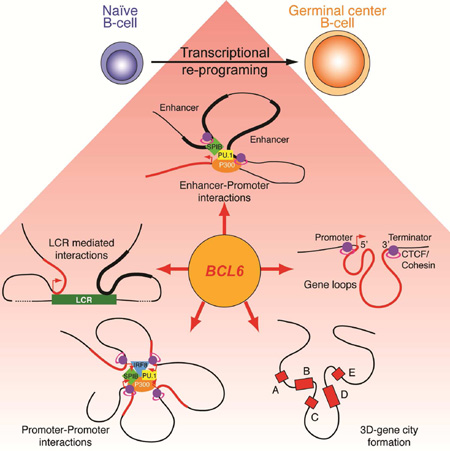
SUMMARY
During the humoral immune response B-cells undergo a dramatic change in phenotype to enable antibody affinity maturation in germinal centers (GCs). Using genome-wide chromosomal conformation capture (Hi-C), we found that GCB-cells undergo massive reorganization of the genomic architecture that encodes the GCB-cell transcriptome. Coordinate expression of genes that specify the GCB-cell phenotype – most prominently, BCL6 – was achieved through a multilayered chromatin reorganization process involving i) increased promoter connectivity, ii) formation of enhancer networks, iii) 5’ to 3’ gene looping, and iv) merging of gene neighborhoods that share active epigenetic marks. BCL6, was an anchor point for the formation of GC-specific gene and enhancer loops on chromosome 3. Deletion of a GC specific, highly interactive locus control region, upstream of Bcl6 abrogated GC formation in mice. Thus, large-scale and multi-tiered genomic three-dimensional reorganization is required for coordinate expression of phenotype-driving gene sets that determine the unique characteristics of GCB-cells.
Graphical Abstract
INTRODUCTION
Production of high affinity, antibody-secreting B-cells is essential for the establishment of humoral immunity (reviewed in (Victora and Nussenzweig, 2012)). During the immune response, T-cell-dependent antigen activation induces naïve B-cells to form germinal centers (GCs) within lymphoid follicles wherein they undergo rapid proliferation. At the same time, these cells endure somatic hypermutation, during which their immunoglobulin genes are progressively mutated. The end result of this process is the emergence of B-cell clones that express new, high-affinity antibodies against specific antigens. The dramatic transition in phenotype of naive B-cells to GCB-cells requires rapid and coordinate epigenetic regulation and expression of multiple genes regulating the cell cycle, DNA damage checkpoints, metabolic pathways, and is crucially dependent on the GC master transcriptional regulator, BCL6 (Hatzi and Menick, 2014). Precisely how the GCB-cell transcriptional program is coordinated efficiently at the genome-wide level is unknown.
The establishment of distinct cellular phenotypes during development and differentiation in multicellular organisms requires coordinated and large-scale changes in transcriptional programming (Cantone and Fisher, 2013; Natoli, 2010; Spitz and Furlong, 2012). Alterations in histone modifications are one example of mechanisms that regulate the transcriptional state of individual genes (Zhou et al., 2011). However, the genomes of higher organisms are large and highly complex, and in a single cell, the chromatin status and transcription of many thousands of genes must be altered simultaneously in a highly efficient and coordinated manner to enable phenotypic change. One way that the genome overcomes this complexity is by large-scale folding and looping. Recent studies using chromosome conformation capture (3C) techniques and three-dimensional (3D) DNA-fluorescence in situ hybridization (FISH) provide evidence that the genomes of higher organisms are physically organized and compartmentalized, and that the 3D folding of specific loci in terminally differentiated cells can help to control gene expression (Bickmore and van Steensel, 2013; Cavalli and Misteli, 2013; Fabre et al., 2015). Genome-wide 3C approaches have further shown that genome compartmentalization and folding are crucial to the way that genes are reprogrammed in Drosophila, and mammalian cells (Sexton et al., 2012, Dixon et al., 2012; Lieberman-Aiden et al., 2009; Sanyal et al., 2012; Dixon et al., 2015; Baù et al., 2011). Here we examined whether physical reorganization of the B-cell genome is linked to and required to enable the dramatic and coordinated changes in expression and epigenetic regulation of genes that drive the phenotypic transition of resting B-cells into GCB-cells.
RESULTS
Extensive reorganization of the genome of GCB-cells is linked to cell morphology and specific chromosomal territories
The GC reaction involves a profound phenotypic shift from resting, antigen-naïve B-cells to highly proliferative GCB-cells (Figure 1A) a process that requires large-scale transcriptional and epigenetic changes. To examine the three-dimensional organization of the B-cell genome during the GC reaction we performed genome-wide chromosomal conformation capture (Hi-C) in purified primary human tonsillar naïve B-cells (IgD+CD38lo; naive B) and GCB-cells (CD77+CD38hi; GCB) (Figure S1A and B, Table S1). Naïve B and GCB-cell Hi-C biological replicates showed high reproducibility (Spearman correlation 0.97 and 0.88 respectively; Figure S1C). Examination of intra-chromosome DNA:DNA interaction maps by principal component analysis (PCA) revealed a significant loss of interactions between the short (p) and long (q) arms of the longest chromosomes in GCB-cells (e.g. Figure 1B; p<10−300; Fisher’s exact test). This large scale remodeling of chromosomal arm contacts revealed the architectural basis of chromosomal de-compaction in GCB-cells, reflecting the increased size and chromatic laxity of GCB-cell nuclei, compared to the smaller and denser morphology of naïve B-cells (Figure S1D). Closer examination uncovered 135,156 chromatin interactions connecting 1 megabase (Mb) blocks of DNA that were significantly different between naïve B and GCB-cells, of which 84,684 interactions were gained in GCB-cells (FDR=0.05; BH corrected Fisher’s exact test; Figure S1E). For example, a region from chromosome 3, 3q27, which encodes the GC master regulator gene, BCL6, showed substantially higher numbers of chromatin interactions in GCB-cells compared to naïve B-cells (Figure 1C; p<0.005; binomial exact test).
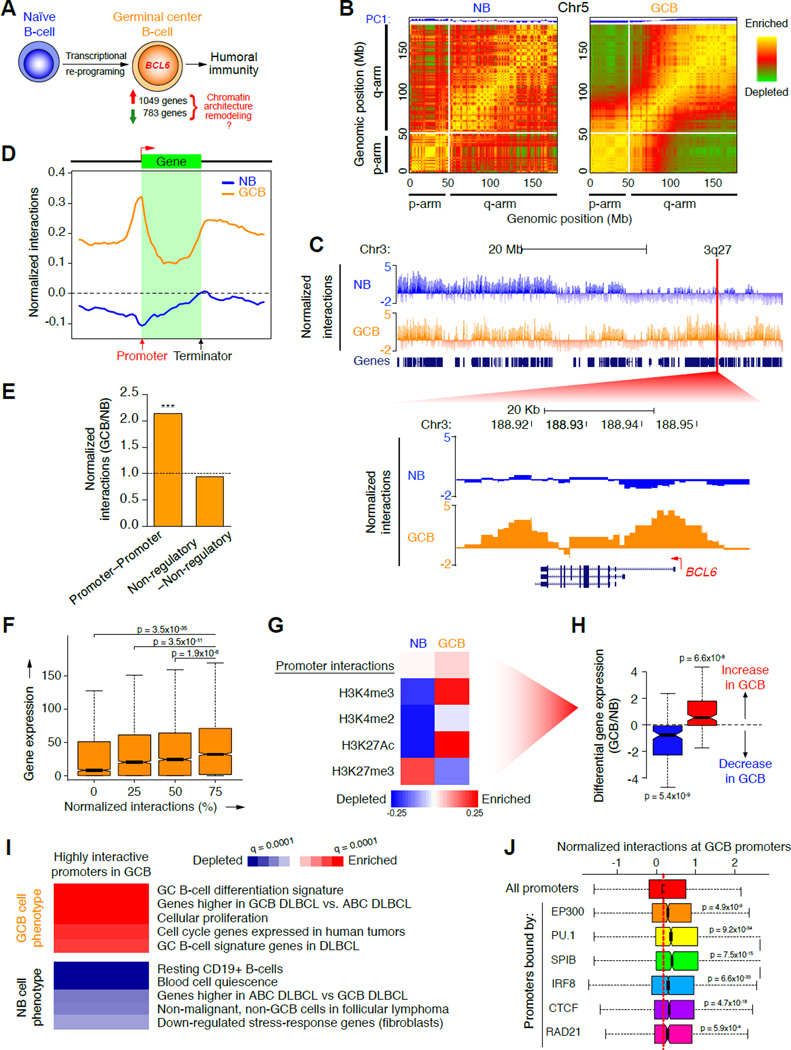
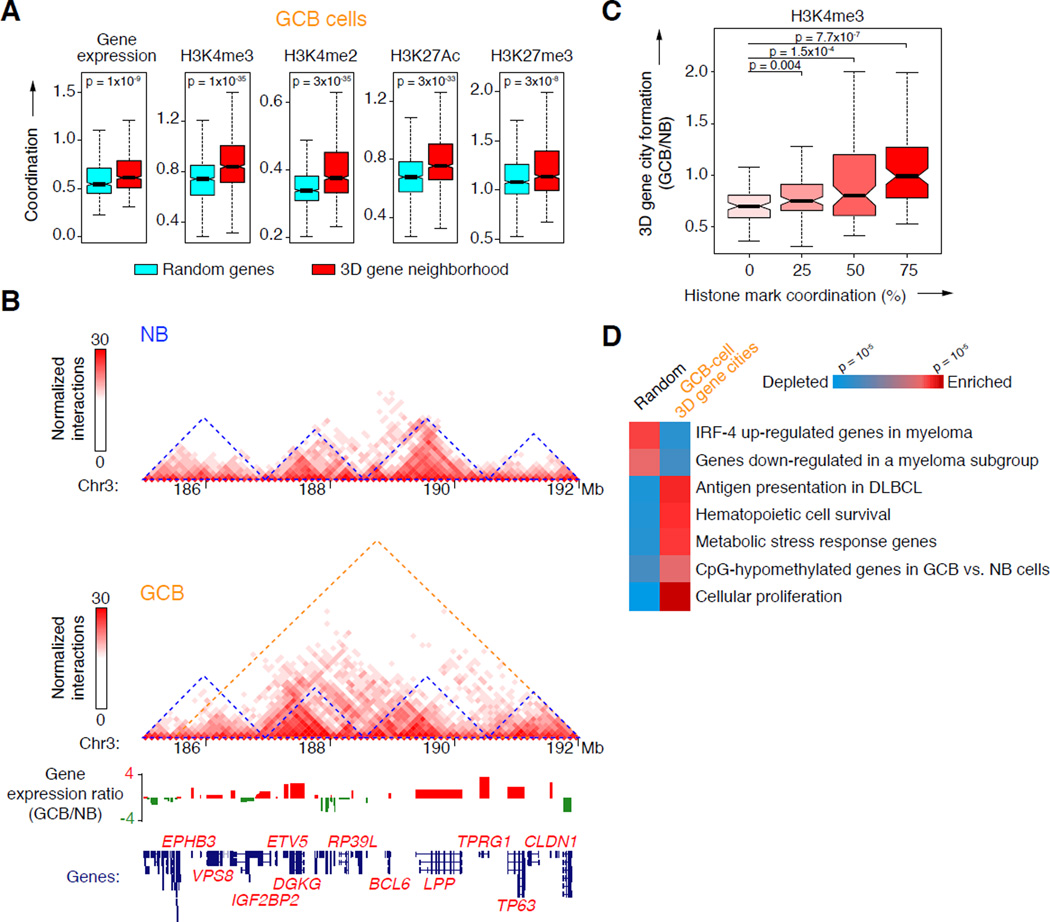
Figure 1. Topological shifts in GCB-cells involve chromosome unpacking and increased promoter interactivity.
A, Illustration of transcriptional changes during germinal center B-cell differentiation. 1,049 and 783 genes are up and down-regulated respectively, as determined by RNA-seq. B, Pearson correlation matrix plots of intra-chromosomal interactions across chromosome 5 at 1 Mb scale in NB and GCB-cells, showing PC1 and loss of interactions between the p (short) and q (long) arms of chromosomes in GCB-cells. The degree of correlation is indicated by yellow (enrichment), green (depletion) or red (no correlation). Centromere positions are indicated by the lack of DNA mappability (white). C, UCSC Genome Browser tracks showing normalized interaction frequencies, as measured by Hi-C, for naïve B and GCB-cells across a region of chromosome 3, with a zoomed in view of a 3q27 locus encoding BCL6. D, Profiles of the abundance of genome-wide interactions (normalized) across regions encoding genes (± 50 Kb) in naïve B and GCB-cells. E, Relative enrichment of promoter–promoter and non-regulatory–non-regulatory interactions, as a fraction of all interactions in GCB versus naïve B-cells. ***p<10−15, χ2 test. F, Correlation between gene expression (RPKM) and normalized interaction frequency percentiles (quartiles) in naïve B and GCB-cells. Median gene expression was compared between the 25% most interactive promoters and all other quartiles, using non-parametric Wilcoxon tests. G, Principal component analysis of promoter interactions (Hi-C) and histone modification peaks (ChIP-seq), in naïve and GCB-cells. Degree of correlation is indicated by the color key showing enrichment (red) and depletion (blue) and represents normalized read counts (for Hi-C and ChIP-seq data). Only genes in this principal component were differentially expressed as shown in: H, Differential gene expression (log2 ratio) of genes enriched (red) or depleted (blue) in the principal component promoter set shown in G. p-values were calculated using a threshold of >20% expression change between naïve B and GCB-cells. I, The heatmap represents GSEA of genes with highly interactive promoters in GCB-cells showing gene signatures that are positively or negatively enriched (FDR = 0.01). J, Correlations between normalized promoter interaction frequencies and enrichment of the listed regulatory factors as determined by ChIP-chip or ChIP-seq in GCB-cells (compared to all promoters). Significance was determined using Mann-Whitney’s test. n.s., non-significant. See also figure S1.
Induction of promoter interactivity is coordinated with activated chromatin and transcription in GCB-cells
We next performed an unbiased, gene-centered analysis of chromatin interactions in naïve B and GCB-cells. We discovered that DNA:DNA interactions involving gene promoters were highly enriched in GCB-cells, compared with naïve B-cells (p<10−16; Fisher’s exact test; Figure 1D) as was the fraction of Hi-C interactions corresponding to promoter–promoter interactions (p<10−15; χ2 test; Figure 1E. Interactive promoters were associated with increased transcription, as determined by RNA-seq in the same specimens (p<10−34; Wilcoxon test; Figure 1F). Promoter interactions were highly correlated with active chromatin marks, as determined by ChIP-seq for H3K4me3, H3K4me2, H3K27Ac in primary human naïve B and GCB-cells (p<10−47, <10−59, <10−39, respectively; Figure S1F). The most highly interactive promoters in GCB-cells were specifically enriched for genes linked to the GCB-cell transcriptional program. These included transcriptional regulators BCL6, MTA3, LMO2 and BCL7A, and components of the B-cell receptor signaling pathway, TEC, PIK3CG and CAMK2B (Figure S1G and H). Enrichment of GCB-cell phenotype-driving gene sets among highly interactive promoters in GCB-cells was confirmed using gene set enrichment analysis (GSEA, FDR=0.10; Figure S1I).
To further explore how gene promoter interactions and epigenetic marks might be linked to the GCB transcriptional program, we performed an unbiased, multidimensional principal component analysis (PCA). This approach identified a set of highly interactive gene promoters in GCB-cells (principal component 1) with increased H3K4me3 and H3K27Ac activating marks in GCB-cells and reduced H3K27me3 (Figure 1G). These promoters showed diametrically opposite features in naïve B-cells: low numbers of interactions, depletion of active and enrichment of repressive chromatin (Figure 1G). Only this specific combination of promoter interactions and epigenetic marks was associated with significant gene upregulation in GCB vs. naïve B-cells (p<10−7; Figure 1H and data not shown). These promoters were enriched in gene sets corresponding to the GCB-cells, GCB-type diffuse large B-cell lymphomas (DLBCLs), cellular proliferation and cell cycle genes (FDR=0.001), and were depleted in gene sets linked with resting B-cells and non-GCB DLBCLs (FDR=0.01; Figure 1I). Hence, differential expression of GC phenotype-driving genes requires an increase in both promoter interactions and active epigenetic marks.
Integrating Hi-C and ChIP-seq we observed that the most highly interactive promoters in GCB-cells but not naïve B-cells (Figure 1J and S1J) were strongly linked to binding by the EP300 histone acetyl-transferase (p<10−8), as well as transcription factors, PU.1 and SPIB (p<10−33 and <10−14, respectively; Mann-Whitney’s test). These factors play important regulatory roles in GCs (Klein and Dalla-Favera, 2008) and interact with EP300 (Bai et al., 2005; Yamamoto et al., 2002). IRF8, which plays an important role in GC formation (Lee et al., 2006), was associated with promoters engaged in interactions in both GCB and naïve B-cells (p<10−32 and <10−105 respectively; Figure 1J and S1J). CTCF and RAD21 (cohesin), which are linked to formation of cis-interacting chromatin (Merkenschlager and Odom, 2013), were significantly associated with highly interactive promoters in GCB-cells (p<10−17 and <10−3, respectively; Figure 1J)
Spatial reorganization and coordination of a GCB-cell specific enhancer network
Cell-type specific gene expression patterns are governed in large part through tissue specific gene enhancer elements, which contact and positively regulate distally located gene promoters (Chepelev et al., 2012; Sanyal et al., 2012; Shen et al., 2012). Using ChIP-seq, each cell type was observed to have 20,173 and 21,807 H3K4me2posH3K4me3neg enhancer elements, respectively. Naïve B-cells contained 5,167 enhancers <50 Kb away from genes; of which 2,818 were unique to naïve B-cells and linked to genes involved in immune homeostasis and lymph nodes (p<10−2; Figure 2A and B). GCB-cells contained 5,339 enhancers <50 Kb from genes, among which 2,990 were unique to GCB-cells and enriched for GCB-cell differentiation, proliferation and GCB-type DLBCL genes, including BCL6, CKS2, MCM2 and 5, RFC3, SPRED2, PRC1 and TIAM2 (p<10−2; Figure 2A and B). Genes associated with active enhancers (H3K4me2posH3K27Acpos) in GCB-cells manifested higher expression than genes associated with poised enhancers (p<10−90; Mann-Whitney test; Figure 2C). Enhancers were significantly more interactive in GCB compared to naïve B-cells (p<10−300; Figure S2A), with active GCB-cell enhancers being significantly more interactive than poised GCB-cell enhancers (p=2.1×10−32, Mann Whitney’s test; Figure 2D). Likewise, enhancers bound by PU1, SPIB, and EP300 (which mediates H3K27 acetylation) were more highly interactive with other loci in GCB-cells (p<10−3, <10−4, and <10−7; respectively, Mann Whitney’s test; Figure 2E). CTCF and RAD21 were also enriched at highly interactive enhancers in GCB-cells (p<10−2 and <10−3, respectively; Figure 2E). The fraction of Hi-C interactions corresponding to enhancer–promoter and enhancer–enhancer interactions was highly increased in GCB-cells versus naïve B-cells (p<10−15; χ2 test; Figure 2F). In contrast interactions between non-regulatory loci were not enriched in GCB-cells, suggesting that interaction of DNA that encodes regulatory information is most relevant to the re-programming of these cells.
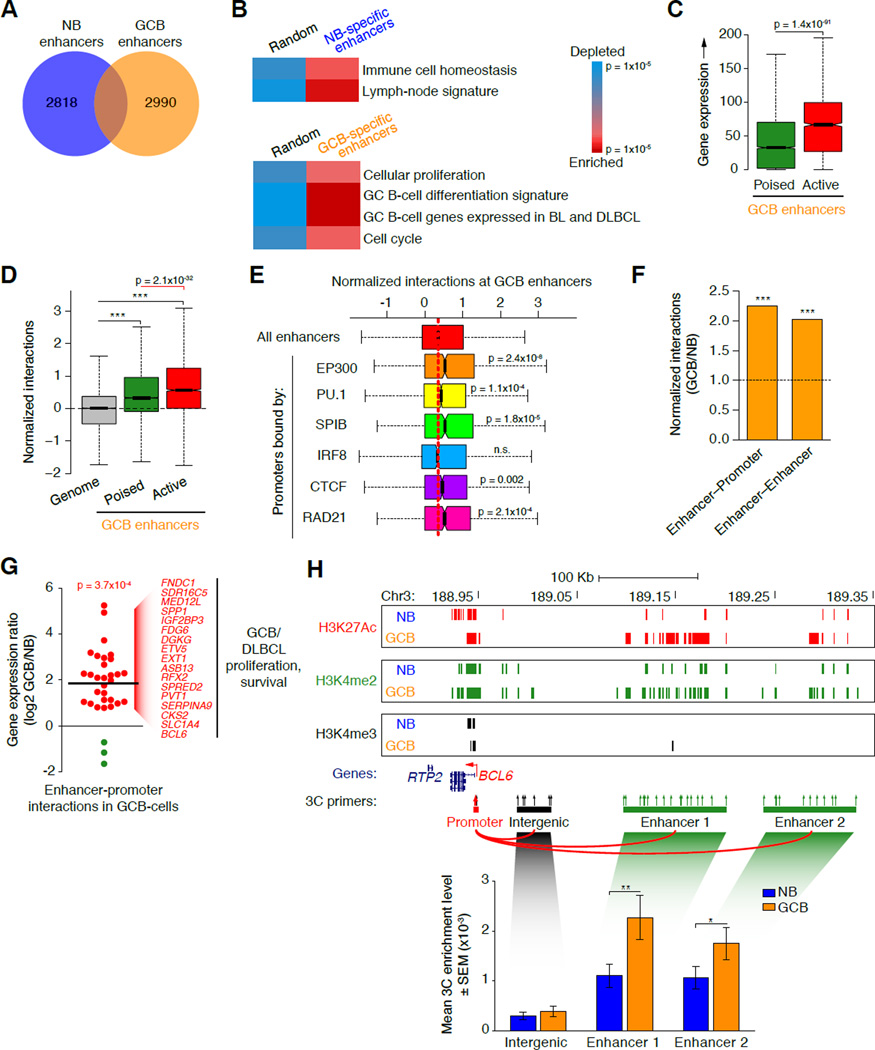
Figure 2. GCB-cells manifest widespread reprogramming of enhancer interactions.
A, Venn diagram showing numbers of unique and overlapping naïve B and GCB-cell specific enhancer regions (H3K4me2pos H3K4me3neg). B, Relative enrichment and depletion for genes <50 Kb away from naïve B and GCB-cell-specific enhancers (versus a random set of genes), showing enrichment of naïve B and GCB-cell signature genes, respectively. C, Expression (RPKM) of genes <50 Kb away from poised (H3K27Acneg H3K4me2pos H3K4me3neg) versus active (H3K27Acpos H3K4me2pos H3K4me3neg) GCB-specific enhancers. D, Normalized interaction frequencies (as measured by Hi-C) at poised versus active GCB-specific enhancers versus the whole genome. E, Correlations between normalized interaction frequencies (measured by HiC) and factor binding, as determined by ChIP-seq, at GCB-cell specific enhancers (compared to all enhancers). Significance was determined using Mann-Whitney’s test. n.s., non-significant. F, Relative enrichment of enhancer–promoter and enhancer–enhancer interactions, as a fraction of all interactions in GCB versus naïve B-cells. ***p<10−15, χ2 test. G, Log2 normalized ratio of gene expression in GCB versus naïve B-cells for 32 genes with highest enhancer-promoter interactivity in GCB-cells (FDR = 0.05, Fisher’s exact test with BH correction). Genes up or downregulated in GCB-cells are colored red or green, respectively. Line indicates the average gene expression ratio for the 32 genes H, UCSC Genome Browser tracks in naïve B and GCB-cells showing chromatin marks and locations of 3C primers (arrows) across a region upstream of BCL6. The red arrow denotes the anchor point for all 3C PCRs. Bar graphs show the mean 3C enrichment in naïve B and GCB-cell, 3C DNA templates (n=3) at the regions indicated. Primer pairs were averaged across each region. Asterisk indicates significance using a one-tailed, unpaired t-test (**: p<0.05; *: p<0.1). See also figure S2.
We next identified genes with substantial increase in interactions between their promoter and distal enhancer in GCB-cells (FDR=0.05; Fisher’s exact test with BH correction; Figure S2B) and differential expression. We identified 32 such genes; the majority of which (90%) were up-regulated in GCB-cells (p<10−3; Fisher’s exact test; Figure 2G) and corresponded to important GCB-cell regulators such as BCL6, CKS2, DGKG, ETV5, PVT1, SERPINA9, and SPP1. We validated GC-specific enhancer–promoter looping using the 3C assay at the BCL6 and CKS2 loci (Figure 2H and Figure S2C), which have enhancers located ~100–380 Kb away from their transcription start sites. Using their proximal promoter regions as anchors, we confirmed that interactions were significantly higher at distal enhancer regions (H3K4me2posH3K4me3neg), compared with nearby (<76 Kb) inter- and intragenic regions either lacking or with low-density enhancer marks (Figure 2H and Figure S2C). Establishment of the GC-specific transcriptional program is therefore linked to enhancer reorganization and connectivity, including direct looping to proximal promoters of critical GC genes such as BCL6.
GC genes feature gene looping linking promoters and downstream regions
Examination of interactions at gene regions showed enrichment not only at promoters, but also around the 3’ end of gene bodies in GCB-cells (Figure 1C), suggesting the potential for 5’ to 3’ gene loop formation. Accordingly we identified 70 individual genes with significantly increased 5’ to 3’ looping in GCB versus naïve B-cells (FDR=0.05; Fisher’s exact test with BH correction; Figure S3A). Notably, genes that showed looping had increased expression (≥1.5-fold) in GCB-cells (p<0.001; Fisher’s exact test; Figure 3A) and were linked to GCB-cell proliferation, cell cycle, stress responses and/or DLBCL survival (FGFR1, ECT2, RICH2, SGPP2, TIAM2, EPHA4, BCL6; Figure 3A and Figure S3A). We confirmed these observations using 3C with an anchor upstream of the BCL6 promoter (Upstream; Figure 3B), demonstrating increased interactions with three regions immediately downstream of the BCL6 transcript end in GCB-cells. In contrast, there were no differences in 3C enrichment at control regions within the BCL6 gene body (Figure 3B). Genes with increased 5’ to 3’ looping also exhibited significantly greater binding of CTCF and RAD21 to their up- and downstream regions, compared to genes without such interactions (p<0.05 and <0.01 for CTCF and RAD21, respectively; Fisher’s exact test; Figure 3C). CTCF binding, with or without RAD21, was evident by ChIP-seq at looped genes (Figure 3D). There was also enrichment of H3K4me3, H3K4me2 and H3K27Ac (p<0.05 for all marks; Fisher’s exact test) but not H3K27me3 (p=0.27) at the downstream regions of looped genes (Figure S3B), suggesting that activated chromatin is a marker for genes that engage in 5’ to 3’ looping. 5’ to 3’ gene looping, potentially mediated by CTCF and/or RAD21, represents a novel layer of mammalian gene reorganization that may facilitate transcription of GCB-cell-specific genes. We speculate that this could occur through efficient recycling of RNA polymerase II (Shandilya and Roberts, 2012), since, using ChIP, we detected RNA polymerase II binding to the 5’ and 3’ ends of looped genes, including BCL6 (Figure S3C).
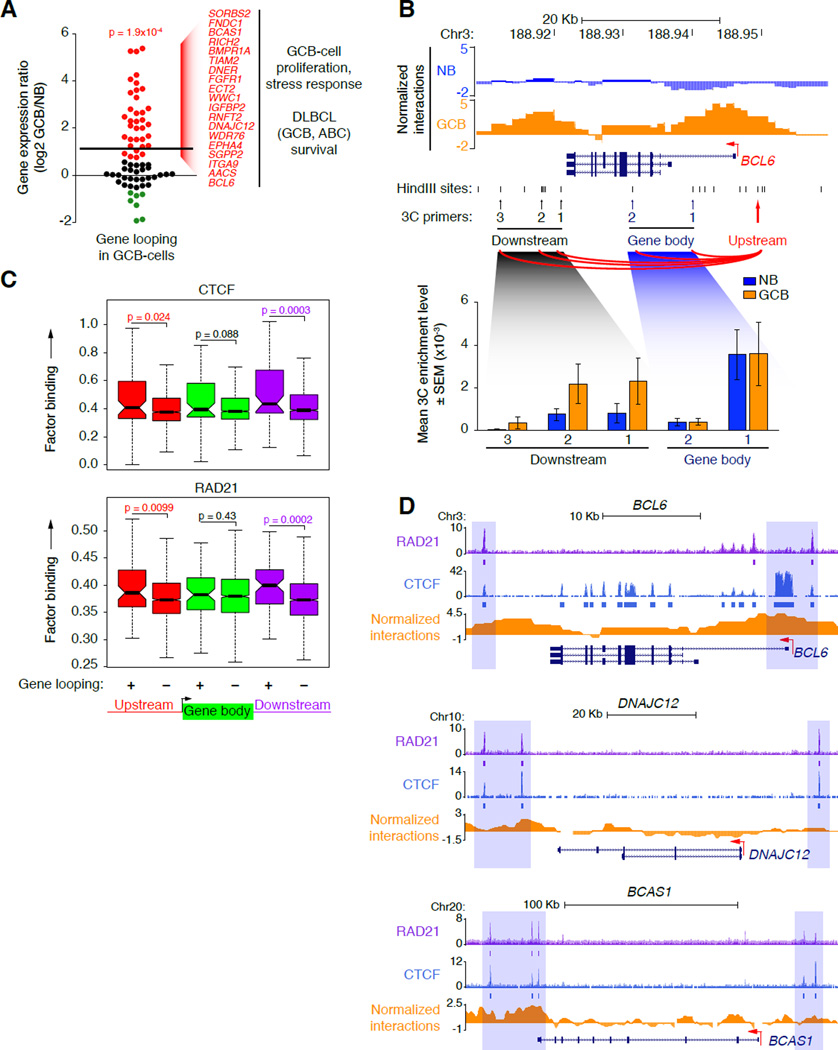
Figure 3. GCB-cell genes form gene loops in concert with CTCF and cohesin binding.
A, Log2 normalized ratio of gene expression in GCB versus naïve B-cells for 70 genes with increased 5’ to 3’ gene looping in GCB-cells (FDR = 0.05, Fisher’s exact test with BH correction). Genes up or downregulated in GCB-cells are colored red or green, respectively. The upper line indicates the average gene expression ratio of the 70 genes. B, UCSC Genome Browser tracks in naïve B and GCB-cells showing normalized interaction frequency, HindIII sites and locations of 3C primers (arrows) across the BCL6 gene region. The red arrow denotes the upstream anchor point. Bar graphs show the mean 3C enrichment in naïve B and GCB-cell 3C DNA templates (n=3) at the locations indicated. C, Average binding frequency of CTCF and RAD21 as determined by ChIP-seq, within the gene body or upstream or downstream regions (± 50 Kb) of genes with increased or decreased gene looping in GCB-cells (+ and −, respectively). Significance was determined using Mann-Whitney’s test. D, UCSC Genome Browser tracks showing normalized Hi-C interactions and locations of CTCF and RAD21 binding (normalized read counts) at representative genes with increased 5’ to 3’ looping in GCB-cells. ChIP-seeqer-defined peaks (ChIP enrichment over input) at these 5’ to 3’ sites are indicated by boxes below the tracks of normalized read counts and are highlighted by blue shading. See also Figure S3.
Remodeling of gene neighborhoods enables coordinated expression of GC-specific gene sets
Groups of genes are separated into boundary-delimited “neighborhoods” that are highly conserved between cell types and mammalian species (Dixon et al., 2012; Vietri Rudan et al., 2015). Using the Hi-C data and a Hidden Markov Model to infer the domain structure of chromosomes down to 100 Kb resolution (Dixon et al, 2012), we identified many cohorts of genes organized into discrete “3D gene neighborhoods” (n=1,525 and 1,587 in naïve B and GCB-cells respectively; 1.35 Mb average size), the majority of which (76.4%) were conserved between naïve B and GCB-cells and enriched for CTCF and cohesin binding at their boundaries (p<10−9; Mann Whitney’s test; Figure S4A). By definition, genes within these 3D neighborhoods displayed abundant interactions with each other. Expression of genes residing within 3D gene neighborhoods was more coordinated (i.e. similarly expressed) in both naïve B and GCB-cells, compared to randomly positioned sets of genes (p=0.001 and <10−9, respectively; Fisher’s exact test; Figure 4A and Figure S4B). Gene promoters within 3D neighborhoods showed greater epigenetic coordination in their enrichment of active (H3K4me3, H3K4me2, H3K27Ac) or repressive (H3K27me3) chromatin marks (p<10−5; Fisher’s exact test; Figure 4A and Figure S4B). 3D gene neighborhoods in B-cells thus correspond to spatially determined regulatory units where genes tend to share regulatory and epigenetic information (Figure S4C).
Figure 4. Altered 3D gene neighborhoods coordinate changes in chromatin and gene expression in GCB-cells.
A, Degree of coordination of gene expression as detected by RNA-seq or histone modifications by ChIP-seq within 3D gene neighborhoods (red) or among randomly selected genes (blue) in GCB-cells. B, Two-dimensional heat maps of normalized interaction frequencies in naïve B and GCB-cells and log2 gene expression ratio (GCB/naïve B) across a chromosome 3 region. 3D gene neighborhoods in naïve B and GCB-cells are indicated (blue and orange triangles, respectively). Gene expression ratios between GCB vs. naïve B-cells determined by RNA-seq is shown below, along with location of each gene. C, Ratio of 3D gene city formation versus H3K4me3 coordination in GCB versus naïve B-cells. Significance was determined using Mann-Whitney’s test. D, Gene set enrichment and depletion among genes located within newly formed 3D gene cities in GCB-cells (>80% increase in interaction frequency) versus a random set of genes. The color scale indicates statistical significance of enrichment and depletion. See also Figure S4.
We observed many instances (n=171) where adjacent 3D gene neighborhoods in naïve B-cells merged together to form larger, de novo 3D gene “cities” in GCB-cells. This is exemplified by a 6 Mb region on chromosome 3, comprised of four individual 3D gene neighborhoods that were physically partitioned in naïve B-cells, but merge together in GCB-cells (Figure 4B). The genes newly connected to one another across former neighborhood boundaries included genes specifically up-regulated in GCB-cells, share common chromatin marks and play important roles in GCB-cell specification, such as BCL6, LPP, DGKG, TP63 and ETV5 (Figure 4B). At the chromosome-wide level, the formation of 3D gene cities in GCB-cells was accompanied by the de novo acquisition of coordinated gene expression between groups of genes that were formerly isolated from each other in naïve B-cells (p=0.009; Spearman correlation). This was also true of epigenetic marks, such as H3K4me3 (p<10−6; Mann-Whitney’s test; Figure 4C). Furthermore, genes within these newly formed 3D gene cities were significantly enriched for GCB-cell phenotype-driving processes, including proliferation, MHC class II-mediated antigen presentation and metabolic stress, and have hypomethylated DNA at their promoter regions in GCB versus naïve B-cells (Shaknovich et al., 2011) (p<10−2; >80% increase in interactions; Figure 4D). 3D gene neighborhood expansion in GCB-cells is thus a chromosome-wide phenomenon that enables regulatory coordination between genes that were formerly isolated and disparately regulated in naïve B-cells, and was consistent with the observed increase in connectivity between promoters and enhancers in GCB-cells. At the macro scale, this was reflected by a genome-wide expansion of chromosomal territories (Hi-C compartments) associated with active chromatin in GCB-cells (e.g. Chr3, including the BCL6 gene; Figure S4D; p<10−10; Mann-Whitney’s test). Indeed, 7.4% of all RefSeq genes shifted from a repressive chromatin compartment in naïve B-cells to an active compartment in GCB-cells (compared with 2.3% of genes that shifted to a repressive compartment in GCB-cells). Notably, these genes play key roles in cell proliferation, cell cycle regulation and chromosome organization, including, PCNA, MCM4, E2F7, CENPN, and CTCFL, and have up-regulated expression in GCB-cells. Hence, remodeling of gene neighborhoods may enable more efficient coordination between sets of genes that play key roles in mediating the GCB-phenotype.
BCL6, the GC master regulator, is the centerpiece of a GCB-specific promoter-enhancer network
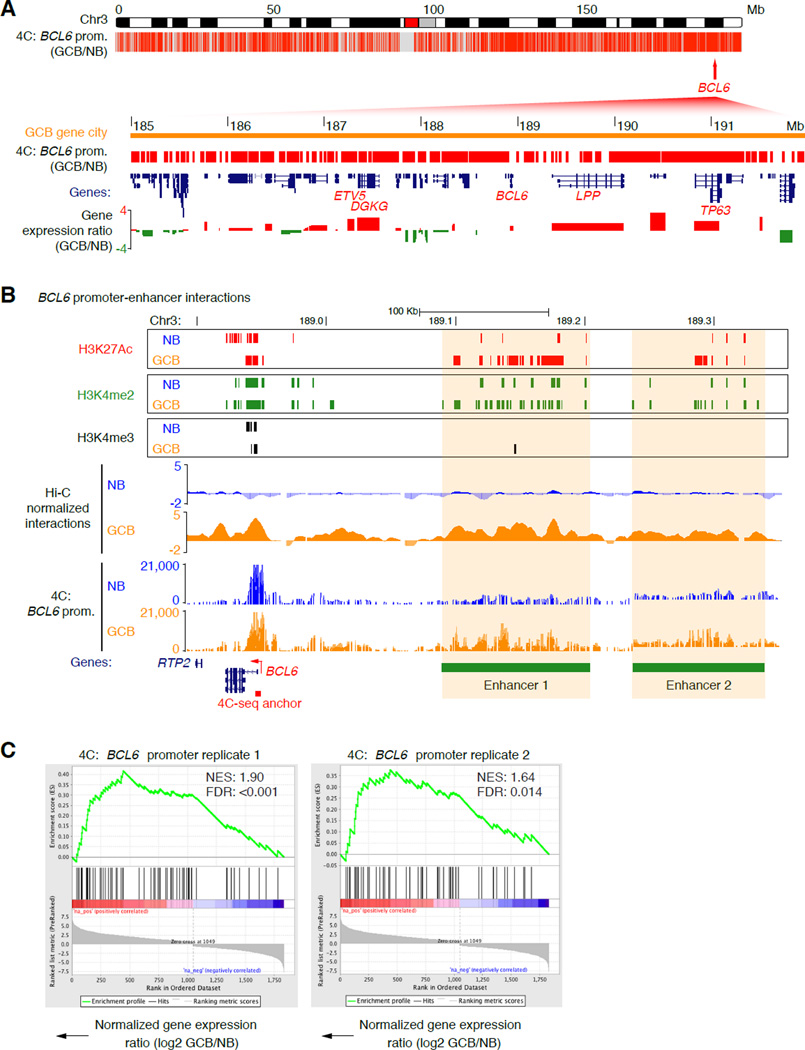
The BCL6 gene was subject to multiple layers of physical reorganization, suggesting that it might play a key role in organizing the topology of the GCB-cell genome. To explore the BCL6 gene interactome in more detail, we employed a targeted 4C-seq approach in naïve B and GCB-cells. First, using 4C bait primers specific for the BCL6 gene promoter region we uncovered ~3,000 intra-chromosomal interactions that were significantly gained in GCB compared with naïve B-cells (p<0.05, Fisher’s exact test with Bonferroni correction; Table S2), the majority of which localized to the GCB-specific 3D gene city identified in Figure 4B (Figure 5A). Enriched BCL6 promoter contacts included interactions with two distal enhancer regions, and between the 5’ and 3’ ends of the BCL6 gene (Figure 5B and S5A). Using GSEA, we discovered significant association between genes that gained contacts with the BCL6 promoter in GCB-cells on chromosome 3 and their up-regulated expression in GCB-cells (FDR<0.015 for both replicates; Figure 5C). The BCL6 interactome on chromosome 3 was also significantly enriched with genes that specify the GCB-cell phenotype and survival of GCB-DLBCLs (p=0.0014), including GCET2, LPP, CADPS, MME, PRKCD, CD80 and MED12L (Figure S5B).
Figure 5. The BCL6 promoter interacts extensively with other GC expressed genes.
A, Plot showing locations of BCL6 promoter contacts made in GCB-cells (versus NB-cells), as detected by 4C-seq across chromosome 3 (p<0.05, Fisher’s exact test). (Below) Zoomed-in view of the GCB-cell 3D gene city showing BCL6 gene contacts, locations of genes, histone modifications and log2 gene expression ratio (GCB/NB). GCB-cell-specific genes are highlighted. B, Genome visualization tracks showing H3K4me3 H3K4me2 and H3K27Ac histone modification peaks, normalized Hi-C interaction frequencies (merged), and normalized read counts of 4C contacts made with the BCL6 gene promoter in NB and GCB-cells (representative of 2 independent experiments) across the BCL6 gene region on chromosome 3. Increased interactions between the BCL6 promoter and two upstream enhancer elements (1 and 2) are evident in GCB-cells (highlighted). C, GSEA showing a positive correlation between up-regulated gene expression and genes in contact with the BCL6 promoter in GCB-cells. NES = normalized enrichment score. See also Figure S5.
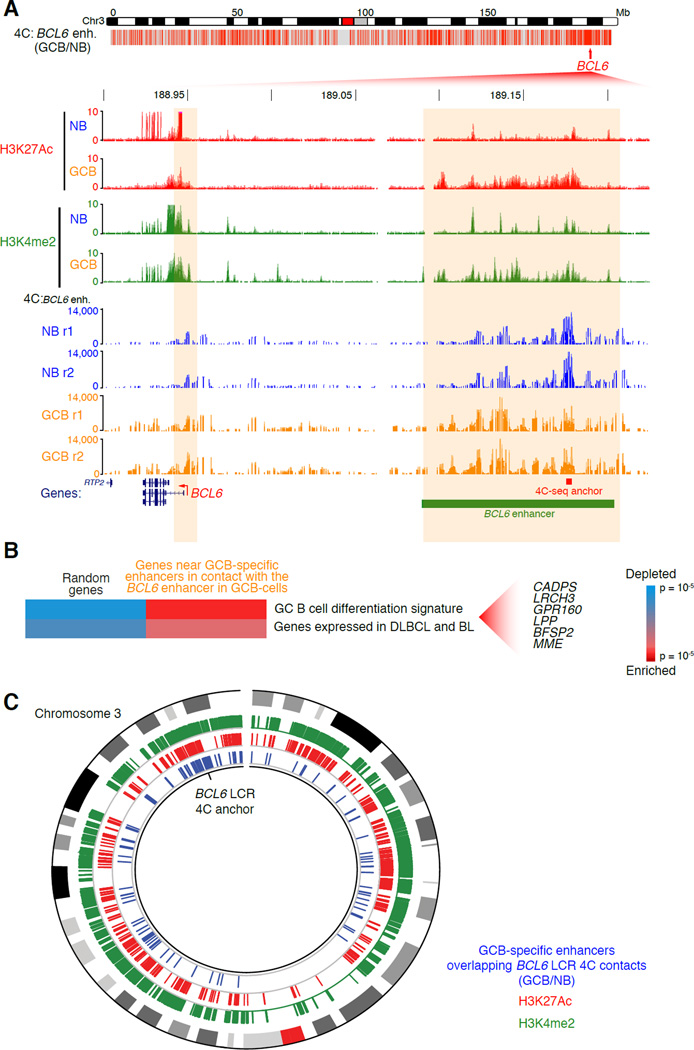
Our Hi-C, 3C and ChIP-seq data revealed a large (~114 Kb) GCB-specific, active enhancer region (H3K4me2posH3K27Acpos) distal to, and in contact with the BCL6 gene promoter as shown in Figure 2. We hypothesized that this region could be a locus control region (LCR) for other GC-specifying genes on chromosome 3. We performed 4C using an anchor within this putative BCL6 LCR, which identified between 2,000 and 3,000 intra-chromosomal interactions gained in GCB compared with naïve B-cells (p<0.05, Fisher’s exact test with Bonferroni correction; Figure 6A, Table S2). The BCL6 promoter was among the loci that gained interactions with the LCR. The BCL6 LCR was also physically connected to 400–600 other genes on chromosome 3 (Table S2). These genes were significantly up-regulated in GCB-cells (p=0.012; Fisher’s exact test), and were significantly enriched for GC signature genes (p<10−4). Notably, the BCL6 LCR was connected to 120 other GCB-cell-specific enhancers on chromosome 3 (Figure 6C). These looped enhancers were in close proximity (< 50 Kb) to genes that are enriched in GC and DLBCL survival pathways, including CADPS, LRCH3, LPP and MME (p<0.002; Figure 6B). A signature feature of these looped enhancers was active chromatin, marked by H3K27Ac (Figure 6C). Indeed, we saw a trend toward increased expression of genes proximal to enhancers looped to the BCL6 enhancer (p<0.09; Fisher’s exact test). Hence the BCL6 enhancer functions as an LCR controlling a large network of GC enhancers and genes.
Figure 6. A BCL6 upstream LCR forms an extensive GC-specific contact network.
A, (Top) Plot showing locations of BCL6 LCR contacts made in GCB-cells (versus NB-cells), as detected by 4C-seq across chromosome 3 (p<0.05, Fisher’s exact test). (Below) Zoomed-in view of the BCL6 gene and LCR showing histone modifications as detected by ChIPseq, and normalized read counts of contacts made with the BCL6 enhancer bait sequence, as detected by replicate 4C-seq, in NB and GCB-cell replicates. B, Gene set enrichment and depletion among genes near to GCB-specific enhancers that form contacts with the putative GCB-cell specific LCR upstream of BCL6 in GCB-cells. Significant genes are listed (p<0.002). C, Circos plot showing (in blue) chromosomal locations of the enhancer-enhancer contacts made between the BCL6 LCR (4C anchor) and other GCB-specific enhancers in GCB-cells (but not in naïve B-cells), as detected by 4C-seq across chromosome 3 (p<0.05, Fisher’s exact test). Regions of enrichment of histone modifications (H3K4me2 and H3K27Ac) as measured by ChIP-seq are shown in green and red, respectively. The outermost ring of the plot is an ideogram of chromosome 3 with cytogenetic bands represented in gray scale and the centromere in red.
The LCR upstream of BCL6 is required for formation of GCs
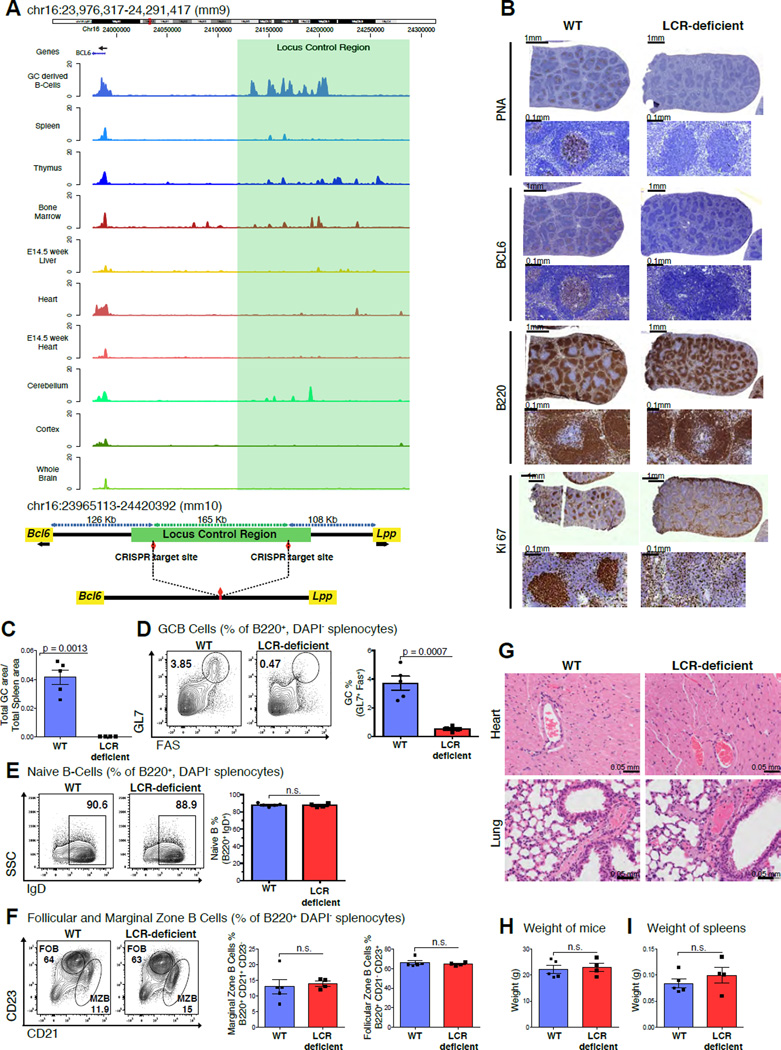
To determine whether activation of this BCL6 LCR is specific to GCB-cells we compared its H3K27Ac profiles with multiple tissues that express BCL6 including T-follicular helper cells using available human H3K27Ac ChIP-seq. This analysis showed that the LCR is exclusively highly H3K27 acetylated only in GCB-cells but not other tissues (Figure S6A). To evaluate whether this LCR might be evolutionarily conserved we mapped the syntenic region in mice and observed that it localized at chromosome 16, 130.4 Kb upstream from the transcriptional start site of the murine Bcl6 locus. Using H3K27Ac ChIP-seq we demonstrated massive enrichment of H3K27Ac in the syntenic LCR in murine GC derived B-cells, upstream of the Bcl6 locus (shaded green; Figure 7A). In contrast, other murine tissues lacked H3K27Ac enrichment at this region (Yue et al., 2014).
Figure 7. Disruption of the LCR leads to GC formation defects.
A, Tracks for H3K27Ac read densities (top panel) normalized to input for murine tissues including GC derived malignant B-cells and other Bcl6 expressing tissues. The region shaded in green represents the location of the syntenic LCR. Schematic illustrating the locus upstream of Bcl6 in the mouse genome before and after CRISPR-mediated deletion (bottom panel). B, Immunohistochemistry images of mouse spleens stained with antibodies against PNA, BCL6, B220 and Ki67. C, Bar plot representing the splenic area occupied by GCs based on PNA-IHC in WT vs. LCR-deficient mice. D, Representative flow cytometry plots of WT and LCR-deficient mice (left panel), quantification for GCB (GL7+FAS+) (right panel). E, F, Representative flow cytometry plots of naïve B-cells (B220+, IgD+), follicular (B220+, CD23high) and marginal zone B-cells (B220+, CD21high). G, Representative images of H&E stained Heart and Lung sections from the WT and LCR-deficient mice. H, I, Bar plots depicting average weight of the mice and spleens in the WT and LCR-deficient groups. Data in B, C, D, E, F, G, H and I are representative from one of two replicate experiments with 5 WT littermates and 4 LCR-deficient mice. The data is shown as mean ± SEM. Significance is calculated by performing a two-tailed, unpaired t-test. P-values are listed wherever the difference is significant. See also Figure S6.
To investigate the functionality of the putative LCR, we used CRISPR-mediated DNA editing in murine oocytes to delete the syntenic 165 Kb region upstream of the Bcl6 gene in mice (Figure 7A) and generated independent founder lines that were bred into the C57BL/6 strain. We immunized LCR-deficient and wild-type (WT) littermate control mice with T-cell dependent antigen and examined lymphoid tissues ten days later to assess GC formation. Immunohistochemical (IHC) analysis of spleen sections using peanut agglutinin (a GCB marker), BCL6 and Ki-67 revealed a profound defect in the formation of GCs in LCR-deficient mice compared to WT mice (p=0.0013; two tailed, unpaired t-test, Figure 7B and C), whereas total B-cells were unchanged between the LCR-deficient and WT mice (Figure S6B). Flow cytometry confirmed the loss of B220+, FAS+, GL7+ (along with B220+, FAS+, CD38−) GCB-cells (p=0.0007; two tailed, unpaired t-test, Figure 7D and S6C). There was no significant difference in naïve B-cell numbers (Figure 7E). Additionally, marginal zone B-cell (B220+, CD21high), follicular zone B-cell (B220+, CD23high) and monocyte (B220−, CD3−, CD11b+, GR1+) numbers were normal (Figure 7F and S6D). These results were confirmed in an independent founder strain (data not shown).
Constitutive BCL6 deficiency causes a severe and lethal phenotype in addition to loss of GCs. Bcl6−/− pups either die in utero or are born runted and die shortly after due to a severe inflammatory syndrome due to BCL6 loss of function in T-cells and macrophages (Dent et al., 1997). In addition, BCL6 plays a role in cortical neuronal differentiation in mice (Tiberi et al., 2012). Compared to Bcl6−/− mice, LCR-deficient mice were born at Mendelian ratios, did not demonstrate any discernible health problems, nor any evidence of inflammation (Figure 7G, 7H and 7I). Notably, Bcl6 nuclear staining in the neurons of the subiculum (a BCL6 expressing region next to the hippocampus) was comparable in LCR-deficient and WT mice (Figure S6E). Altogether, these results support the notion that this GC-specific highly interactive region is required for GC formation and functions as a GC-specific LCR. These results provide evidence of a role for a non-coding region in the regulation of the GC program during B-cell affinity maturation.
DISCUSSION
Our integrative study comparing 3D chromosomal organization in human naïve B-cells and GCB-cells, reveals a number of mechanisms through which reorganization of chromosomal architecture may accommodate and coordinate changes in transcriptional programs. The loss of inter-arm chromosomal looping in GCB-cells reveals a topological basis for nuclear decompaction in GCB-cells. At higher resolution we observed a process whereby 3D gene neighborhoods merge to form interconnected “cities”. Merging of 3D gene neighborhoods in GCB-cells is accompanied by the sharing of information in the form of chromatin epigenetic marks, and de novo coordination of transcription between genes in interacting units. This represents a potentially economical manner through which cells can control, or mix and match, cohorts of genes to more efficiently coordinate changes in their transcriptional programs. For example, BCL6, which represses cell cycle checkpoint genes and is specifically up-regulated in GC GCB-cells, becomes coordinately expressed with genes that facilitate proliferation through the merging of 3D gene neighborhoods on chromosome 3. Similarly, 3D gene neighborhoods encompassing the Hoxd gene cluster undergo a conformational switch to control limb development in mouse embryos (Andery, et al., 2013).
An overarching mechanism described herein is the de novo increase of interactions and connectivity between promoters in activated B-cells, linked to binding of B-cell transcriptional activators (PU.1, SPIB and IRF8), activated chromatin (H3K4me3, H3K4me2 and H3K27Ac) and recruitment of the architectural remodelers CTCF and cohesin, whose long-range actions have been documented during immunoglobulin V(D)J recombination and MHC II gene selection in B-cells (Degner et al., 2011; Guo et al., 2011; Majumder and Boss, 2011). Importantly the differential expression of genes in GCB-cells was most strongly associated with both increased promoter interactivity and coordinated activating and repressing histone marks. Hence the GCB-cell transcriptional program is synchronized through de novo assembly of cell type-specific promoter networks, thereby facilitating an efficient exchange of regulatory information. This is consistent with models proposed in the literature (Sutherland and Bickmore, 2009), and recent genome-wide studies of interactions in cancer cell lines, ESCs, differentiating T helper cells and during pre to pro-B-cell transition (Apostolou et al., 2013; Lin et al., 2012; Hakim et al., 2013; Li et al., 2012). Activated genes in GCB-cells more extensively utilize distal enhancer regions marked by H3K27Ac and binding of EP300, PU.1 and SPIB, whereas genome-wide interactions less frequently engage poised enhancers. Active enhancers became connected to promoters in GCB-cells for cell type-specific gene expression. Finally, our high-resolution analysis has enabled the identification of a genome-wide role for 5’ to 3’ gene looping in the regulation of cell type-specific transcription. This may aid the re-loading of RNA polymerase II to the promoters of genes that are highly expressed, as postulated in studies of gene loops in yeast and in mammalian cell lines (Laine et al., 2009; Tan-Wong et al., 2008; Tan-Wong et al., 2009), and may require activated chromatin, as well as CTCF and RAD21 binding, to direct 5’ to 3’ gene folding.
The requirement for such mechanisms to efficiently coordinate transcription is consistent with the profound phenotypic change that B-cells undergo as they transition from the resting state to the GC reaction. The master regulator of GCB-cell differentiation, BCL6, which is subject to remodeling by each of these different architectural layers exemplifies this model and supports the functional importance of synchronous 3D conformational changes in the control of cell type specification. The most prominent feature of the BCL6-centric changes in chromatin organization was identification of a GCB-cell-specific LCR upstream of the BCL6 gene, which displayed all of the hallmark features of a cell-specifying regulatory element: (i) GCB-cell-specific enrichment for H3K27Ac, (ii) looping to connect the promoters and enhancers of GC-specifying genes, and (iii) is specifically required for the formation of functional GCs. Loss of this LCR likely results in failure to induce and coordinate expression of multiple GC related genes including BCL6.
Altogether, these different layers of architectural reorganization fit into a dynamic and cooperative model of cellular activation whereby genes that require fast and efficient transcription in GCB-cells, such as those required for BCR signaling, cell proliferation and repression of DNA repair checkpoints, undergo coordinated structural remodeling upon differentiation signals in the GC. Chromatin tethering at an enhancer region upstream of the BCL6 gene represents a mechanism by which GC-specific genes are looped together for coordinate expression on chromosome 3. Although this study was conducted in mature human B-cells, we propose that the architecture of the genome is also likely to be critical for specification of other cellular phenotypes, especially when major shifts in transcriptional programming are required, and that epigenetic and transcriptional reprogramming is in this way functionally linked to the structural organization of genes in the nucleus.
EXPERIMENTAL PROCEDURES
Chromatin immunoprecipitation (ChIP)
ChIP was performed in 1% formaldehyde-fixed naïve B and GCB-cells, or a mature B-cell line, OCI-Ly1, as previously described (Ci et al., 2009), using the following antibodies: H3K4me3 (Abcam, catalogue no. ab8580), H3K4me2 (Abcam, ab32356), H3K27Ac (Abcam, ab4729), H3K27me3 (Millipore, 17-622), PU.1 (Santa Cruz, sc-352), EP300 (Santa Cruz, sc-585x), CTCF (Millipore, 07-729), RAD21 (Abcam, ab992), RNA polymerase II (Santa Cruz, sc-899). Quantitative ChIP was performed using SYBER Green-based QPCR and the primer sequences in Table S3.
Chromosome conformation capture (3C), Hi-C and 4C
3C DNA templates were generated as previously described (van Berkum and Dekker, 2009) using HindIII digestion. 3C enrichment was determined using 50 ng of 3C DNA and SYBR Green-based QPCR and the 3C primer sequences in Table S4 and S5. Primer pairs were normalized to enrichment at adjacent HindIII fragments within the promoters of each gene. BAC plasmids encompassing the genomic regions of interest and used as controls for primer efficiency were as follows: RP11-632M13, CTD-2527P7, RP11-464D9 (BCL6 region); CTD-3120O5, RP11-1066F16 (CKS2 region). Hi-C DNA templates were generated as previously described (Lieberman-Aiden et al., 2009). 4C DNA templates were generated from 50 µg of HindIII-digested and ligated 3C DNA templates using a 4 basepair (bp) cutter, NlaIII, according to a previously published protocol (Gheldof et al., 2012).
ChIP-seq, RNA-seq, Hi-C and 4C-seq Library Preparation and Illumina Sequencing Processing
ChIP-seq and RNA-seq libraries were prepared using the Illumina ChIP-Seq and TruSeq RNA sample kits, respectively. 4C-seq libraries were generated using a set of 16 uniquely indexed, Illumina sequencing-based bait primers anchored at a NlaIII-HindIII fragment encompassing exon 1 and the promoter region of the BCL6 gene, or at a region within the distal enhancer region (enhancer 1) upstream of the BCL6 gene (see Table S7 for 4C-seq primer sequences). Hi-C DNA sequencing libraries were generated as previously described (Lieberman-Aiden et al., 2009) with 20–50 µg of Hi-C DNA template. Libraries were validated using the Agilent Technologies 2100 Bioanalyzer and Quant-iT™ dsDNA HS Assay (Life Technologies), and 8–10 pM sequenced on an Illumina GAIIx or HiSeq2000 sequencer as follows: ChIP-seq, 1 × 36; RNA-seq, 2 × 50; Hi-C, 2 × 50 cycles. 6.5 pM of the 4C-seq libraries were sequenced on an Illumina HiSeq2000 sequencer using 1 × 75 cycles, with 20–30% of a PhiX control library spiked into each lane, to account for low sample complexity. ChIP-seq data was aligned to the hg18 genome using BWA (Li and Durbin, 2009), clonal reads discarded, and regions of enrichment (peaks) identified using ChIPseeqer (Giannopoulou and Elemento, 2011). RNA-seq data was aligned to transcripts using TopHat (Trapnell et al., 2009), gene expression quantified in RPKM (Mortazavi et al., 2008) and differentially expressed genes identified (Wang et al., 2012). Hi-C reads were aligned to the hg18 genome using BWA and ambiguous reads and clonal reads discarded. Hi-C interactions were derived from read-pairs with consistent placement relative to HindIII sites (<500 bp away). Accordingly, read-pairs were filtered to remove background ligation products (non-ligated, self-ligated, non-digested or re-ligated DNA). The raw numbers of Hi-C read-pairs that were sequenced on each lane, uniquely aligned, and which represent intra-chromosomal interactions post-filtering, are summarized for each of the naïve B and GCB-cell replicates in Table S1. 4C-seq reads that contained the BCL6 promoter bait sequence or either of the BCL6 enhancer bait sequences (A, B or C) (>95% of total reads) were filtered using FASTX (http://hannonlab.cshl.edu/fastx_toolkit), aligned to the hg18 genome using BWA and ambiguous reads discarded. The numbers of uniquely aligned 4C-seq reads that were sequenced for each cell type and replicate are summarized in Table S2.
Generation of LCR-deficient mice using CRISPR mediated genome editing
Guide RNA sequences targeting the regions flanking the 165 Kb LCR in mice were selected with the CRISPR design tool at http://crispr.mit.edu/. Complementary oligos were ordered from integrated DNA technologies (Table S6), annealed and cloned into the BbsI restriction enzyme digestion site of the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid. PCR was performed to amplify template DNA coding for the sgRNA sequence with an upstream T7 promoter sequence (Table S6). In-vitro transcription was performed using the Ambion MEGAshortscript T7 Transcription kit (Life Technologies). The sgRNA was subsequently purified using the Ambion MEGAclear kit (Life Technologies). The embryonic microinjection of the sgRNA pair along with Cas9 mRNA was in F1 hybrid zygotes from FVB/NJ and B6(Cg)-Tyrc-26/J mice. Microinjections were performed as described in (Singh, Schimenti, & Bolcun-filas, 2014). 81 of the 117 one-cell embryos progressed to the two-cell stage and were transferred into 4 pseudo-pregnant mice. A total of 15 pups were born from the resulting pregnancy. Pups were transferred to the animal facility at Weill Cornell Medical College where they were bred and all experiments were performed with the approval of the institutional animal care and use committee.
Immunohistochemistry
Mouse organs were fixed in 4% formaldehyde and embedded in paraffin. Sections were dewaxed in xylene and hydrated into graded alcohols. Endogenous peroxidase activity was blocked with 1% hydrogen peroxide in PBS for 15 minutes. Pretreatment was performed in a steamer using 10mM citrate buffer, pH6.0 for 30 minutes. Sections were incubated overnight with primary antibodies Bcl-6 (Rabbit polyclonal antibody from Santa Cruz Biotechnology, Cat. No. sc-858) diluted at 1:750; Biotinylated PNA (Vector Laboratories, Cat. No. B1075) diluted at 1:1000. Sections were washed with PBS and incubated with the appropriate secondary antibody followed by avidin-biotin complexes (Vector Laboratories, Burlingame, CA, Cat. No. PK-6100). Antibody reaction was visualized with 3-3’ Diaminobenzidine (Sigma, Cat. No. D8001) and counterstained with hematoxylin. Tissue sections were dehydrated in graded alcohols, cleared in xylene and mounted. Immunohistochemistry of the formalin or PFA-fixed paraffin-embedded sections for CD3, Ki67 and B220 was performed on a Leica Bond™ RX using the Bond™ Polymer Refine Detection Kit (Cat. No. DS9800). The sections were pre-treated using heat mediated antigen retrieval with EDTA pH9, (Leica Biosystem Epitope Retrieval Solution 2, Cat. No. AR9640) for 20mins. The sections were then incubated with CD3 (Abcam, Cat. No. ab16669) diluted at 1:100; Ki67 (Abcam, Cat. No. ab16667) diluted at 1:100; and B220 (BD Bioscience, Cat. No. 550286) diluted at 1:200 for 30mins. DAB was used as the chromogen. The sections were then counterstained with hematoxylin and mounted.. ImageJ 1.44o software (NIH) was used for IHC quantification. H&E and IHC stained sections were examined by a board certified veterinary pathologists.
Germinal center formation analysis using flow cytometry
For assessment of GC formation defects, LCR-deficient mice were intraperitoneally injected with 500ul of 1:10 diluted (in D-PBS) sheep red blood cells. On day 10 post-immunization mice were euthanized with CO2 to collect spleens. The splenic mononuclear cells were separated using Fico/Lite-LM (Atlanta Biologicals). Cells were stained for GCB-cells, naïve B-cells, follicular zone B-cells, marginal zone B-cells and monocytes and analyzed on a BD FACS Canto II flow cytometer.
Supplementary Material
Acknowledgments
We thank the Epigenomics Core Facility (WCMC); the Dekker lab and Nynke van-Berkum, for advice on Hi-C; Jiao Maria and Dr. Sebastien Monette from the WCMC and MSKCC lab of comparative pathology; Dr. John Schimenti from Cornell University (Ithaca); the Cornell Stem Cell and Transgenic Core facility, and Hao Shen. A.M.M. and O.E. are funded through NCI R01 CA104348. O.E. is funded by a US National Science Foundation CAREER grant 1054964 and NCI R01 CA194547. K.L.B. was supported by an American Society of Hematology Scholar Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.L.B., A.M.M. and O.E. designed the studies and interpreted the data with T.D.S. and R.S. Experiments were performed as follows: K.L.B., D.W.P. and B.L.S., B-cell isolation, flow cytometry, microscopy, 3C; K.L.B. and D.W.P., 4C-seq; K.L.B., Hi-C-seq, RNA-seq, H3K27me3 and RAD21 ChIP-seq; R.S., design, generation and analysis of LCR-deficient mice; Y.J., H3K4me2, H3K4me3, H3K27Ac, CTCF, and PU.1 ChIP-seq; K.H., EP300 ChIP-seq; R.S., RNA PolII QChIP; W.R., surgical isolation of tonsil specimens; W.B. flow cytometry and IHC for LCR-deficient mice; T.D.S. developed Hi-C data algorithms and integrated computational analyses, with input from O.E.; K.L.B. contributed to development of the Hi-C data filtering pipeline and analyzed 4C-seq data, with input from O.E. M.T. contributed to data interpretation. K.L.B. wrote and prepared the manuscript with A.M.M., O.E., T.D.S., and R.S.
Accession Number
The Gene Expression Omnibus (GEO) accession number for the Hi-C and RNA-seq data sets reported in this paper is GSE84022.
REFERENCES
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. Genome-wide Chromatin Interactions of the Nanog Locus in Pluripotency, Differentiation, and Reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Srinivasan L, Perkins L, Atchison ML. Protein acetylation regulates both PU.1 transactivation and Ig kappa 3' enhancer activity. J Immunol. 2005;175:5160–5169. doi: 10.4049/jimmunol.175.8.5160. [DOI] [PubMed] [Google Scholar]
- Baù D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W, Polo JM, Cerchietti L, Shaknovich R, Wang L, Yang SN, Ye K, Farinha P, Horsman DE, Gascoyne RD, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276(5312):589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, … Ren B. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PJ, Benke A, Joye E, Nguyen Huynh TH, Manley S, Duboule D. Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc of the Natl Acad Sci USA. 2015;112:3964–13969. doi: 10.1073/pnas.1517972112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof N, Leleu M, Noordermeer D, Rougemont J, Reymond A. Detecting long range chromatin interactions using the chromosome conformation capture sequencing (4C-seq) method. Methods Mol Biol. 2012;786:211–225. doi: 10.1007/978-1-61779-292-2_13. [DOI] [PubMed] [Google Scholar]
- Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng H, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Nakayamada S, Voss TC, Baek S, Hager GL. Spatial congregation of STAT binding directs selective nuclear architecture during T-cell functional differentiation. Genome Res. 2013;23:462–472. doi: 10.1101/gr.147652.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Melnick A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends in Molecular Medicine. 2014;20(6):343–352. doi: 10.1016/j.molmed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centers: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Laine J, Singh B, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes & Development. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Boss JM. Cohesin Regulates MHC Class II Genes through Interactions with MHC Class II Insulators. J Immunol. 2011;187:4236–4244. doi: 10.4049/jimmunol.1100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams B, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012;48:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, Ballon G, Yang SN, Weinhold N, Reimers M, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118:3559–3569. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya J, Roberts SG. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim Biophys Acta. 2012;1819:391–400. doi: 10.1016/j.bbagrm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, Mccleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-filas E. A Mouse Geneticist ’ s practical guide to CRISPR applications. Genetics: Early Online. 2014 Jan;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci USA. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes & Development. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberi L, van den Ameele J, Dimidschstein J, Piccirilli J, Gall D, Herpoel A, Vanderhaeghen P. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nature Neuroscience. 2012;15(12):1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum NL, Dekker J. Determining spatial chromatin organization of large genomic regions using 5C technology. Methods Mol Biol. 2009;567:189–213. doi: 10.1007/978-1-60327-414-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C Reveals that CTCF Underlies Evolution of Chromosomal Domain Architecture. Cell Reports. 2015;10(8):1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kang DD, Shen K, Song C, Lu S, Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al. An R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics. 2012;28:2534–2536. doi: 10.1093/bioinformatics/bts485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kihara-Negishi F, Yamada T, Suzuki M, Nakano T, Oikawa T. Interaction between the hematopoietic Ets transcription factor Spi-B and the coactivator CREBbinding protein associated with negative cross-talk with c-Myb. Cell Growth Differ. 2002;13:69–75. [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.