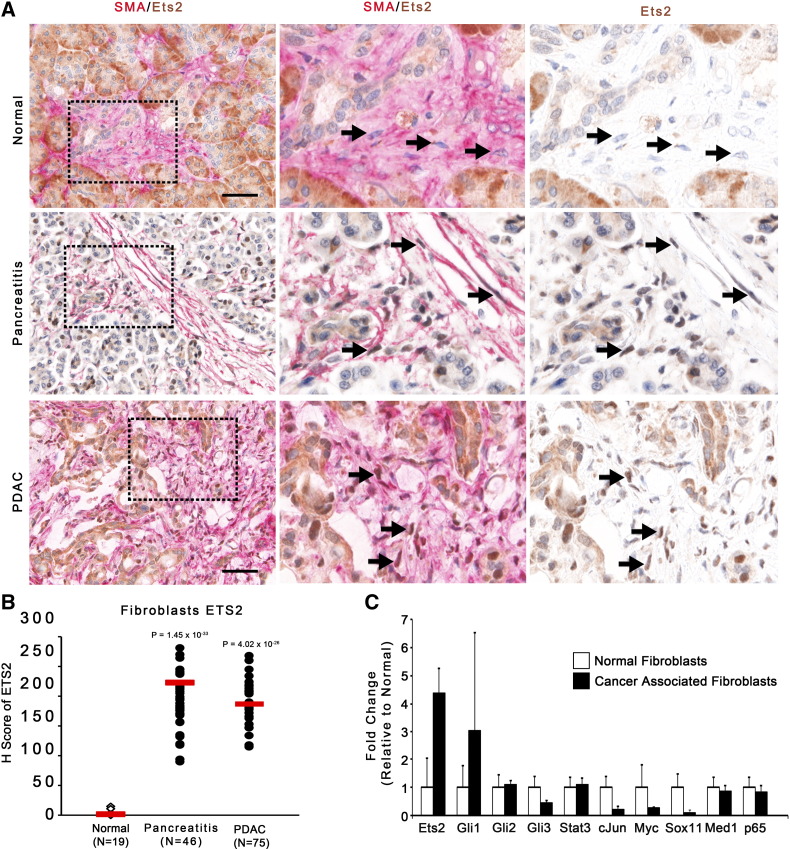
Previous studies have shown that ETS2 levels increase in ductal tumor cells with PDAC staging, but fibroblast ETS2 levels have yet to be explored [51]. Dual-color IHC staining of normal pancreatic tissue, pancreatitis tissue, and PDAC patient tissue for alpha-SMA and ETS2 was analyzed using the Vectra multispectral imaging system (Figure 1A, see Materials And Methods). The analysis demonstrated that nuclear ETS2 staining was significantly enriched in fibroblasts from both pancreatitis and PDAC patient samples relative to normal pancreatic controls (Figure 1B). Robust staining within acinar and ductal epithelial cells in both normal and PDAC patient samples was also observed, consistent with previous reports [51]. To further test the hypothesis that ETS2 is activated in pancreatic fibroblasts, primary fibroblasts from resected PDAC patient tumors were isolated, and mRNA expression of Ets-2 was compared relative to other proto-oncogenic transcription factors. Among the 10 transcription factors analyzed, only Ets-2 and Gli1 showed significantly higher levels of mRNA in cancer-associated fibroblasts relative to control pancreatic fibroblasts (Figure 1C). Gli1, the nuclear effector of Hedgehog receptor signaling, has been previously shown to be expressed by pancreatic cancer–associated fibroblasts [52], [53].
Figure 1.
Human pancreatic cancer associated fibroblasts activate Ets-2 signaling.
(A) Dual-color IHC (ETS2, brown; SMA, red) of human normal or PDAC Tissue Microarray. Insets show higher magnified view of representative areas. Black arrows indicate ETS2+/SMA+ fibroblasts. Scale bars 25 μm. (B) Quantification of ETS2 staining in SMA+ positive cells shown in A (red bar indicates mean). (C) qRT-PCR analysis of proto-oncogenic transcription factors in pancreatic fibroblasts isolated from normal or PDAC patients (normalized to Rpl4, represented as fold-change relative to normal fibroblast expression; n = 3; bars represent means ± Standard Deviation [SD]).
Fibroblast Ets-2 Is Upregulated Early and Late in Pancreatic Tumorigenesis
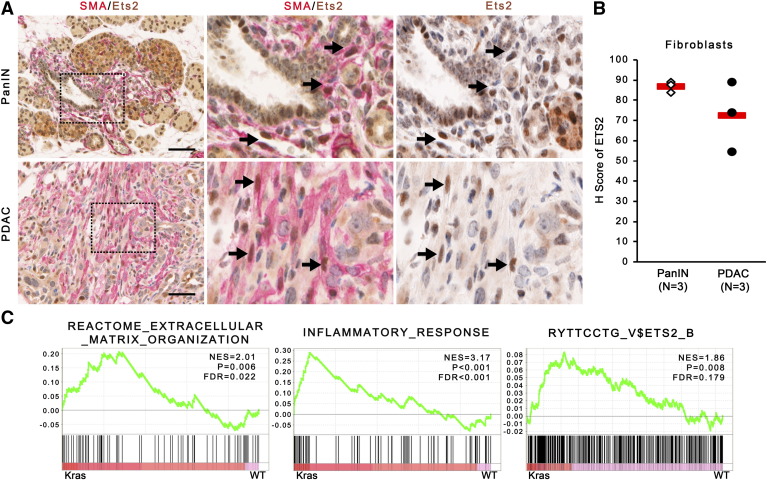
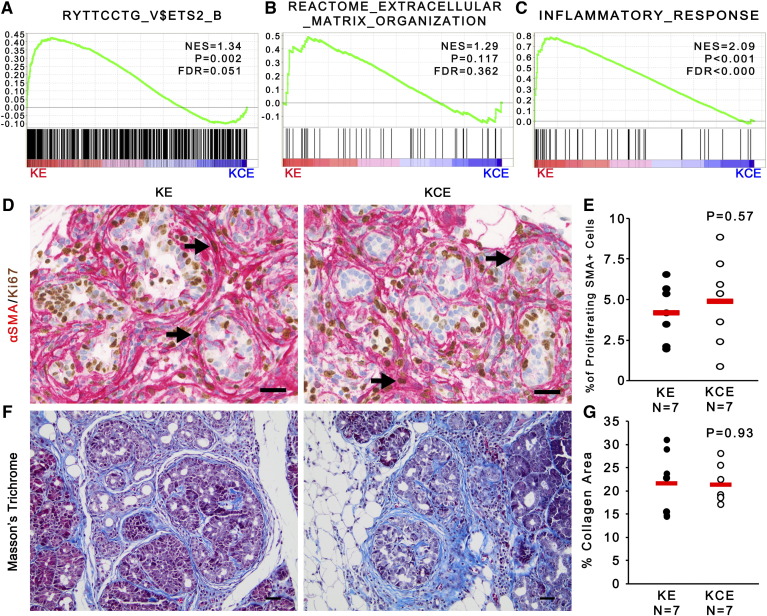
SMA-ETS2 dual IHC was repeated in a cohort of LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre (KPC) mice, demonstrating that ETS2 is activated in fibroblasts during early ADM and PanIN stages (2 weeks of age) as well as in PDAC (4 months of age, Figure 2, A and B). Gene set enrichment analysis (GSEA) of RNA sequencing of fibroblast mRNA isolated from WT or KPC pancreata at the ADM stage demonstrated that extracellular matrix remodeling and inflammatory response pathways were highly enriched in KPC fibroblasts relative to WT fibroblasts (Figure 2C). GSEA also identified ETS2-regulated genes as an altered process in fibroblasts isolated from KPC mice undergoing ADM (Figure 2C). This same ETS2 GSEA pathway (“RYTTCCTG_V$ETS2_B” Broad Institute MSigDB gene set) was significantly altered (P = 1.68 × 10−4) in the “activated stroma” signature relative to “normal stroma” in the recent analysis of stromal gene expression from 145 PDAC patients [33].
Figure 2.
ETS2 is upregulated in PanIN and PDAC fibroblasts.
(A) Dual-color IHC (ETS2, brown; SMA, red) of pancreata from KPC mice at PanIN or PDAC stage. Insets show higher magnified view of representative areas. Black arrows indicate ETS2+/SMA+ fibroblasts. Scale bars 25 μm. (B) Quantification of ETS2 staining in SMA+ positive cells shown in A (red bar indicates mean). (C) GSEA enrichment plots for ECM remodeling, inflammation, and ETS2-regulated gene pathways in mouse WT or KPC pancreatic fibroblast cultures (n = 3).
Fibroblast-Specific Deletion of Ets-2 Decreases KrasG12D-Driven ADM
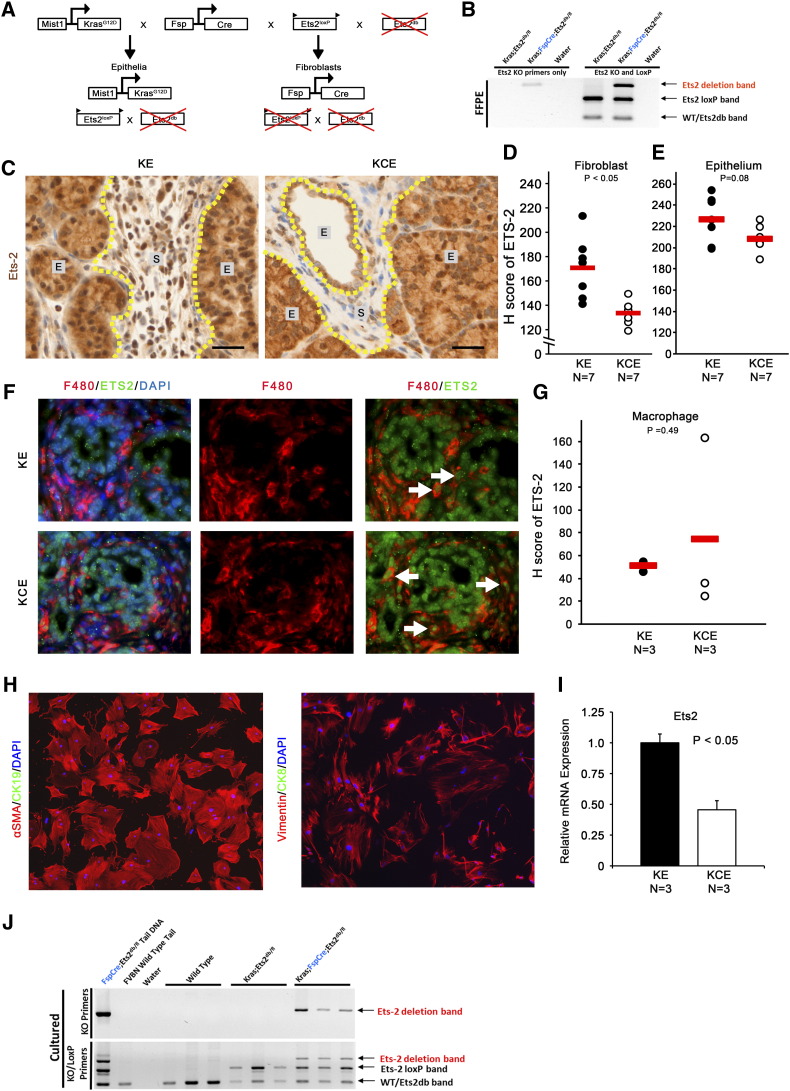
To determine the role of Ets2 signaling in epithelial-stromal cross talk during ADM formation, we utilized the Mist1KrasG12D knock-in allele in which oncogene expression originates in the acinar cell compartment [10]. This model was combined with fibroblast-specific Fsp-Cre, which was previously created and characterized extensively by our group [41], [42], and Ets2db/loxP alleles to achieve efficient fibroblast-specific conditional deletion of Ets-2. Mist1-KrasG12D; Ets2db/loxP (KE) control mice and Mist1-KrasG12D; Fsp-Cre; Ets2db/loxP (KCE) experimental mice were generated (Supplemental Figure 1A, see Materials and Methods). Recombination of the Ets2loxP allele was confirmed in DNA isolated from formalin-fixed, paraffin-embedded tissue sections by genotyping PCR (Supplemental Figure 1B). IHC staining and quantification using Vectra multispectral imaging of ETS2 in pancreatic tissue sections from KE and KCE mice confirmed in vivo deletion exclusively in the fibroblast compartment (Supplemental Figure 1, C and D), whereas ETS2 levels in the pancreatic epithelium remained unchanged (Supplemental Figure 1E). Pancreatic fibroblasts were isolated from KE and KCE mice and shown to be positive for fibroblast markers SMA and vimentin and negative for epithelial markers cytokeratin 8 (CK8) or cytokeratin 19 (CK19) by co-IF (Supplemental Figure 1H). KCE fibroblast cultures had lower Ets-2 mRNA relative to KE (Supplemental Figure 1I), and genotyping PCR was performed to show deletion of Ets-2loxP exclusively in cultures isolated from Fsp-Cre–bearing mice (Supplemental Figure 1J).
Supplemental Figure 1.
(A) Graphic of inherited alleles in epithelial and fibroblast cellular lineages. (B) Ets2 genotyping PCR of DNA derived from formalin-fixed, paraffin-embedded tissue of indicated genotypes. (C) Dual-color IHC (ETS2, brown; SMA, red) of KE and KCE pancreata (E = epithelium; S = stroma). Scale bars 25 μm. (D–E) Quantification of ETS2 staining in SMA+ positive fibroblasts or pancreatic epithelium shown in A (red bar indicates mean; n = 7). (F) Co-IF for F480/ETS2; white arrows indicate F480/ETS2 dual-positive cells. (G) Quantification of ETS2 staining in F4/80+ positive cells shown in F (red bar indicates mean; n = 3). (H) Co-IF of pancreatic fibroblast cultures stained for SMA/CK19 or vimentin/CK8. (I) qRT-PCR of Ets-2 in KE and KCE mouse fibroblast cultures (normalized to Rpl4, represented as fold-change relative to ETS2WT expression; n = 3; bars represent means ± SD). (J) Ets2 genotyping PCR of DNA derived from cultured fibroblasts of indicated genotypes.
To further confirm the fibroblast specificity of the Fsp1-Cre in the pancreas, as previously demonstrated in normal mammary gland and mammary tumors [41], [42], we performed co-IF staining with the macrophage marker F4/80 and ETS2. Consistent with our published reports, there was no difference in ETS2 staining in F4/80 macrophages between KE and KCE mice (Supplemental Figure 1, F and G). These collective results indicate that Ets-2 is efficiently deleted in KCE mice exclusively in the fibroblast compartment and this deletion is maintained under in vitro culture conditions.
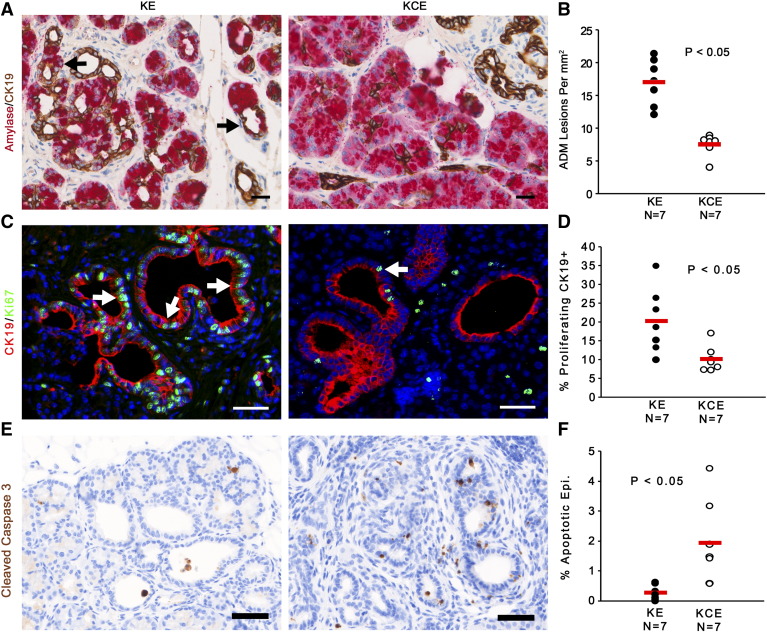
Ets-2 specific deletion in stromal fibroblasts significantly decreased the amount of ADM events, identified by ducts that stain positive for ductal cell marker CK19 and acinar cell marker β-amylase (Figure 3, A and B). CK19/Ki67 co-IF staining showed that ductal cells in KCE mice had two-fold lower proliferation than KE lesions (Figure 3, C and D; matched H&E in Supplemental Figure 2A). Staining with apoptosis marker cleaved caspase 3 showed increased numbers of apoptotic epithelial cells in the preneoplastic ADMs in KCE mice relative to KE (Figure 3, E and F).
Figure 3.
Stromal ETS2 ablation decreases pancreatic ADM.
(A and B) Dual-color IHC and quantitation for β-amylase (red) and CK19 (brown). Black arrows indicate ADM lesions. (C and D) Co-IF for CK19 (red) and Ki67 (green) and quantification of percentage of proliferating CK19+ cells, indicated by white arrows. (E and F) IHC for cleaved caspase 3 (brown) and quantification of percentage of epithelial cells undergoing apoptosis. All scale bars 25 μm (n = 7; red bar indicates mean).
Supplemental Figure 2.
(A) Matched H&E images supplemental to Figure 3E.
Ets-2 Deleted Fibroblasts Are Viable and Maintain an Activated Phenotype
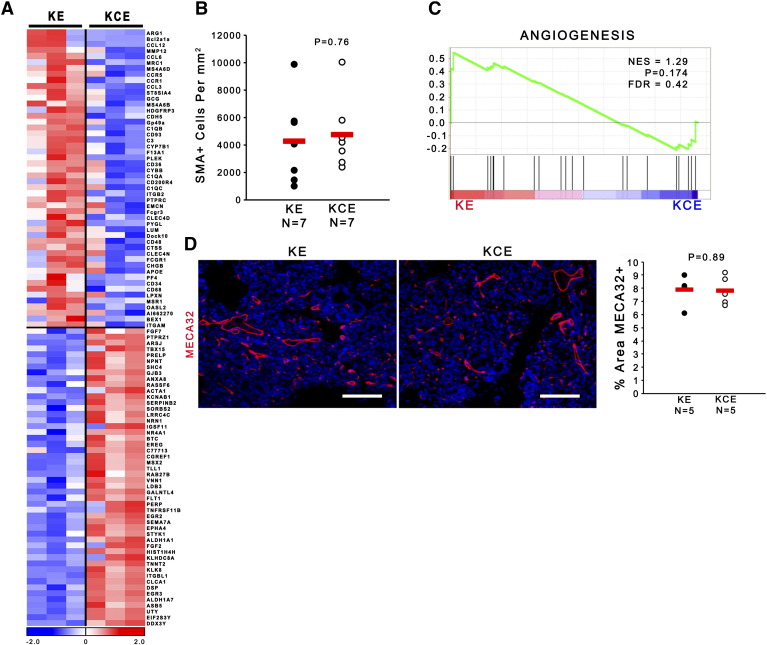
Gene expression analysis of Ets2-intact and Ets2-deleted fibroblast revealed that loss of Ets2 in pancreatic fibroblasts led to significant changes (Supplemental Figure 3A and Supplemental Table 1). GSEA identified the ETS2 target gene signature as significantly downregulated in the KCE versus KE fibroblasts (Figure 4A, P = .002). However, in contrast to our findings in breast cancer [42], GSEA indicated that pancreatic fibroblasts lacking Ets-2 maintained an activated extracellular matrix (ECM) remodeling state, as there was no significant difference between KCE and KE fibroblasts (Figure 4B; P = .117 and false discovery rate [FDR] > 25%). In contrast, GSEA identified that inflammatory responses were significantly affected by Ets2 deletion in fibroblasts (Figure 4C, P < .001).
Supplemental Figure 3.
(A) Heatmap of top 50 up- and downregulated genes in KE and KCE fibroblasts. (B) Total SMA+ cells per mm3 supplemental to Figure 4D. (C) GSEA enrichment plots for angiogenesis pathway in KE versus KCE fibroblasts (n = 3). (D) IF for Meca32 (red) and quantification of percent area Meca32+. Scale bars 25 μm (n = 5; red bar indicates mean).
Figure 4.
Ets-2 deletion does not affect fibroblast proliferation or collagen deposition.
(A–C) GSEA enrichment plots for Ets2-regulated genes, ECM remodeling, and inflammation pathways in KE and KCE fibroblasts (n = 3). (D and E) Dual-color IHC of SMA (red) and Ki67 (brown) and quantification showing the percentage of proliferating SMA+ cells, indicated by black arrows. (F and G) Masson's Trichrome stain and quantification of the percent area of collagen staining (n = 7; red bar indicates mean).
Dual-color IHC staining of SMA and Ki67 showed no difference in the proliferation or total number of SMA-positive fibroblasts in KCE mice compared to KE controls, demonstrating that differences in ADM formation between KE and KCE mice were not due to decreased viability or proliferation of Ets2-null fibroblasts (Figure 4D and Supplemental Figure 3B, respectively). An important physiological function for activated stromal fibroblasts is to form and remodel a collagen-rich extracellular matrix. Masson's Trichrome staining revealed no significant difference in the area of collagen-rich ECM between KCE and KE mice, consistent the GSEA prediction (Figure 4, F and G). Similarly, GSEA showed no difference in angiogenesis pathways between KE and KCE fibroblasts (Supplemental Figure 3C; P = .174 and FDR > 25%), and Meca-32 staining of endothelial cells demonstrated no significant differences between control and ETS2-null tissue (Supplemental Figure 3D).
Ets-2 Deletion In Fibroblasts Alters the Immune Cell Infiltrate during ADM
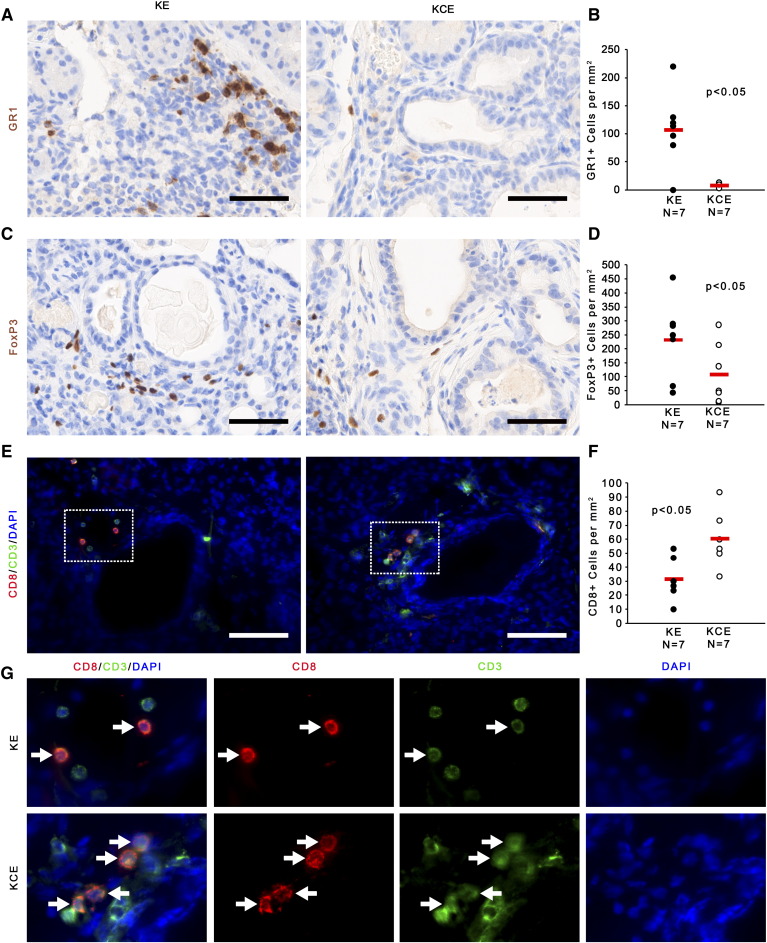
To explore the predicted changes in inflammatory responses upon Ets2 deletion in pancreatic fibroblasts, antibodies for cell-type–specific markers were used to test whether KE and KCE mice had different infiltration of GR1-positive myeloid-derived suppressor cells (MDSCs), F4/80-positive macrophages, FoxP3-positive Tregs, or CD8-positive cytotoxic T cells. The results demonstrated that MDSCs, Tregs, and macrophages were recruited to the stroma in KE mice, and all three cell populations were significantly depleted in the KCE mice with Ets2 ablation (Figure 5, A–D and Supplemental Fig. 4, A and B, respectively). Of note, recruitment of MDSCs into the pancreatic stroma of KCE mice was decreased by 100-fold compared to KE mice (Figure 5A). In contrast, CD8+ cytotoxic T cells were increased two-fold in KCE mice compared to KE controls (Figure 5, E and F). CD8+ cells were confirmed to be dual positive for CD3 (Figure 5G; white arrows).
Figure 5.
Ets-2 fibroblast deletion alters the pancreatic immune microenvironment.
(A and B) IHC for GR1 (brown) and quantification of positive cells per mm2. (C and D) IHC for FoxP3 (brown) and quantification of positive cells per mm2. (E and F) Co-IF for CD8 (red) and CD3 (green) and quantification of CD8-positive cells per mm2. (G) Insets show higher magnified view of representative areas; white arrows indicate CD3+/CD8+ cells. All scale bars 25 μm (n = 7; red bar indicates mean).
Supplemental Figure 4.
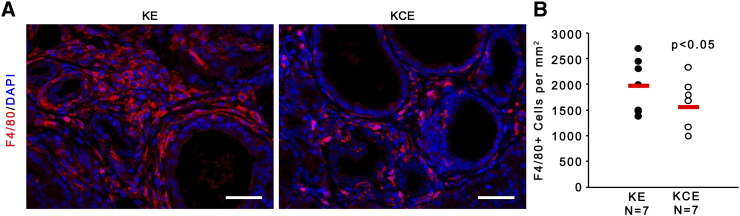
(A) IF for F4/80 (red) and quantification of F4/80+ cells per mm3. Scale bars 25 μm (n = 7; red bar indicates mean).
Ets-2 Directly Regulates Chemokine Expression in Pancreatic Fibroblasts
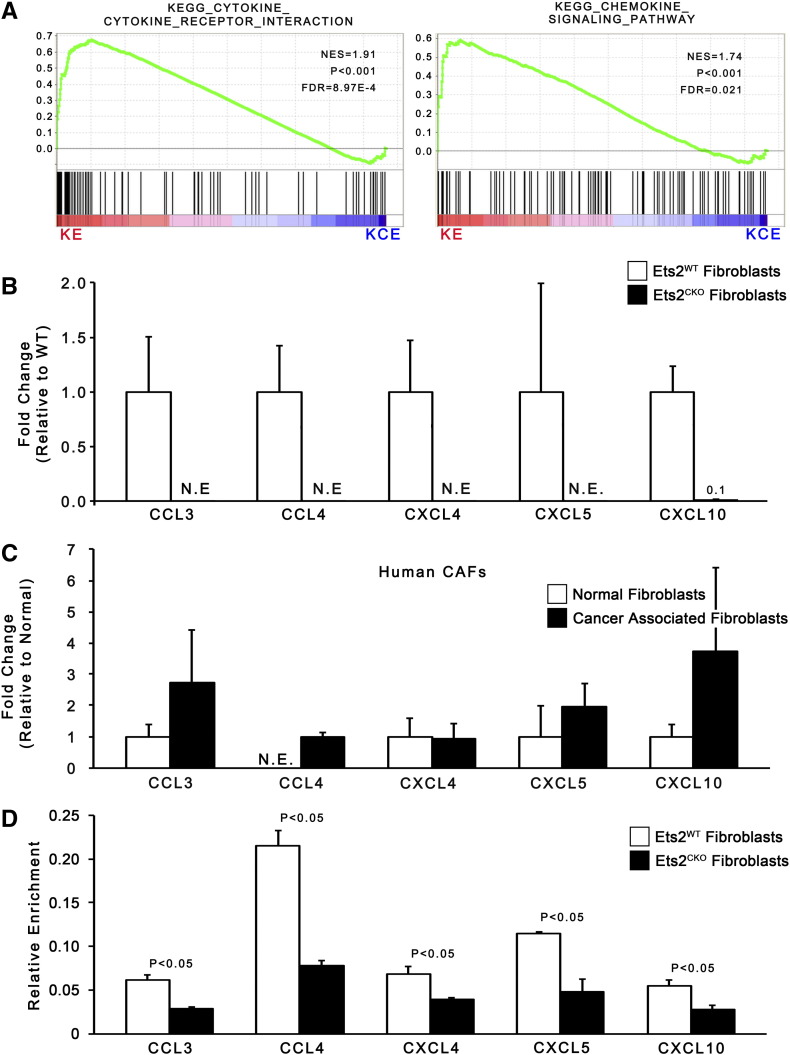
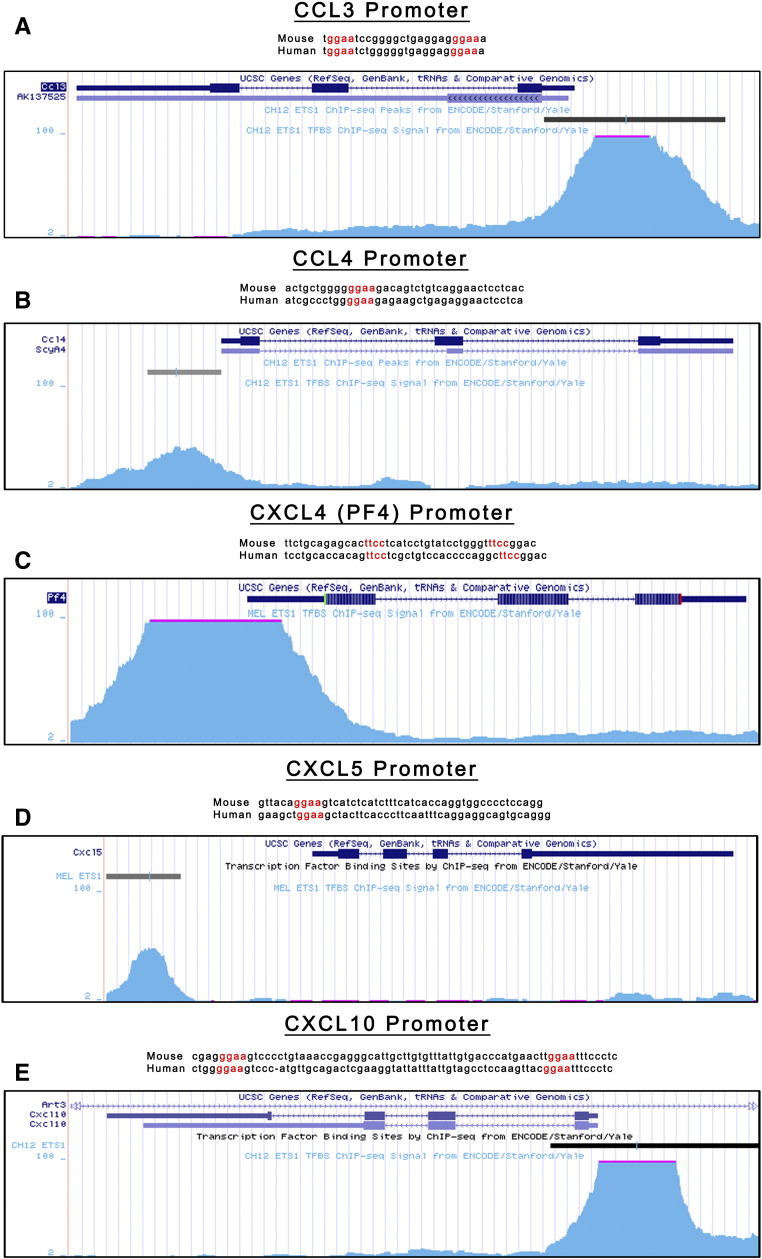
GSEA revealed that chemokine and cytokine pathways were significantly affected by Ets2 deletion (Figure 6A). Further in silico dissection of the chemokine and cytokine GSEA pathways showed that 15 secreted factors were present on the leading edge analysis of the pathways (Supplemental Table 2). The 15 ligands belonged to 3 subgroups: chemokine ligands, interleukins, and TNF superfamily members (Supplemental Table 3). Using publically available data on the Encyclopedia of DNA Elements consortium database, we used ChIP-Seq data available for the closely related transcription factor ETS1 (GEO accession GSM1003774 and GSM1003777) to identify predicted ETS binding sites in the promoters of the 15 ligands. The promoters of 9 of the 15 factors (60%) contained ETS1 ChIP-Seq peaks that are also predicted ETS2 binding sites (Supplemental Table 3). The ETS2 binding sites were conserved in mice and human (Supplemental Figure 5). Out of these nine, five factors (CCL3, CCL4, CXCL4, CXCL5, CXCL10) are known to function in recruiting T cells, macrophages, or MDSCs [54], [55], [56], [57], [58] (Supplemental Table 3).
Figure 6.
Ets-2 directly regulates chemokine and cytokine production.
(A) GSEA enrichment plots for cytokine and chemokine pathways in KE versus KCE fibroblasts (n = 3). (B) qRT-PCR of secreted factors in KE and KCE mouse fibroblasts (normalized to Rpl4, represented as fold-change relative to ETS2WT expression; n = 3; bars represent means ± SD; N.E. = not expressed). (C) qRT-PCR of secreted factors in human PDAC-derived CAF cultures (normalized to Rpl4, represented as fold-change relative to normal fibroblast expression; n = 3; bars represent means ± SD). (D) Relative enrichment for ETS2 binding to the promoters of indicated genes in KE and KCE mouse fibroblasts (n = 5; bars represent means ± SD).
Supplemental Figure 5.
(A–E) UCSC Genome Browser visualization of the promoter regions of CCL3 (A), CCL4 (B), CXCL4 (C), CXCL5 (D), and CXCL10 (E) overlaid with ETS1 ChIP-Seq data from Encyclopedia of DNA Elements (GEO accession GSM1003774 and GSM1003777). For each gene, mouse and human DNA sequences showing predicted ETS2 binding site conservation.
Strikingly, real-time RT-PCR confirmed that mRNAs for all five ligands were expressed in KE fibroblasts, whereas expression was not detectable in cells with Ets2 deletion (Figure 6B). Furthermore, all five ligands were expressed in human PDAC patient-derived cancer associated fibroblasts (Figure 6C). ETS2 ChIP on chromatin from KE and KCE pancreatic fibroblasts showed significant ETS2 enrichment in ETS2-intact versus deleted fibroblasts (Figure 6D).
Discussion
The results presented here identify ETS signaling in fibroblasts as a means of immune cell recruitment to ADM. ETS2 directly coordinates a chemokine signature that alters the immune contexture at the early ADM stage. Loss of ETS2 signaling in the pancreatic stroma abrogates production of key chemokines in fibroblasts, resulting in decreased infiltration of macrophage, MDSC, and Treg populations while increasing CD8 T cells.
The stroma undergoes a dramatic expansion in concert with the stepwise development of pancreatic cancer, suggesting that the stroma is an active partner in disease initiation and progression. However, characterizing the complex interactions between the pancreatic epithelium and the adjacent stroma has proved difficult, as recent reports have highlighted the degree of heterogeneity that exists in the pancreatic stroma. In particular, depletion of SMA-positive cells accelerated pancreatic cancer, whereas depletion of FAP-positive cells slowed tumor growth [29], [31]. Interestingly, both studies identified immune cell subpopulations that dynamically interact with stromal fibroblasts. Tumors depleted for SMA-positive cells were responsive to α-CTLA-4 therapy, which functions by altering T-cell activation [31]. Tumors depleted for FAP-positive cells were dependent on both CD4 and CD8 T-cell populations, and fibroblast depletion synergized with α-PD-1 and α-CTLA-4 immune checkpoint therapy [29]. These studies indicate that fibroblast-immune cell cross talk is complex, and our results establish that ETS2 is responsible for the production of several key immunomodulatory chemokines and cytokines. We propose that ETS2 is activated in fibroblasts early during epithelial transformation of the pancreas and helps to establish an immune microenvironment that is conducive to increased ADM formation.
The function of ETS2-mediated signaling in pancreatic fibroblasts is to modulate the presence of immune cells in the preneoplastic stroma and is a phenotype that is distinct from its role in mammary fibroblasts. Our previous studies in mammary tumor growth have shown that the primary function of ETS2 in the fibroblast compartment is to promote angiogenesis, which was not observed in the pancreas [41], [42]. Furthermore, differences in chemokine and cytokine gene expression and changes in immune cell populations identified in this work were not observed with Ets2 loss in the breast tumor stroma [41], [42]. Thus, ETS2 alters the fibroblast transcriptome in a tissue-specific fashion and subsequently modulates the communication between the epithelial cell and its microenvironment in distinct manner. This is consistent with the observation that fibroblasts maintain a complex positional memory that drives specific gene expression patterns based on their topographical location [59]. These results also highlight the diversity of stromal fibroblasts and show that a more detailed understanding of the signaling networks within fibroblasts derived from cancers of different origins is required to more precisely understand epithelial-fibroblast cross talk.
The most striking change in immune cell recruitment upon loss of ETS2 signaling from the fibroblasts was in the immunosuppressive MDSC population. These cells are recruited early during pancreatic tumorigenesis, as they are present at the ADM/PanIN stage and progressively increase during malignant progression [22], [60]. Additionally, MDSC levels positively correlate with FoxP3-positive Treg numbers in pancreatic cancer, consistent with our results at the ADM stage [61]. Further functional studies into MDSCs in pancreatic cancer has revealed that pancreatic cancer–associated stellate cells secrete factors that drive MDSC differentiation, which in turn suppresses CD8-positive T-cell proliferation [62]. It has also been observed that MDSC and CD8-positive populations are present in a nearly mutually exclusive relationship in both PanIN and PDAC, suggesting that early host immunosuppressive responses are able to impede T-cell infiltration [22]. The decrease in MDSCs in Ets2-deleted pancreata may open the door for recruitment of CD8-positive T cells. These cells have previously been characterized for their ability to eliminate tumor cells during pancreatic cancer development and progression [63], and patients who have increased CD8-positive populations have improved prognosis [64], [65]. The genetic data presented here suggest that ETS2 is involved in the interactions between fibroblasts and immunosuppressive cells that may contribute to KrasG12D epithelial cell proliferation indirectly.
KrasG12D epithelial cell proliferation may also be directly affected by ETS2 in fibroblasts, as chemokines are a major conduit of epithelial-fibroblast communication in the pancreas [63]. The ETS2 targets CCL3, CCL4, CXCL5, and CXCL10 have all been linked to increased tumor cell proliferation in a variety of cancers [66], [67], [68], [69], [70]. ETS2 signaling may also play an important signal transduction role from within the epithelial cell, given its role downstream of RAS-MAPK and mutant P53 signaling pathways [38], [39], [71], [72], [73]. Thus, ETS2 could present an overlap between KRAS and p53 signaling, but further biochemical work will be required to pin down the interworkings of this signaling network within the pancreatic tumor cell. Targeting ETS2 signaling may be beneficial to inhibit both epithelial cell intrinsic oncogenic mechanisms as well as the fibroblast-driven immune-modulatory effects defined herein.
The following are the supplementary data related to this article.
Table of top 50 up- and downregulated genes in pancreatic fibroblast cultures isolated from KE or KCE animals. All values are presented as RMA.
GSEA summary files showing genes in each gene set. Green indicated that gene is present on leading edge when comparing KE versus KCE pancreatic fibroblast culture gene expression.
Table summarizing subfamilies of ETS2-regulated genes showing ETS1 ChIP-Seq peaks, known receptors, and chemoattractive abilities. Red indicates gene was selected for further analysis. See Supplemental References for citations.
Supplementary material.
Conflict of Interest
The authors report no conflicts of interest.
Acknowledgements
We acknowledge Jason Bice, Daphne Bryant, Nicole Drummond, and Lisa Rawahneh from the OSU Solid Tumor Biology Histology Core for their technical support. We also thank Chelsea K. Martin for assisting in initial analysis of pancreatic tumor histology. We acknowledge the OSUCCC Genomics, Microscopy, Transgenic/Knockout, Target Validation, Analytic Cytometry, Bioinformatics, and Biostatistics Shared Resources. This study was supported by National Institutes of Health grants PO1 CA097189 (M. C. O. and G. L.), NRSA F31 CA189757 (J. R. P.), and R01 CA124586 (S. F. K.). This work was also supported by the Pelotonia Fellowship Program (J. R. P.). Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the Pelotonia Fellowship Program.
Footnotes
This study was supported by National Institutes of Health grants PO1 CA097189 (M. C. O. and G. L.), NRSA F31 CA189757 (J. R. P.), and R01 CA124586 (S. F. K.). This work was also supported by the Pelotonia Fellowship Program (J. R. P.). Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect those of the Pelotonia Fellowship Program.
References
- 1.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z. National Cancer Institute; Bethesda, MD: 2015. SEER Cancer Statistics Review, 1975-2012. [ http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, posted to the SEER web site] [Google Scholar]
- 3.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53(4):549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 4.Barton CM, Staddon SL, Hughes CM, Hall PA, O'Sullivan C, Kloppel G, Theis B, Russell RC, Neoptolemos J, Williamson RC. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64(6):1076–1082. doi: 10.1038/bjc.1991.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarpa A, Capelli P, Mukai K, Zamboni G, Oda T, Iacono C, Hirohashi S. Pancreatic adenocarcinomas frequently show p53 gene mutations. Am J Pathol. 1993;142(5):1534–1543. [PMC free article] [PubMed] [Google Scholar]
- 6.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733. doi: 10.1053/j.gastro.2011.12.042. [e9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 8.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS, Sandgren EP. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63(9):2016–2019. [PubMed] [Google Scholar]
- 9.Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105(48):18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuveson DA, Zhu L, Gopinathan A, Willis NA, Kachatrian L, Grochow R, Pin CL, Mitin NY, Taparowski EJ, Gimotty PA. Mist1-KrasG12D knock-in mice develop mixed differentiation metastatic exocrine pancreatic carcinoma and hepatocellular carcinoma. Cancer Res. 2006;66(1):242–247. doi: 10.1158/0008-5472.CAN-05-2305. [DOI] [PubMed] [Google Scholar]
- 11.De La OJ, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105(48):18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP, Pan FC, Akiyama H, Wright CV, Jensen K. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(6):737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, Hieber M, Arbeiter A, Klein S, Kong B. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23(3):406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Morris JP, Yan W, Schofield HK, Gurney A, Simeone DM, Millar SE, Hoey T, Hebrok M, Pasca di Magliano M. Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res. 2013;73(15):4909–4922. doi: 10.1158/0008-5472.CAN-12-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liou GY, Doppler H, Necela B, Krishna M, Crawford HC, Raimondo M, Storz P. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol. 2013;202(3):563–577. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi G, DiRenzo D, Qu C, Barney D, Miley D, Konieczny SF. Maintenance of acinar cell organization is critical to preventing Kras-induced acinar-ductal metaplasia. Oncogene. 2013;32(15):1950–1958. doi: 10.1038/onc.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Nandakumar N, Shi Y, Manzano M, Smith A, Graham G, Gupta S, Vietsch EE, Laughlin SZ, Wadhwa M. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7(324):ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12(2):81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 19.Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171(1):263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsa I, Longnecker DS, Scarpelli DG, Pour P, Reddy JK, Lefkowitz M. Ductal metaplasia of human exocrine pancreas and its association with carcinoma. Cancer Res. 1985;45(3):1285–1290. [PubMed] [Google Scholar]
- 21.Remmers N, Anderson JM, Linde EM, DiMaio DJ, Lazenby AJ, Wandall HH, Mandel U, Clausen H, Yu F, Hollingsworth MA. Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 2013;19(8):1981–1993. doi: 10.1158/1078-0432.CCR-12-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67(19):9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 23.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korc M. Pancreatic cancer–associated stroma production. Am J Surg. 2007;194(4 Suppl.):S84–S86. doi: 10.1016/j.amjsurg.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18(16):4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neesse A, Algul H, Tuveson DA, Gress TM. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut. 2015;64(9):1476–1484. doi: 10.1136/gutjnl-2015-309304. [DOI] [PubMed] [Google Scholar]
- 27.Erkan M, Hausmann S, Michalski CW, Fingerle AA, Dobritz M, Kleeff J, Friess H. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat Rev Gastroenterol Hepatol. 2012;9(8):454–467. doi: 10.1038/nrgastro.2012.115. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What we have learned about pancreatic cancer from mouse models. Gastroenterology. 2012;142(5):1079–1092. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti–PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110(50):20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita H, Ohuchida K, Mizumoto K, Nakata K, Yu J, Kayashima T, Cui L, Manabe T, Ohtsuka, Tanaka M. alpha-Smooth muscle actin expressing stroma promotes an aggressive tumor biology in pancreatic ductal adenocarcinoma. Pancreas. 2010 doi: 10.1097/MPA.0b013e3181dbf647. [DOI] [PubMed] [Google Scholar]
- 33.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollingsworth MA, Yeh JJ, Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stavey KJ, Hume DA, Maki RA, Ostrowski MC. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Nat Genet. 1996;16(2):538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patton SE, Martin ML, Nelsen LL, Fang X, Mills GB, Bast RC, Jr., OStrowski MC., Jr Activation of the ras-mitogen–activated protein kinase pathway and phosphorylation of ets-2 at position threonine 72 in human ovarian cancer cell lines. Cancer Res. 1998;58(10):2253–2259. [PubMed] [Google Scholar]
- 36.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24(24):10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabbout M, Dakhlallah D, Sharma S, Bronisz A, Srinivasan R, Piper M, Marsh CB, Ostrowski MC. MicroRNA 17-92 cluster mediates ETS1 and ETS2-dependent RAS-oncogenic transformation. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, Chauhan K, Daniels SR, Boccetta M, Garrett MR. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26(8):830–845. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, Jackson JG, Suh YA, Elizondo-Fraire AC, Yang P. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci U S A. 2014;111(30):11145–11150. doi: 10.1073/pnas.1404139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollareddy M, Dimitrova E, Vallabhaneni KC, Chan A, Le T, Chauhan KM, Carrero ZI, Ramakrishnan G, Watabe K, Haupt Y. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat Commun. 2015;6:7389. doi: 10.1038/ncomms8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, Thompson JC, Caserta E, Wang H, Chong JL. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461(7267):1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace JA, Li F, Balakrishnan S, Cantemir-Stone CZ, Pecot T, Martin C, Kladney RD, Sharma SM, Trimboli AJ, Fernandez SA. Ets2 in tumor fibroblasts promotes angiogenesis in breast cancer. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ, Martin CK, Li F. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol. 2012;14(2):159–167. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Man AK, Young LJ, Tynan JA, Lesperance J, Egeblad M, Werb Z, Hauser CA, Muller WJ, CArdiff RD, Oshima RG. Ets2-dependent stromal regulation of mouse mammary tumors. Mol Cell Biol. 2003;23(23):8614–8625. doi: 10.1128/MCB.23.23.8614-8625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behrens P, Mathiak M, Mangold E, Kirdorf S, Wellmann A, Fogt F, Rothe M, Florin A, Wernert N. Stromal expression of invasion-promoting, matrix-degrading proteases MMP-1 and -9 and the Ets 1 transcription factor in HNPCC carcinomas and sporadic colorectal cancers. Int J Cancer. 2003;107(2):183–188. doi: 10.1002/ijc.11336. [DOI] [PubMed] [Google Scholar]
- 46.Behrens P, Rothe M, Wellmann A, Krischler J, Wernert N. The Ets-1 transcription factor is up-regulated together with MMP 1 and MMP 9 in the stroma of pre-invasive breast cancer. J Pathol. 2001;194(1):43–50. doi: 10.1002/path.844. [DOI] [PubMed] [Google Scholar]
- 47.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, Cresap N, Rosol TJ, Robinson ML, Eng C. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68(3):937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 48.Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, Muthusamy N, Man AK, Oshima RG, Leone G. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114(5):1123–1130. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12(9):1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu L, Gulati P, Fernandez S, Pennell M, Kirschner L, Jarjoura D. Fully moderated T-statistic for small sample size gene expression arrays. Stat Appl Genet Mol Biol. 2011;10(1) doi: 10.2202/1544-6115.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito Y, Miyoshi E, Takeda T, Sakon M, Ihara S, Tsujimoto M, Matsuura N. Ets-2 overexpression contributes to progression of pancreatic adenocarcinoma. Oncol Rep. 2002;9(4):853–857. [PubMed] [Google Scholar]
- 52.Walter K, Omura N, Hong SM, Griffith M, Vincent A, Borges M, Goggins M. Overexpression of smoothened activates the sonic hedgehog signaling pathway in pancreatic cancer-associated fibroblasts. Clin Cancer Res. 2010;16(6):1781–1789. doi: 10.1158/1078-0432.CCR-09-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455(7211):406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 54.Ajuebor MN, Hogaboam CM, Le T, Proudfoot AE, Swain MG. CCL3/MIP-1alpha is pro-inflammatory in murine T cell–mediated hepatitis by recruiting CCR1-expressing CD4(+) T cells to the liver. Eur J Immunol. 2004;34(10):2907–2918. doi: 10.1002/eji.200425071. [DOI] [PubMed] [Google Scholar]
- 55.Burger JA, Quiroga MP, Hartmann E, Burkle A, Wierda WG, Keating MJ, Rosenwald A. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009;113(13):3050–3058. doi: 10.1182/blood-2008-07-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balamayooran G, Batra S, Cai S, Mei J, Worthen GS, Penn AL, Jeyaseelan S. Role of CXCL5 in leukocyte recruitment to the lungs during secondhand smoke exposure. Am J Respir Cell Mol Biol. 2012;47(1):104–111. doi: 10.1165/rcmb.2011-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrovic-Djergovic D, Popovic M, Chittiprol S, Cortado H, Ransom RF, Partida-Sanchez S. CXCL10 induces the recruitment of monocyte-derived macrophages into kidney, which aggravate puromycin aminonucleoside nephrosis. Clin Exp Immunol. 2015;180(2):305–315. doi: 10.1111/cei.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivastava K, Cockburn IA, Swaim A, Thompson LE, Tripathi A, Fletcher CA, Shirk EM, Sun H, Kowalska MA, Fox-Talbot K. Platelet factor 4 mediates inflammation in experimental cerebral malaria. Cell Host Microbe. 2008;4(2):179–187. doi: 10.1016/j.chom.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, Brown PO. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99(20):12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21(6):836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60(10):1419–1430. doi: 10.1007/s00262-011-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mace TA, Ameen Z, Collins A, Wojcik S, Mair M, Young GS, Fuchs JR, Eubank TD, Frankel WL, Bekaii-Saab T. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013;73(10):3007–3018. doi: 10.1158/0008-5472.CAN-12-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wormann SM, Diakopoulos KN, Lesina M, Algul H. The immune network in pancreatic cancer development and progression. Oncogene. 2014;33(23):2956–2967. doi: 10.1038/onc.2013.257. [DOI] [PubMed] [Google Scholar]
- 64.Ademmer K, Ebert M, Muller-Ostermeyer F, Friess H, Buchler MW, Schubert W, Malfertheiner P. Effector T lymphocyte subsets in human pancreatic cancer: detection of CD8 + CD18+ cells and CD8 + CD103+ cells by multi-epitope imaging. Clin Exp Immunol. 1998;112(1):21–26. doi: 10.1046/j.1365-2249.1998.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukunaga A, Miyamoto M, Cho Y, Murakami S, Kawarada Y, Oshikiri T, Kato K, Kurokawa T, Suzuoki M, Nakakubo Y. CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas. 2004;28(1):e26–e31. doi: 10.1097/00006676-200401000-00023. [DOI] [PubMed] [Google Scholar]
- 66.Tsai SC, Lin SJ, Lin CJ, Chou YC, Lin JH, Yeh TH, Chen MR, Huang LM, Lu MY, Huang YC. Autocrine CCL3 and CCL4 induced by the oncoprotein LMP1 promote Epstein-Barr virus–triggered B cell proliferation. J Virol. 2013;87(16):9041–9052. doi: 10.1128/JVI.00541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lentzsch S, Gries M, Janz M, Bargou R, Dorken B, Mapara MY. Macrophage inflammatory protein 1-alpha (MIP-1 alpha) triggers migration and signaling cascades mediating survival and proliferation in multiple myeloma (MM) cells. Blood. 2003;101(9):3568–3573. doi: 10.1182/blood-2002-08-2383. [DOI] [PubMed] [Google Scholar]
- 68.Zheng J, Zhu X, Zhang J. CXCL5 knockdown expression inhibits human bladder cancer T24 cells proliferation and migration. Biochem Biophys Res Commun. 2014;446(1):18–24. doi: 10.1016/j.bbrc.2014.01.172. [DOI] [PubMed] [Google Scholar]
- 69.Kawamura M, Toiyama Y, Tanaka K, Saigusa S, Okugawa Y, Hiro J, Uchida K, Mohri Y, Inoue Y, Kusunoki M. CXCL5, a promoter of cell proliferation, migration and invasion, is a novel serum prognostic marker in patients with colorectal cancer. Eur J Cancer. 2012;48(14):2244–2251. doi: 10.1016/j.ejca.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 70.Maru SV, Holloway KA, Flynn G, Lancashire CL, Loughlin AJ, Male DK, Romero IA. Chemokine production and chemokine receptor expression by human glioma cells: role of CXCL10 in tumour cell proliferation. J Neuroimmunol. 2008;199(1–2):35–45. doi: 10.1016/j.jneuroim.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 71.Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23(6):213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- 72.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2(11):827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 73.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Alawar R, Katona BW, Shilatifard A. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525(7568):206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of top 50 up- and downregulated genes in pancreatic fibroblast cultures isolated from KE or KCE animals. All values are presented as RMA.
GSEA summary files showing genes in each gene set. Green indicated that gene is present on leading edge when comparing KE versus KCE pancreatic fibroblast culture gene expression.
Table summarizing subfamilies of ETS2-regulated genes showing ETS1 ChIP-Seq peaks, known receptors, and chemoattractive abilities. Red indicates gene was selected for further analysis. See Supplemental References for citations.
Supplementary material.