Abstract
BACKGROUND
Bone metastasis from primary prostate cancer leads to markedly diminished quality of life with poor long-term survival. Bone seeking treatment options are limited with adverse consequences on rapidly proliferating tissues such as bone marrow. In the present study, we seek to determine the bone-enriching capabilities of monomethyl auristatin E phosphate (MMAEp), a derivative of the potent antimitotic monomethyl auristatin E (MMAE).
METHODS
The in vitro actions and mechanisms of cytotoxicity were assessed using cell viability, immunofluorescence, flow cytometry, and Western blot analysis. In vivo efficacy was determined using an intratibial xenograft mouse model of human prostate cancer and live animal imaging.
RESULTS
The half maximal inhibitory concentration (IC50) of MMAE and MMAEp was determined to be approximately 2 nM and 48 nM, respectively, in PC-3 and C4-2B cell lines. MMAEp retained the mechanism of action of MMAE in reducing microtubule polymerization and stalling cell cycle progression at the G2/M transition. MMAEp was able to bind hydroxyapatite in in vitro assays. MMAEp significantly reduced intratibial tumor growth compared to the vehicle control treatment.
CONCLUSIONS
MMAEp is an antimitotic compound that binds to calcium ions in the bone and inhibits prostate tumor growth in the bone.
Keywords: Prostate cancer, Therapeutics, MMAEp, Apoptosis, Cell cycle
INTRODUCTION
Bone metastases from primary prostate cancer (PCa) are a significant public health burden. According to the most recent data from the American Cancer Society, more than 26,000 men will die of prostate cancer in 2016 [1]. Sixty five percent of these patients demonstrate bone metastasis at autopsy [2]. Bone metastasis can cause painful skeletal related events (SREs) such as vertebral compression and femoral neck fractures [3].
There are six FDA-approved treatment options for patients with metastatic castration resistant PCa (mCRPC): docetaxel, cabazitaxel, enzalutamide, abiraterone, radium-223, and sipuleucel-T [4]. These treatments increase overall survival in the range of 2-5 months, but not without side effects. Docetaxel and cabazitaxel are members of the taxane family of compounds that induce microtubule stabilization [5]. Both drugs can also compromise tissue function that relies on rapid cellular proliferation, such as intestinal epithelium and bone marrow [6]. Enzalutamide (an androgen receptor antagonist) and abiraterone (a 17α-hydroxylase inhibitor) act to disrupt androgen's ability to signal to its cognate receptor [7,8]. Anti-androgen therapy resistance has become a serious concern (9). Radium-223 is a α-particle emitting radionuclide that mimics cations (e.g., calcium2+) in their uptake and enrichment within the bone matrix [10]. This is in addition to the observation that phosphorus-32 can also enrich in the bone matrix via a similar mechanism [11-13]. Radium-223 is currently the preferred radiopharmaceutical treatment over phosphorus-32 as the latter demonstrates severe myelosuppression due to its β-emissions acting over a longer, millimeter-length distance [13]. This is in contrast to radium-223's shorter micrometer-length distance of emission which demonstrates diminished, but still apparent, bone marrow suppression [14,15]. Aminobisphosphonates (NBPs) are another class of bone enriching compounds currently in use for the treatment of painful SREs. NBPs work by mimicking pyrophosphate to enrich within the bone matrix [16]. Once enriched, they are taken up by bone-resorbing osteoclasts and inhibit the enzyme farnesyl pyrophosphate synthase [17]. In the context of PCa bone metastasis, this disrupts the “vicious cycle” of osteoclast-mediated release of growth factors from the bone stroma, thus reducing SREs in patients with advanced PCa [18]. Germaine to the present study is the observation that the literature has documented cases of bone seeking compounds for the treatment of bone metastasis.
Monomethyl auristatin E (MMAE) is a synthetic derivative of dolastatin-10 developed in the lab of George Pettit [19,20]. MMAE is similar to taxanes (i.e. docetaxel and cabazitaxel) in that it disrupts microtubule dynamics, albeit through inhibition of tubulin polymerization [19]. MMAE is characterized as a linear depsipeptide consisting of five derivative amino acids with methyl and methoxy-substituted side chains. MMAE is chiefly being investigated as the main cytotoxic component of molecules called antibody directed conjugates (ADCs) to treat several different cancer types [21-24]. Previous investigations have revealed that MMAE induces apoptosis through a mechanism of cell death called mitotic catastrophe [25-27]. In the present study, we generated a custom modification of MMAE by adding a phosphate to the carboxy terminal to create a potentially bone-enriching compound that could induce apoptosis in bone metastatic cells. The objective of this study was to determine the bone seeking ability of a phosphate modification of MMAE named monomethyl auristatin E phosphate (MMAEp) in order to determine MMAEp's suitability in the treatment of cancer bone metastases. We found that MMAEp caused cell growth arrest and cell death via apoptosis, and that MMAEp bound to calcium ions in hydroxyapatite in vitro and reduced tumor burden in an intratibial xenograft mouse model of human prostate cancer.
MATERIALS AND METHODS
Cell Culture
PC-3 and C4-2B cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and Leland WK Chung (Cedars-Sinai Medical Center, Los Angeles, CA, USA) [28], respectively. PC-3 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM with 4.5g/L glucose, L-glutamine, and sodium pyruvate, Mediatech Inc., Manassas, VA, USA; catalogue number 10-013-CV) containing 10% fetal bovine serum (FBS, JR Scientific Inc., Woodland, CA, USA; catalogue number 43603-500), 100 IU/ml penicillin/streptomycin cocktail (Mediatech, Inc., Manassas, VA, USA; catalogue number 30-004-CI). C4-2B cells were cultured in HyClone™ RPMI-1640 medium (with 2.05 mM L-glutamine, GE Healthcare Life Science, Logan, UT, USA) with 10% FBS and 100 IU/ml penicillin/streptomycin cocktail. Cultures were maintained in an incubator set to 5% CO2 atmosphere at 37°C (Thermo Scientific, Forma Series II Water Jacketed CO2 Incubator, Thermo Fisher Scientific, Waltham, MA, USA).
Cell Viability Assay
The number of living cells was determined using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega Corp., Fitchburg, WI, USA). Cells were plated in 96-well opaque Costar plates as previously described [29]. In brief, 4,000 cells in 100 μL media were added to each well in triplicate and allowed to adhere overnight. Medium was removed the next day and cells were re-fed with media containing dimethylsulfoxide (DMSO) vehicle, MMAE (dose range: 0.1-5 nM), or MMAEp (dose range: 1-100 nM). MMAEp was developed based on Dr. Zongbing You's original idea to put a phosphate group onto MMAE, in order to make it able to bind to calcium ions. MMAEp was custom-synthesized by Concortis/Levena Biopharma, San Diego, CA, USA. Cell-free wells were established as control for determining background. After 72 hours, 70 μL of CellTiter-Glo® viability reagent was added to each well and placed on an orbital shaker in the dark for 2 minutes. The plate was allowed to sit at room temperature for 10 minutes before being read on a FLUOstar OPTIMA (BMG Labtech GmbH, Ortenberg, Germany) microplate reader. Viability was determined using the formula [(Luminescence of the treatment group – Background luminescence) ÷ (Luminescence of the control group – Background luminescence)] × 100%. Data are presented as the mean and standard error of the mean (SEM) of three independent experiments. Half maximal inhibitory concentrations (IC50) values were calculated using linear regression on the dataset.
Immunofluorescence
Polylysine-coated slides were sterilized with 70% ethanol prior to plating with either PC-3 or C4-2B cells. Two hundred microliters of cells at a concentration of 5×105 cells/ml were applied to the slides in an area outlined by a wax pen. Cells were allowed to adhere to the slides overnight while being kept in a 150-mm dish. The following day, medium was aspirated and cells were re-fed with media containing either DMSO vehicle, MMAE (4 nM), or MMAEp (96 nM) for 24 h. Cells were fixed in methanol and extensively washed with phosphate-buffered saline (PBS). The slides were then blocked in goat IgG for 1 h. The slides were probed with mouse anti-α-tubulin antibody (working titer: 0.5 μg/ml, catalogue number 322588, Invitrogen) for 3 h at room temperature and counterstained for 10 minutes with Hoechst stain (0.2 μg/ml) at room temperature. Both stains were applied in the dark with extensive PBS washes in between applications of stain. Cells were imaged using a Nikon Eclipse 80i microscope connected to a Photometrics CoolSnap EZ camera. The image acquisition software utilized for this study was NIS Elements (Nikon Instruments Inc., Melville, NY, USA). Quantification of immunofluorescence signal was accomplished using the CellProfiler program [30,31].
Hydroxyapatite Chromatography
Approximately 120 mg of Bio-Rad CHT™ ceramic hydroxyapatite type I crystals (lot number S400892, Bio-Rad Laboratories, Hercules, CA) were loaded in a spin column and equilibrated with 500 μl of 150 mM Tris NaCl (25 mM Tris base and 125 mM sodium chloride) via spinning down in a microcentrifuge. An equivalent volume of Tris NaCl containing 1 μM MMAE or MMAEp was then loaded onto the equilibrated column and incubated for 30 seconds. The columns were then centrifuged for 10 seconds. This procedure was repeated a total of 5 times per sample. Column eluents were then passed through a 0.22 μm filter for sterilization. Stock concentrations of 150 mM Tris NaCl (negative control), 2 nM MMAE in 150 mM Tris NaCl (positive control), and 48 nM MMAEp in 150 mM Tris NaCl were also passed through a 0.22 μm filter for sterilization. Negative and positive controls were diluted in 1 ml DMEM for application to PC-3 cells. Column eluents were diluted in the same volume of DMEM with the assumption that both MMAE and MMAEp solutions contained their initial 1 μM concentration of the agents. PC-3 cells were treated using the abovementioned cell viability assay and measured on a BioTek Synergy H1 microplate reader (BioTek Instruments Inc., Winooksi, VT).
Cell Cycle Analysis
PC-3 and C4-2B cells were plated in 60-mm dishes at a concentration of 5×105 cells/dish in triplicate. After overnight adhesion, cells were re-fed with media containing either DMSO vehicle, MMAE (4 nM), or MMAEp (96 nM), and incubated for 24 hours. Cells were then trypsinized and spun down for 5 minutes at 1,500 rounds per minute (rpm) at 4°C. The supernatant was aspirated and each sample was suspended in 500 μL PBS to achieve single cell suspension. Cells were pelleted again at 1,500 rpm for 5 minutes at 4°C and suspended in 70% ice-cold ethanol. Samples were kept at −20°C overnight. Cells were stained with 500 μL of propidium iodide solution containing 0.1% Triton™ X-100, 0.2 mg/mL DNase-free RNase A, and 20 μg/mL propidium iodide for 30 minutes in the dark. The number of cells in G1/G0, S, G2/M phases of the cell cycle were determined using flow cytometry and DNA content frequency histogram deconvolution software (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Data are presented as the mean and standard deviation (SD) of a representative of triplicate experiments.
Cell Apoptosis
Apoptosis was determined by performing sub-G1 analysis on the samples treated for cell cycle analysis as described above. The ModFit software package was used to gate the number of cells in the sub-G1 region. The line on each panel represents the gating used to count the number of cells in the sub-G1 region. Data are presented as the mean and standard deviation (SD) of a representative of three independently performed experiments.
Western Blot Analysis
Approximately 5×105 PC-3 and C4-2B cells were plated in 60-mm dishes in 3 ml medium. After overnight adherence, cells were treated with DMSO, 2 nM MMAE, or 48 nM MMAEp for 48 and 72 hours. Protein samples were collected in RIPA lysis buffer (50nM sodium fluoride, 0.5% Igepal CA-630 (NP-40), 10 mM sodium phosphate, 150 mM sodium chloride, 25 mM Tris (pH 8.0), 1 mM phenylmethylsulfonyl fluoride, 2 mM ethylenediaminetetraacetic acid, 1.2 mM sodium vanadate). Protein concentration was assessed using 200 μl Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, Hercules, CA) and a BioTek Synergy H1 microplate reader (BioTek Instruments Inc., Winooksi, VT). Approximately 50 μg of protein per sample was loaded onto a 10% sodium dodecyl sulfate -polyacrylamide gel and subjected to electrophoresis for 3 hours at 200 volts. The protein was then transferred to a polyvinylidene difluoride membrane in a semi-dry transfer chamber for 30 minutes at 20 volts. The membrane was then blocked for one hour at room temperature with 5% dry milk in TBST buffer (25 mM Tris-HCl, 125 mM sodium chloride, 0.1% Tween-20). The membrane was then probed with primary antibody overnight at 4°C and then incubated with IRDye® 800CW and IRDye® 680RD secondary antibodies (LI-COR Biosciences, Lincoln, NE). Results were generated using an Odyssey Infrared Imager (LI-COR Biosciences). Antibodies used include the following: mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and rabbit anti-cleaved poly ADP ribose polymerase (PARP) (Millipore, Billerica, MA).
Animal Study
Animal study was approved by the Animal Care and Use Committee of Tulane University. Athymic, Nu/Nu, 7-8-week-old male mice were purchased from Charles River Laboratories (Germantown, MD). Mice were anesthetized with isofluorane and placed in a position with the knee joint flexed to an approximate 75° angle to visualize the patellar tendon. The knee joint was sterilized with 70% ethanol and betadine solutions. The tibial marrow space was accessed with a 21-gauge needle as a trocar (guide) using a firm twisting motion through the top of the tibial plateau. Once the 21-gauge needle had burrowed through the cortical bone into the marrow, a 26-gauge needle attached to a Hamilton 50-μL syringe was used to introduce an inoculum of 2×105 PC-3-Luc-LacZ cells (stably expressing luciferase and β-galactosidase) in 10 μL of a PBS/Matrigel® mix (1:1 volume ratio) into the bone marrow space. Animals were then placed back in the cage in a ventral position under a warming light and observed for 15-30 minutes following the procedure and 0.1 mg/kg Buprenex analgesic was administered subcutaneously. Animals were randomized into either control (n = 10) or treatment group (n = 10) by flipping a coin. Treatment began immediately with intraperitoneal administration of either MMAEp at 1.75 mg/kg body weight or DMSO vehicle at an equivalent volume in 200 μL PBS. This was repeated once every four days for a total of six doses over a twenty-four day period. Starting seven days post tumor cell implantation, animals were imaged using an in vivo imaging system (IVIS® Lumina XRMS Series III, PerkinElmer, Waltham, MA, USA) for tumor luminescence via intraperitoneal administration of 80 μL of 40 mg/ml D-luciferin (Cayman Chemical Company, Ann Arbor, MI, USA) as described previously [29]. Animals were imaged once per week. The IVIS® imager settings were set to collect a minimum of 600 events per animal. The tumor sizes were represented as average radiance (log p/s/cm2/sr). Data are presented as the mean and standard error of the mean (SEM).
Histology
Mouse bone tumors were isolated subsequent to euthanasia and fixed overnight in 4% paraformaldehyde at 4°C and kept in 70% ethanol thereafter. Since bone was present in the samples, the tumors were demineralized for 5 days at room temperature on an orbital shaker in the Immunocal reagent (Decal Chemical Corp., Tallman, NY). Once samples were demineralized, they were serially dehydrated in increasing percentages of ethanol (75%, 85%, 95%, and 100%) for approximately one hour at each percentage. Tissue samples were then cleared in 100% xylene for 1 – 2 hours. Samples were then incubated in a 50/50 mix of xylene and paraffin wax for 10 minutes at 60°C and embedded in 100% paraffin wax for 30 minutes at 60°C and left to cool at room temperature overnight. Five-μm tissue sections were cut and processed through two washes in 100% xylene and then rehydrated in decreasing concentrations of ethanol (100% twice, 95% twice, 85% once, and 75% once) for 5 minutes each. Sections were washed extensively in tap water before staining with either hematoxylin and eosin or toluidine blue.
Statistical analysis
The in vitro experiments were replicated three times. Figures were developed using Graphpad Prism® 5.0 (Graphpad Software, La Jolla, CA). Statistical analysis was performed using a two-tailed Student's t-test with α < 0.05 as the threshold for significance.
RESULTS
MMAE and MMAEp inhibit proliferation of human prostate cancer cells
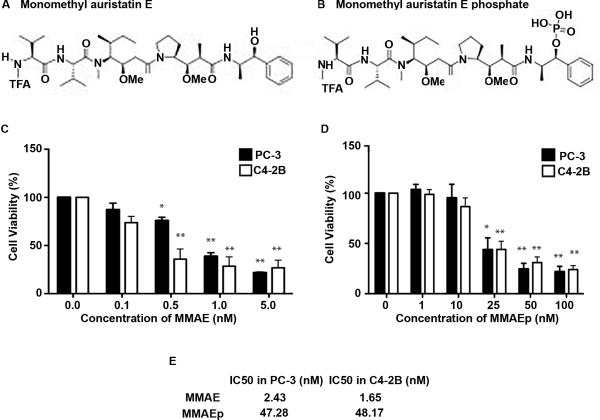
The chemical structures of both MMAE and MMAEp are depicted in Figures 1A and 1B, respectively. In order to determine the half maximal inhibitory concentration (IC50), a common parameter of drug efficacy, we treated prostate cancer cell lines in a dose-dependent manner over a 72-hour period and subjected these lines to a cell viability assay. Treatment of PC-3 and C4-2B cell lines showed significantly reduced viability starting at 0.5 nM MMAE and 25 nM MMAEp, respectively (Fig. 1C and 1D). Using linear regression, we calculated IC50 values of 2.43 nM (in PC-3 cells) and 1.65 nM (in C4-2B cells) for MMAE and 47.28 nM (in PC-3 cells) and 48.17 nM (in C4-2B cells) for MMAEp (Fig. 1E).
Fig. 1.
Chemical structures and cell viability assay to determine IC50 values. A and B: Chemical structures of MMAE and MMAEp, respectively. C and D: Cell viability assay used to calculate the IC50 values of both compounds. Data are presented as mean ± SEM of three independently performed experiments. * p<0.01, ** p<0.001 compared to the control (0 nM). E: Calculated IC50 values for both compounds in PC-3 and C4-2B cell lines using linear regression analysis.
MMAE and MMAEp inhibit microtubule polymerization and MMAEp binds to calcium ions
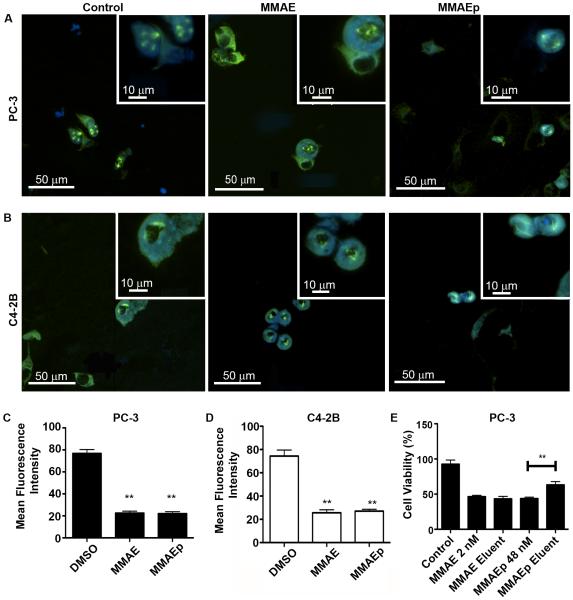
Having established the IC50 values of both compounds, we next sought to establish the mechanism of action using MMAE as the positive control. In order to assess whether MMAEp retained the mechanism of action of MMAE, we treated PC-3 and C4-2B cell lines for 24 hours with DMSO, 2× IC50 (4 nM) MMAE, or 2× IC50 (96 nM) MMAEp. Representative mitotic figures are depicted in Figures 2A and 2B. Using a human cell pipeline in the CellProfiler program, we were able to calculate a significant reduction in microtubule fluorescent signal in both human prostate cell lines (Fig. 2C and 2D). In order to assess the ability of MMAEp to bind to calcium ions, we passed MMAE and MMAEp through a hydroxyapatite column. We tried to directly quantify the concentration of the compounds in the eluents using mass spectrometry and other spectrometers, however, we could not obtain consistent data due to lack of reliable quantitative measurements of the new compounds. As an alternative approach, we paired hydroxyapatite chromatography with our cell viability assay to determine if binding occurred between MMAEp and the calcium charges found in hydroxyapatite crystals. Hydroxyapatite crystals are the main mineral component of bone and are often found in regular crystalline structures and amorphous deposits in living bone tissue [32]. When we compared MMAE eluent against an MMAE IC50 positive control, we found no significant differences in PC-3 cell viability. In contrast, we found that MMAEp eluent permitted significantly higher PC-3 cell viability rate when compared against an MMAEp IC50 positive control (Fig. 2E), suggesting that MMAEp was partially absorbed by the hydroxyapatite column.
Fig. 2.
Mechanisms of actions of MMAE and MMAEp. A and B: Representative immunofluorescent images of PC-3 and C4-2B cells treated with either DMSO control, 4 nM MMAE, or 96 nM MMAEp for 24 hours and stained for α-tubulin and Hoechst. Original magnification, large image: ×200; insets: ×400; C and D: Measured fluorescence intensity of α-tubulin staining signals. Data are presented as mean ± SD of 20 mitotic figures for each group. ** p<0.01 compared to control. E: Hydroxyapatite chromatography paired with cell viability assay to detect binding of MMAEp to calcium ions. Data are presented as the mean ± SD of a representative of three independently performed experiments. ** p<0.01 compared to MMAEp control.
MMAE and MMAEp induce G2/M cell cycle arrest and increase the sub-G1 population
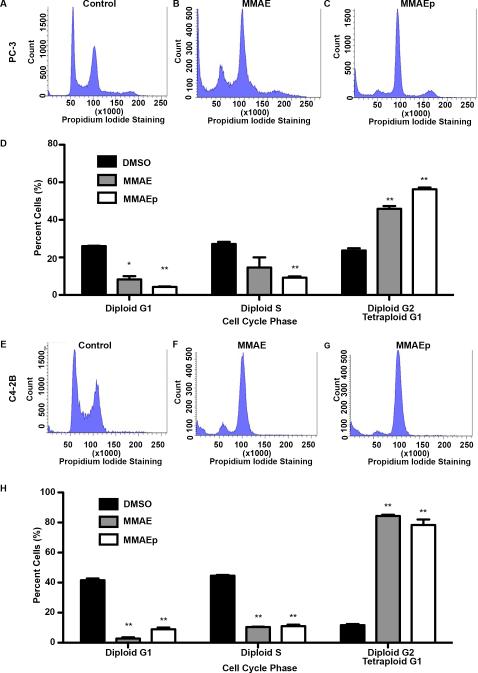
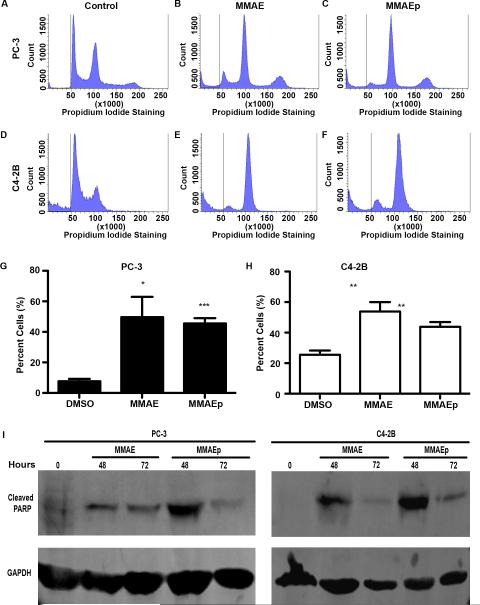
We next performed cell cycle analysis to assess whether or not MMAEp causes cell cycle arrest due to disruption of microtubule dynamics. We found that MMAEp stalled cell cycle progression at G2/M in PC-3 and C4-2B cells, in concordance with MMAE (Fig. 3A to 3H). Additionally, we analyzed the sub-G1 population for differences in apoptosis at the cellular level. We found that both MMAE and MMAEp significantly increased the sub-G1 population (Fig.4A to 4H). Western blot analysis demonstrated increased cleaved PARP expression, a caspase-dependent marker of apoptosis, after 48 and 72 hours of treatment (Fig. 4I).
Fig. 3.
MMAE and MMAEp cause cell cycle arrest at G2/M. A to C and E to G: Representative histograms of cell cycle analysis performed in PC-3 and C4-2B cell lines. Cells were treated for 24 hours with either DMSO control, 4 nM MMAE, or 96 nM MMAEp. D and H: Quantification of cell cycle analysis in PC-3 and C4-2B cell lines. Data are presented as representative mean ± SD of three independently performed experiments. * p<0.05, ** p<0.01 compared to DMSO control.
Fig. 4.
MMAE and MMAEp induce apoptosis. A to F: Representative histograms of sub-G1 (representing apoptotic cells) analysis performed on PC-3 and C4-2B cell lines. Cells were treated for 24 hours with either DMSO control, 4 nM MMAE, or 96 nM MMAEp. G and H: Quantification of sub-G1 populations in PC-3 and C4-2B cell lines. Data are presented as representative mean ± SD of three independently performed experiments. * p<0.05, ** p<0.01 compared to control. I: Western blot analysis of cleaved PARP (an apoptotic marker) in PC-3 and C4-2B cells. Cells were treated with 2 nM MMAE and 48 nM MMAEp at the indicated time points, respectively. Blots are presented as representative images of three independently performed experiments.
MMAEp significantly reduces tumor burden in a mouse model
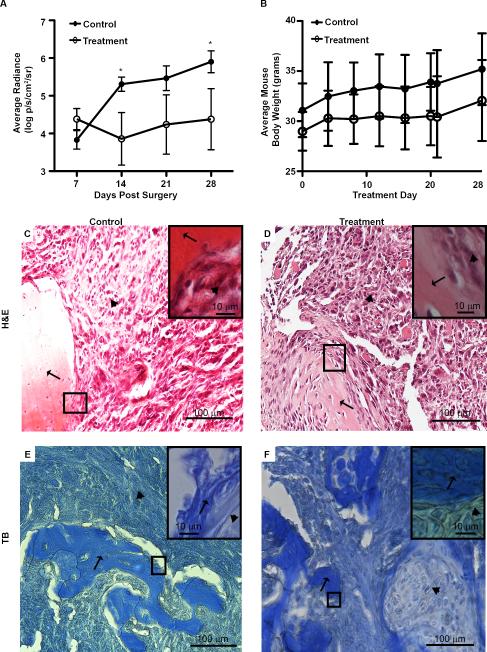
We implemented an intratibial cancer cell injection model to assess MMAEp's in vivo efficacy. Non-invasive in vivo imaging of tumor burden in mice showed that MMAEp was able to significantly reduce tumor growth after 14 and 28 days of treatment (Fig. 5A), without significantly affecting animal body weight, a surrogate for toxicity (Fig. 5B). We harvested bone samples after sacrifice and stained tumor tissue sections with H&E and toluidine blue to confirm tumor growth in the bone marrow space in both control and treatment mice (Fig. 5C to 5F).
Fig. 5.
In vivo efficacy of MMAEp. A: Bone tumor growth curves. Luminescence values are log transformed. Data are presented as mean ± SEM of n=10 mice per group. * p<0.05 compared to control. B: Average mouse body weight over the course of experiment. Data are presented as mean ± SD of n=10 mice per group. C to F: Representative hematoxylin and eosin staining and toluidine blue staining of mouse bone tumors. Black arrows indicate areas of the bone; arrowheads indicate areas of the tumor. H&E: hematoxylin and eosin; TB: toluidine blue.
DISCUSSION
MMAE is not able to be approved as a monotherapy due to its toxicity. We found that MMAE at 1 mg/kg body weight killed all mice (data not shown). MMAE's utility in the current study was solely as a positive control in revealing the mechanisms of drug action. Its utility as the cytotoxic component of the antibody conjugate complex has been well established [21]. Once the antibody portion of the conjugate binds to its cognate antigen on the cancer cell, the complex is internalized and MMAE is released via a cleavable linker peptide [22]. Because of the success of brentuximab vedotin (Adcetris®, Seattle Genetics, Bothell, WA), several new compounds are currently undergoing clinical trials against a range of cancer types [23-24]. In Francisco et al's initial characterization of brentuximab, then known as cAC10-vcMMAE, the published IC50 values of MMAE in several different lymphoma cell lines were in the mid-picomolar to low nanomolar range [21]. The apparent discrepancy in toxicity between Francisco et al's work and this study may be attributable to the fact that lymphoma cell lines are cultured in suspension, whereas PC-3 and C4-2B lines are adherent cells, deriving survival signaling from contact with the culture dishes.
Another study on MMAE conjugate was done by Buckel and colleagues [33]. They were able to establish that MMAE had high picomolar to low nanomolar IC50 values in HCT-116, PANC-1 and 779E cell lines. This is again in concordance with the toxicity seen in our present study. Buckel and colleague's work is also relevant as they used MMAE in a targeted manner, conjugating it to an activatable cell penetrating peptide containing several arginine-glycineglutamate domains. These domains, the cleavage substrates of matrix metalloproteinases, are digested in the tumor microenvironment, allowing the cell penetrating peptide to deliver MMAE to the cytosol of cancer cells and induce apoptosis.
Based on the data presented in the present study, both MMAE and MMAEp cause cell cycle arrest and induce apoptosis, similar to that demonstrated by Buckel and colleagues. Our initial in vitro findings demonstrated a 24-fold reduction in cytotoxicity of MMAEp in comparison to MMAE, most likely due to the negative charge of the phosphate group preventing trafficking across the cell membrane. We believe that this allows MMAEp to be better tolerated in vivo than MMAE that caused all treated animals to die after two doses at 1mg/kg body weight. More importantly, we found that MMAEp binds to calcium ions as evidenced by a reduction in cytotoxicity after exposure to hydroxyapatite crystals. This finding, coupled with our in vivo data, provides evidence to support our hypothesis that MMAEp binds to calcium ions found in bone matrix and prevents growth of bone metastasis. We believe that our findings support the contention that MMAEp is a unique modification of MMAE that utilizes this potent antimitotic in a targeted manner.
Although the present study provides some proof-of-principle evidence to support the potential usefulness of MMAEp, more studies are warranted to investigate the pharmacokinetics and pharmacodynamics. Trafficking of the drug across vascular and membranous barriers in living tissue represents the next challenge in establishing MMAEp's efficacy. A comparison of the effects (e.g., on cellular proliferation and apoptosis) of MMAEp in vivo in bone environment immediately following MMAEp administration is needed to validate the notion that MMAEp is more effective in bone targeting. Also pertinent to the study of MMAEp as a viable bone metastasis treatment is to determine whether or not this compound is toxic to proliferative cells found in bone marrow and bone tissue, such as lymphoid/myeloid precursors and osteoblasts. Alternatively, we will test our ideas to put more than one phosphate group to MMAE or to use other osteophilic compounds to conjugate with MMAE, in order to increase its bone-enriching capacity.
CONCLUSIONS
Our findings suggest that MMAEp induces cell cycle arrest and apoptosis. MMAEp retains the mechanism of action of MMAE by preventing microtubule polymerization and has the ability to bind to calcium ions. MMAEp is a bone-seeking compound that can inhibit tumor growth in a mouse intratibial xenograft model of human prostate cancer.
ACKNOWLEDGEMENTS
The authors thank Mary Price at The Louisiana Cancer Research Consortium FACS Core supported by National Institute of General Medical Sciences of the National Institutes of Health (Award Number P20GM103518).
Grant sponsor: National Institutes of Health; Grant numbers: R01CA174714, P20GM103518, and G12MD007595; Grant sponsor: Department of Defense; Grant numbers: W81XWH-14-1-0050, W81XWH-14-1-0149, W81XWH-14-1-0458, and W81XWH-15-1-0444; Grant sponsor: The Developmental Fund of Tulane Cancer Center (TCC) and Louisiana Cancer Research Consortium (LCRC) Fund; the content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Department of Defense.
Footnotes
Disclosure statement: The authors declare no conflict of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Sartor O, Reid RH, Hoskin PJ, Quick DP, Ell PJ, Coleman RE, Kotler JA, Freeman LM, Olivier P, Group QSS. Samarium-153-Lexidronam complex for treatment of painful bone metastases in hormone-refractory prostate cancer. Urology. 2004;63(5):940–945. doi: 10.1016/j.urology.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 3.Coleman RE. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res. 2006;12(20 Pt 2):6243s–6249s. doi: 10.1158/1078-0432.CCR-06-0931. [DOI] [PubMed] [Google Scholar]
- 4.Crawford ED, Higano CS, Shore ND, Hussain M, Petrylak DP. Treating Patients with Metastatic Castration Resistant Prostate Cancer: A Comprehensive Review of Available Therapies. J Urol. 2015 doi: 10.1016/j.juro.2015.06.106. [DOI] [PubMed] [Google Scholar]
- 5.Fitzpatrick JM, de Wit R. Taxane mechanisms of action: potential implications for treatment sequencing in metastatic castration-resistant prostate cancer. Eur Urol. 2014;65(6):1198–1204. doi: 10.1016/j.eururo.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 6.Doyle-Lindrud S. Managing side effects of the novel taxane cabazitaxel in castrate-resistant prostate cancer. Clin J Oncol Nurs. 2012;16(3):286–291. doi: 10.1188/12.CJON.286-291. [DOI] [PubMed] [Google Scholar]
- 7.Patel NK, Finianos A, Whitaker KD, Aragon-Ching JB. Advanced prostate cancer - patient survival and potential impact of enzalutamide and other emerging therapies. Ther Clin Risk Manag. 2014;10:651–664. doi: 10.2147/TCRM.S57509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zobniw CM, Causebrook A, Fong MK. Clinical use of abiraterone in the treatment of metastatic castration-resistant prostate cancer. Res Rep Urol. 2014;6:97–105. doi: 10.2147/RRU.S29003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santer FR, Erb HH, McNeill RV. Therapy escape mechanisms in the malignant prostate. Semin Cancer Biol. 2015;35:133–144. doi: 10.1016/j.semcancer.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Parker C, Nilsson S, Heinrich D, Helle SI, O'Sullivan JM, Fosså SD, Chodacki A, Wiechno P, Logue J, Seke M, Widmark A, Johannessen DC, Hoskin P, Bottomley D, James ND, Solberg A, Syndikus I, Kliment J, Wedel S, Boehmer S, Dall'Oglio M, Franzén L, Coleman R, Vogelzang NJ, O'Bryan-Tear CG, Staudacher K, Garcia-Vargas J, Shan M, Bruland Ø , Sartor O, Investigators A Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369(3):213–223. doi: 10.1056/NEJMoa1213755. [DOI] [PubMed] [Google Scholar]
- 11.Friedell H, Storaasli J. The use of radioactive phosphorus in the treatment of carcinoma of the breast and widespread metastases to the bone. American Journal of Roentgenology and Radium Therapy. 1950;64:559–575. [PubMed] [Google Scholar]
- 12.Maxfield J, Maxfield J, Maxfield W. The use of radioactive phosphorus and testosterone in metastatic bone lesions from breast and prostate. Southern Medical Journal. 1958;51:320–328. doi: 10.1097/00007611-195803000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Rubini G, Nicoletti A, Rubini D, Asabella AN. Radiometabolic treatment of bone-metastasizing cancer: from 186rhenium to 223radium. Cancer Biother Radiopharm. 2014;29(1):1–11. doi: 10.1089/cbr.2013.1549. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson S, Larsen RH, Fosså SD, Balteskard L, Borch KW, Westlin JE, Salberg G, Bruland OS. First clinical experience with alpha-emitting radium-223 in the treatment of skeletal metastases. Clin Cancer Res. 2005;11(12):4451–4459. doi: 10.1158/1078-0432.CCR-04-2244. [DOI] [PubMed] [Google Scholar]
- 15.Dan TD, Eldredge-Hindy HB, Hoffman-Censits J, Lin J, Kelly WK, Gomella LG, Lallas CD, Trabulsi EJ, Hurwitz MD, Dicker AP, Den RB. Hematologic Toxicity of Concurrent Administration of Radium-223 and Next-generation Antiandrogen Therapies. Am J Clin Oncol. 2015;00:000–000. doi: 10.1097/COC.0000000000000181. DOI: 10.1097/COC0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake MT, Clarke BL, Khosla S. Bisphosphonates: mechanism of action and role in clinical practice. Mayo Clin Proc. 2008;83(9):1032–1045. doi: 10.4065/83.9.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tricarico PM, Girardelli M, Kleiner G, Knowles A, Valencic E, Crovella S, Marcuzzi A. Alendronate, a double-edged sword acting in the mevalonate pathway. Mol Med Rep. 2015;12(3):4238–4242. doi: 10.3892/mmr.2015.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidle UH, Birzele F, Kollmorgen G, Rüger R. Molecular Mechanisms of Bone Metastasis. Cancer Genomics Proteomics. 2016;13(1):1–12. [PubMed] [Google Scholar]
- 19.Bai R, Pettit GR, Hamel E. Dolastatin 10, a powerful cytostatic peptide derived from a marine animal. Inhibition of tubulin polymerization mediated through the vinca alkaloid binding domain. Biochem Pharmacol. 1990;39(12):1941–1949. doi: 10.1016/0006-2952(90)90613-p. [DOI] [PubMed] [Google Scholar]
- 20.Pettit GR, Srirangam JK, Barkoczy J, Williams MD, Durkin KP, Boyd MR, Bai R, Hamel E, Schmidt JM, Chapuis JC. Antineoplastic agents 337. Synthesis of dolastatin 10 structural modifications. Anticancer Drug Des. 1995;10(7):529–544. [PubMed] [Google Scholar]
- 21.Francisco JA, Cerveny CG, Meyer DL, Mixan BJ, Klussman K, Chace DF, Rejniak SX, Gordon KA, DeBlanc R, Toki BE, Law CL, Doronina SO, Siegall CB, Senter PD, Wahl AF. cAC10-vcMMAE, an anti-CD30-monomethyl auristatin E conjugate with potent and selective antitumor activity. Blood. 2003;102(4):1458–1465. doi: 10.1182/blood-2003-01-0039. [DOI] [PubMed] [Google Scholar]
- 22.Furtado M, Rule S. Emerging Pharmacotherapy for Relapsed or Refractory Hodgkin's Lymphoma: Focus on Brentuximab Vedotin. Clin Med Insights Oncol. 2012;6:31–39. doi: 10.4137/CMO.S6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yardley DA, Weaver R, Melisko ME, Saleh MN, Arena FP, Forero A, Cigler T, Stopeck A, Citrin D, Oliff I, Bechhold R, Loutfi R, Garcia AA, Cruickshank S, Crowley E, Green J, Hawthorne T, Yellin MJ, Davis TA, Vahdat LT. EMERGE: A Randomized Phase II Study of the Antibody-Drug Conjugate Glembatumumab Vedotin in Advanced Glycoprotein NMB-Expressing Breast Cancer. J Clin Oncol. 2015;33(14):1609–1619. doi: 10.1200/JCO.2014.56.2959. [DOI] [PubMed] [Google Scholar]
- 24.Burris HA, Gordon MS, Gerber DE, Spigel DR, Mendelson DS, Schiller JH, Wang Y, Choi Y, Kahn RS, Wood K, Maslyar DJ, Infante JR. In: A phase I study of DNIB0600A, an antibody-drug conjugate (ADC) targeting NaPi2b, in patients (pts) with non-small cell lung cancer (NSCLC) or platinum-resistant ovarian cancer (OC) Cannistra SA, editor. American Society of Clinical Oncology; Chicago, IL: 2014. Supplemental Abstract 2504. [Google Scholar]
- 25.Portugal J, Mansilla S, Bataller M. Mechanisms of drug-induced mitotic catastrophe in cancer cells. Curr Pharm Des. 2010;16(1):69–78. doi: 10.2174/138161210789941801. [DOI] [PubMed] [Google Scholar]
- 26.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15(7):1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 27.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12(6):385–392. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 28.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, Pathak S, von Eschenbach AC, Chung LW. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54(10):2577–2581. [PubMed] [Google Scholar]
- 29.Parajuli KR, Zhang Q, Liu S, You Z. Aminomethylphosphonic acid inhibits growth and metastasis of human prostate cancer in an orthotopic xenograft mouse model. Oncotarget. 2016;7(9):10616–10626. doi: 10.18632/oncotarget.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27(8):1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratnayake JT, Mucalo M, Dias GJ. Substituted hydroxyapatites for bone regeneration: A review of current trends. J Biomed Mater Res B Appl Biomater. 2016;00:000–000. doi: 10.1002/jbm.b.33651. DOI: 10.1002/jbm.b.33651. [DOI] [PubMed] [Google Scholar]
- 33.Buckel L, Savariar EN, Crisp JL, Jones KA, Mier Hicks A, Scanderbeg DJ, Nguyen QT, Sicklick JK, Lowy AM, Tsien RY, Advani SJ. Tumor radiosensitization by monomethyl auristatin E: mechanism of action and targeted delivery. Cancer Res. 2015;75(7):1376–1387. doi: 10.1158/0008-5472.CAN-14-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]