Abstract
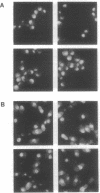
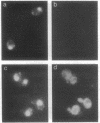
Cytochrome c with a nuclear localization signal added at the N terminus was mistargeted to the nucleus, resulting in a yeast strain deficient in mitochondrial cytochrome c. Reversion of this strain allowed the isolation of temperature-conditional mutants defective in nuclear transport, as demonstrated with one of these mutants, nip1-1, that was shown to be defective in nuclear accumulation of a LacZ protein containing a nuclear localization signal of the yeast ribosomal protein L29. The NIP1+ gene was cloned and shown to encode a 93,143-Da protein. Furthermore, an epitope-labeled NIP1 protein migrated in SDS/polyacrylamide gels with a mass of approximately 100,000 Da and was shown by immunofluorescence to localize mainly in the cytoplasm. NIP1+ was shown to be an essential gene by gene disruption experiments. Intriguingly, NIP1 has a serine-rich acidic N-terminal region that is similar in this regard to the N-terminal region of a previously described nuclear localization signal-binding protein, NSR1.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991 Sep 6;66(5):837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Baim S. B., Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988 Apr;8(4):1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer M., Goldfarb D. S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990 Mar 23;60(6):999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M. D., Mathews A. J., Nall B. T., Baim S. B., Eustice D. C., Sherman F. Differential stability of two apo-isocytochromes c in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990 Feb 15;265(5):2733–2739. [PubMed] [Google Scholar]
- Dumont M. E., Cardillo T. S., Hayes M. K., Sherman F. Role of cytochrome c heme lyase in mitochondrial import and accumulation of cytochrome c in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Nov;11(11):5487–5496. doi: 10.1128/mcb.11.11.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M. N. Nuclear protein localization. Biochim Biophys Acta. 1991 Mar 7;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Lee W. C., Mélèse T. Identification and characterization of a nuclear localization sequence-binding protein in yeast. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8808–8812. doi: 10.1073/pnas.86.22.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., Xue Z. X., Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991 Apr;113(1):1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moerschell R. P., Tsunasawa S., Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1988 Jan;85(2):524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Silver P. Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer D. D., Forbes D. J. Nuclear import can be separated into distinct steps in vitro: nuclear pore binding and translocation. Cell. 1988 Mar 11;52(5):641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- Reichelt R., Holzenburg A., Buhle E. L., Jr, Jarnik M., Engel A., Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990 Apr;110(4):883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. D., Mills A. D., Dilworth S. M., Laskey R. A., Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988 Mar 11;52(5):655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Russo P., Li W. Z., Hampsey D. M., Zaret K. S., Sherman F. Distinct cis-acting signals enhance 3' endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991 Mar;10(3):563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERMAN F., SLONIMSKI P. P. RESPIRATION-DEFICIENT MUTANTS OF YEAST. II. BIOCHEMISTRY. Biochim Biophys Acta. 1964 Jul 15;90:1–15. doi: 10.1016/0304-4165(64)90113-8. [DOI] [PubMed] [Google Scholar]
- Sadler I., Chiang A., Kurihara T., Rothblatt J., Way J., Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J Cell Biol. 1989 Dec;109(6 Pt 1):2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sherman F., Jackson M., Liebman S. W., Schweingruber A. M., Stewart J. W. A deletion map of cyc1 mutants and its correspondence to mutationally altered iso-1-cytochromes c of yeast. Genetics. 1975 Sep;81(1):51–73. doi: 10.1093/genetics/81.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A. How proteins enter the nucleus. Cell. 1991 Feb 8;64(3):489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- Underwood M. R., Fried H. M. Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J. 1990 Jan;9(1):91–99. doi: 10.1002/j.1460-2075.1990.tb08084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]