Abstract
RNA sequencing has allowed high-throughput screening of differential gene expression in many tissues and organisms. Xenopus laevis is a classical embryological and cell-free extract model system, but its genomic sequence had been lacking due to difficulties arising from allotetraploidy. There is currently much excitement surrounding the release of the completed X. laevis genome (version 9.1) by the Joint Genome Institute (JGI), which provides a platform for genome-wide studies. Here we present a deep RNA-seq dataset of transcripts expressed in dorsal and ventral lips of the early Xenopus gastrula embryo using the new genomic information, which was further annotated by blast searches against the human proteome. Overall, our findings confirm previous results from differential screenings using other methods that uncovered classical dorsal genes such as Chordin, Noggin and Cerberus, as well as ventral genes such as Sizzled, Ventx, Wnt8 and Bambi. Complete transcriptome-wide tables of mRNAs suitable for data mining are presented, which include many novel dorsal- and ventral-specific genes. RNA-seq was very quantitative and reproducible, and allowed us to define dorsal and ventral signatures useful for gene set expression analyses (GSEA). As an example of a new gene, we present here data on an organizer-specific secreted protein tyrosine kinase known as Pkdcc (protein kinase domain containing, cytoplasmic) or Vlk (vertebrate lonesome kinase). Overexpression experiments indicate that Pkdcc can act as a negative regulator of Wnt/ β-catenin signaling independently of its kinase activity. We conclude that RNA-Seq in combination with the Xenopus laevis complete genome now available provides a powerful tool for unravelling cell-cell signaling pathways during embryonic induction.
Keywords: RNA-Seq, Xenopus laevis, Dorsal-Ventral patterning, Spemann organizer, Secreted Tyrosine kinase, Vlk
Introduction
Xenopus laevis is a favorite model system in developmental biology. From its embryos we have learned the molecular basis of fundamental processes such as egg cytoplasmic determinants, mesoderm induction, Hox genes, signaling by Spemann’s organizer, induction of the central nervous system, and many other processes that are conserved among all vertebrates (Harland and Gerhart, 1997; De Robertis and Kuroda, 2004; Bier and De Robertis, 2015). With its large and numerous eggs, Xenopus laevis has provided an essential source for cell-free extracts to study cell biological aspects of the cell cycle, cell division and cytoskeletal assembly (Wuhr et al, 2014). Xenopus may be considered an ideal post-genomic model organism for those interested in the biochemistry of early development. However, progress has been hampered by the lack of a complete X. laevis genome. The recent completion of this genome by Harland and colleagues (Session et al., 2016) therefore opens an exciting new era in Xenopus embryology. We initiated this study to test the potential of this new genomic information for improved RNA-seq analysis of early embryos. The Joint Genome Institute Xenopus laevis 9.1 sequence (JGI9.1, also known as XENLAv9.1) made public through Xenbase (Bowes et. al, 2010) was utilized as the reference in these gene expression explorations.
The sequence of the related frog Xenopus tropicalis has been available for some time (Hellsten, 2010). However, X. tropicalis eggs are less abundant and smaller than those of X. laevis, making the latter species the favorite for cut-and-paste embryological experiments as well as preparation of cell extracts. The problem with Xenopus laevis is that it is an allotetraploid species (Kobel, 1996). This means that matings between two different Xenopus species resulted in one with double the number of chromosomes than its tropicalis-like diploid ancestors. Allopolyploidy is an effective method for generating new species of fish, amphibians and plants because if the two species differ sufficiently, chromosome pairs will still form bivalents during meiosis instead of forming tetravalents that cause aneuploidy (Weiler and Ohno, 1962). The recent Xenopus laevis genomic sequencing has revealed that the species hybridization event took place only 17 to 18 million years ago. One of the precursor subgenomes had longer chromosomes (L-subgenome) and was domineering in the sense that it retained many more active genes than the shorter S-subgenome, which underwent more gene loss by deletion, pseudogene formation and intrachromosmal rearrangements (Session et al, 2016). Duplicated transcripts are therefore annotated as L or S forms. However, we found that the annotation of transcripts by comparison to human gene symbols in JGI9.1 left room for improvement.
Our laboratory has focused on Dorsal-Ventral (D-V) cell differentiation in Xenopus laevis. From dorsal to ventral, ectoderm gives rise to neural plate, neural crest and epidermis, while mesoderm differentiates into prechordal plate, notochord, somite, kidney, lateral plate mesoderm and blood islands (De Robertis and Kuroda, 2004). A signaling center known as the Spemann organizer has long been known to play a key role in D-V patterning. The organizer appears at the beginning of gastrulation and corresponds to the dorsal blastopore lip, the site where dorsal mesoderm starts to involute. The Spemann organizer has proven to be a very productive fishing ground for discovery of novel genes involved in cell signaling (reviewed in De Robertis, 2009). A plethora of organizer-specific genes, many of which encode secreted growth factor antagonists of the Nodal, BMP and Wnt signaling pathways, have been isolated. D-V patterning relies on the formation of a gradient of BMP activity, in which low BMP levels promote the formation of dorsal structures such as the neural plate, notochord or somites while high BMP activity induces tissues with ventral characteristics like the lateral plate mesoderm and blood islands (Bier and De Robertis, 2015). In this regard, BMP antagonists such as Noggin (Nog) (Smith and Harland, 1992), Follistatin (Fst) (Hemmati-Brivanlou et al., 1994) and Chordin (Chrd) (Sasai et al., 1994; Piccolo et al., 1996) play an essential role in establishing the D-V BMP morphogen gradient (Khokha et al., 2005; Plouhinec et al., 2013).
A ventral signaling center develops 180° from the Spemann organizer, and secretes many proteins – such as Sizzled (Szl), Xolloid-related (Xlr), Crossveinless 2 (Cv2), and Twisted gastrulation (Tsg) - that are involved in a biochemical pathway that regulate Chordin/BMP complexes diffusing from the dorsal side (Reversade and De Robertis, 2005; Plouhinec et al., 2013). Dorsal and ventral genes are under opposite transcriptional regulation by BMP signaling (Reversade and De Robertis, 2005). These two signaling centers communicate with each other over long distances through protein-protein interactions with the Chordin morphogen, as well as through transcriptional feedback loops (Plouhinec et al., 2013). Each action of the organizer elicits a corresponding reaction from the ventral center (Bier and De Robertis, 2015). These regulatory mechanisms are thought to ensure the self-organizing and resilient properties of the embryo, which has the remarkable property of regenerating the missing half after surgical bisection (De Robertis, 2009).
The isolation and identification of organizer- or ventral-specific genes has relied on the classical techniques of expression cloning (Smith and Harland, 1992), differential cDNA radioactive screening (Bouwmeester et al., 1996), secretion cloning (Pera and De Robertis, 2000), cDNA macroarrays (Wessely et al., 2004), and oligonucleotide microarrays (Hufton et al., 2006). The advent of the Xenopus laevis genome now holds promise for revitalizing the field when used in conjunction with high-throughput methods such as RNA-seq, which provides a powerful method for transcriptomic analysis in many organisms (Wang et al., 2009; Yanai et al., 2011; Collart et al., 2014; Wuhr et al., 2014; Dichmann et al., 2015; Peshkin et al., 2015; Reuter et al., 2015).
Here we report an RNA-seq screen for genes expressed in dorsal and ventral blastopore lips of Xenopus laevis gastrula embryos taking advantage of the newly available reference genome. The results confirm the classical dorsal and ventral genes and identify many novel ones. We provide validation for one of the dorsal genes, Pkdcc/Vlk, a secreted tyrosine kinase (Bordoli et al., 2014) whose role in Xenopus development had remained unnoticed until very recently (Vitorino et al., 2015). We present complete lists of genes correlated with D-V patterning, which provide new avenues for understanding the signaling pathways and gene signatures involved in early vertebrate development.
Materials and methods
Libraries preparation and Illumina sequencing
RNA was isolated with an Absolutely RNA miniprep Kit (Agilent). Libraries were constructed with the Illumina TruSeq RNA Library Preparation Kit V2 according to manufacturer’s protocol. Briefly, 1 µg total RNA from stage 10.5 wild type embryos, dorsal lips or ventral lips was used for input RNA. Then mRNA was purified through polyA+ selection. After fragmentation of the mRNA, first strand and second strand synthesis, double strand cDNA (ds cDNA) was obtained. The ds cDNA was then further processed for A-tailing, end repair and ligation of appropriate adaptors. Finally, the ds cDNA with adaptors was amplified by PCR to generate libraries. After measurement of the length and concentration of the libraries by Bioanalyser and Qubit, the libraries were sent for sequencing on Illumina Hi-Seq 2000 using standard methods to generate 100-bp single end reads by the Broad Stem Cell Research Center at UCLA.
Sequencing data processing
After sequencing, adaptors were first trimmed from the RNA reads and RNA reads of low quality filtered. The clean RNA reads were mapped using Tophat against the Xenopus laevis JGI9.1 (XENLAv9.1) transcript sequences in Xenbase. Transcripts counts were normalized using DESeq package in R (Anders and Huber, 2010; Love et al., 2014). RPKM (Reads Per Kilobase per Million mapped reads) for transcripts was calculated in R. Human gene names were assigned to transcripts using an annotation file obtained by blasting the JGI9.1 peptide sequences to human reference protein sequences. Differential gene expression was calculated with DESeq. Heatmap was generated in R. Gene Ontology (GO) analysis was performed in the DAVID database (Huang et al., 2009). The RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE75278).
Gene Set Enrichment Analysis
The correlations between the dorsal and ventral signature gene sets with Chordin, Bambi, SMC1A and Pkdcc were analyzed by the Gene Set Enrichment Analysis (GSEA) approach (Subramanian et al., 2005). Normalized gene expression data for 26 independent libraries was obtained by RNA-seq from stage 10 to 12 Xenopus laevis embryos. The Pearson correlation coefficient between Chordin, Bambi, SMC1A (a uniform and highly expressed control gene) or Pkdcc and all 44,501 transcripts in each library was calculated using Excel. Genes were ranked according to their Pearson correlation coefficient with Chordin, Bambi, SMC1A or Pkdcc throughout all 26 different conditions using Excel. The ranked lists were then analyzed for enrichment of the dorsal or ventral gene set using the GSEA software from the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp) (Mootha et al., 2003; Subramanian, et al., 2005). Statistical significance was measured with a permutation based Kolmogorov–Smirnoff nonparametric rank test (1,000 permutations).
Embryo manipulations, mRNA injections and whole-mount in situ hybridization
Xenopus laevis were purchased from Nasco and embryos generated through in vitro fertilization. The embryos were cultured in 0.1× Marc's Modified Ringers (MMR) and staged according to Nieuwkoop and Faber (1967). Dorsal and ventral lips were dissected at stage 10.5 in 1× MMR solution and immediately lysed for RNA preparation. Stage 10.5 wild-type embryos were lysed for RNA preparation at the same time as dorsal and ventral lips; 2 whole embryos, 16 dorsal lips, or 20 ventral lips were used for each library.
Analysis of EST and genome sequences available on Xenbase revealed the presence of two Pkdcc homologs in the frog Xenopus laevis, similarly to X. tropicalis and the fish Danio rerio. While a full length Pkdcc1 clone was available, we did not find any deposited full length Pkdcc2 clone. EST and genomic sequences from Xenbase were therefore aligned to design 5’ and 3’ primers containing the predicted start and the stop codon, respectively. These primers allowed amplification of a specific band of approximately 1.5 kb from cDNA derived from late gastrula (stage 12) cDNA. Primers and cloning details are available upon request.
For in vitro mRNA synthesis, pCS2-xPkdcc1-Flag, pCS2-xPkdcc2-Flag, pCS2-xPkdcc2-myc, pCS2-eGFP, pCS2-nLacZ and pCS2-xWnt8 were linearized with NotI and transcribed with Sp6 RNA polymerase using the Ambion® mMessage mMachine. Synthetic mRNA was injected in the animal blastomeres of 4-cell stage Xenopus embryos, in the following total amounts: Pkdcc1/2 = 0.5–1 ng total; eGFP = 1 ng total; xWnt8 = 4 pg. A total of 24 ng of Morpholino against β-catenin (Heasman et al., 2000) was injected radially at the 2-cell stage.
For quantitative and semiquantitative RT-PCR analyses of injected embryos, whole embryos and animal caps excised at stage 9 were processed for RNA extraction, cDNA synthesis and PCR as previously described (Colozza and De Robertis, 2014). The following primer pairs were used for Pkdcc: Pkdcc1, Fw: ctcttggacattatgaatgccac, Rv: gcctgtccatgttctttgatga. Pkdcc2, Fw: tttcacgtatcttctgccacac, Rv: gtgaatggctcctgcacaca. Additional RT-PCR primer sets and PCR conditions used can be found at http://www.hhmi.ucla.edu/derobertis.
For hybridization probe synthesis, pCS2-xPkdcc1 and pCS2-xPkdcc2 were linearized with BamH1 and transcribed with T3 RNA polymerase. Other probes used in this study were Otx2 (forebrain and cement gland marker), BF1 (telencephalon marker) and Rx2a (eye field and forebrain marker). Whole-mount in situ hybridization and Red-Gal staining were performed as described at http://www.hhmi.ucla.edu/derobertis.
For protein biochemistry of Pkdcc1 and 2, please see Supplemental material and methods.
Results
RNA sequencing of Xenopus laevis
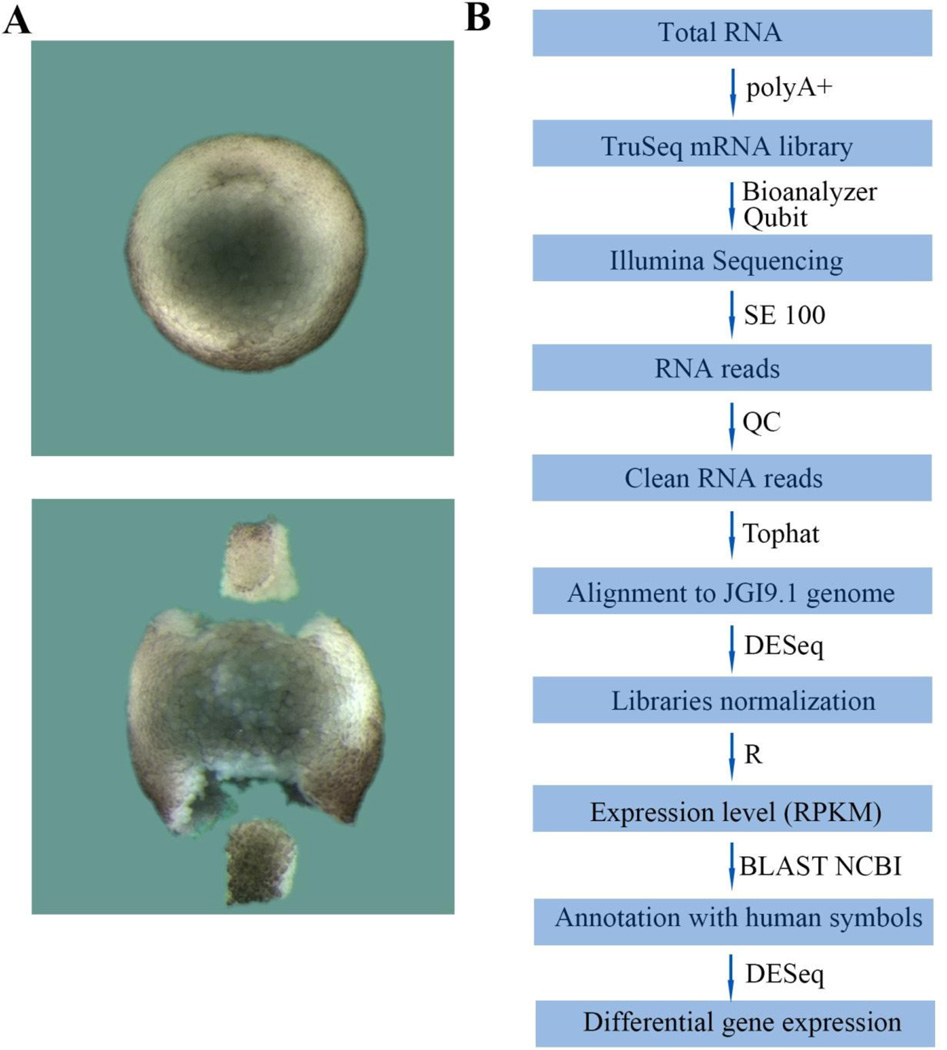
To screen for differentially expressed dorsal and ventral genes by RNA-seq, libraries were made from dorsal or ventral dissected fragments or whole embryos at early gastrula (stage 10.5) (Fig. 1A). We used a single-end 100 bp sequencing method which produced long reads of high quality that were analyzed by the bioinformatics pipeline shown in Fig. 1B. Sequencing reads were mapped to the X. laevis JGI9.1 (XENLAv9.1) transcripts in Xenbase. We observed that many genes - such as Chordin, Nodal and many others - were annotated only by their numbered ID location. Therefore, we introduced an additional step by which all sequences were blasted to the human proteome in NCBI. This step yielded a greatly increased number of gene annotations, and in all our tables we include, in addition to the JGI9.1 ID, a column with the human protein ID and one with the human gene symbols (Supplemental Table S1). The availability of human gene symbols makes it possible to relate the Xenopus laevis RNA-seq information to human gene expression sets.
Fig. 1.
D-V dissections and experimental flowchart. (A) Dorsal and ventral lip dissection. The upper panel shows a wild-type stage 10.5 Xenopus embryo, while the lower panel shows dorsal (Dlip, top) and ventral lips (Vlip, bottom) dissected from a sibling stage 10.5 embryo. (B) Outline of the RNA-seq analysis pipeline described in materials and methods.
Transcriptomic analysis of early gastrula wild-type embryos, dorsal lips and ventral lips
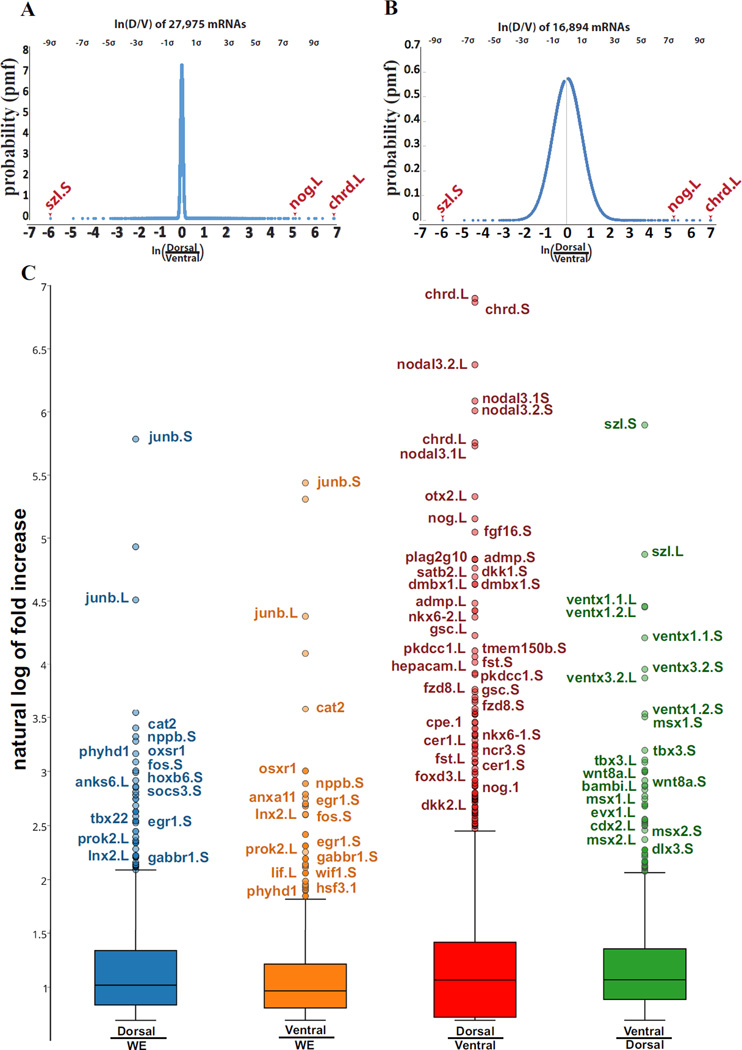
RNA-seq was very quantitative, and the vast majority of transcripts were equally expressed in dorsal and ventral lips. In Fig. 2A, the natural logarithm of the ratio of dorsal over ventral transcripts per million reads (TPM) formed a very sharp peak centered on genes that were equally expressed. When we removed the 11,000 transcripts that were most similar in both samples, the curve adopted a typical Gaussian shape (Fig. 2B). From this curve the standard deviation could be calculated. The gene most highly enriched in the dorsal lip was Chordin with a remarkable 9 standard deviations over the mean. At the ventral end of the spectrum, 8 standard deviations below the mean, the most differentially expressed gene was Sizzled, a protein that functions in the Chordin pathway through the inhibition of the chordinase Xolloid (Lee et al., 2006) (Fig. 2B).
Fig. 2.
Distribution of transcripts sequenced from dorsal and ventral lips. (A) Curve showing normal distribution of fold change of dorsal transcripts. The curve was created using 27,594 mRNAs obtained from the first experiment after selecting for abundance of expression; the ordinate indicates the probability that a gene is located in the curve by chance (probability mass function). Genes were organized by the dorsal/ventral natural logarithm, providing dorsal genes with positive values, while assigning negative numbers to ventral genes. (B) A normal Gaussian curve was obtained by artificially removing about 11,000 transcripts expressed at similar levels in both dorsal and ventral lips. The graph indicates that the data obtained was evenly distributed throughout both dorsal and ventral lips and defined the standard deviation. The most dorsal gene was Chordin, 9 standard deviations above the mean, while the most ventrally expressed gene was Sizzled, 8 standard deviations above the mean. (C) Box and whisker plots reveal transcripts upregulated upon bisection of dorsal (Dorsal/WE) or ventral (Ventral/WE) lips compared to uncut wild type whole embryos (WE), and dorsal and ventral transcriptomes (Dorsal/Ventral and Ventral/Dorsal). Only some of the outliers above the whisker are annotated, with Short on the right and Long subgenome genes on the left. Chordin is referred to by its abbreviation chrd in this Figure and throughout the Supplementary Tables and Xnr3 in the main text is referred to as nodal 3 here. Note that cutting the embryo triggers the expression of injury response genes in both fragments.
We next compared transcripts from dorsal lips to those of wild-type whole embryos. Unexpectedly, the most upregulated genes were JunB, Cat2, NppB, Phyhd1, Oxsr1, Fos and Egr1 (Fig. 2C). Similarly, comparing ventral lips to whole embryos the most upregulated genes included JunB, Cat2, Oxsr1, NppB, Egr1 and Fos (Fig. 2C). Some of these genes are involved in wound healing processes, such as JunB and Fos that can be activated by the JNK and MAPK pathways (Eming et al., 2014; Werner and Grose, 2003). It seems likely that these genes were activated in the dorsal and ventral explants as an injury response to surgery. It only took around 20 minutes to excise and harvest the lips, and it came as a surprise to us that these wound healing genes could be so strongly activated in such a short time period. The entire list of injury response genes activated in both dorsal and ventral regions is provided in Table S2.
When transcripts of dorsal lips were divided over those of ventral lips, many of the familiar dorsal genes emerged, and were indicated as outliers in the box plot of Fig. 2C. Most of the well-known dorsal genes, such as Chordin, Nodal, Noggin and Goosecoid, as well as many novel genes were included among the dorsally enriched genes. Conversely, the ventral gene outliers included previously characterized ventral genes such as Szl, Wnt8, Ventx and Msx, as well as new genes (Fig. 2C). In conclusion, the comparison of dorsal and ventral explants circumvents transcripts triggered by injury response, and many D-V genes could be identified.
Reproducibility of D-V transcriptomes
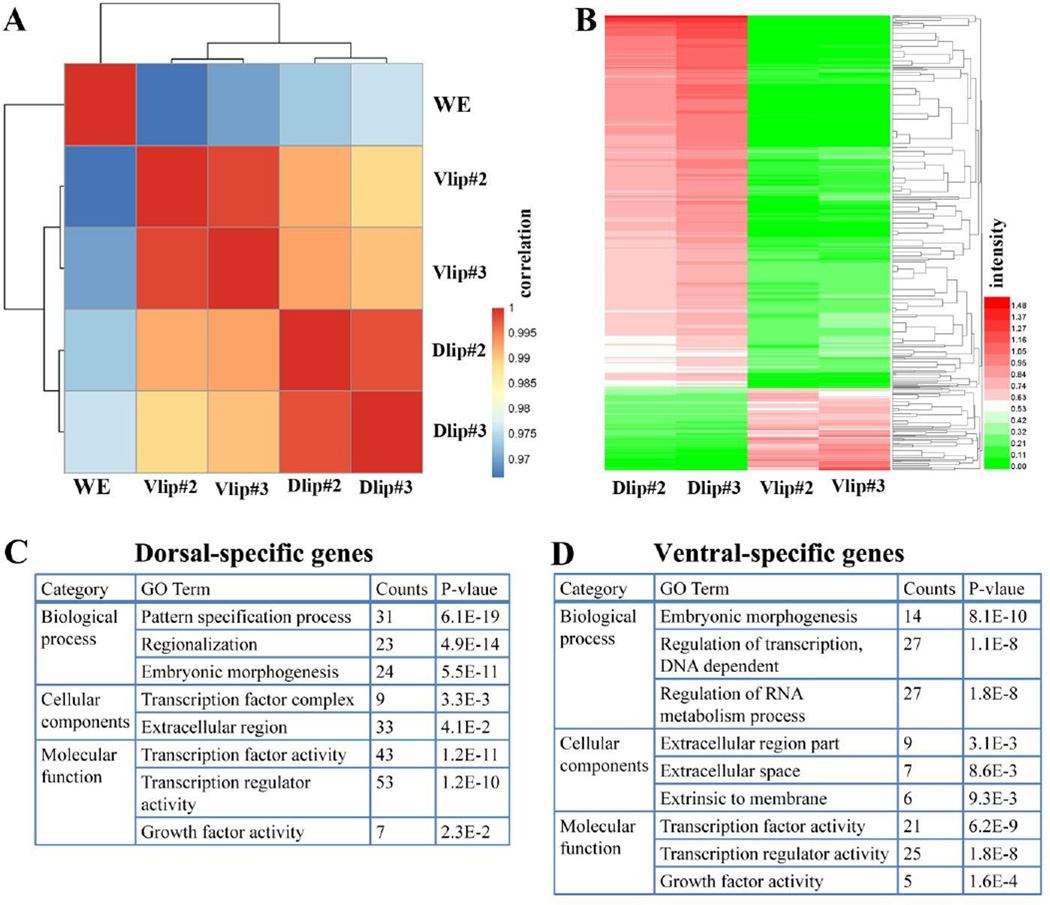
We next tested the reproducibility of our transcriptome analysis by sequencing two independent dorsal and ventral lip samples. To optimize reproducibility these were sequenced in the same lane. We performed the clustering analysis based on the correlation of samples, and a correlation plot showed that the sequencing results from the duplicate ventral lip libraries clustered together and, similarly, that the dorsal lip sequences correlated with each other (Fig. 3A). To ascertain reproducibility in a quantitative way, we prepared an excel table in which all 44,000 transcripts matching the JGI9.1 genomic sequence were ranked according to the p-value of the variation in their RPKM in the four dorsal and ventral lip samples from experiments 2 and 3 (Table S1). This table also includes the transcripts from the first experiment shown in Fig. 2. Heatmap analysis of the top dorsal and ventral transcripts (see below) showed that transcripts were reproducibly enriched in the duplicate samples (Fig. 3B). Taken together, these results demonstrated that RNA-seq D-V results were extremely reproducible.
Fig. 3.
The D-V transcriptomes obtained by RNA-seq are highly reproducible. (A) Correlation plot of duplicate dorsal and ventral lip RNA-seq experiments from different egg clutches shows high reproducibility of the method. The correlation scores were calculated as the Pearson Correlation Coefficient (PCC) and color-coded as shown in the scale bar on the right of the panel. Note that Vlip and Dlip samples clustered into groups based on PCC. (B) Heatmap of differentially expressed genes between dorsal and ventral lip libraries. The expression data (RPKM) of each gene across different samples was scaled and represented as z-scores. The expression levels (expressed as logarithm of RPKM) are indicated in the scale bar on the right panel. (C) Table showing the top gene ontology (GO) terms associated with dorsal-specific genes. (D) Table of GO terms for ventral-specific transcripts.
Analysis of dorsal genes and ventral genes
In Tables S3 and S4 we present ranked lists of 316 dorsal-most and 95 ventral-most enriched transcripts. Genes that fulfilled three criteria were chosen using the data from experiments 2 and 3. For dorsal transcripts: the adjusted p-value had to be lower than 0.05, RPKM had to be above 1 in dorsal lips, and RPKM had to be as least 2-fold higher in dorsal lips than in ventral lips. For ventral transcripts: the adjusted p-value had to be lower than 0.1, RPKM had to be above 1 in ventral lips, and RPKM had to be at least 2-fold higher in ventral lips than in dorsal lips. Among the 316 dorsal transcripts (out of 44,424 total transcripts) we found most of the classic Spemann organizer genes (Table S3). The gene with highest fold induction was the secreted BMP antagonist Chordin which was increased 1480-fold in dorsal lips compared to ventral lips. This was in keeping with the key role of Chordin for the inducing properties of the Spemann organizer and its high abundance in the dorsal lip. Additionally, another classic Spemann organizer gene, Xnr3 (Xenopus nodal-related 3), a target gene of the early Wnt pathway, was also significantly enriched in the dorsal lip compared to ventral lip, with a 1396-fold induction. Other Spemann organizer secreted growth factor antagonists, such as Cerberus, ADMP, Noggin, Dkk and Frzb, were also greatly enriched in the dorsal lip (Table S3). All of these genes were expressed at high RPKM levels, indicating that the previous Xenopus differential screens have been almost saturating for abundant transcripts.
Among the ventral transcripts, the two most highly induced genes were Wnt8a and Sizzled which were, respectively, 32 and 29 times higher in ventral lips than dorsal lips. Wnt8a is a ventral ligand for the canonical Wnt signaling pathway and Sizzled serves as an inhibitor of Xolloid proteases that cleave Chordin in the extracellular space. Other abundant ventral genes, such as the BMP targets Bambi, Ventx, ID3 and Msx1, were also enriched in the ventral lip (Table S4). Gene ontology analyses revealed that the dorsal genes and ventral genes identified in this way were mainly involved in pattern specification, embryonic morphogenesis, transcriptional complexes, and growth factor activity (Fig. 3C and D). The D-V gene expression lists making use of the recently completed Xenopus laevis genome represent a rich resource for Xenopus embryologists.
Gene Set Expression Analysis of D-V genes
A goal of these genome-wide analyses is to identify the signaling pathways that operate in multiple experimental conditions, as opposed to just comparing only two alternatives at a time. Gene Set Enrichment Analysis (GSEA) allows one to compare many RNA-seq samples to a particular set of genes (Subramanian et al., 2005). Including the dorsal and ventral lip samples described in detail here, we sequenced a total of 26 different libraries of gastrula embryos (stages 10, 11, 12). These comprised whole embryos, dorsal and ventral lips, β-catenin MO, LiCl-treated embryos, and regenerating sagittal, dorsal and ventral halves, which will be analyzed in detail in future publications. All sequenced transcripts (representing about 1.3 billion reads) were ranked according their Pearson correlation coefficient with Chordin or Bambi throughout the 26 libraries. We then analyzed this list, containing 40,501 transcripts, for enrichment of dorsal or ventral gene signatures. These signatures were built under stringent conditions, requiring that D-V genes fulfill the criteria detailed in Tables S5 and S6 in each of the three independent dorsal and ventral lip experiments.
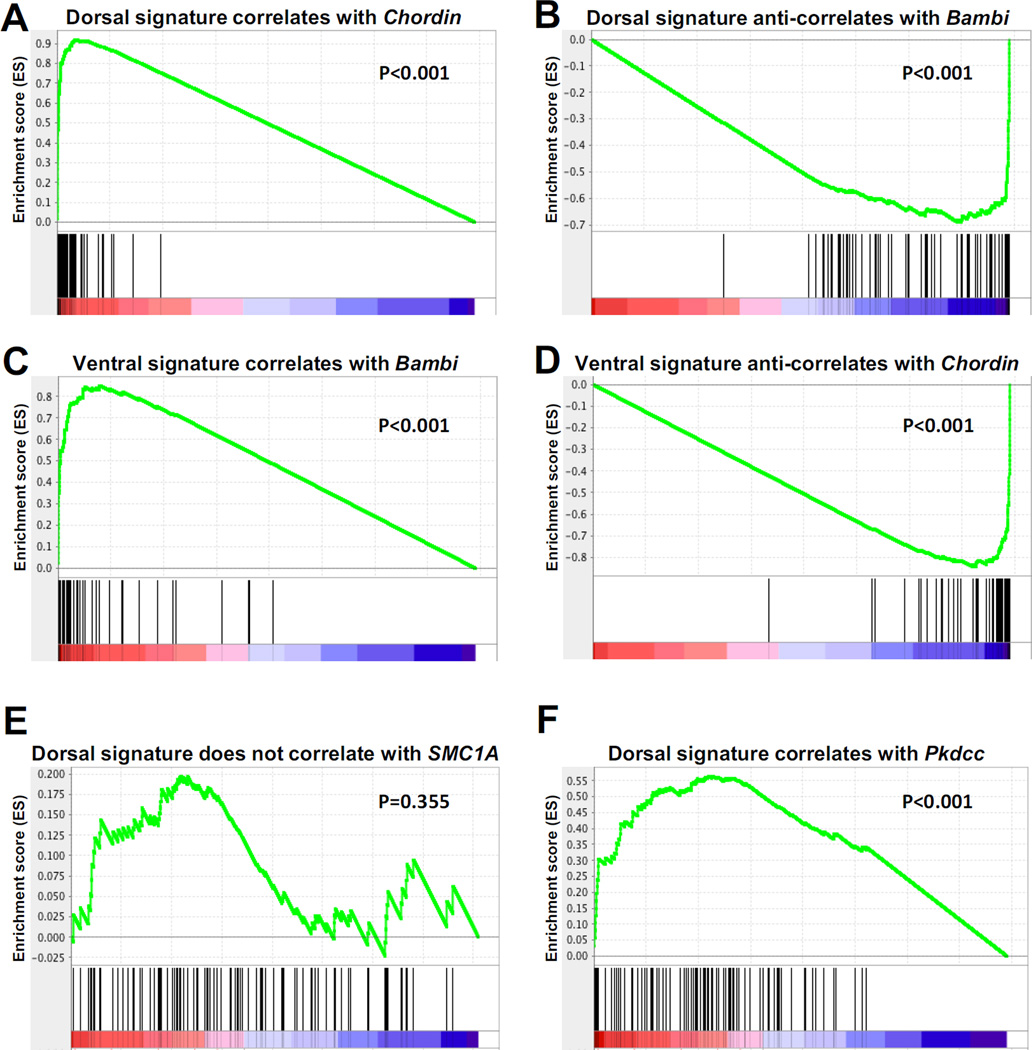
When all transcripts were ranked by their correlation to Chordin, the dorsal signature of 107 genes was significantly (P<0.001) enriched at the top of the ranked list, as expected (Fig. 4A). Conversely, when all transcripts were ranked by their correlation to ventrally expressed Bambi, the dorsal signature gene set was significantly (P<0.001) enriched at the bottom of this list (Fig. 4B). Likewise, the 70-gene ventral signature significantly correlated with Bambi (Fig. 4C) and was anti-correlated with Chordin (Fig. 4D). As a control, when all transcripts were ranked by their correlation to the uniformly expressed gene Structural Maintenance of Chromosomes 1A (SMC1A) no statistically significant enrichment was observed (Fig, 4E).
Fig. 4.
Gene Set Enrichment Analysis (GSEA) of dorsal and ventral gene signatures. (A–B) The dorsal gene signature (comprised of 107 genes) positively correlated with Chordin (P<0.001), a Spemann organizer marker, and negatively correlated (or anti-correlated) with BAMBI (P<0.001), a marker of ventral tissues. (C–D) The ventral signature (which contains 70 genes) positively correlated with BAMBI (P<0.001), while it negatively correlated with Chordin (P<0.001). (E) SMC1A, used here as a negative control, did not correlate with the dorsal (P=0.355) nor the ventral (not shown) signature. (F) Pkdcc, a novel dorsal gene described here, significantly correlated with the dorsal gene signature (P<0.001).
To our surprise, the ranked lists of genes correlated to Chordin or Bambi from the 26 libraries were an excellent predictor of dorsal or ventral genes (Tables S7 and S8). For example, immediately after Chordin one finds Noggin, and within the top genes ADMP, Goosecoid, Hhex, Xnr3, Dkk1 and Follistatin (Table S7). When genes were ranked according to their correlation with Bambi, within the top ten genes, out of 40,501 transcripts, one finds Smad7, HAS1 (Hyaluronan synthase 1), Sizzled, Msx1, ID3 and Twisted gastrulation, all of which are known ventral genes (Table S8). The results suggest that, by analyzing large numbers of experimental conditions using GSEA or Pearson correlations (Tables S7 and S8), it will be possible to define the signaling pathways and new gene signatures involved in embryonic induction during early development. These analyses would not have been possible without the complete genomic sequencing of Xenopus laevis.
Pkdcc1 and 2 are Spemann Organizer-specific secreted tyrosine kinases
Among the new dorsally enriched genes, a prominent one was Pkdcc (Table S3). We present here its characterization as a validation that our gene lists indeed identify true organizer-specific genes. While this work was in preparation a study of Pkdcc in Xenopus and its function in non-canonical Wnt signaling was reported (Vitorino et al., 2015). Pkdcc (also known as sgk493, adtk1, AW548124) represents the first secreted tyrosine kinase identified (Bordoli et al., 2014) and belongs to a family of related kinases that also comprises Fam69A-C, Deleted in Autism 1 (DIA1) and DIA1-Related (DIA1R), which share a common subcellular localization inside the Golgi apparatus (Dudkiewicz et al., 2013; Tagliabracci et al., 2015). Work in mouse and zebrafish has shown that Pkdcc was expressed in regions homologous to dorsal organizer (Goncalves et al., 2011; Kinoshita et al., 2009; Imuta et al., 2009).
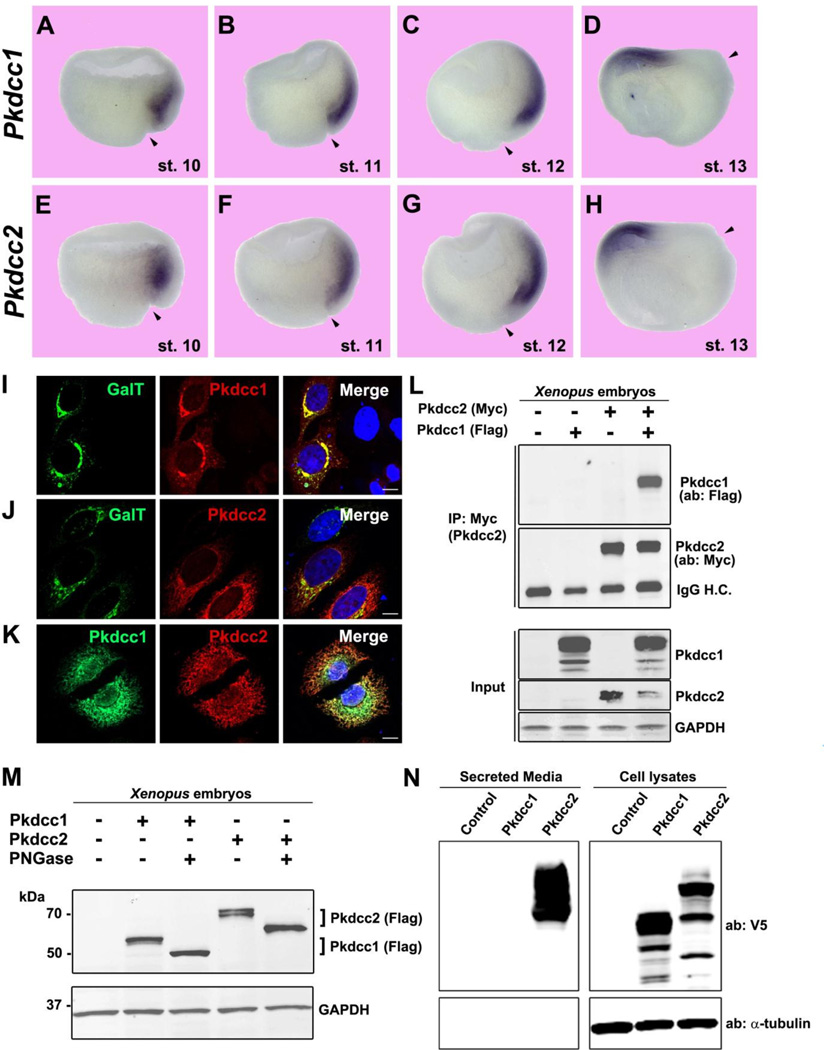
Xenopus laevis has two Pkdcc homologs, as do Xenopus tropicalis and zebrafish (Bordoli et al., 2014; Melvin et al., 2013). Amino acid sequence comparisons revealed that Xenopus Pkdcc2 shared 60% homology with human Pkdcc/Vlk at the protein level, while Pkdcc1 was less conserved, with a 40% homology (Fig. S1). Between each other, Xenopus Pkdcc1 and 2 shared 40% similarity (Fig. S1). Both genes were expressed form stage 10.5 onwards (Fig. S2A). By in situ hybridization both were found to be expressed in the Spemann organizer, with Pkdcc2 extending more anteriorly (Fig. 5A and E). Both genes correlated with the dorsal gene signature in GSEA analysis (Fig. 4F). At later gastrulation stages, Pkdcc1 and 2 presented similar and overlapping expression patterns, comprising in the anterior of the involuting dorsal mesendoderm (Fig. 5B–H). At later stages, both mRNAs were expressed in the eye and in lateral plate mesoderm, with Pkdcc1 extending more posteriorly in comparison to Pkdcc2 (Fig. S2B–I).
Fig. 5.
Pkdcc1 and 2 are expressed in the Spemann Organizer and localize to the Golgi apparatus. (A–H) Whole-mount in situ hybridization on hemisectioned Xenopus gastrula embryos showing Pkdcc1 and Pkdcc2 expression pattern from early to late gastrula (stage 10–13). Note the more anterior localization (in anterior endomesoderm) of Pkdcc2 compared to Pkdcc1 at stage 10. By the end of gastrulation Pkdcc1/2 expression was restricted to the prechordal plate. Arrowheads indicate dorsal lip position. (I–K) Immunofluorescence on HeLa cells overexpressing Pkdcc1-Flag and Pkdcc2-myc shows that the two kinases co-localize with the Golgi marker Galactosyl-Transferase (GalT). The two kinases also showed an overlapping subcellular localization. Nuclei were stained with DAPI (merge). Scale bars represent 10 µm. (L) Immunoprecipitation experiment showing physical interaction between Pkdcc1 and Pkdcc2 in Xenopus embryos. Pkdcc1-flag and/or Pkdcc2-myc mRNAs were injected at 4-cell stage and embryos were lysed at stage 12. One third of the total protein extract was saved and used as input. (M) Pkdcc1 and 2 are glycosylated, as shown by the electrophoretic mobility shift caused by PNGase F treatment. (N) Analysis of V5-tagged Pkdcc1 and 2 secretion in HEK293T cells showing that Pkdcc2 was found in the culture medium, indicating extensive secretion.
We found that Pkdcc transcription is induced by Wnt8 mRNA and blocked by β-catenin morpholino (Fig. S3A–C). In mammals, Pkdcc/Vlk protein is glycosylated and known to transit the Golgi before being secreted (Bordoli et al., 2014). To determine whether Xenopus Pkdcc1 and 2 share similar subcellular localization and biochemical properties, we first performed immunofluorescence in HeLa cells overexpressing Pkdcc1 or 2. Both co-localized with the Golgi marker Galactosyl-Transferase (GalT) (Storrie et al., 1998) and with each other (Fig. 5I–K). In mRNA co-injection experiments in Xenopus embryos, Pkdcc1 and 2 were able to bind each other (Fig. 5L). The proteins were glycosylated, since removal of N-linked oligosaccharides with PNGase F, caused faster electrophoretic mobility (Fig. 5M). To determine whether both enzymes were secreted, we analyzed the extracellular medium of HEK293T cells overexpressing V5-tagged forms of Pkdcc1 and 2. Surprisingly, only Pkdcc2 was secreted in abundance into the culture medium (Fig. 5N). Altogether, these data indicate that Xenopus Pkdcc1 and 2 are related organizer glycoproteins that bind to each other, and that at least one of which can be secreted extracellularly, presumably to phosphorylate tyrosines in organizer secreted factors.
Pkdcc1 or 2 overexpression induces expansion of head structures and inhibits canonical Wnt
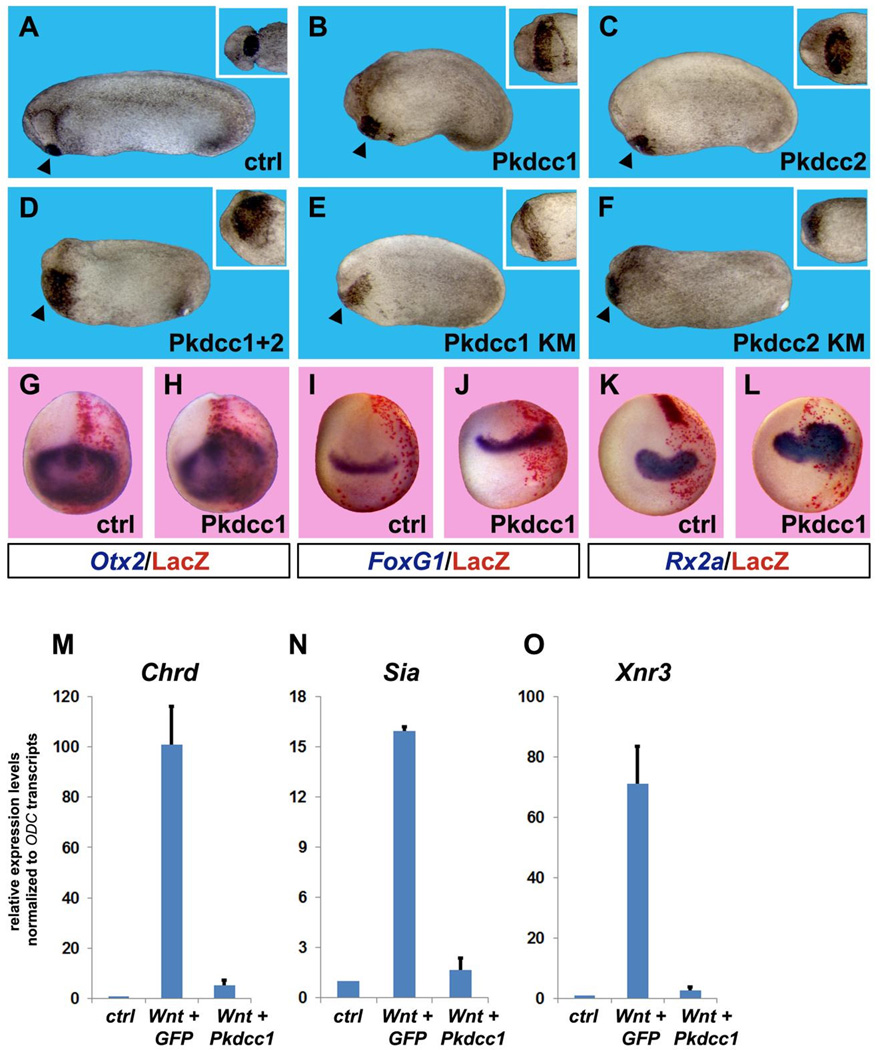
Mouse knock-out studies show that Pkdcc is required for craniofacial development and long bone elongation (Goncalves et al., 2011; Imuta et al., 2009; Kinoshita et al., 2009; Melvin et al., 2013; Probst et al., 2013). We investigated the effects of Pkdcc1 and 2 overexpression by microinjecting mRNA encoding these kinases in the animal blastomeres of 4-cell stage Xenopus embryos and assessed the phenotype at early tailbud stage. Overexpression of Pkdcc1 or 2 led to similar phenotypes, consisting in a prominent expansion of the cement gland and head region (Fig. 6A–C). Co-expressing both enzymes together led to an even stronger effect, suggesting possible synergy (Fig. 6D). Although enzymatic kinase activity was not tested directly in this study, the conservation of the clones with its mammalian, zebrafish and Xenopus homologues (Bordoli et al., 2014; Kinoshita et al., 2009; Melvin et al., 2013; Vitorino et al., 2015) suggests that the microinjected mRNA should be active. To test whether the anterior-promoting effect of Pkdcc required kinase activity, we mutated a Lys residue to Met (KM) in the known ATP binding site of both Pkdcc1 and 2, which has been shown to inactivate kinase activity (Bordoli et al., 2014). To our surprise, Pkdcc1 and 2 kinase mutants still induced cement gland enlargement when overexpressed in Xenopus embryos (Fig. 6E and F). This result indicated that the Tyrosine kinase activity is not involved in the induction of the cement gland.
Fig. 6.
Overexpression of Pkdcc enlarges anterior tissues and inhibits canonical Wnt signaling. (A–C) Overexpression of 1 ng of Pkdcc1 or Pkdcc2 mRNA caused expansion of anterior structures such as the cement gland (see insets). Percent of embryos showing enlarged cement gland and head were 0% n=60 for controls; 81% n=54 for Pkdcc1; 84% n=38 for Pkdcc2. (D) This effect is synergistic, since injection of 500 pg each of Pkddc1 and 2 caused an even greater enlargement of the cement gland (98% of the embryos showed this phenotype, n=50). (E–F) Kinase mutant (KM) forms of Pkdcc1 and 2 also caused an increase of cement gland tissues, suggesting that the kinase activity is dispensable for Pkdcc-mediated anteriorizing effects. Percent of embryos showing enlarged cement glands were: 82% n=27 for Pkdcc1-KM; 83% n=24 for Pkdcc2-KM. All embryos are shown from lateral views, insets show ventral view of the cement gland from the same embryos. Arrowheads point to cement gland. (G–L) Whole-mount in situ hybridizations for anterior neural markers show that Pkdcc1 mRNA overexpression expands Otx2, FoxG1 (also known as BF1) and Rx2a. The injected side was identified through Red Gal staining of the lineage tracer nuclear Lacz (nLacz) coinjected with Pkdcc1. Control embryos were injected with nLacz only. Percentage of Pkdcc1-injected embryos showing expanded neural markers: Otx2 66% n=27; FoxG1 76% n=47; Rx2a 92% n=38. (M–O) Pkdcc1 abolishes canonical Wnt signaling. Quantitative RT-PCR analysis performed on animal cap explants co-injected with xWnt8 and Pkdcc1 mRNAs shows that Pkdcc1 greatly reduces induction of canonical Wnt target genes Chordin (Chrd), Siamois (Sia) and Nodal related 3 (Xnr3). GFP mRNA was used as a negative control. Transcript levels of the housekeeping gene ODC were used for normalization. Error bars indicate standard deviations derived from three independent experiments.
Next, we asked whether cement gland enlargement was part of a more general anteriorizing effect. Embryos injected unilaterally with Pkdcc1 mRNA and the lineage tracer LacZ were harvested at neurula (stage 16), fixed, and processed for in situ hybridization for anterior neural markers. The results showed that Pkdcc1 induced an expansion of the forebrain/cement gland marker Otx2 (Pannese et al., 1995), the telencephalic marker FoxG1 (also known as Bf1) (Bourguignon et al., 1998) and the eye-field marker Rx2a (Andreazzoli et al., 1999; Casarosa et al., 1997) (Fig. 6G–L). In addition, quantitative RT-PCR data from Pkdcc1 mRNA injected embryos also showed an increase in anterior markers (Fig. S3D).
In Xenopus, head and anterior development requires suppression of canonical Wnt signaling by several inhibitors, such as Cerberus, Dkk1, Frzb-1, Notum, Shisa and Tiki, while increased late Wnt signaling leads to the reduction of head structures (Bouwmeester et al., 1996; Glinka et al., 1997, 1998; Yamamoto et al., 2005; Zhang et al., 2012, 2015). Therefore, we hypothesized that the anteriorization effect of Pkdcc might be explained by inhibition of canonical Wnt signaling. To test this, we took advantage of the Xenopus animal cap system, which is very amenable for quantitative analyses of Wnt-induced genes. Interestingly, while Wnt8 mRNA overexpression was able to upregulate Wnt target genes such as Chordin, Siamois and Xnr3, coexpression of Pkdcc1 inhibited their induction almost completely (Fig. 6M–O). Although the molecular mechanism of Wnt inhibition by Pkdcc overexpression is unclear, it might be related to the regulation of protein trafficking through the secretory pathway. In NIH3T3 cultured cells, overexpression of Pkdcc/Vlk, or its kinase-inactive mutant, has been shown to inhibit transport from the Golgi to the plasma membrane (Kinoshita et al., 2009). The overexpression results presented here suggest that Pkdcc1 may act as a Wnt antagonist.
Discussion
In this study we took advantage of the recently completed genome sequence of Xenopus laevis to explore the transcriptome of the early gastrula embryo using high-throughput RNA-seq. The allotetraploidy of X. laevis arising from the hybridization between two different species had been an obstacle in previous research (Kobel, 1996). Initially, we used as reference the X. tropicalis genome (Hellsten, 2010) and a X. laevis egg and embryo reference transcriptome (Peshkin et al., 2015). Many more sequences were identified in our RNA-seq analyses using the new Xenopus laevis JGI 9.1 genome. However, many of the transcripts in JGI9.1 were annotated only with ID numbers. For example, Chordin and many other genes could not be identified. We improved the annotation by making a blast search of our gene lists to the human proteome. Two additional columns, indicating human protein ID and human gene symbol, were introduced in our Supplemental Tables. The annotation of Xenopus laevis with human gene symbols greatly helps one to correlate frog early development studies with human signaling pathway gene sets. In future, it may even allow the identification of existing permanent human cell lines that most resemble embryonic signaling centers.
In this first study, we concentrated on genes expressed dorsally or ventrally in the early gastrula. We generated lists of genes ranked according to their enrichment from dorsal to ventral and vice versa. The most differentially expressed genes were the BMP antagonist Chordin and the Xolloid inhibitor Sizzled, both of which are key players of the D-V patterning biochemical pathway (Plouhinec et al., 2013). The classical Spemann organizer genes – such as Xnr3, Cerberus, ADMP, Goosecoid, Noggin, Follistatin and Crescent - were also identified (Table S3). Similarly, the well-known ventral genes – Wnt8, BAMBI, Ventx, ID3 and Msx1 - were readily detected (Table S4). All these genes had high RPKM values, indicating that the previous organizer screens had been essentially saturating for highly expressed genes. This is in a sense analogous to the work in Drosophila, in which screening for zygotic cuticle mutants was saturating (Nüsslein-Volhard and Wieschaus, 1980), but in the case of Xenopus this was applicable only to abundant genes. The use of the complete genome permitted the identification of many additional D-V genes than were even possible in the elegant previous study by Hufton et al. (2006) using microarrays. A plethora of novel genes are listed in our Supplemental Tables 3 and 4. To construct these lists we chose an arbitrary cut-off of genes expressed at above 1 RPKM, but it is possible that low-level expressed genes could have developmental roles if they had catalytic or signaling activities. In Table S1 we present all data for 44,000 genes expressed from three independent dorsal and ventral lip gastrula libraries. The expression ranking for all genes, even the low-expressed ones, can be reconstructed readily from the Excel platform presented in Table S1.
As a proof of principle of the ability of RNA-seq to identify novel genes, we provided data on Pkdcc/Vlk 1 and 2. These are secreted Tyrosine kinases (Bordoli et al., 2014), and we thought they would be interesting to study because they in principle could phosphorylate a plethora of organizer-specific secreted factors. When overexpressed, these protein showed canonical Wnt inhibitor activity but, to our disappointment, this property was not dependent on their Tyrosine kinase activity. While this manuscript was in preparation, Vitorino et al., (2015) published a similar expression pattern in Xenopus laevis, now confirmed by this work. They also demonstrated by morpholino loss-of-function experiments that Pkdcc1 and 2 regulate Wnt/PCP signaling (Vitorino et al., 2015). Their results are extended here by showing an additional role for these genes as inhibitors of canonical Wnt signaling in microinjected animal cap explants.
Because of its allotetraploid genome, Xenopus laevis has many duplicated genes, and in general both copies are expressed (Session et al., 2016; Peshkin et al., 2015). This drawback is mitigated by the many advantages of Xenopus laevis for embryology. The availability of the sequenced genome and transcriptome will be invaluable for the design of morpholinos and gene editing loss-of-function reagents that have proven essential to uncover fundamental developmental mechanisms (Blum et al., 2015). Multiple morpholinos can be injected simultaneously to cause knockdowns (Khokha et al., 2005; Reversade and De Robertis, 2005), indicating that the duplicated gene problem can be overcome if required. Although we have not analyzed the levels in detail, we confirm that many alloalleles are expressed at the gastrula stage and will require double knockdowns.
We defined dorsal and ventral gene expression signatures that were suitable for gene set expression analysis (GSEA) (Subramanian et al., 2005). We sequenced the total of 26 gastrula libraries in different experimental conditions, and found that our signatures correlated with D-V genes (Fig. 4). The Pearson correlation coefficient indicates the degree to which gene expression levels vary with respect to one another. When transcripts in all these libraries are ranked according to the Pearson correlation coefficient, for example, to Chordin or BAMBI, we were surprised to find that correlated genes were excellent predictors of D-V enriched transcripts, including some that had been overlooked in other analyses (Tables S7 and S8).
We found it particularly interesting that the gene that correlated the most highly with Chordin was Noggin, and vice versa. These two BMP antagonists synergize with each other in mouse knockouts (Bachiller et al., 2000) and in Xenopus (Khokha et al., 2005). The close correlation indicates that they share very similar transcriptional regulatory mechanisms. It may be worthwhile in the future to explore whether Chordin and Noggin interact with each other or with other components of the D-V biochemical pathway at the protein level.
The Xenopus gastrula is one of the best understood systems for long-range signaling by morphogen gradients (Bier and De Robertis, 2015). Our next step will be to define gene-response signatures for the Wnt, BMP, Nodal and FGF pathways during gastrulation. As more transcriptomes in different experimental conditions are obtained, we hope this will further illuminate the molecular mechanisms of embryonic induction, and in particular how half-embryos can regenerate the missing part. In the meantime, the gene lists in the Supplemental Tables presented here represent a rich resource for Xenopus embryologists.
Supplementary Material
Highlights.
Genome-based RNA-seq is a powerful tool to detect differential gene expression.
RNA-seq was used to screen for dorsal and ventral genes in Xenopus.
Classic and novel dorsal/ventral genes were reported.
Dorsal and ventral gene signatures provide a rich resource for Xenopus embryology.
Pkdcc/Vlk is a secreted tyrosine kinase expressed in the Spemann Organizer.
Acknowledgments
We thank L. Peshkin and M. Kirshner for access to their RNA-seq database before publication, and Prof. G. Fan from UCLA for guiding us in library preparation, B. Storrie for GalT-GFP Golgi marker, and J. Belo and R. Harland for sharing information prior to publication. E.A.S. and M.D.J.B. were supported by the NIH MARC program. This work was supported by the Norman Sprague endowment and the Howard Hughes Medical Institute, of which E.M.D.R. is an investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Angeloni D, Menna E, Barsacchi G. Role of Xrx1 in Xenopus eye and anterior brain development. Development. 1999;126:2451–2460. doi: 10.1242/dev.126.11.2451. [DOI] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- Bier E, De Robertis EM. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science. 2015;348:aaa5838. doi: 10.1126/science.aaa5838. [DOI] [PubMed] [Google Scholar]
- Blum M, De Robertis EM, Wallingford JB, Niehrs C. Morpholinos: antisense and sensibility. Dev. Cell. 2015;35:145–149. doi: 10.1016/j.devcel.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Bordoli MR, Yum J, Breitkopf SB, Thon JN, Italiano JE, Jr, Xiao J, Worby C, Wong SK, Lin G, Edenius M, Keller TL, Asara JM, Dixon JE, Yeo CY, Whitman M. A secreted tyrosine kinase acts in the extracellular environment. Cell. 2014;158:1033–1044. doi: 10.1016/j.cell.2014.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125:4889–4900. doi: 10.1242/dev.125.24.4889. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a headinducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature. 1996;15:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38:D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. Xrx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech. Dev. 1997;61:187–198. doi: 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141:1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colozza G, De Robertis EM. Maternal syntabulin is required for dorsal axis formation and is a germ plasm component in Xenopus. Differentiation. 2014;88(1):17–26. doi: 10.1016/j.diff.2014.03.002. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Spemann's organizer and the self-regulation of embryonic fields. Mech. Dev. 2009;126:925–941. doi: 10.1016/j.mod.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell. Dev. Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann DS, Walentek P, Harland RM. The alternative splicing regulator Tra2b is required for somitogenesis and regulates splicing of an inhibitory Wnt11b isoform. Cell Rep. 2015;10:527–536. doi: 10.1016/j.celrep.2014.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkiewicz M, Lenart A, Pawlowski K. A novel predicted calcium-regulated kinase family implicated in neurological disorders. PLoS One. 2013;8:e66427. doi: 10.1371/journal.pone.0066427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med. 2014;6:265sr266. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Onichtchouk D, Blumenstock C, Niehrs C. Head induction by simultaneous repression of Bmp and Wnt signaling in Xenopus. Nature. 1997;389:517–519. doi: 10.1038/39092. [DOI] [PubMed] [Google Scholar]
- Goncalves L, Filipe M, Marques S, Salgueiro AM, Becker JD, Belo JA. Identification and functional analysis of novel genes expressed in the Anterior Visceral Endoderm. Int. J. Dev. Biol. 2011;55:281–295. doi: 10.1387/ijdb.103273lg. [DOI] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann’s organizer. Annu. Rev. Cell Dev. Biol. 1997;13:611–617. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. β-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev. Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neutralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hufton AL, Vinayagam A, Suhai S, Baker JC. Genomic analysis of Xenopus organizer function. BMC Dev. Biol. 2006;6:27. doi: 10.1186/1471-213X-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imuta Y, Nishioka N, Kiyonari H, Sasaki H. Short limbs, cleft palate, and delayed formation of flat proliferative chondrocytes in mice with targeted disruption of a putative protein kinase gene, Pkdcc (AW548124) Dev. Dyn. 2009;238:210–222. doi: 10.1002/dvdy.21822. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann’s organizer leads to a catastrophic loss of dorsal structures. Dev. Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Era T, Jakt LM, Nishikawa S. The novel protein kinase Vlk is essential for stromal function of mesenchymal cells. Development. 2009;136:2069–2079. doi: 10.1242/dev.026435. [DOI] [PubMed] [Google Scholar]
- Kobel HR. Allopolyploidy speciation. In: Tinsley RC, Kobel HR, editors. The biology of Xenopus. Oxford: Clarendon Press; 1996. pp. 391–401. [Google Scholar]
- Lee HX, Ambrosio AL, Reversade B, De Robertis EM. Embryonic dorsal-ventral signaling: secreted Frizzled-related proteins as inhibitors of Tolloid proteinases. Cell. 2006;124:147–159. doi: 10.1016/j.cell.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin VS, Feng W, Hernandez-Lagunas L, Artinger KB, Williams T. A morpholino-based screen to identify novel genes involved in craniofacial morphogenesis. Dev. Dyn. 2013;242:817–831. doi: 10.1002/dvdy.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1a-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis. Amsterdam, The Netherlands: North Holland Publishing Co.; 1967. [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. The Xenopus homologue of Otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development. 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- Pera EM, De Robertis EM. A direct screen for secreted proteins in Xenopus embryos identifies distinct activities for the Wnt antagonists Crescent and Frzb-1. Mech. Dev. 2000;96:183–195. doi: 10.1016/s0925-4773(00)00394-4. [DOI] [PubMed] [Google Scholar]
- Peshkin L, Wühr M, Pearl E, Haas W, Freeman RM, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouhinec JL, Zakin L, Moriyama Y, De Robertis EM. Chordin forms a self-organizing morphogen gradient in the extracellular space between ectoderm and mesoderm in the Xenopus embryo. Proc. Natl. Acad. Sci. USA. 2013;110:20372–20379. doi: 10.1073/pnas.1319745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst S, Zeller R, Zuniga A. The hedgehog target Vlk genetically interacts with Gli3 to regulate chondrocyte differentiation during mouse long bone development. Differentiation. 2013;85:121–130. doi: 10.1016/j.diff.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123:1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session A, Uno Y, Kwon T, Chapman J, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Putnam N, Stites J, van Heeringen S, Quigley I, Heinz S, Hellsten U, Lyons JB, Suzuki A, Kondo M, Ogino H, Ochi H, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Mozaffari S, Shu S, Schmutz J, Jenkins J, Grimwood J, Carlson J, Mitros T, Simakov O, Heald R, Miller K, Haudenschild C, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Kinoshita T, Ohta Y, Mawaribuchi S, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Kitzman J, Shendure J, Nakayama T, Izutsu Y, Robert J, Dichmann D, Flajnik M, Houston DW, Marcotte E, Wallingford J, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra G, Fujiyama A, Harland RM, Taira M, Rokhsar D. Genome evolution in the allotetraploid frog Xenopus laevis. Manuscript submitted. 2016 doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- Storrie B, White J, Röttger S, Stelzer EH, Suganuma T, Nilsson T. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 1998;143(6):1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci VS, Wiley SE, Guo X, Kinch LN, Durrant E, Wen J, Xiao J, Cui J, Nguyen KB, Engel JL, Coon JJ, Grishin N, Pinna LA, Pagliarini DJ, Dixon JE. A single kinase generates the majority of the secreted phosphoproteome. Cell. 2015;161:1619–1632. doi: 10.1016/j.cell.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitorino M, Silva AC, Inacio JM, Ramalho JS, Gur M, Fainsod A, Steinbeisser H, Belo JA. Xenopus Pkdcc1 and Pkdcc2 Are Two New Tyrosine Kinases Involved in the Regulation of JNK Dependent Wnt/PCP Signaling Pathway. PLoS One. 2015;10:e0135504. doi: 10.1371/journal.pone.0135504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler C, Ohno S. Cytological confirmation of female heterogamety in the African water frog (Xenopus laevis) Cytogenetics. 1962;1:217–223. doi: 10.1159/000129731. [DOI] [PubMed] [Google Scholar]
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- Wessely O, Kim JI, Geissert D, Tran U, De Robertis EM. Analysis of Spemann organizer formation in Xenopus embryos by cDNA macroarrays. Dev. Biol. 2004;269:552–566. doi: 10.1016/j.ydbio.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Wuhr M, Freeman RM, Jr, Presler M, Horb ME, Peshkin L, Gygi SP, Kirschner MW. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr. Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors. Wnt and FGF. Cell. 2005;120:223–235. doi: 10.1016/j.cell.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev. Cell. 2011;19:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Abreu JG, Yokota C, MacDonald BT, Singh S, Coburn KL, Cheong SM, Zhang MM, Ye QZ, Hang HC, Steen H, He X. Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell. 2012;149:1565–1577. doi: 10.1016/j.cell.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cheong SM, Amado NG, Reis AH, MacDonald BT, Zebisch M, Jones EY, Abreu JG, He X. Notum is required for neural and head induction via Wnt deacylation, oxidation, and inactivation. Dev. Cell. 2015;32:719–730. doi: 10.1016/j.devcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.