Abstract
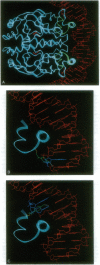
We have developed a straightforward biochemical method to determine the orientation of the DNA binding motif of a sequence-specific DNA binding protein relative to the DNA site in the protein-DNA complex. The method involves incorporation of a photoactivatable crosslinking agent at a single site within the DNA binding motif of the sequence-specific DNA binding protein, formation of the derivatized protein-DNA complex, UV-irradiation of the derivatized protein-DNA complex, and determination of the nucleotide(s) at which crosslinking occurs. We have applied the method to catabolite gene activator protein (CAP). We have constructed and analyzed two derivatives of CAP: one having a phenyl azide photoactivatable crosslinking agent at amino acid 2 of the helix-turn-helix motif of CAP, and one having a phenyl azide photoactivatable crosslinking agent at amino acid 10 of the helix-turn-helix motif of CAP. The results indicate that amino acid 2 of the helix-turn-helix motif is close to the top-strand nucleotides of base pairs 3 and 4 of the DNA half site in the CAP-DNA complex, and that amino acid 10 of the helix-turn-helix motif is close to the bottom-strand nucleotide of base pair 10 of the DNA half site in the CAP-DNA complex. The results define unambiguously the orientation of the helix-turn-helix motif relative to the DNA half site in the CAP-DNA complex. Comparison of the results to the crystallographic structure of the CAP-DNA complex [Schultz, S., Shields, S. & Steitz, T. (1991) Science 253, 1001-1007] indicates that the method provides accurate, high-resolution proximity and orientation information.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- D'Andrea A. D., Haseltine W. A. Modification of DNA by aflatoxin B1 creates alkali-labile lesions in DNA at positions of guanine and adenine. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4120–4124. doi: 10.1073/pnas.75.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Characterization of protein-DNA complexes by affinity cleaving. Methods Enzymol. 1991;208:497–515. doi: 10.1016/0076-6879(91)08026-e. [DOI] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989 Dec 25;17(24):10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Pendergrast P. S., Gunasekera A. Conversion of a helix-turn-helix motif sequence-specific DNA binding protein into a site-specific DNA cleavage agent. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2882–2886. doi: 10.1073/pnas.87.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Kolb A., Buc H., Kunkel T. A., Krakow J. S., Beckwith J. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K. S., Dervan P. B. Structural motif of the DNA binding domain of gamma delta-resolvase characterized by affinity cleaving. J Biol Chem. 1990 Sep 25;265(27):16534–16540. [PubMed] [Google Scholar]
- Gunasekera A., Ebright Y. W., Ebright R. H. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem. 1992 Jul 25;267(21):14713–14720. [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hixson S. H., Hixson S. S. P-Azidophenacyl bromide, a versatile photolabile bifunctional reagent. Reaction with glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1975 Sep 23;14(19):4251–4254. doi: 10.1021/bi00690a016. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Bebenek K., McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Mack D. P., Sluka J. P., Shin J. A., Griffin J. H., Simon M. I., Dervan P. B. Orientation of the putative recognition helix in the DNA-binding domain of Hin recombinase complexed with the hix site. Biochemistry. 1990 Jul 17;29(28):6561–6567. doi: 10.1021/bi00480a003. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Bavykin S. G., Belyavsky A. V., Karpov V. L., Preobrazhenskaya O. V., Shick V. V., Ebralidse K. K. Mapping DNA-protein interactions by cross-linking. Methods Enzymol. 1989;170:386–408. doi: 10.1016/0076-6879(89)70058-6. [DOI] [PubMed] [Google Scholar]
- Oakley M. G., Dervan P. B. Structural motif of the GCN4 DNA binding domain characterized by affinity cleaving. Science. 1990 May 18;248(4957):847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Shin J. A., Ebright R. H., Dervan P. B. Orientation of the Lac repressor DNA binding domain in complex with the left lac operator half site characterized by affinity cleaving. Nucleic Acids Res. 1991 Oct 11;19(19):5233–5236. doi: 10.1093/nar/19.19.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Bruist M. F., Simon M. I., Dervan P. B. Synthesis of a sequence-specific DNA-cleaving peptide. Science. 1987 Nov 20;238(4830):1129–1132. doi: 10.1126/science.3120311. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987 Nov 20;198(2):311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Zhang X. P., Ebright R. H. Identification of a contact between arginine-180 of the catabolite gene activator protein (CAP) and base pair 5 of the DNA site in the CAP-DNA complex. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4717–4721. doi: 10.1073/pnas.87.12.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. P., Gunasekera A., Ebright Y. W., Ebright R. H. Derivatives of CAP having no solvent-accessible cysteine residues, or having a unique solvent-accessible cysteine residue at amino acid 2 of the helix-turn-helix motif. J Biomol Struct Dyn. 1991 Dec;9(3):463–473. doi: 10.1080/07391102.1991.10507929. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]