Abstract
Antibiotic resistance is increasingly widespread, largely due to human influence. Here, we explore the relationship between antibiotic resistance genes and the antimicrobial chemicals triclosan, triclocarban, and methyl-, ethyl-, propyl-, and butylparaben in the dust microbiome. Dust samples from a mixed-use athletic and educational facility were subjected to microbial and chemical analyses using a combination of 16S rRNA amplicon sequencing, shotgun metagenome sequencing, and liquid chromatography tandem mass spectrometry. The dust resistome was characterized by identifying antibiotic resistance genes annotated in the Comprehensive Antibiotic Resistance Database (CARD) from the metagenomes of each sample using the Short, Better Representative Extract Data set (ShortBRED). The three most highly abundant antibiotic resistance genes were tet(W), blaSRT-1, and erm(B). The complete dust resistome was then compared against the measured concentrations of antimicrobial chemicals, which for triclosan ranged from 0.5 to 1970 ng/g dust. We observed six significant positive associations between the concentration of an antimicrobial chemical and the relative abundance of an antibiotic resistance gene, including one between the ubiquitous antimicrobial triclosan and erm(X), a 23S rRNA methyltransferase implicated in resistance to several antibiotics. This study is the first to look for an association between antibiotic resistance genes and antimicrobial chemicals in dust.
Introduction
With whole metagenome sequencing, it is now possible to monitor the development of antibiotic resistance in entire microbial communities. This technology has made it possible to study the collection of antibiotic resistance genes – the resistome – in a wide array of biological systems. Researchers are discovering that antibiotic resistance is ubiquitous and interconnected with microbial community structure and dynamics. Antibiotic resistance genes naturally occur in the human infant gut during the first days of microbial colonization, even in the absence of antibiotic drugs.1 Antibiotic resistance genes are also increasingly present in bacteria associated with livestock, driven by the widespread application of veterinary antibiotics, as well as in the outdoor environment.2
Antibiotic resistance is also prevalent in the indoor environment, where humans spend up to 90% of their time. Microbes in the indoor environment, referred to as the indoor microbiome, have been identified as an important source of antibiotic-resistant infections. For example, cultivable antibiotic-resistant Staphylococcus aureus isolates are more concentrated in indoor air than outdoor air,3 and nonhuman factors (i.e., elements in the house excluding its residents) in the household environment can contribute to the recurrence of antibiotic-resistant infections in the inhabitants of certain homes.4 Despite the increase in knowledge of antibiotic resistance in host-associated microbiomes and the existence of high-profile antibiotic-resistant pathogens in the built environment, we still know very little about the indoor resistome.
The spread of antibiotic resistance genes is exacerbated by the widespread use of antimicrobial chemicals, which are prevalent in building materials and personal care products.5 Unlike antibiotic drugs, antimicrobials leave longlasting residues and can accumulate in the environment. Triclosan, a commonly used antimicrobial, can promote the development of resistance itself.6 Some strategies for triclosan resistance, such as alterations to the outer membrane or expression of nonspecific efflux pumps, give bacteria cross-resistance to antibiotic drugs, as was observed in S. aureus, Pseudomonas aeruginosa, Escherichia coli, Salmonella enterica, and Rhodospirillum rubrum.6 Efflux pumps are not commonly subject to horizontal transfer, limiting the spread of efflux-mediated cross-resistance between species; nevertheless, all bacteria have genes encoding efflux pumps,7 suggesting that triclosan could elicit multidrug resistance in many species independently. Like antibiotics, triclosan can have dramatic impacts on the structure of host-associated gut microbiomes, at least in zebrafish.8 In the environment, triclosan resistance increases in microbial communities that are exposed to higher concentrations of the chemical.9
Antimicrobial chemicals are prevalent in indoor dust. Surveys of household dust from Belgium, Canada, China, and Spain found triclosan in 137 of 138 samples collected, with median values in the ppb range.10 The concentration of triclosan in household dust is thus comparable to that found in wastewater, where triclosan is an emerging contaminant of concern. Another antimicrobial, methylparaben, was detected in ≥90% of samples collected in the US, China, Korea, Japan, Spain, and Canada, with median values ranging from 320 to 1470 ppb,10c,11 exceeding concentrations found in municipal wastewater by approximately 4 orders of magnitude.12 However, the relationship between antimicrobial chemicals on the indoor dust microbiome and resistome have not been examined.
This study is the first to report a relationship between antibiotic resistance genes and antimicrobial chemicals in the indoor environment. We tested the following hypotheses: 1) the composition of the resistome and/or the microbiome correlate with the concentration of antimicrobial chemicals and 2) the abundance of specific antibiotic resistance genes correlates with the concentration of antimicrobial chemicals. To test these hypotheses, we collected 44 dust samples from a mixed-use athletic and educational facility. Samples were subdivided for chemical and DNA extraction and examined using liquid chromatography tandem mass spectrometry (LC-MS/MS) and next-generation nucleic acid sequencing. The dust in this building, which is characterized by natural ventilation and high infiltration, contained slightly less triclosan than found in previous studies (median concentration of 200 ppb in this study compared to 220–702 ppb elsewhere10) and low concentrations of antibiotic resistance genes (median reads per kilobase per million reads (RPKM) per sample 23.24 compared to 47.71 in other studies13). Nevertheless, a positive association was observed between the concentration of antimicrobials and the abundance of multiple antibiotic resistance genes, including the 23S rRNA methyltransferases erm(C), erm(X), and erm(33) and the efflux pumps tet(K) and vga(A). These results suggest that antimicrobial chemicals and antibiotic resistance genes are positively correlated in the built environment.
Experimental Section
Sample Collection
The building to be sampled was selected for its space types (i.e., rooms with different patterns of occupancy and usage) to capture a large diversity of indoor microbial communities.14 Dust was collected using a vacuum fitted with Dustream collectors (Indoor Biotechnologies, Charlottesville, VA). Dust was vacuumed from each space until at least two collectors had been filled or no further apparent dust was available. Samples were stored in sterile plastic bags in the dark at room temperature until further processing. Dust was aliquoted for chemical or DNA extraction by mixing the collected sample and distributing the desired mass using sterile forceps and spatulas in a sterile hood. In total, 44 samples were collected (Table S1).
Chemical Analysis
Samples with sufficient mass (≥0.75 g of dust; Table S1) were subsampled, and 100 mg aliquots were placed in 10 mL centrifuge tubes for chemical extraction. Aliquots were spiked with 100 μL of 100 ppb isotope-labeled internal standards, extracted twice using 3 mL of 1:1 (v/v) methanol:acetone with 10 mM acetic acid, centrifuged, and pooled. Samples were dried to approximately 1 mL and mixed with 5 mL of 10 mM acetic acid for loading onto an Oasis HLB 3 cc Flangeless Vac Cartridge solid phase extraction column (Waters) through 60 mg sorbent per cartridge of 30 μm particle size. After solid phase extraction, the samples were reconstituted in 1 mL of methanol and diluted 1:1 (v/v) with water. Ten microliters of each sample was injected onto a 2100 HPLC (Shimadzu) coupled with an API 4000 triple quadrupole mass spectrometer (MS/MS AB Sciex) operating in electrospray ionization (ESI) mode. The analytes were separated on an X-Bridge C8 column (2.1 × 100 mm, 3.5 μm particle size; Waters) preceded by an equivalent guard column using a gradient LC protocol. The flow rate was 0.2 mL/min, and the gradient was as follows: starting at 50% methanol for 1 min, then ramped to 95% methanol over 5 min, held at 95% methanol for 3 min, then decreased back to 50% methanol over 1 min, and held at 50% methanol for 1 min. A switching valve allowed sample to flow to the MS/MS between 0.5 and 10.5 min of each 11 min run. The ESI was operated in negative mode, and the source parameters were set as follows: curtain gas = 25 psi, gas 1 = 70 psi, gas 2 = 50 psi, IS = −4500 eV, source temperature = 500 °C, entrance potential = −10 eV, and collision activated dissociation gas = 12 psi. Target times and masses are listed in Table S2. The average relative recoveries for all the analytes ranged from 79% to 97%, with standard deviation varying between 5% to 31%. Each sample was aliquoted and extracted in two independent experiments; duplicates were averaged for statistical analysis. All reagents were mass spectrometry grade.
DNA Analysis
For samples containing ≥0.25 g of dust (Table S1), 0.25 g was aliquoted for DNA extraction.15 For the remaining samples the entirety of the sample was extracted, and the mass of dust was recorded. Genomic DNA was extracted as previously.14b Libraries for 16S rRNA gene amplicon sequencing were prepared at the University of Oregon Genomics Core Facility using 1 μL of template DNA from all extractions and the 515F and 806R primers targeting the V4 region of the 16S rRNA gene.16 Samples were sequenced on an Illumina HiSeq in paired-end 150 bp mode. Sequence data were processed using Another Automated Data Analysis Management Application (AnADAMA; https://huttenhower.sph.harvard.edu/anadama) using the Usearch16SPipeline pipeline.
AnADAMA automates the following steps: paired-end 16S amplicon sequencing reads were demultiplexed using QIIME v1.8 (split_libraries_fastq.py), stitched using ea-utils command line tools (https://expressionanalysis.github.io/ea-utils/), and clustered into operational taxonomic units (OTUs) using UPARSE for its low false positive rate for OTU reporting relative to other identification pipelines. Reads were quality-filtered using the UPARSE17 quality-filtering threshold of Emax = 1, at which the most probable number of base errors per read is zero for filtered reads.18 Filtered reads were trimmed to a fixed length, and singletons were removed and clustered de novo into OTUs, followed by chimera filtering. Taxonomic classification and quantification of OTUs were then performed against the Greengenes19 version 13.5 16S rRNA database.20
For shotgun metagenome sequencing, libraries were prepared using the Nextera XT DNA Sample Preparation Kit (Illumina) from all extractions containing ≥15 ng/mL DNA. Sequencing was performed on an Illumina NextSeq in paired-end mode at the University of Oregon Genomics Core Facility. Metagenomic data were processed using the AnADAMA WGSPipeline pipeline, per the following automated steps. Whole genome shotgun sequencing reads were as follows: 1) concatenated into a single fastq file, 2) filtered for low quality reads and human sequence contaminants using Knead v0.3 (http://huttenhower.sph.harvard.edu/kneaddata), and 3) taxonomically classified and quantified using MetaPhlAn221 (http://huttenhower.sph.harvard.edu/metaphlan2).
All sequence data are available in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/; BioProject ID PRJNA321035).
Operational taxonomic units (OTUs) derived from the 16S rRNA sequencing data identified as chloroplasts and mitochondria, as well as likely contaminants that were prevalent in both the reagent controls and the samples, were removed as per Meadow et al.22 The OTUs removed in this step were identified as Halomonadaceae and Shewanella. One sample with a low number of reads (<4000) after contaminant removal was excluded from downstream analysis.
Architectural Analysis
For each space sampled, the following data were collected: space type; number, type, and operational state of windows; and crack area for all windows and doors. All architectural metadata and the mapping file for further analysis are presented in Tables S1 and S3.
Data Analysis
Bayesian source tracking was performed using the SourceTracker23 algorithm in MacQIIME (v1.9.1) to identify putative sources of indoor microbes. Human Microbiome Project data from five body sites, included with the SourceTracker tutorial, were supplied as the source data. In addition to data from the present study, we analyzed data from two recently published indoor microbiome studies on athletic facilities24 and homes.25 All three data sets were processed using the same USEARCH-based OTU picking pipeline prior to SourceTracker analysis.
Antibiotic resistance genes were quantified using the Short, Better Representative Extract Data set (ShortBRED).26 Briefly, ShortBRED comprises two parts, ShortBRED-identify and ShortBRED-quantify. ShortBRED-identify generates unique peptide markers for a set of protein sequences. Specifically, ShortBRED-identify used an 85% amino acid identity threshold to cluster the protein homologue model gene set of the CARD database into nonredundant representative sequences. ShortBRED-quantify then maps translated reads to protein markers at ≥95% amino acid identity across ≥95% of the marker length. The use of markers, rather than full protein sequences, ensures quicker and more accurate mapping, leading to fewer false positive RPKM counts. Moreover, reads are required to map with more specificity to markers than markers are required to align with one another. For comparison with results from previous studies,13 antibiotic resistance genes were also identified using ShortBRED with markers from the Antibiotic Resistance Database (ARDB).27 Because the ARDB is no longer maintained, results using CARD as the reference database are likely to more accurately reflect the current understanding of antibiotic resistance genes.
Statistical analyses were performed using R or Hierarchical All-against-All significance testing (HAllA; http://huttenhower.sph.harvard.edu/halla) as appropriate. Community-level analyses of the 16S rRNA data were performed in R on samples rarefied to 16,000 sequence reads. Taxonomic beta diversity was calculated using the Canberra dissimilarity metric. Dissimilarity among chemical profiles (triclosan, triclocarban, methylparaben, ethylparaben, propylparaben, and butylparaben) was determined by calculating pairwise Euclidean distances on each sample’s suite of chemical concentrations (normalized by log transformation). To determine whether microbial community composition and total chemical profiles were correlated, the taxonomic and chemical dissimilarity/distance matrices were compared using a Mantel test in the R vegan package. Additionally, permutational analysis of variance (PERMANOVA) testing was performed to determine whether variation in community composition was significantly explained by concentrations of individual chemicals (log-transformed) using the vegan package in R. Relationships between individual chemical concentrations and antibiotic resistance gene family relative abundances were tested using HAllA. Input files for HAllA were filtered to remove all elements that were not detected in more than one sample. Antibiotic resistance genes were analyzed using Pearson correlation for individual comparisons and a Benjamini-Hochberg q-value threshold of 0.2 to control the false discovery rate for multiple testing.
Data analysis and visualization was performed in R primarily using functions from R packages vegan28 and labdsv.29 Data and R code necessary to reproduce the analyses are found at https://github.com/uo-green-lab/dust-2015.
Results and Discussion
To explore the relationship between antimicrobial chemicals and antibiotic resistance genes in dust, we collected 44 dust samples from 31 spaces in a mixed-use athletic and educational facility. We analyzed these samples for antimicrobial chemicals, microbial community profiles, and antibiotic resistance gene profiles. We obtained chemical data from 23 samples (the remainder had insufficient dust mass for chemical analysis). After quality control, we obtained microbial community data from 42 samples and antibiotic resistance gene data from 42 samples.
Correlation of Architectural Design with Microbial Community Composition
From the 16S ribosomal gene amplicon sequencing, we observed a median of 23,980 reads per sample (minimum 16,450, maximum 37,460), corresponding to 853 operational taxonomic units after filtering and quality control. Of the 44 samples, 41 yielded populations where no single OTU accounted for over 50% based on the relative abundances (Figure S1). Two samples were 52% Enterobacteriaceae, and one was 65% Pseudomonas.
Prior research has shown that indoor microbial community composition is linked to building design.14,30 With regards to design, space type has been a demonstrated significant factor.14b For this reason, we intentionally selected a building with a variety of space types to capture a wide swath of microbiome compositions. We observed a significant correlation between space type and community dissimilarity (PERMANOVA, R2 = 0.22885, p = 0.006; Figure S2a).
Another factor that has been shown to correlate with dust microbiome composition is building ventilation type.30 Our study building was constructed in 1921 and was designed to supply air through the windows via natural ventilation. Additionally, the building envelope is not tightly sealed, resulting in high infiltration via cracks. On average, the widths of the cracks were 70, 101, 730, and 413 mm (top, sides, middle, and bottom, respectively) around exterior doors and 105, 168, 226, and 152 mm around double-hung windows. Crack area, normalized by space floor area, averaged 0.4 in.2/sq ft or 2788 mm2/sq m. This building can be characterized as “leaky,” and the indoor microbiome is likely more influenced by the influx of outdoor air than in buildings that are tighter.14,30
Infiltration, like natural ventilation, increases the influx of environmental microbes.30 The large crack areas we observed likely influence the composition of the indoor microbiomes, although this effect could be exacerbated or mitigated by other factors, such as wind or stack-driven effects, occupant behavior, site-specific wind patterns, or mechanical system operation patterns. Approaching the question with microbiological methodology, we used the Bayesian SourceTracker algorithm to examine the potential influence of environmental versus human-derived sources on the dust microbiome.
The majority of the bacterial community in most samples was predicted to be of putative environmental (i.e., nonhuman) origin (Figure 1a). Twenty-five percent or less was predicted to be of human origin, of which the majority was associated with human skin (Figure 1a). Overall, we observed lower proportions of bacteria of human origin in our samples compared to previous studies of surfaces in athletic facilities and household indoor dust (Figure 1b). The indoor microbiome in our study system thus appears to be heavily influenced microbes that are not human-associated, more so than previously studied built environments.
Figure 1.
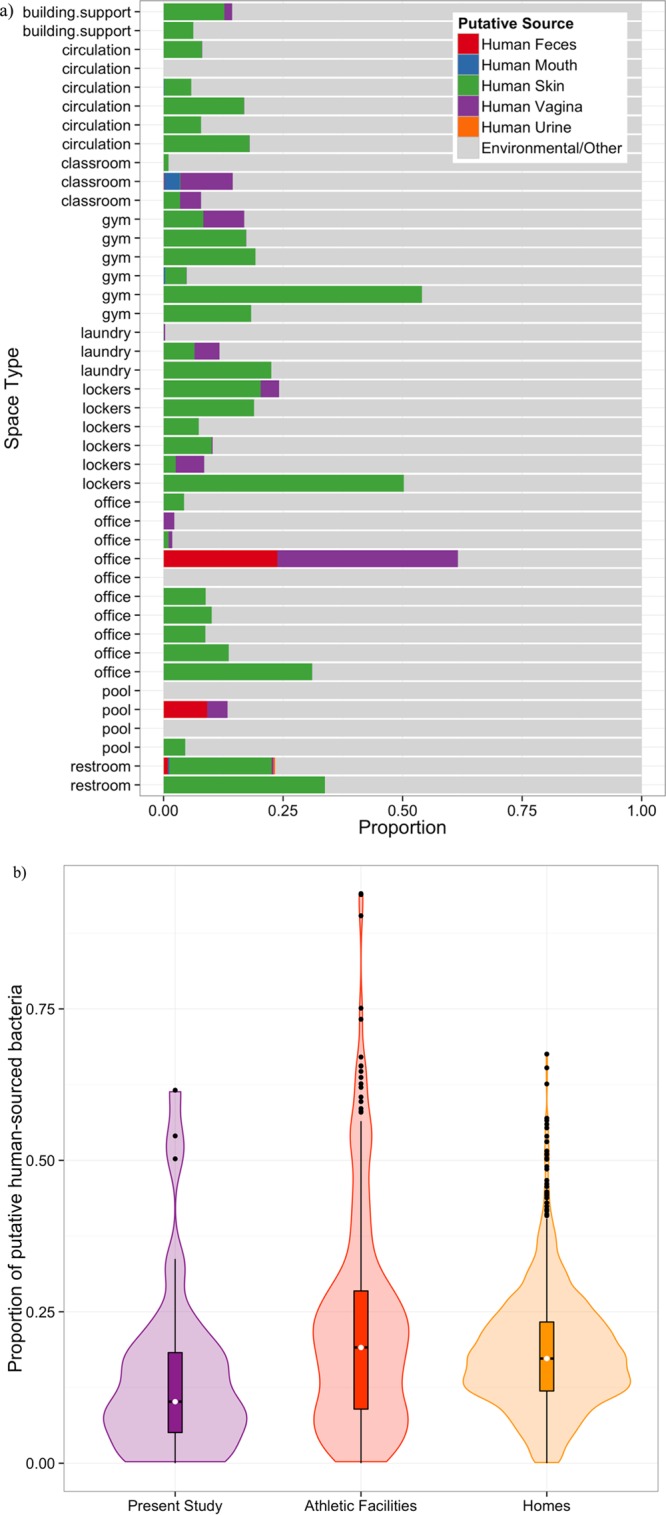
Influx of outdoor air, and thus outdoor microbes, likely had a large influence on the microbiome of the building in this study. a) The proportion of bacteria attributed to a human source using SourceTracker by sample. b) Violin plot, showing the median (white dot) and interquartile ranges (box and whiskers) as well as the density distribution (shaded area) of the proportion of bacteria of putative human origin in the present study (purple, n = 42), other athletic facilities (red, n = 308), and private homes (yellow, n = 1366).
Correlation of Antimicrobial Chemicals with Microbial Community Composition
We surveyed our samples for several antimicrobial chemicals that have reliably been detected in previous studies of indoor dust. Triclosan, triclocarban, and methyl-, ethyl-, propyl-, and butylparaben were quantified in all samples where sufficient dust mass was available (≥0.75 g). While these antimicrobials were detected at varying concentrations throughout the building (Tables 1 and S3), they were comparable to values previously observed in built environments. Other studies of indoor dust reported median values ranging from 320–1470, 11–276, 182–800, and 2–212 ng/g for methyl-, ethyl-, propyl-, and butylparaben, respectively;10a,10c,11 the median values from our study are 1020, 60, 380, and 60 ng/g, respectively. Median concentrations of triclosan in household dust ranged from 220 to 702 ng/g in other studies;10 in ours, the median concentration is 200 ng/g.
Table 1. Concentrations in ng/g Dust of Antimicrobial Chemicals Detected in This Studya.
| space type | triclosan | triclocarban | methylparaben | ethylparaben | propylparaben | butylparaben |
|---|---|---|---|---|---|---|
| classroom | 968 | 598 | 1695 | 200 | 996 | 111 |
| 1003 | 460 | 673 | 123 | 684 | 75 | |
| gym | 310 | 251 | 1559 | 112 | 519 | 88 |
| 132 | 88 | 526 | 50 | 130 | 20 | |
| office | 164 | 348 | 846 | 64 | 305 | 68 |
| 90 | 323 | 415 | 34 | 233 | 46 | |
| circulation | 302 | 274 | 1708 | 162 | 674 | 95 |
| 37 | 42 | 513 | 99 | 228 | 29 | |
| laundry | 356 | 162 | 421 | 18 | 124 | 39 |
| lockers | 326 | 2629 | 1603 | 51 | 502 | 65 |
| 191 | 3347 | 777 | 21 | 426 | 32 | |
| building support | 20 | 115 | 244 | 9 | 36 | 4 |
| pool | 85 | 156 | 399 | 13 | 58 | 18 |
| 165 | 130 | 348 | 22 | 73 | 29 |
Upper values are the average measurement for all samples collected in a given space type; lower values are one standard deviation. Where no standard deviation is given, only replicates were collected.
The correlation between the concentration of antimicrobial chemicals and the microbial community composition was not significant (p = 0.086; Supplementary Figure 2b) as determined from a Mantel test. Individual PERMANOVA tests revealed that the concentration of triclosan and all of the parabens correlate weakly yet significantly with microbial community dissimilarity (R2 between 0.07 and 0.08, p < 0.03). HAllA did not reveal any individual taxa that were significantly associated with any antimicrobial chemicals (data not shown).
Correlation of Antimicrobial Chemicals with Antibiotic Resistance Genes
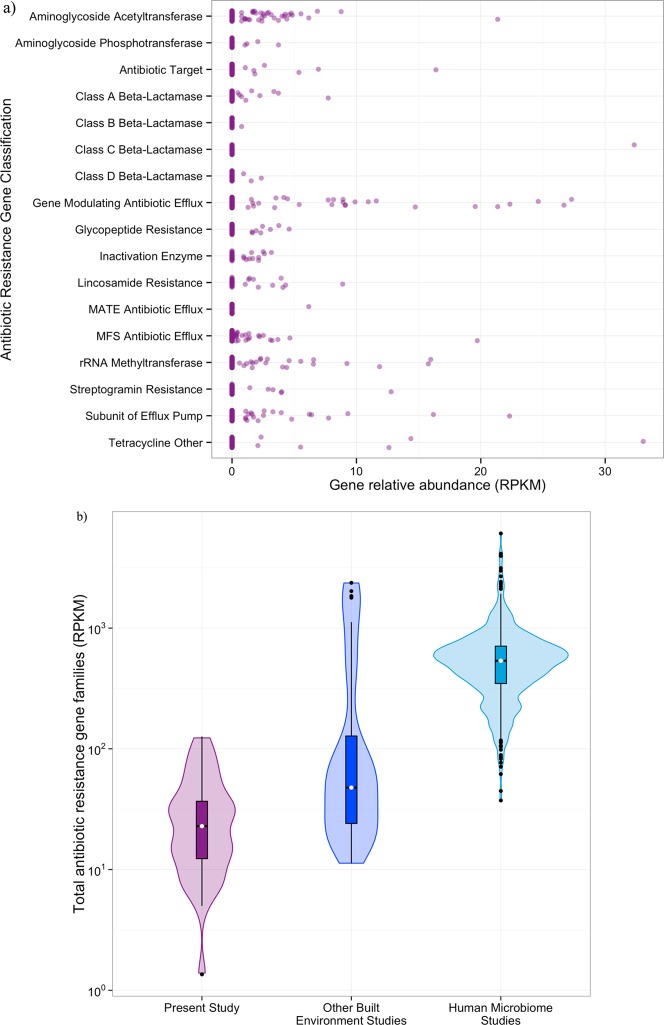
We identified antibiotic resistance gene families from the shotgun metagenomic data using markers generated from the CARD31 database. We identified 17 antibiotic resistance gene families throughout the building (Figure 2a, Table S4). The spaces had on average 21 RPKM attributed to an antibiotic resistance gene, although the standard deviation was quite large (27 RPKM). The three most highly abundant antibiotic resistance genes encoded the tetracycline-resistance gene tet(W), the extended-spectrum beta-lactamase blaSRT-1, and the macrolide-resistance gene erm(B) (Table S4). While present at low abundances, together these genes cover resistance to a wide spectrum of antibiotics.
Figure 2.
Antibiotic resistance gene families (identified in ARDB) were identified throughout the building but at lower relative abundances than observed elsewhere. a) Dot plot of the relative abundance (in RPKM) of each antibiotic gene class by sample. b) Violin plot, showing the median (white dot) and quartile ranges (box and whiskers) as well as the density distribution (shaded area) of the relative abundance of all antibiotic resistance gene families (in RPKM) from the present study (purple, n = 36), other built environment microbiomes (blue, n = 40), and the human microbiome (teal, n = 552).
Compared to previous investigations of the built environment resistome, our study system has fewer antibiotic resistance genes (median RPKM per sample 23.24 compared to 47.71 in other studies;13Figure 2b). The built environments included in this analysis are homes, offices, a hospital, a pier, and a metropolitan transit system. Data from a mixed-use athletic and educational facility was not available for comparison. Overall, the relative abundance of antibiotic resistance genes in all built environments is an order of magnitude lower than in the human gut (536.30 RPKM; Figure 2b). This finding is consistent with observations that the human gut has the highest abundance of antibiotic resistance genes of any surveyed biological system.32
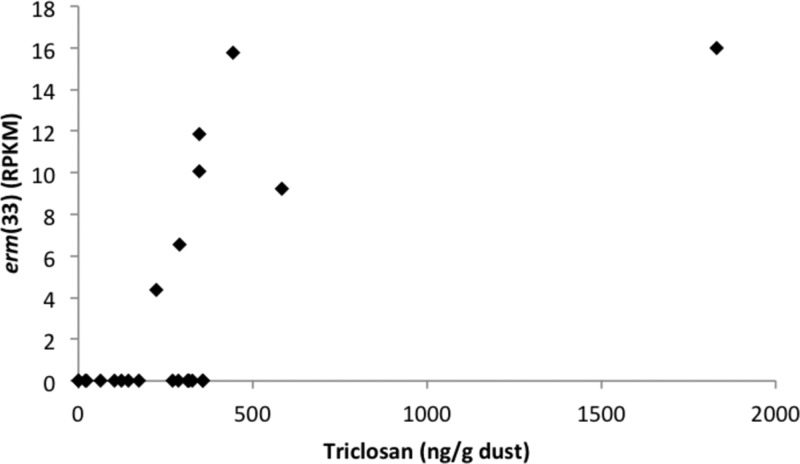
Using HAllA, we identified several antibiotic resistance genes that are significantly positively associated (FDR q < 0.05, similarity score >0.5) with triclosan or methylparaben (Table 2). Specifically, higher relative abundances of the 23S rRNA methyltransferase gene erm(X) were associated with higher concentrations of triclosan, and higher relative abundances of erm(33), which is a hybrid between erm(A) and erm(C) that is specific to Staphylococcus sp., and erm(C) were associated with higher concentrations of methylparaben (Figure 3). Ribosomal RNA methyltransferases, such as these erm genes, can confer resistance to antibiotics that act by targeting the peptidyltransferase center in bacteria, thereby inhibiting protein synthesis.33
Table 2. Select Antibiotic Resistance Genes Associated with Antimicrobial Chemicalsa.
| identifier | description | similarity score | p-value | q-value | |
|---|---|---|---|---|---|
| triclosan | erm(X) | 23S rRNA transferase | 0.86 | 0.001 | 0.036 |
| tet(K) | tetracycline resistance efflux pump | 0.83 | 0.001 | 0.036 | |
| vga(A) | streptogramin resistance efflux pump | 0.65 | 0.001 | 0.036 | |
| methylparaben | erm(33) | 23S rRNA transferase | 0.73 | 0.001 | 0.036 |
| erm(C) | 23S rRNA transferase | 0.68 | 0.001 | 0.036 | |
| cmr | resistance efflux pump | 0.58 | 0.002 | 0.048 |
Adjusted p-values (q-values) were calculated to account for multiple testing. Only associations with similarity scores >0.5 and adjusted p-values <0.05 are shown.
Figure 3.
A 23S ribosomal RNA methyltransferase (erm(33)) was significantly associated with triclosan.
In addition, relative abundances of two genes encoding efflux pumps (tet(K) and vga(A)) were associated with triclosan concentration, and relative abundances of the cmr efflux pump were associated with methylparaben concentration. tet(K) encodes an efflux pump with specific activity toward tetracyline and reduced activity toward derivatives of tetracycline lacking the 6-hydroxy group.34vga(A) encodes an ATP-binding cassette transporter that confers resistance to lincosamides and streptogramin A, as well as pleuromutilins,35 which have only been approved for use in humans since 2007.36 While triclosan has not previously been associated with erm methyltransferases or the tet(K) or vga(A) efflux pumps, it has been associated with an increased expression of multiple efflux pump genes in other settings.37
Of the six associations we identified (Table 2), three involve rRNA methyltransferases, a class of antibiotic resistance genes known for macrolide resistance. However, neither triclosan nor methylparaben is structurally or functionally related to macrolides. The remaining associations each involve an efflux pump specific to tetracycline, streptogramin, or chloramphenicol. The targets of these efflux pumps all represent different classes of antibiotics, and again none are structurally or functionally related to triclosan or methylparaben.
While our data show that higher relative abundances of antibiotic resistance genes correlate with higher concentrations of antimicrobial chemicals in indoor dust, further studies are needed to determine if this relationship is causative.
Implications and Limitations
Antimicrobials are widely distributed, high-production volume chemicals, so they are routinely found in many different environments including surface waters, fish, and human bodily fluids.5,38 Because they are so commonly found to bioaccumulate in these environments, the effects of triclosan on microbial communities have mostly been studied in aquatic systems and wastewater treatment. Triclosan was identified as a contaminant of emerging concern in drinking water and found to accumulate in wastewater at concentrations reaching parts per billion.38 These concentrations are comparable to those found in indoor dust. In microbial communities seeded from wastewater treatment plants, the addition of triclosan caused an increase in the copy number of the multidrug efflux pump gene mexB,39 which also confers resistance to beta-lactams, quinolones, tetracyclines, and trimethoprim.40 In benthic microbial communities, where typical triclosan concentrations are lower than in sewage sludge, triclosan was not associated with any changes in the microbial community composition or diversity, but communities exposed to higher concentrations of triclosan yielded higher proportions of cultivable triclosan-resistant isolates.9 Ours is thus not the first study to identify a link between the antimicrobial triclosan and antibiotic resistance; it is, however, the first to do so in indoor dust.
Our observations are correlative and do not give evidence of a causal relationship between the presence of antimicrobial chemicals in dust and an increase in antibiotic resistance genes in the dust microbiome. However, they do raise questions regarding how antibiotic resistance gene selection occurs and whether antimicrobial chemicals also affect microbial viability. Other studies investigating the effect of triclosan in various situations similarly found that the chemical has little effect on the microbial community composition,9,41 possibly due to insufficient exposure time42 or concentration.41 The median concentrations of triclosan we and others observed in dust is 3 orders of magnitude lower than the concentration of triclosan when used as an active ingredient in toothpaste (0.3–2 mg/g).38,43 Unfortunately, questions centered on bacterial gene selection and viability cannot be directly addressed using the current sequencing methods. Because we collected dust in situ, we cannot assess whether the relationship between antimicrobials and antibiotic resistance genes reflects events occurring before or after the chemicals and microbes are deposited in the dust.
Based on our data, microbial community composition correlates significantly with space type but not with antimicrobial chemical concentration in indoor dust; however, the concentration of individual antimicrobials strongly correlate with the relative abundance of individual antibiotic resistance genes. This observation suggests that these antimicrobials do little to affect the assembly of microbial communities in dust but they may have a strong influence on the promotion and retention of antibiotic resistance genes. This finding is similar to what was found in benthic microbial communities.9 As the indoor microbiome plays an important role in human health,4,15 the effect of antimicrobials on the accumulation of antibiotic resistance genes in the indoor microbiome deserves more attention.
Acknowledgments
The authors would like to thank Ali Rahnavard, Koji Yasuda, Eric Franzosa, Lauren McIver, Dale Northcutt, and Brendan Bohannan for their help. We are also grateful to Rodney Bloom and everyone who graciously allowed us to vacuum their offices. This work was funded by the Sloan Foundation (G-2013-6-04 & 2012-5-39 MBRP), the NSF (CAREER DBI-1053486), and the NIEHS (R01ES020889). The content does not necessarily represent the views of the NIEHS or NIH.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.6b00262.
Heatmap of the 25 most abundant genera, as determined by 16S rRNA amplicon sequencing and ecological analyses of the observed microbial communities (PDF)
Metadata, including total mass collected, for all samples (XLSX)
Target times and masses for analytes (XLSX)
Mapping file containing architectural metadata and all measured chemical concentrations (ng/g) used in sequence data analysis (XLSX)
Listing of all antibiotic resistance gene markers identified in this study and their ontologies (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Gibson M. K.; Crofts T. S.; Dantas G. Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 2015, 27, 51–56. 10.1016/j.mib.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.; Ward M.; van Bunnik B.; Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc., B 2015, 370 (1670), 20140083. 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandara A.; Mota L. C.; Flores C.; Perez H. R.; Green C. F.; Gibbs S. G. Isolation of Staphylococcus aureus and Antibiotic Resistant Staphylococcus aureus from Residential Indoor Bioaerosols. Environ. Health Perspect. 2006, 114 (12), 1859–1864. 10.1289/ehp.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. F.; Iverson S. A.; Baron P.; Vasse A.; Silbergeld E. K.; Lautenbach E.; Morris D. O. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect. Dis. 2012, 12, 703–16. 10.1016/S1473-3099(12)70156-1. [DOI] [PubMed] [Google Scholar]
- Halden R. U. On the need and speed of regulating triclosan and triclocarban in the United States. Environ. Sci. Technol. 2014, 48 (7), 3603–11. 10.1021/es500495p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey D. E.; McNamara P. J. The impact of triclosan on the spread of antibiotic resistance in the environment. Front. Microbiol. 2015, 5, 780. 10.3389/fmicb.2014.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L.; Sanchez M. B.; Martinez-Solano L.; Hernandez A.; Garmendia L.; Fajardo A.; Alvarez-Ortega C. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 2009, 33 (2), 430–49. 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- Gaulke C. A.; Barton C. L.; Proffitt S.; Tanguay R. L.; Sharpton T. J. Triclosan Exposure Is Associated with Rapid Restructuring of the Microbiome in Adult Zebrafish. PLoS One 2016, 11 (5), e0154632. 10.1371/journal.pone.0154632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury B.; Scott J.; Rosi-Marshall E. J.; Kelly J. J. Triclosan exposure increases triclosan resistance and influences taxonomic composition of benthic bacterial communities. Environ. Sci. Technol. 2013, 47 (15), 8923–30. 10.1021/es401919k. [DOI] [PubMed] [Google Scholar]
- a Fan X.; Kubwabo C.; Rasmussen P.; Jones-Otazo H. Simultaneous quantitation of parabens, triclosan, and methyl triclosan in indoor house dust using solid phase extraction and gas chromatography-mass spectrometry. J. Environ. Monit. 2010, 12 (10), 1891–7. 10.1039/c0em00189a. [DOI] [PubMed] [Google Scholar]; b Lu X.; Chen M.; Zhang X.; Sun Y.; Zhu D.; Zhang Q.; Wang B.; Zhang Z. Simultaneous quantification of five phenols in settled house dust using ultra-high performance liquid chromatography-tandem mass spectrometry. Anal. Methods 2013, 5 (19), 5339. 10.1039/c3ay40602d. [DOI] [Google Scholar]; c Canosa P.; Rodríguez I.; Rubí E.; Cela R. Determination of parabens and triclosan in indoor dust using matrix solid-phase dispersion and gas chromatography with tandem mass spectrometry. Anal. Chem. 2007, 79, 1675–81. 10.1021/ac061896e. [DOI] [PubMed] [Google Scholar]; d Geens T.; Roosens L.; Neels H.; Covaci A. Assessment of human exposure to Bisphenol-A, Triclosan and Tetrabromobisphenol-A through indoor dust intake in Belgium. Chemosphere 2009, 76 (6), 755–60. 10.1016/j.chemosphere.2009.05.024. [DOI] [PubMed] [Google Scholar]
- a Wang L.; Liao C.; Liu F.; Wu Q.; Guo Y.; Moon H.-B.; Nakata H.; Kannan K. Occurrence and Human Exposure of p-Hydroxybenzoic Acid Esters (Parabens), Bisphenol A Diglycidyl Ether (BADGE), and Their Hydrolysis Products in Indoor Dust from the United States and Three East Asian Countries. Environ. Sci. Technol. 2012, 46 (21), 11584–11593. 10.1021/es303516u. [DOI] [PubMed] [Google Scholar]; b Ramírez N.; Marcé R. M.; Borrull F. Determination of parabens in house dust by pressurised hot water extraction followed by stir bar sorptive extraction and thermal desorption–gas chromatography–mass spectrometry. J. Chromatogr. A 2011, 1218 (37), 6226–6231. 10.1016/j.chroma.2011.05.098. [DOI] [PubMed] [Google Scholar]; c Canosa P.; Pérez-Palacios D.; Garrido-López A.; Tena M. T.; Rodríguez I.; Rubí E.; Cela R. Pressurized liquid extraction with in-cell clean-up followed by gas chromatography–tandem mass spectrometry for the selective determination of parabens and triclosan in indoor dust. J. Chromatogr. A 2007, 1161 (1–2), 105–112. 10.1016/j.chroma.2007.05.089. [DOI] [PubMed] [Google Scholar]; d Rudel R. A.; Camann D. E.; Spengler J. D.; Korn L. R.; Brody J. G. Phthalates, Alkylphenols, Pesticides, Polybrominated Diphenyl Ethers, and Other Endocrine-Disrupting Compounds in Indoor Air and Dust. Environ. Sci. Technol. 2003, 37 (20), 4543–4553. 10.1021/es0264596. [DOI] [PubMed] [Google Scholar]
- a Selvaraj K. K.; Sivakumar S.; Sampath S.; Shanmugam G.; Sundaresan U.; Ramaswamy B. R. Paraben resistance in bacteria from sewage treatment plant effluents in India. Water Sci. Technol. 2013, 68 (9), 2067–2073. 10.2166/wst.2013.447. [DOI] [PubMed] [Google Scholar]; b Wells M. J. M.; Bell K. Y.; Traexler K. A.; Pellegrin M.-L.; Morse A. Emerging Pollutants. Water Environ. Res. 2010, 82 (10), 2095–2170. 10.2175/106143010X12756668802292. [DOI] [Google Scholar]
- Hsu T.; Joice R.; Vallarino J.; Abu-Ali G.; Hartmann E. M.; Shafquat A.; DuLong C.; Baranowski C.; Gevers D.; Green J. L.; Morgan X. C.; Spengler J. D.; Huttenhower C. Urban Transit System Microbial Communities Differ by Surface Type and Interaction with Humans and the Environment. mSystems 2016, 1 (3), e00018-16. 10.1128/mSystems.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kembel S. W.; Jones E.; Kline J.; Northcutt D.; Stenson J.; Womack A. M.; Bohannan B. J.; Brown G. Z.; Green J. L. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012, 6 (8), 1469–79. 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kembel S. W.; Meadow J. F.; O’Connor T. K.; Mhuireach G.; Northcutt D.; Kline J.; Moriyama M.; Brown G. Z.; Bohannan B. J.; Green J. L. Architectural design drives the biogeography of indoor bacterial communities. PLoS One 2014, 9 (1), e87093. 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K. E.; Rauch M.; Matsui E.; Iwai S.; Calatroni A.; Lynn H.; Mitchell H.; Johnson C. C.; Gern J. E.; Togias A.; Boushey H. A.; Kennedy S.; Lynch S. V. Development of a Standardized Approach for Environmental Microbiota Investigations related to Asthma Development in Children. J. Microbiol. Methods 2012, 91 (2), 231–239. 10.1016/j.mimet.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G.; Lauber C. L.; Walters W. A.; Berg-Lyons D.; Huntley J.; Fierer N.; Owens S. M.; Betley J.; Fraser L.; Bauer M.; Gormley N.; Gilbert J. A.; Smith G.; Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6 (8), 1621–4. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10 (10), 996–8. 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Edgar R. C.; Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31 (21), 3476–82. 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- McDonald D.; Price M. N.; Goodrich J.; Nawrocki E. P.; DeSantis T. Z.; Probst A.; Andersen G. L.; Knight R.; Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6 (3), 610–8. 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G.; Zaneveld J.; Caporaso J. G.; McDonald D.; Knights D.; Reyes J. A.; Clemente J. C.; Burkepile D. E.; Vega Thurber R. L.; Knight R.; Beiko R. G.; Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31 (9), 814–21. 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N.; Waldron L.; Ballarini A.; Narasimhan V.; Jousson O.; Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9 (8), 811–4. 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow J. F.; Altrichter A. E.; Bateman A. C.; Stenson J.; Brown G. Z.; Green J. L.; Bohannan B. J. Humans differ in their personal microbial cloud. PeerJ 2015, 3, e1258. 10.7717/peerj.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D.; Kuczynski J.; Charlson E. S.; Zaneveld J.; Mozer M. C.; Collman R. G.; Bushman F. D.; Knight R.; Kelley S. T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8 (9), 761–3. 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.; Gibbons S. M.; Lax S.; Eshoo-Anton T. W.; Owens S. M.; Kennedy S.; Gilbert J. A.; Hampton-Marcell J. T. Athletic equipment microbiota are shaped by interactions with human skin. Microbiome 2015, 3, 25. 10.1186/s40168-015-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan A.; Dunn R. R.; Reich B. J.; Pacifici K.; Laber E. B.; Menninger H. L.; Morton J. M.; Henley J. B.; Leff J. W.; Miller S. L.; Fierer N. The ecology of microscopic life in household dust. Proc. R. Soc. London, Ser. B 2015, 282, 1814. 10.1098/rspb.2015.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski J.; Gibson M. K.; Franzosa E. A.; Segata N.; Dantas G.; Huttenhower C. High-Specificity Targeted Functional Profiling in Microbial Communities with ShortBRED. PLoS Comput. Biol. 2015, 11 (12), e1004557. 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37 (Database issue), D443–7. 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J.; Blanchet F. G.; Kindt R.; Legendre P.; Minchin P. R.; O’Hara R. B.; Simpson G. L.; Solymos P.; Stevens M. H.; Wagner H.. vegan: Community Ecology Package. R package version 2.3-1; 2015.
- Roberts D. W.labdsv: Ordination and Multivariate Analysis for Ecology. R package version 1.7-0; 2015.
- Meadow J. F.; Altrichter A. E.; Kembel S. W.; Kline J.; Mhuireach G.; Moriyama M.; Northcutt D.; O’Connor T. K.; Womack A. M.; Brown G. Z.; Green J. L.; Bohannan B. J. M. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air 2014, 24 (1), 41–48. 10.1111/ina.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. G.; Waglechner N.; Nizam F.; Yan A.; Azad M. A.; Baylay A. J.; Bhullar K.; Canova M. J.; De Pascale G.; Ejim L.; Kalan L.; King A. M.; Koteva K.; Morar M.; Mulvey M. R.; O’Brien J. S.; Pawlowski A. C.; Piddock L. J. V.; Spanogiannopoulos P.; Sutherland A. D.; Tang I.; Taylor P. L.; Thaker M.; Wang W.; Yan M.; Yu T.; Wright G. D. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57 (7), 3348–3357. 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D.; Walsh F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol. Ecol. 2016, 92 (2), fiv168. 10.1093/femsec/fiv168. [DOI] [PubMed] [Google Scholar]
- Vester B.; Long K. S.. Antibiotic Resistance in Bacteria Caused by Modified Nucleosides in 23S Ribosomal RNA; Landes Bioscience: Austin, TX, 2000. [Google Scholar]
- Guay G. G.; Rothstein D. M. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob. Agents Chemother. 1993, 37 (2), 191–198. 10.1128/AAC.37.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry D. R.; McCloskey L.; Gwynn M. N.; Rittenhouse S. F.; Scangarella N.; Shawar R.; Holmes D. J. Genetic Characterization of Vga ABC Proteins Conferring Reduced Susceptibility to Pleuromutilins in Staphylococcus aureus. Antimicrob. Agents Chemother. 2008, 52 (12), 4507–4509. 10.1128/AAC.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak R.; Shlaes D. M. The pleuromutilin antibiotics: a new class for human use. Curr. Opin. Invest. Drugs (London, England: 2000) 2010, 11 (2), 182–91. [PubMed] [Google Scholar]
- a Chuanchuen R.; Beinlich K.; Hoang T. T.; Becher A.; Karkhoff-Schweizer R. R.; Schweizer H. P. Cross-Resistance between Triclosan and Antibiotics in Pseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects nfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45 (2), 428–432. 10.1128/AAC.45.2.428-432.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b McMurry L. M.; Oethinger M.; Levy S. B. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 1998, 166 (2), 305–9. 10.1111/j.1574-6968.1998.tb13905.x. [DOI] [PubMed] [Google Scholar]; c Srinivasan V. B.; Singh B. B.; Priyadarshi N.; Chauhan N. K.; Rajamohan G. Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PLoS One 2014, 9 (5), e96288. 10.1371/journal.pone.0096288. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005, 56 (1), 20–51. 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Bedoux G.; Roig B.; Thomas O.; Dupont V.; Le Bot B. Occurrence and toxicity of antimicrobial triclosan and by-products in the environment. Environ. Sci. Pollut. Res. 2012, 19 (4), 1044–65. 10.1007/s11356-011-0632-z. [DOI] [PubMed] [Google Scholar]
- McNamara P. J.; LaPara T. M.; Novak P. J. The impacts of triclosan on anaerobic community structures, function, and antimicrobial resistance. Environ. Sci. Technol. 2014, 48 (13), 7393–400. 10.1021/es501388v. [DOI] [PubMed] [Google Scholar]
- a Schweizer H. P. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob. Agents Chemother. 1998, 42 (2), 394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ziha-Zarifi I.; Llanes C.; Köhler T.; Pechere J.-C.; Plesiat P. In Vivo Emergence of Multidrug-Resistant Mutants ofPseudomonas aeruginosa Overexpressing the Active Efflux System MexA-MexB-OprM. Antimicrob. Agents Chemother. 1999, 43 (2), 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretro T.; Sonerud T.; Mangelrod E.; Langsrud S. Evaluation of the antibacterial effect of a triclosan-containing floor used in the food industry. J. Food Prot. 2006, 69, 627–633. [DOI] [PubMed] [Google Scholar]
- Kampf G.; Kramer A. Epidemiologic Background of Hand Hygiene and Evaluation of the Most Important Agents for Scrubs and Rubs. Clin. Microbiol. Rev. 2004, 17 (4), 863–893. 10.1128/CMR.17.4.863-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S.; Chandrasekhar S.; Shashikumar K. V.; Payne D.; Maclure R.; Kapadiya B.; Schafer F.; Adams S. Plaque triclosan concentration and antimicrobial efficacy of a new calcium carbonate toothpaste with 0.3% triclosan compared to a marketed 0.3% triclosan toothpaste. Int. J. Clin. Dent. 2013, 24 (2), 68–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.