Abstract
Reassortment of 2009 (H1N1) pandemic influenza virus (pdH1N1) with other strains may produce more virulent and pathogenic forms, detection and their rapid characterization is critical. In this study, we reported a “one-size-fits-all” approach using a next-generation sequencing (NGS) detection platform to extensively identify influenza viral genomes for diagnosis and determination of novel virulence and drug resistance markers. A de novo module and other bioinformatics tools were used to generate contiguous sequence and identify influenza types/subtypes. Of 162 archived influenza-positive patient specimens, 161(99.4%) were positive for either influenza A or B viruses determined using the NGS assay. Among these, 135(83.3%) were A(H3N2), 14(8.6%) were A(pdH1N1), 2(1.2%) were A(H3N2) and A(pdH1N1) virus co-infections and 10(6.2%) were influenza B viruses. Of the influenza A viruses, 66.7% of A(H3N2) viruses tested had a E627K mutation in the PB2 protein, and 87.8% of the influenza A viruses contained the S31N mutation in the M2 protein. Further studies demonstrated that the NGS assay could achieve a high level of sensitivity and reveal adequate genetic information for final laboratory confirmation. The current diagnostic platform allows for simultaneous identification of a broad range of influenza viruses, monitoring emerging influenza strains with pandemic potential that facilitating diagnostics and antiviral treatment in the clinical setting and protection of the public health.
Introduction
Influenza viruses belonging to the family Orthomyxoviridae, are classified into influenzavirus A, B, and C. While influenza A and C viruses infect humans and many other species, influenza B viruses only infect humans and seals [1,2,3]. The Influenza virus genome consists of eight negative single-stranded RNA segments encoding eleven proteins: hemagglutinin (HA), neuraminidase (NA), nucleoprotein (NP), polymerase proteins (PB1, PB2, and PA), matrix proteins (M1 and M2), and nonstructural proteins (NS1 and NS2) [4]. Based on reactivity with surface glycoproteins HA and NA, influenza A viruses are classified into eighteen HA subtypes (H1-H18) and eleven NA subtypes (N1-N11) with a total of 144 subtypes possible, while influenza B virus has no subtypes [4,5,6,7,8]. New influenza viral variants emerge either due to mutations in the virus surface glycoproteins (antigenic drift) or reassortment between viral gene segments from different strains (antigenic shift). While, seasonal influenza outbreaks are thought to arise primarily due to antigenic drift, pandemics are thought to result from complex reassortment events among swine, human, and avian reservoirs. Some of them have been reported to directly infect humans, or transmit avian influenza A viruses from domestic poultry to human [4,9,10]. In addition, influenza viruses normally circulate in pigs are called “variant” viruses when found in people. There was a large outbreak of influenza A(H3N2) variant virus (H3N2v) during the 2010–13 seasons in U.S. and a highly pathogenic avian influenza (HPAI) H5N1 virus was identified in early 2014 in North America [10]. According to a USDA report in January 21 2015, HPAI H5N1, H5N2, and H5N8 viruses, which can be potentially transmitted to domestic poultry population leading to an outbreak [11], have been detected in U.S. wild birds in the winter of 2015 [12]. Identification of H7N9, H10N8 and other viruses in humans outside the U.S. has also been reported [9,13]. Reassortment of a 2009 H1N1 pandemic influenza A virus (pdH1N1) and influenza A(H3N2) variant virus (H3N2v) with other circulating seasonal strains in the U.S. can produce virus variants with transmissibility and altered virulence for humans [14,15]. Multiple influenza types/subtypes usually circulate during each season, for example, pdH1N1 were the predominant viruses circulating in December 2013 with fewer influenza B viruses, but influenza B virus became the predominant circulating virus by late March 2014, while seasonal A(H3N2), novel H3N2v, and mixed A/B viruses infections were also identified during the season in the U.S. [16].
It was reported that rapid influenza diagnostic tests showed low and variable sensitivity [17,18] depending on the influenza A subtype [19], while some FDA-cleared diagnostic tests did not detect the novel H3N2v viruses [20]. Diagnosis of influenza infection from an unknown risk respiratory specimen involves multiple assays [17,21]. Identification of the first case of A(H5N1) virus infection in North America involved multiple laboratories [10]. Current methods that evaluate antiviral resistance require two independent tests for the S31N substitutions conferring amantadine-resistance [22,23] and the H274Y substitution conferring oseltamivir-resistance [22,24]. Conventional methods used by diagnostic or reference laboratories for characterization of novel influenza viruses are ineffective since the tests are based on a small conserved region of each HA, NA, and M genes for detection and subtyping, requires the use of many sets of primers, steps and multiple reactions. In addition, this approach may be ineffective for newly emerging influenza subtypes due to mismatch of PCR primers and culture-based genome sequencing which requires high biosafety levels, preventing its application in clinics. Next-generation sequencing (NGS) analysis has been previously reported to greatly improve manipulation procedure for identification of influenza virus strains, these studies focused on examining influenza A [25,26,27] or B [28] viruses using cultured virus [27], random [27], universal [25], and multiple [28] primers. None of these studies reported the use of a paired set of degenerate universal primers to simultaneously identify whole-genome sequences of influenza A and B viruses from a large set of clinical specimens using an NGS assay. We previously reported development of a genomic nanomicroarray and NGS combination approach for influenza virus screening and laboratory confirmation [29]. Here, we extended the NGS approach to potential application for influenza virus surveillance, whole-genome characterization of putative antiviral resistance markers and virulence factors that may confer high virulence in human hosts. We developed a novel universal RT-PCR-NGS platform by testing 162 clinical influenza-positive specimens and demonstrated a “one-size-fits-all” approach to simultaneously identify unknown influenza infections and co-infections in a single tube reaction per sample. The analytical sensitivity of the NGS platform was also evaluated in our studies.
Materials and Methods
Viruses, Clinical Specimens and Tests
Pre-titrated stocks of reference strains of influenza viruses were procured from Dr. Stephen Lindstrom (Centers for Disease Control and Prevention, CDC, Atlanta, GA) or cultured in embryonated chicken eggs at US Food and Drug Administration (FDA). Clinical nasopharyngeal swab specimens were collected from patients with symptoms of influenza-like illness, submitted during the 2010–11 and 2012–13 influenza seasons to the Clinical Virology Laboratory (CVL), Yale New Haven Hospital, New Haven, CT. These specimens were tested as requested by the patients’ physicians using a direct fluorescent antibody (DFA) immunoassay (SimulFluor Respiratory Virus Screen Reagent, Millipore, Billerica, MA) and/or by quantitative RT-PCR (RT-qPCR), as previously described [21]. A total of 162 nasopharyngeal swab specimens that tested as influenza A- and/or B-positive were de-identified and sent to the Laboratory of Molecular Virology (LMV) at FDA (Institutional Review Board (IRB) approval# Research Involving Human Subjects Committee (RIHSC):13-048B) for sequencing (Fig 1). All participants, or their parents or guardians, provided written informed consent when their specimens collected in Yale New Haven Hospital.
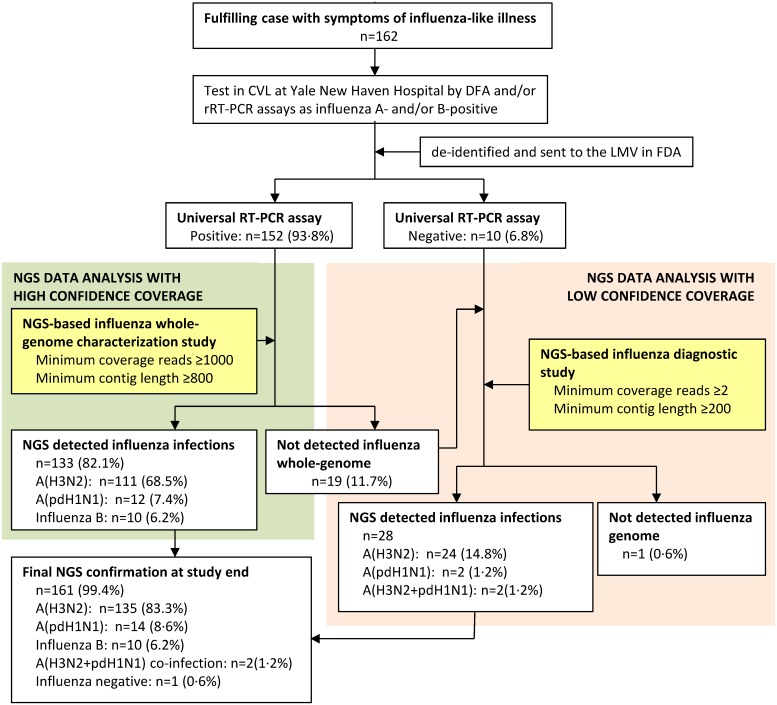
Fig 1. Study profile.
Enrollment of 162 patients aged 20 months to 89 years with symptoms of influenza-like illness during February 19, 2011 to March 8, 2013 from over twenty cities and towns in Connecticut, and submitted to the Clinical Virology Laboratory (CVL) at Yale New Haven Hospital, New Haven, Connecticut prior to detect by universal RT-PCR and NGS assays at the Laboratory of Molecular Virology (LMV) in FDA. A total of 381 genome sequences identified from 55 specimens including A(H3N2), A(pdH1N1), and Influenza B viruses were submitted into GenBank. DFA, direct fluorescent antigen test; RT-qPCR, quantitative RT-PCR; NGS, next-generation sequencing.
RNA Extraction and Universal RT-PCR
The RNA extraction, some primers, and Reverse Transcription-PCR (RT-PCR) procedures were described previously [29] and in S1 File. According to previous reports and our testing algorithm using multiple alternative options [29,30,31,32], a new set of degenerate universal primers for RT (uniflu, 5’-IAGCARAAGC -3’) and fusion primers for PCR (unifluF, 5’-ACGACGGGCGACAIAGCARAAGC-3’; unifluR, 5’-ACGACGGGCGACAAGTAGWAACA-3’) were designed and tested. The 13bp flanking sequences italicized were added at the 5’ end of each PCR primer to enhance the annealing temperature and achieve high fidelity and yield in PCR amplification. These primers target highly conserved regions at the 5'- and 3'- terminus of each segment of influenza virus to amplify the whole-genome concomitantly, and have coverage of sequence variants for influenza A, B, and C viruses to achieve improvements in analytical sensitivity and inclusivity.
MiSeq Sequencing
Sample preparation and bioinformatics data analysis was performed as described [29]. Briefly, the concentration of mega-amplicons were measured by using the Qubit dsDNA BR Assay System (Covaris, Woburn, MA, USA), and 1 ng of DNA product was processed for NGS sample preparation by using a Illumina Nextera XT DNA Sample Preparation Kit. Mega-amplicons for each specimen were internally marked with dual-index primers (S2 Fig). Up to 96 specimens were barcoded and run in one lane of the sequencer. NGS was performed using a MiSeq v2 kit (500 cycles) to produce 2x250 paired-end reads (Illumina, San Diego, CA). After automated cluster generation in MiSeq, the sequencing was processed and genomic sequence reads obtained.
De novo Assembly Strategy
Sequencing reads of approximately 250bp in length were trimmed and the sequence data verified using FastQC software prior to de novo assembly. Contigs for individual gene segments from influenza viruses were generated using the de novo assembly module on the CLC bio software (v6.0.6, CLC bio, Cambridge, MA). The parameter for mapping reads back to contiguous was set on as similarity fraction 0.9; length fraction 0.5; mismatch cost 2; insertion cost 3 and deletion cost 3. For characterization of influenza whole-genomes with high confidence coverage, minimum contig length was set at 800bp to assemble the consensus sequences and minimum coverage was 1000 reads [33]. For diagnosis of influenza virus infections, minimum contig length used was 200bp and higher lengths for 2x reads per base in de novo assembly (low confidence coverage, Fig 1 and S3 Fig) [34].
Bioinformatics Data Analysis
A master file containing all unique contigous sequences of each mega-amplicon was generated and used to perform an all-by-all blast search in Influenza Research Database (IRD, www.fludb.org), the Global Initiative on Sharing All Influenza Data (GISAID, platform.gisaid.org) and NCBI database. The match with the highest score was retained to identify the specific influenza genome. Assembled sequences were aligned by the CLUSTAL W program, and phylogenetic analysis was performed using MEGA 5 and the neighbor-joining method [35]. The amino acid sequence for each segment identified was generated and aligned using the Vector NTI Advance package (v·11·5·2, Life Technologies, Grand Island, NY). The serotype and genotype according to the genetic lineage of influenza A viruses were determined using FluGenome [36].
Coverage of Depth and Breadth
The depth of sequence coverage (DOC) was calculated using formulae LN/G, where L is the read length, N is the number of reads and G is the theoretical genome length. The breadth of sequence coverage (BOC) was calculated as percentage of actual testing contig length (≥1000 reads or ≥30x of DOC) divided by the theoretical genome length [34,37]. Total average length of influenza A and B virus genomes was 13,588bp and 13,559bp, respectively. Representative lengths of individual gene segments from influenza A virus (A/Puerto Rico/8/1934(H1N1)) were as follows; 2341bp (PB2), 2341bp (PB1), 2233bp (PA), 1778bp (HA), 1565bp (NP), 1413bp (NA), 1027bp (M) and 890bp (NS).
Sequence Accession Numbers
The newly reported influenza genome sequences are available in GenBank under the following accession numbers: KM654599-KM654931, and KM654933-KM654980.
Results
NGS Detects Influenza Whole-Genome Segments
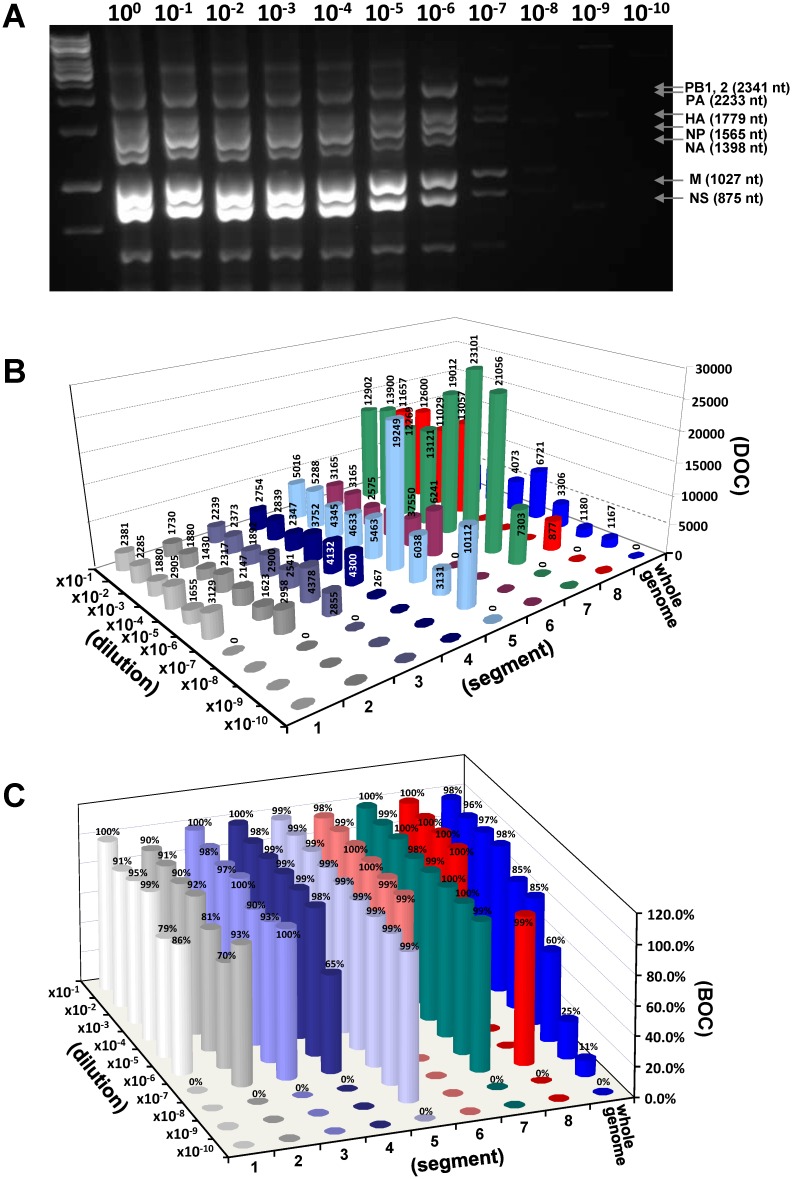
To determine the sensitivity of NGS detection, ten-fold serial dilutions of H5N1 viral RNA (1.8x109 TCID50/mL) were first produced followed by RT-PCR exponential amplification for whole-genome segments. As shown in Fig 2A, 10−7 dilution of A(H5N1) viral RNA (1.8x103 TCID50/mL) was detected in the RT-PCR assay shown as multiple faint visible bands. A total of ten PCR mega-amplicons from the dilutions were next tested using MiSeq, multiple contigs were finally automatically generated for each mega-amplicons, and all contig sequences were correctly identified as genome of A/Viet Nam/1203/2004(H5N1) virus. The overall average of DOC was 4,715. The average of BOC for each identified contig was 96% (≥30x depth) indicating that the near full-length genome sequence was covered (Table 1). Analysis of reads at each dilution level showed that the whole-genome DOC decreases from 4,073 in 10−1 dilution to 1,167 in 10−9 dilution (Fig 2B), and the whole-genome BOC decreases from 98% to 11% at the 10−9 dilution. We observed that the NP gene (segment 5) represented 99% of BOC at all detected titration points with the lowest detectable level at 10−9 dilution (1.8x101 TCID50/mL) with high confidence coverage (Fig 2C).
Fig 2.
(A) Agarose gel illustrating the sensitivity of the universal RT-PCR assay. The picture shows PCR mega-amplicons in 2% agarose gel representing the whole-genome segments of A/Viet Nam/1203/2004(H5N1) virus in dilution series, and detection of viral RNA in a dilution of 10−7 (Table 1). (B) Illustrating average depth of coverage (DOC). (C) average breadth of coverage (BOC) in NGS characterization of the influenza genome.
Table 1. Length, reads and coverage from serial dilutions of influenza A(H5N1) virus.
| gene segment (bp) | serial dilutions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | 10−7 | 10−8 | 10−9 | 10−10 | ||
| PB2 (2341) | length (bp) | 2341 | 2132 | 2230 | 2323 | 1854 | 2024 | 0 | 0 | 0 | 0 |
| reads (x103) | 22·3 | 21·4 | 17·6 | 27·2 | 15·5 | 29·3 | 0 | 0 | 0 | 0 | |
| depth | 2381 | 2285 | 1880 | 2905 | 1655 | 3129 | 0 | 0 | 0 | 0 | |
| breadth | 100% | 91% | 95% | 99% | 79% | 86% | 0% | 0% | 0% | 0% | |
| PB1 (2341) | length (bp) | 2109 | 2123 | 2102 | 2158 | 1895 | 1648 | 2174 | 0 | 0 | 0 |
| reads (x103) | 16·2 | 17·6 | 13·4 | 21·7 | 20·1 | 15·2 | 27·7 | 0 | 0 | 0 | |
| depth | 1730 | 1880 | 1430 | 2317 | 2147 | 1623 | 2958 | 0 | 0 | 0 | |
| breadth | 90% | 91% | 90% | 92% | 81% | 70% | 93% | 0% | 0% | 0% | |
| PA (2233) | length (bp) | 2233 | 2192 | 2171 | 2233 | 2016 | 2085 | 2224 | 0 | 0 | 0 |
| reads (x103) | 20 | 21·2 | 16·9 | 25·9 | 22·7 | 39·1 | 25·5 | 0 | 0 | 0 | |
| depth | 2239 | 2373 | 1892 | 2900 | 2541 | 4378 | 2855 | 0 | 0 | 0 | |
| breadth | 100% | 98% | 97% | 100% | 90% | 93% | 100% | 0% | 0% | 0% | |
| HA (1779) | length (bp) | 1772 | 1747 | 1753 | 1757 | 1760 | 1751 | 1160 | 0 | 0 | 0 |
| reads (x103) | 19·6 | 20·2 | 16·7 | 26·7 | 29·4 | 30·6 | 1·9 | 0 | 0 | 0 | |
| depth | 2754 | 2839 | 2347 | 3752 | 4132 | 4300 | 267 | 0 | 0 | 0 | |
| breadth | 100% | 98% | 99% | 99% | 99% | 98% | 65% | 0% | 0% | 0% | |
| NP (1565) | length (bp) | 1556 | 1556 | 1556 | 1556 | 1556 | 1554 | 1556 | 1556 | 1543 | 0 |
| reads (x103) | 31·4 | 33·1 | 27·2 | 29 | 34·2 | 120·5 | 37·8 | 19·6 | 63·3 | 0 | |
| depth | 5016 | 5288 | 4345 | 4633 | 5463 | 19249 | 6038 | 3131 | 10112 | 0 | |
| breadth | 99% | 99% | 99% | 99% | 99% | 99% | 99% | 99% | 99% | 0% | |
| NA (1398) | length (bp) | 1371 | 1389 | 1398 | 1398 | 1389 | 1389 | 0 | 0 | 0 | 0 |
| reads (x103) | 17·7 | 17·7 | 14·4 | 19·3 | 21 | 34·9 | 0 | 0 | 0 | 0 | |
| depth | 3165 | 3165 | 2575 | 3451 | 3755 | 6241 | 0 | 0 | 0 | 0 | |
| breadth | 98% | 99% | 100% | 100% | 99% | 99% | 0% | 0% | 0% | 0% | |
| M (1027) | length (bp) | 1027 | 1016 | 1027 | 1003 | 1016 | 1027 | 1027 | 1018 | 0 | 0 |
| reads (x103) | 53 | 57·1 | 50·4 | 53·9 | 78·1 | 94·9 | 86·5 | 30 | 0 | 0 | |
| depth | 12902 | 13900 | 12269 | 13121 | 19012 | 23101 | 21056 | 7303 | 0 | 0 | |
| breadth | 100% | 99% | 100% | 98% | 99% | 100% | 100% | 99% | 0% | 0% | |
| NS (875) | length (bp) | 884 | 883 | 889 | 891 | 0 | 0 | 0 | 877 | 0 | 0 |
| reads (x103) | 40·8 | 44·1 | 38·6 | 45·7 | 0 | 0 | 0 | 14·4 | 0 | 0 | |
| depth | 11657 | 12600 | 11029 | 13057 | 0 | 0 | 0 | 4103 | 0 | 0 | |
| breadth | 100% | 100% | 100% | 100% | 0% | 0% | 0% | 100% | 0% | 0% | |
| Whole Genome (13559) | length (bp) | 13293 | 13038 | 13126 | 13319 | 11486 | 11478 | 8141 | 3451 | 1543 | 0 |
| reads (x103) | 221 | 232·5 | 195·1 | 249·3 | 220·9 | 364·5 | 179·3 | 64 | 63·3 | 0 | |
| depth* | 4073 | 4287 | 3597 | 4597 | 4073 | 6721 | 3306 | 1180 | 1167 | 0 | |
| breadth* | 98% | 96% | 97% | 98% | 85% | 85% | 60% | 25% | 11% | 0% | |
| No. of genes. | 8 | 8 | 8 | 8 | 7 | 7 | 5 | 3 | 1 | 0 | |
Ten-fold serial dilutions were used in titrations of influenza A(H5N1) virus to establish the analytical sensitivity by universal RT-PCR and NGS assays. Minimum contiguous length sets at 800bp to assemble the consensus sequences for characterization of each segment full-length sequence and minimum coverage is 1000 reads. A calculated whole-genome base of A/Viet Nam/1203/2004 virus is 13559bp.
* calculation for virus whole-genome.
Molecular Confirmation of Simultaneous Detection of Influenza A and B Viruses in a Single Specimen/Test
Since some influenza related hospitalized cases were found to be associated with mixed A and B virus co-infections [1,16], analytical inclusivity studies were performed next by using a new set of degenerate universal primers to achieve improved coverage for varieties of both influenza A and B viruses. We tested A/Fujian Gulou/1896/09(H1N1), A/Perth/16/2009(H3N2), and B/Wisconsin/01/2010 reference viruses. Total RNA was extracted from spiked samples containing either individual or a mixture of two strains 1:1, and RT-PCR was performed. After the NGS test and following de novo assembly, eight contigs were generated for A(H1N1), A (H3N2), and B viruses, respectively. Fifteen and sixteen contigs were generated from the mixed A(H1N1), and A(H3N2) with B viruses supported by high confidence coverage, respectively (S1 Table). A validation assessment for these contigs from mixed viruses confirmed that the genome sequences are identical to the gene segments of the A(H1N1), A(H3N2), or B viruses, respectively. This is the first report of analytical validation of degenerate universal primers that can simultaneously amplify influenza A and B viruses for NGS identification and discrimination of influenza viruses in a single sample/test.
Detection of Influenza Virus in Nasopharyngeal Swab Specimens
By using the aforementioned strategy, we next tested 162 clinical nasopharyngeal swab specimens (Fig 1) which previous detection in CVL showed 74%(120/162) DFA test positive, 60%(98/162) RT-qPCR test positive, and 6.2%(10/162) for influenza B viruses. The range of Ct values for the 162 specimens is between 17.4 and 39.2. The PCR mega-amplicons prepared for each of the clinical specimens were analyzed using MiSeq. 99.4%(161/162) specimens detected influenza viral genome (see description below, S4 Table). To simplify the table, we reported only 57 isolates with whole-genome segments including 35 A(H3N2) isolates, twelve A(pdH1N1), and ten influenza B viruses identified in NGS data analysis. The results from four different detection methods including DFA, RT-qPCR, universal RT-PCR and NGS assays were summarized in Table 2 to represent the types/subtypes for influenza viruses. Results of NGS data analysis representing full-length sequences were listed in S2 Table. The average whole-genome DOC for these viruses was between 0.1x103 and 13.7x103, and average whole-genome BOC was between 13% and 100% supporting high confidence coverage.
Table 2. Detection of influenza A and B viruses in nasopharyngeal swab specimens from naturally infected patients collected in the 2010–11 and 2012–13 seasons in Connecticut.
| Patient ID | Age | Gender | Collected Date | City, State | Detection Methods | |||
|---|---|---|---|---|---|---|---|---|
| DFA | RT-qPCR(Ct) | Univ·PCR | NGS Verified | |||||
| Flu003 | 9 | M | 12/29/2012 | Hamden, CT | B | B(22·9) | ++? | B |
| Flu053 | 28m. | M | 12/30/2012 | New Haven, CT | B | not done | +? | B |
| Flu096 | 9 | F | 2/5/2013 | New Haven, CT | B | not done | +? | B |
| Flu101 | 72 | M | 3/5/2013 | West Haven, CT | B | B(21·2) | ++? | B |
| Flu103 | 33 | F | 3/7/2013 | New Haven, CT | B | not done | +++? | B |
| Flu107 | 23m. | M | 3/5/2013 | New Haven, CT | B | not done | +? | B |
| Flu108 | 15 | M | 3/4/2013 | Hamden, CT | B | B(18·7) | ++? | B |
| Flu110 | 33 | F | 3/8/2013 | New Haven, CT | B | B(21·4) | ++? | B |
| Flu111 | 11 | F | 3/8/2013 | East Haddam, CT | B | not done | +? | B |
| Flu118 | 3 | M | 1/27/2013 | West Haven, CT | B | not done | ++? | B |
| Flu042 | 76 | M | 12/28/2012 | Cheshire, CT | A | not done | +++ | H3N2 |
| Flu050 | 79 | F | 12/29/2012 | Essex, CT | A | A(21·1) | ++++ | H3N2 |
| Flu051 | 36 | F | 12/30/2012 | New Haven, CT | A | not done | ++++ | H3N2 |
| Flu055 | 26 | F | 12/27/2012 | East Haven, CT | A | not done | ++++ | H3N2 |
| Flu056 | 62 | F | 12/30/2012 | Clinton, CT | A | A(24·4) | ++++ | H3N2 |
| Flu060 | 68 | F | 12/30/2012 | New Haven, CT | A | not done | ++++ | H3N2 |
| Flu066 | 29 | F | 12/30/2012 | East Haven, CT | A | not done | ++ | H3N2 |
| Flu068 | 84 | M | 12/29/2012 | Hamden, CT | A | A(17·9) | ++++ | H3N2 |
| Flu072 | 28 | M | 12/29/2012 | New Haven, CT | A | not done | +++ | H3N2 |
| Flu074 | 89 | M | 12/29/2012 | West Haven, CT | not done | A(18·0) | ++++ | H3N2 |
| Flu075 | 47 | M | 12/30/2012 | Killingworth, CT | A | A(19·2) | ++++ | H3N2 |
| Flu085 | 71 | M | 12/31/2012 | Branford, CT | A | not done | ++++ | H3N2 |
| Flu086 | 71 | M | 12/31/2012 | Branford, CT | A | not done | +++ | H3N2 |
| Flu090 | 49 | M | 1/1/2013 | Wallingford, CT | A | A(17·4) | ++++ | H3N2 |
| Flu098 | 82 | M | 2/4/2013 | Milford, CT | A | A(21·8) | +++ | H3N2 |
| Flu099 | 44 | F | 1/3/2013 | Madison, CT | A | A(17·9) | ++++ | H3N2 |
| Flu105 | 20m. | F | 3/4/2013 | Hamden, CT | A | not done | ++++ | H3N2 |
| Flu117 | 61 | F | 1/25/2013 | New Haven, CT | A | not done | ++++ | H3N2 |
| Flu119 | 6 | M | 1/24/2013 | Shelton, CT | not done | A(20·2) | ++++ | H3N2 |
| Flu122 | 85 | F | 1/27/2013 | Bridgeport, CT | A | not done | ++++ | H3N2 |
| Flu124 | 56 | F | 1/25/2013 | New Haven, CT | A | not done | +++ | H3N2 |
| Flu135 | 14 | F | 1/28/2013 | North Haven, CT | A | not done | ++++ | H3N2 |
| Flu138 | 22m. | M | 1/30/2013 | New Haven, CT | A | A(19·5) | ++++ | H3N2 |
| Flu139 | 7 | M | 2/1/2013 | Derby, CT | A | not done | ++++ | H3N2 |
| Flu140 | 16 | M | 1/29/2013 | New Haven, CT | A | not done | ++++ | H3N2 |
| Flu141 | 56 | M | 1/28/2013 | New Haven, CT | A | not done | ++++ | H3N2 |
| Flu146 | 44 | F | 2/8/2013 | New Haven, CT | A | A(23·1) | ++++ | H3N2 |
| Flu147 | 6 | F | 2/1/2013 | New Haven, CT | A | not done | ++++ | H3N2 |
| Flu159 | 84 | M | 2/10/2013 | Hamden, CT | A | A(18·0) | ++++ | H3N2 |
| Flu166 | 37 | F | 1/26/2013 | New Haven, CT | A | A(28·8) | ++++ | H3N2 |
| Flu169 | 24 | F | 1/25/2013 | West Haven, CT | A | A(25·4) | ++++ | H3N2 |
| Flu170 | 66 | M | 1/25/2013 | West Haven, CT | not done | A(19·4) | ++++ | H3N2 |
| Flu175 | 39 | M | 1/25/2013 | New Haven, CT | A | A(28·5) | - | H3N2 |
| Flu177 | 84 | F | 1/27/2013 | New Haven, CT | A | A(27·5) | - | H3N2 |
| Flu178 | 39 | M | 1/31/2013 | Hamden, CT | A | not done | ++++ | H3N2 |
| Flu100 | 39 | M | 2/5/2013 | Meriden, CT | not done | A(23·9) | +++ | pdH1N1 |
| Flu113 | 42 | F | 1/25/2013 | West Haven, CT | A | not done | ++++ | pdH1N1 |
| Flu150 | 39 | M | 2/6/2013 | Meriden, CT | A | not done | ++++ | pdH1N1 |
| Flu161 | 61 | F | 1/31/2013 | Hamden, CT | A | not done | ++++ | pdH1N1 |
| Flu165 | 8 | M | 1/30/2013 | North Haven, CT | A | not done | ++++ | pdH1N1 |
| Flu182 | 55 | M | 4/7/2011 | New Haven, CT | A | A | ++++ | pdH1N1 |
| Flu183 | 76 | F | 3/7/2011 | West Haven, CT | not done | A | +++ | pdH1N1 |
| Flu184 | 30 | F | 2/22/2011 | Old Lyme, CT | not done | A | +++ | pdH1N1 |
| Flu186 | 55 | M | 2/19/2011 | Naugatuck, CT | not done | A | +++ | pdH1N1 |
| Flu187 | 81 | F | 2/23/2011 | New Haven, CT | A | A | ++++ | pdH1N1 |
| Flu188 | 10 | M | 2/28/2011 | Stamford, CT | A | A | ++++ | pdH1N1 |
| Flu189 | 46 | M | 3/6/2011 | New Haven, CT | A | A | ++++ | pdH1N1 |
Abbreviations: M, male; F, female; m., months; DFA, direct fluorescent antigen test; RT-qPCR, quantitative RT-PCR; Ct, cycle threshold; Inad, inadequate cells for DFA; A, influenza A virus; B, influenza B virus; “+”, universal PCR positive in agarose gel assay for influenza A virus; “-”, universal PCR negative in agarose gel assay; “?”, PCR positive with different pattern from influenza A virus.
NGS Assay Shows High Detection Sensitivity
When we evaluated analytical sensitivity of the NGS method and compared it with other clinical tests (S3 Table), the criteria of minimum coverage length of 200bp and more than 2 reads was used to assemble contigs mapping to the influenza genome. Both M (278bp/810 reads) and NS (716bp/16 reads) genes were identified at the 10−10 dilution level in the reference H5N1 strain indicating highly sensitive detection. Likewise, we further re-analyzed ten universal RT-PCR negative specimens and 19 universal RT-PCR positive but NGS negative specimens when using the criteria of minimum coverage length of 800bp and 1000 reads (Fig 1). The NGS data showed that multiple partial influenza genomic sequences were identified in each of 28 specimens representing different influenza genomic profiles (S3 Table) and confirmed the presence of 24 A(H3N2) and two A(pdH1N1) infections. There were two cases identified as A(H3N2) and A(pdH1N1) viruses co-infections (Flu087 and Flu185). The specimen Flu175 contained full-length segments for M (1033bp/3033 reads) and NS genes (920bp/1203 reads), while Flu177 contained full-length gene sequence for the M gene (1031bp/3645 reads).
Overall Reporting of Influenza A and B Virus Infections
A total of 27.4 million sequence reads representing a total length of 1.73 million base-pairs were obtained from 162 clinical specimens that were processed (S4 Table). Of these, approximately 26 million reads (95%) matched the influenza virus genome representing 78% of total read length with an average of 30,000 reads per contig from a total of 867 contigs. 99.4% (161/162) of specimens were identified to contain influenza genomes and their types/subtypes were determined. Among the influenza positive specimens, 83.3% (135/162) were seasonal A(H3N2), 8.6% (14/162) were A(pdH1N1), and 6.2% (10/162) were influenza B viruses. Two cases (1.2%) were identified as A(H3N2) and A(pdH1N1) virus co-infections, and influenza genome was not found in specimen Flu199. Among 123 influenza A viruses with whole-genome sequence data supported by high confidence reads (S5 Table), we obtained 83.6% (823/8 x123) contigs in total. The highest DOC and BOCs were observed for the HA, NP, NA, M, and NS genes with each demonstrating 99–100% coverage. The minimum average DOC observed for the PB1 gene was 1,467. The frequency of being detected by the RT-PCR-NGS assay was highest for gene segment M, followed by NS, NP, PA, NA, HA, PB1, and PB2. Genotype results and representative phylogenetic trees for HA, NA, and M genes are described in S1 Fig. The details of key signature amino-acid mutations from 47 selected influenza A viruses are summarized in Table 3, The molecular biomarkers for pandemic risk, drug-resistant-, transmission-associated signature mutations, and virulence factors were assessed and further described in the discussion section. We concluded that we have simultaneously and correctly identified genetically distinct lineages of influenza A(H3N2 and pdH1N1) and B viruses in clinical specimens.
Table 3. Comparison of the amino-acid substitutions in the proteins of 47 influenza A(H3N2, and pdH1N1) viruses.
| Strain | Subtype | PB2 | PB1 | PA | NP | NA | M2 | NS1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 199 | 475 | 627 | 327 | 100 | 356 | 409 | 100 | 119 | 152 | 11 | 20 | 31 | 92 | ||
| Flu042 | H3N2 | S | M | K | K | A | R | N | V | V | Y | I | N | N | D |
| Flu050 | H3N2 | S | M | K | K | A | K | S | I | V | Y | I | N | N | D |
| Flu051 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu055 | H3N2 | S | M | K | K | A | R | N | V | V | Y | I | N | N | D |
| Flu056 | H3N2 | S | M | K | K | A | K | S | I | V | Y | I | N | N | D |
| Flu060 | H3N2 | S | L | K | K | A | R | S | I | V | Y | I | N | N | D |
| Flu066 | H3N2 | S | L | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu068 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu072 | H3N2 | S | L | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu074 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu075 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu085 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu086 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu090 | H3N2 | S | L | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu098 | H3N2 | S | M | K | K | A | R | S | I | V | Y | I | N | N | D |
| Flu099 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu105 | H3N2 | S | L | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu117 | H3N2 | S | L | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu119 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu122 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu124 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu135 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu138 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu139 | H3N2 | S | M | K | K | A | R | N | V | V | Y | I | N | N | D |
| Flu140 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu141 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu146 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu147 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu159 | H3N2 | S | M | K | K | A | R | N | V | V | Y | I | N | N | D |
| Flu166 | H3N2 | S | L | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu169 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu170 | H3N2 | S | M | K | K | A | K | S | I | V | Y | I | N | N | D |
| Flu175 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu177 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu178 | H3N2 | S | M | K | K | A | R | S | V | V | Y | I | N | N | D |
| Flu100 | pdH1N1 | A | L | E | R | - | R | N | I | S | R | T | S | N | D |
| Flu113 | pdH1N1 | A | L | E | R | I | R | N | I | S | R | T | S | N | D |
| Flu150 | pdH1N1 | A | L | E | R | V | R | N | I | S | R | T | S | N | D |
| Flu161 | pdH1N1 | A | L | E | R | I | R | N | I | S | R | T | S | N | D |
| Flu165 | pdH1N1 | A | L | E | K | - | R | N | I | S | R | T | S | N | D |
| Flu182 | pdH1N1 | A | L | E | R | V | R | N | I | S | R | T | N | N | D |
| Flu183 | pdH1N1 | A | L | E | R | V | R | N | I | S | R | T | S | N | D |
| Flu184 | pdH1N1 | A | L | E | R | V | R | N | - | S | R | T | S | N | D |
| Flu186 | pdH1N1 | A | L | E | R | V | R | N | I | S | R | T | S | N | D |
| Flu187 | pdH1N1 | A | L | E | R | - | R | N | I | S | R | T | S | N | D |
| Flu188 | pdH1N1 | A | L | E | R | - | R | N | I | S | R | T | N | N | D |
| Flu189 | pdH1N1 | A | L | E | R | V | R | N | I | S | R | T | S | N | D |
| A/California/04/09 | pdH1N1 | A | L | E | R | V | R | N | V | S | R | T | S | N | D |
| A/Panama/07/99 | H3N2 | S | M | K | K | A | R | N | V | V | Y | I | N | S | D |
| A/Alberta/01/2014 | H5N1 | A | L | E | R | V | K | S | R | S | R | T | S | S | E |
The reference sequences are based on the inspection of amino-acids found in the majority of human pdH1N1, H3N2, and H5N1 viruses. The human-specific substitution residues were highlighted in bold and italic. A dashed line indicates that a space was inserted in the sequence to preserve the alignment.
Discussion
We performed NGS as part of a new detection platform for diagnosis of influenza virus infections using clinical specimens. A method for identification and discrimination of functional significance of diverse influenza viruses has been established. We report (i) multiplexed detection and simultaneous discrimination of influenza virus infections in nasopharyngeal swabs and identification of genetically distinct lineages of influenza A(H3N2 and pdH1N1) and B viruses in a single sequencing run, (ii) identification of influenza virus co-infections in a single specimen/test, (iii) development of new degenerate universal primer pairs for identification of influenza viruses and demonstration of highly sensitive detection of influenza virus infections using the NGS assay. We proposed a “one-size-fits-all” approach using an NGS diagnostic platform for extensive identification of a broad range of influenza virus infections including co-infections, by revealing their comprehensive genetic context and providing matrix reporting information for final sequence-based confirmation.
A total of 162 influenza-positive de-identified nasopharyngeal swab specimens collected from patients in the Connecticut were tested in a blinded fashion using the RT-PCR-NGS platform. No reassortants were found but multiple mutations were detected in the specimens tested. Sequence analysis of 123 influenza A viruses revealed that 66.7%(82/123) of A(H3N2) viruses had a single signature mutation of E627K in the PB2 protein, and 88%(108/123) of influenza A(H3N2 and pdH1N1) viruses contained the S31N mutation in the M2 protein. A mutation of PB2 E627K has been reported to confer high virulence to the virus by enhancing replication efficiency, and increasing polymerase activity and disease severity of avian influenza viruses in mammals [38]. The S31N mutation in the transmembrane region of the M2 protein confers resistance to amantadine [9,39]. The emergence of E627K(PB2) and S31N(M2) mutations raise concerns of increased human disease severity. Influenza viruses contain multi-basic amino-acid motif at the proteolytic cleavage site of HAs, which is associated with broad tissue tropism and organ dissemination and determines viral pathogenicity [40]. Investigation of HAs in tested specimens showed a single arginine that appeared at the site (PSIQSR↓G) in A(pdH1N1) and (PEKQTR↓G) in A(H3N2) viruses, suggesting that these viruses belonged to a low pathogenic strain that poses less risk to humans.
We aligned all protein sequences of 47 influenza A viruses and compared their sequences with those in humans that have been reported as amino-acid signatures in past influenza outbreaks (Table 3). The emergence of zanamivir- and oseltamivir-resistant viruses is facilitated by mutations in the NA protein, which provides a major target for developing anti-influenza drugs [41,42]. Sequence analysis revealed that the E119V signature mutations in these specimens may be susceptible to oseltamivir in A(H3N2) but not in A(pdH1N1) viruses. Three signature residues in the PA protein, PA-100A, PA-356A, and PA-409N, were reported in most H1N1, H2N2, H3N2 or H5N1 strains and could cause pandemics [43]. A mammalian-adapting V100A substitution was identified in most A(H3N2) but not in A(pdH1N1) viruses, and S409N was identified in all A(pdH1N1) and some A(H3N2) viruses, indicating their pandemic potential. NP protein plays an important role in viral RNA replication and cross-species transmission [44]. An amino-acid signature, valine at position NP-100 reported in 2009 pandemic H1N1 strain [45], was found as V100I mutation in all A(pdH1N1) and five A(H3N2) viruses, suggesting genetic changes of influenza A viruses in the 2012–13 seasons in this region. Influenza B virus is typically considered a mild disease and has been causing 20% to 50% of total influenza incidence [46]. An E105K point mutation at the NA tetramer in influenza B virus was reported leading to reduced susceptibility to NA inhibitor drugs [47]. This mutation was not identified in the B viruses tested in current study.
It has been recognized that clinical influenza tests have variable sensitivity and multiple tests may be required for accurate diagnosis. NGS analysis of influenza viruses has been previously reported [25,26,27,28]. However, the sensitivity and simultaneous detection of various influenza viruses from a large set of clinical specimens in a single test has not been reported. Our analytical sensitivity study demonstrated that NGS can identify the full-length NP gene of influenza at a 10−9 dilution level (1.8x101 TCID50/mL) in the characterization study, and detect partial sequences of M and NS genes at 10−10 dilution level in the analytical study (S3 Table). When we performed universal RT-PCR-NGS assays in the analytical study using specimens with low virus concentrations [21], we further confirmed that multiple influenza genomic sequences were identified in each of the 28 human specimens demonstrating that NGS based diagnostics can achieve highly sensitive detection equivalent to the clinical RT-qPCR test at low virus concentrations. To determine whether the current NGS platform could detect influenza virus in different vertebrate species in addition to humans, we tested chicken specimens obtained from Egypt and identified A(H5N1) [48] and A(H9N2) (unpublished data) subtypes. Influenza C virus infection is not routinely tested in clinical settings, therefore, specimens and reference materials were not evaluated in current study.
Since the degenerate universal primers reported here target conserved sequences for influenza A, B and C viruses, it is anticipated that the current NGS based detection platform would enable detection and accurate characterization of influenza infection including novel, emerging strains and reassortants arising during outbreaks. This platform allows multiplex identification and simultaneous discrimination of functional significance in a single test and provides the whole spectrum of the influenza genome. The method described here will have significant implications from the perspective of screening, monitoring, drug resistance, and vaccine development. These features of the assay will facilitate diagnostics and antiviral treatment in the clinical setting and enhance protection of public health. With the aid of bioinformatics, mathematics and epidemiologic studies, a typical genetic matrix composition from a known, and a novel emerging influenza infection can be determined. Development of an automated assembly and analysis pipeline will help uncover new levels of innovation and efficiency of transferring raw reads to specific genomic identification which will facilitate future use of an NGS diagnostics platform for public health surveillance and in clinical microbiology laboratory [49].
Supporting Information
(TIF)
(TIF)
(TIFF)
(PDF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
This work was funded through FDA Center for Biologics Evaluation and Research (CBER) intramural and Medical Countermeasures Initiative funds. We are thankful to FDA, CBER core facility for help with some NGS assays and oligonucleotide synthesis. The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy. We would like to thank Drs. Heike Sichtig and Shyh-Ching Lo for their critical reading of our manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files. Sequences have been deposited into GenBank, GenBank accession numbers are: KM654599-KM654931, and KM654933-KM654980.
Funding Statement
The research was supported by Center for Biologics Evaluation and Research (CBER) intramural and US Food and Drug Administration (FDA) Medical Countermeasures Initiative (MCMi) funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hay AJ, Gregory V, Douglas AR, Lin YP (2001) The evolution of human influenza viruses. 0962–8436 (Print). 1861–1870 p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA (2000) Influenza B virus in seals. Science 288: 1051–1053. [DOI] [PubMed] [Google Scholar]
- 3.Bodewes R, Morick D, de Mutsert G, Osinga N, Bestebroer T, et al. (2013) Recurring influenza B virus infections in seals. Emerg Infect Dis 19: 511–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56: 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, et al. (2005) Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79: 2814–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCauley JW, Hongo S, Kaverin NV, Kochs G, Lamb RA, et al. (2012) Family Orthomyxoviridae Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses,: pp. 749–761. Edited by King A.M.Q., Adams M.J., Carstens E.B. & Lefkowitz E.J.. Amsterdam: Elsevier. [Google Scholar]
- 7.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, et al. (2006) Global patterns of influenza a virus in wild birds. Science 312: 384–388. [DOI] [PubMed] [Google Scholar]
- 8.Tong S, Zhu X, Li Y, Shi M, Zhang J, et al. (2013) New world bats harbor diverse influenza A viruses. PLoS Pathog 9: e1003657 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao R, Cao B, Hu Y, Feng Z, Wang D, et al. (2013) Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368: 1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 10.Pabbaraju K, Tellier R, Wong S, Li Y, Bastien N, et al. (2014) Full-genome analysis of avian influenza A(H5N1) virus from a human, North America, 2013. Emerg Infect Dis 20: 887–891. 10.3201/eid2005.140164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.USDA (2013) Highly pathogenic avian influenza standard operating procedures. Available: http://www.aphis.usda.gov/animal_health/emergency_management/downloads/sop/sop_hpai_e-e.pdf.
- 12.Bevins SN, Dusek RJ, White CL, Gidlewski T, Bodenstein B, et al. (2016) Widespread detection of highly pathogenic H5 influenza viruses in wild birds from the Pacific Flyway of the United States. Sci Rep 6: 28980 10.1038/srep28980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Yuan H, Gao R, Zhang J, Wang D, et al. (2014) Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383: 714–721. 10.1016/S0140-6736(14)60111-2 [DOI] [PubMed] [Google Scholar]
- 14.Lindstrom S, Garten R, Balish A, Shu B, Emery S, et al. (2012) Human infections with novel reassortant influenza A(H3N2)v viruses, United States, 2011. Emerg Infect Dis 18: 834–837. 10.3201/eid1805.111922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, et al. (2010) Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328: 1529 10.1126/science.1189132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epperson S, Blanton L, Kniss K, Mustaquim D, Steffens C, et al. (2014) Influenza activity—United States, 2013–14 season and composition of the 2014–15 influenza vaccines. MMWR Morb Mortal Wkly Rep 63: 483–490. [PMC free article] [PubMed] [Google Scholar]
- 17.Ganzenmueller T, Kluba J, Hilfrich B, Puppe W, Verhagen W, et al. (2010) Comparison of the performance of direct fluorescent antibody staining, a point-of-care rapid antigen test and virus isolation with that of RT-PCR for the detection of novel 2009 influenza A (H1N1) virus in respiratory specimens. J Med Microbiol 59: 713–717. 10.1099/jmm.0.017244-0 [DOI] [PubMed] [Google Scholar]
- 18.Uyeki TM, Prasad R, Vukotich C, Stebbins S, Rinaldo CR, et al. (2009) Low sensitivity of rapid diagnostic test for influenza. Clin Infect Dis 48: e89–92. 10.1086/597828 [DOI] [PubMed] [Google Scholar]
- 19.Faix DJ, Sherman SS, Waterman SH (2009) Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 361: 728–729. 10.1056/NEJMc0904264 [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease C, Prevention (2012) Evaluation of rapid influenza diagnostic tests for influenza A (H3N2)v virus and updated case count—United States, 2012. MMWR Morb Mortal Wkly Rep 61: 619–621. [PubMed] [Google Scholar]
- 21.Landry ML, Ferguson D (2014) Comparison of Simplexa Flu A/B & RSV PCR with cytospin-immunofluorescence and laboratory-developed TaqMan PCR in predominantly adult hospitalized patients. J Clin Microbiol 52: 3057–3059. 10.1128/JCM.00738-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease C, Prevention (2009) Update: drug susceptibility of swine-origin influenza A (H1N1) viruses, April 2009. MMWR Morb Mortal Wkly Rep 58: 433–435. [PubMed] [Google Scholar]
- 23.Masuda H, Suzuki H, Oshitani H, Saito R, Kawasaki S, et al. (2000) Incidence of amantadine-resistant influenza A viruses in sentinel surveillance sites and nursing homes in Niigata, Japan. Microbiol Immunol 44: 833–839. [DOI] [PubMed] [Google Scholar]
- 24.Baranovich T, Saito R, Suzuki Y, Zaraket H, Dapat C, et al. (2010) Emergence of H274Y oseltamivir-resistant A(H1N1) influenza viruses in Japan during the 2008–2009 season. J Clin Virol 47: 23–28. 10.1016/j.jcv.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 25.de la Rosa-Zamboni D, Vazquez-Perez JA, Avila-Rios S, Carranco-Arenas AP, Ormsby CE, et al. (2012) Molecular characterization of the predominant influenza A(H1N1)pdm09 virus in Mexico, December 2011-February 2012. PLoS One 7: e50116 10.1371/journal.pone.0050116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flaherty P, Natsoulis G, Muralidharan O, Winters M, Buenrostro J, et al. (2012) Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res 40: e2 10.1093/nar/gkr861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutvisuttinunt W, Chinnawirotpisan P, Simasathien S, Shrestha SK, Yoon IK, et al. (2013) Simultaneous and complete genome sequencing of influenza A and B with high coverage by Illumina MiSeq Platform. J Virol Methods 193: 394–404. 10.1016/j.jviromet.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 28.Zhou B, Lin X, Wang W, Halpin RA, Bera J, et al. (2014) Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics. J Clin Microbiol 52: 1330–1337. 10.1128/JCM.03265-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, Ragupathy V, Liu J, Wang X, Vemula SV, et al. (2015) Nanomicroarray and multiplex next-generation sequencing for simultaneous identification and characterization of influenza viruses. Emerg Infect Dis 21: 400–408. 10.3201/eid2103.141169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Archives of Virology 146: 2275–2289. [DOI] [PubMed] [Google Scholar]
- 31.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, et al. (2009) Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol 83: 10309–10313. 10.1128/JVI.01109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou S (1997) A practical approach to genetic screening for influenza virus variants. J Clin Microbiol 35: 2623–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treangen TJ, Salzberg SL (2012) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampson J, Jacobs K, Yeager M, Chanock S, Chatterjee N (2011) Efficient study design for next generation sequencing. Genet Epidemiol 35: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 36.Lu G, Rowley T, Garten R, Donis RO (2007) FluGenome: a web tool for genotyping influenza A virus. Nucleic Acids Res 35: W275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP (2014) Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 15: 121–132. 10.1038/nrg3642 [DOI] [PubMed] [Google Scholar]
- 38.Hatta M, Gao P, Halfmann P, Kawaoka Y (2001) Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293: 1840–1842. [DOI] [PubMed] [Google Scholar]
- 39.Zaraket H, Saito R, Suzuki Y, Suzuki Y, Caperig-Dapat I, et al. (2010) Genomic events contributing to the high prevalence of amantadine-resistant influenza A/H3N2. Antivir Ther 15: 307–319. 10.3851/IMP1538 [DOI] [PubMed] [Google Scholar]
- 40.Steinhauer DA (1999) Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258: 1–20. [DOI] [PubMed] [Google Scholar]
- 41.McKimm-Breschkin J, Trivedi T, Hampson A, Hay A, Klimov A, et al. (2003) Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob Agents Chemother 47: 2264–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okomo-Adhiambo M, Demmler-Harrison GJ, Deyde VM, Sheu TG, Xu X, et al. (2010) Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: challenges in diagnosis of oseltamivir resistance. Antimicrob Agents Chemother 54: 1834–1841. 10.1128/AAC.01608-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Lu L, Sun Z, Chen GW, Wen Y, et al. (2013) Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect 15: 432–439. 10.1016/j.micinf.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 44.Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S (2008) Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu Rev Microbiol 62: 403–424. 10.1146/annurev.micro.62.081307.162746 [DOI] [PubMed] [Google Scholar]
- 45.Pan C, Cheung B, Tan S, Li C, Li L, et al. (2010) Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS One 5: e9549 10.1371/journal.pone.0009549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang SS, Banner D, Paquette SG, Leon AJ, Kelvin AA, et al. (2014) Pathogenic influenza B virus in the ferret model establishes lower respiratory tract infection. J Gen Virol 95: 2127–2139. 10.1099/vir.0.064352-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujisaki S, Takashita E, Yokoyama M, Taniwaki T, Xu H, et al. (2012) A single E105K mutation far from the active site of influenza B virus neuraminidase contributes to reduced susceptibility to multiple neuraminidase-inhibitor drugs. Biochem Biophys Res Commun 429: 51–56. 10.1016/j.bbrc.2012.10.095 [DOI] [PubMed] [Google Scholar]
- 48.Amen O, Vemula SV, Zhao J, Ibrahim R, Hussein A, et al. (2015) Identification and Characterization of a Highly Pathogenic H5N1 Avian Influenza A Virus during an Outbreak in Vaccinated Chickens in Egypt. Virus Res 210: 337–343. 10.1016/j.virusres.2015.09.004 [DOI] [PubMed] [Google Scholar]
- 49.Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, et al. (2014) A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 24: 1180–1192. 10.1101/gr.171934.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIFF)
(PDF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Sequences have been deposited into GenBank, GenBank accession numbers are: KM654599-KM654931, and KM654933-KM654980.