Abstract
Gaucher’s disease is a common genetic disease caused by mutations in the β-glucocerebrosidase (GBA1) gene that have been also linked to increased risk of Parkinson’s disease and Lewy Body Dementia. Stabilization of misfolded mutant β-glucocerebrosidase (GCase) represents an important therapeutic strategy in synucleinopathies. Here we report a novel class of GCase quinazoline inhibitors, obtained in a high throughput screening, with moderate potency against wild-type GCase. Rational design and a SAR study of this class of compounds led to a new series of quinazoline derivatives with single digit nanomolar potency. These compounds were shown to selectively stabilize GCase when compared to other lysosomal enzymes and to increase N370S mutant GCase protein concentration and activity in cell assays. To the best of our knowledge, these molecules are the most potent non-iminosugar GCase inhibitors to date that may prove useful for future mechanistic studies and therapeutic approaches in Gaucher’s and Parkinson’s diseases.
Keywords: β-Glucocerebrosidase, Gaucher’s disease, Parkinson’s disease, Quinazoline, Modulator, SAR
Table of Contents Graphic
INTRODUCTION
Gaucher’s disease (GD), the most common lysosomal storage disease, is caused by a recessively inherited deficiency in β-glucocerebrosidase (GCase) and subsequent accumulation of glucoceramides, toxic lipid substrates1,2. Substrate accumulation leads to hepatosplenomegaly, bone marrow suppression, and bone lesions1–3. Many of the GCase mutations are missense mutations4 that result in single amino acid substitutions of the enzyme. Most of these mutations, including the prevalent N370S mutation, are still functional, although with very low residual GCase activity5 due to enzyme misfolding, and proteasome-mediated breakdown5. Current treatments for GD include enzyme replacement therapy (ERT) and substrate reduction therapy (SRT) 2,6. In recent years, mutations in GBA1 were also found to be a major risk factor for Parkinson’s disease (PD) and dementia with Lewy bodies (DLB)7–11. Accumulation of β-glucosylceramide, the substrate of GCase, in neurons promotes the formation of α-synuclein oligomers, which are considered toxic in PD12. Enhancement of GCase activity is thought to be a potential therapeutic strategy for GCase-associated synucleinopathies, including PD13,14.
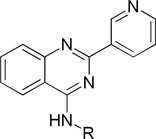
An emerging therapeutic approach involves the restoration of proper folding and lysosome delivery of degradation-prone mutant enzymes using small molecules as pharmacological chaperones (PCs)5. Previous studies have shown that iminosugars increase the cellular activity of the N370S mutant form of GCase15,16, as well as of the wild-type enzyme5,17. Isofagomine (IFG, 1) attracted the most attention in the iminosugar class of compounds (Figure 1)18. However, iminosugars tend to have poor selectivity and relatively short half-lives in cells19. Several different scaffolds of non-iminosugar inhibitors (2 and 3 are examples in Figure 1) have been reported as GCase PCs since 200720–24; however, the binding site and the interaction with GCase of these non-iminosugar PCs remain unknown.
Figure 1.
Structures of GCase inhibitors
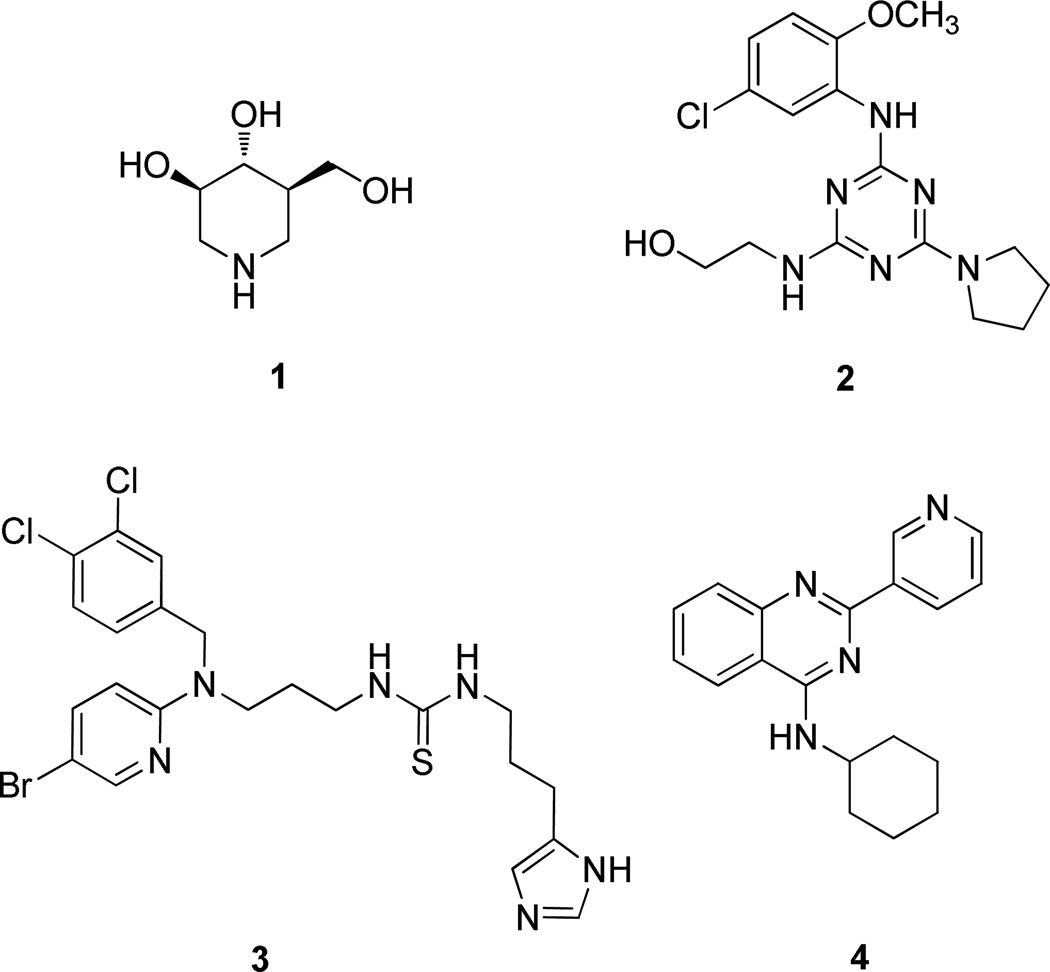
In our high throughput screening effort to discover potent GCase modulators, compound 4 (Figure 1) was identified as a potent GCase inhibitor (IC50 0.177 µM) in a 4-methylumbelliferyl β-D-glucopyranoside (4MU-β-Glc) enzyme activity based high throughput screen. The activity was confirmed with additional synthesized compounds. To further develop potent GCase modulators and use them to explore the properties of the binding site, we carried out a structure activity relationship (SAR) study of a series of quinazoline derivatives, leading to the discovery of single digit nanomolar potency GCase inhibitory modulators.
CHEMISTRY
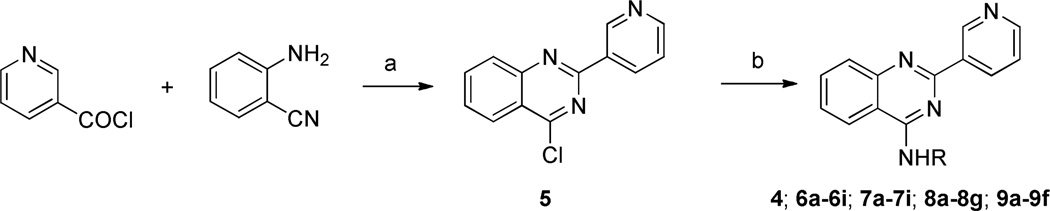
The synthesis of compound 4 and its analogues for SAR exploration is straightforward and is detailed in Scheme 1 and 2. As showed in Scheme 1, 5 was prepared from 2-amino-benzonitrile and nicotinoyl chloride according to a known method25. The reaction of 5 and appropriate amines in the presence of potassium carbonate as a base afforded 4, 6a-6i, 7a-7i, 8a-8g, and 9a-9f.
Scheme 1.
Synthesis of 4, 6a-6i, 7a-7i, 8a-8g, and 9a-9f with substituents on the secondary amine
Reagents and conditions: (a) (i) sulfolane (ii) PCl5; (b) RNH2, K2CO3, DMF
Scheme 2.
Synthesis of 11a-11h with modifications at the 2-position of the quinazoline ring
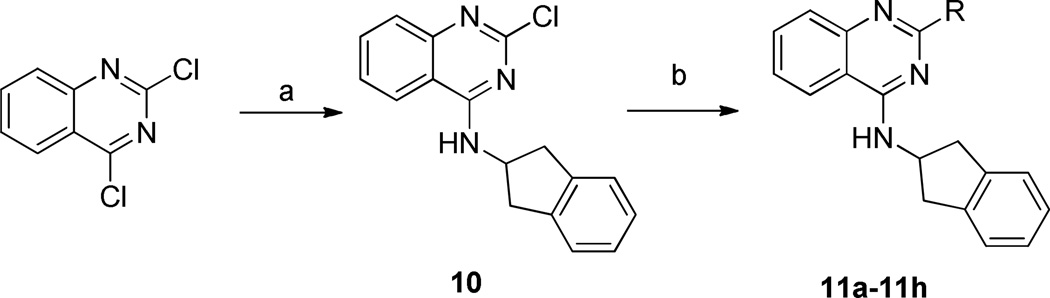
Reagents and conditions: (a) 2,3-dihydro-1H-inden-2-amine, K2CO3, DMF; (b) RB(OH)2, Pd(PPh3)4, K2CO3, 1,4-dioxane, H2O
Additional analogues having modifications at position 2 of the quinazoline ring were synthesized, employing alkylation of 2,3-dihydro-1H-inden-2-amine with 2,4-dichloroquinazoline, followed by Suzuki coupling with appropriate boronic acids to afford 11a-11h (Scheme 2). The structure and purity of all the prepared compounds were confirmed by spectroscopic and analytical techniques.
RESULTS AND DISCUSSION
In our high throughput screening efforts to discover potent GCase inhibitors/activators, we used recombinant wild-type GCase and 4MU-β-Glc as substrate in an optimized pH 5.9 buffer26. In previous studies, high concentrations of taurocholate (4–10 mM) were used to improve the signal in GCase enzyme activity assays16,27,28. We found that taurocholate can interfere with our assay results, which was also reported recently by Berger et al.29; therefore, we excluded taurocholate in our screening assay. Using this approach, we discovered several different scaffolds of GCase inhibitors and activators with moderate activity. Among these, we identified a quinazoline compound (4, Figure 1) as a potent GCase inhibitor. The quinazoline ring had previously been found as the best scaffold for GCase inhibitors among several ring systems assayed21. Here we describe our modifications of the substituents on the quinazoline ring.
To examine the SAR at the amino group, a series of substituents was introduced at the 4-position of the N-cyclohexyl ring of 4 (Table 1). A 4-methyl substituent (6a, cis/trans = 3/2 mixture) resulted in 3-fold higher activity (IC50 56 nM), while 4-ethyl substitution (6b, cis/trans = 3/2 mixture) did not show a significant change of activity, suggesting that a smaller hydrophobic group at this position may be beneficial. Then, a trans-4-methyl compound (6c) was synthesized, which exhibited great improvement of inhibitory activity (IC50 20 nM). Further modification by installation of 4-methoxyl (6d), 4,4-dimethyl (6e), or 4,4-difluoro (6f) groups at the same position decreased the potency; amino substituents were detrimental to activity. Replacement of the cyclohexyl group by tetrahydro-2H-pyran-3-yl (6h), or N-substituted piperidine (6i-6k) afforded weak or inactive compounds, supporting the importance of the lipophilic cyclohexyl ring.
Table 1.
Structure and inhibitory activity of 2-(pyridin-3-yl)quinazoline derivatives with substituted cyclohexyl and related ringsa
 | |||||
|---|---|---|---|---|---|
| Comp. | R | IC50 (µM) | Comp. | R | IC50 (µM) |
| 4 | 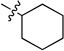 |
0.177 ± 0.012 | 6f | 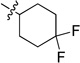 |
0.234 ± 0.017 |
| 6a | 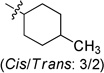 |
0.056 ± 0.005 | 6g | 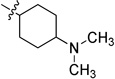 |
Inactive |
| 6b | 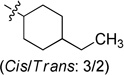 |
0.126 ± 0.007 | 6h | 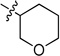 |
0.891 ± 0.023 |
| 6c | 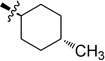 |
0.020 ± 0.002 | 6i | 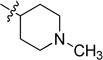 |
Inactive |
| 6d | 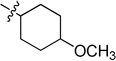 |
0.251 ± 0.018 | 6j | 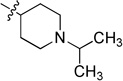 |
33.21 ± 9.71 |
| 6e | 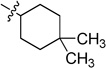 |
0.431 ± 0.042 | 6k | 36.07 ± 8.72 | |
Experiments were performed in triplicate, and the mean ± SD is shown.
To further expand the SAR of the amino group, the cyclohexyl ring of 4 was replaced by a series of saturated carbon rings of different sizes. A dramatic SAR was observed with different carbon rings (7a-7e). As shown in Table 2, the larger cycloalkyl rings were more potent; the compound with a cyclooctyl group (7a, IC50 27 nM) was the most potent. However, when bulk was introduced to the cycloalkyl ring, the potency of the compounds (7f and 7g) decreased, suggesting that the hydrophobic binding pocket may be compact. Introduction of one or two carbons between the cyclohexyl and NH groups (7h and 7i) in 4 decreased the inhibitory activity, again indicating a hydrophobic pocket with limited volume.
Table 2.
Structure and inhibitory activity of 2-(pyridin-3-yl)quinazoline derivatives with saturated alkyl ringsa
 | |||||
|---|---|---|---|---|---|
| Comp. | R | IC50 (µM) | Comp. | R | IC50 (µM) |
| 7a | 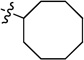 |
0.027 ± 0.002 | 7f | 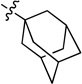 |
0.926 ± 0.08 |
| 7b | 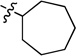 |
0.042 ± 0.003 | 7g | 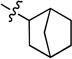 |
0.282 ± 0.043 |
| 7c | 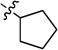 |
0.72 ± 0.05 | 7h | 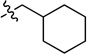 |
0.405 ± 0.040 |
| 7d | 2.84 ± 0.34 | 7i | 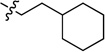 |
0.857 ± 0.033 | |
| 7e | 28.07 ± 3.21 | ||||
Experiments were performed in triplicate, and the mean ± SD is shown.
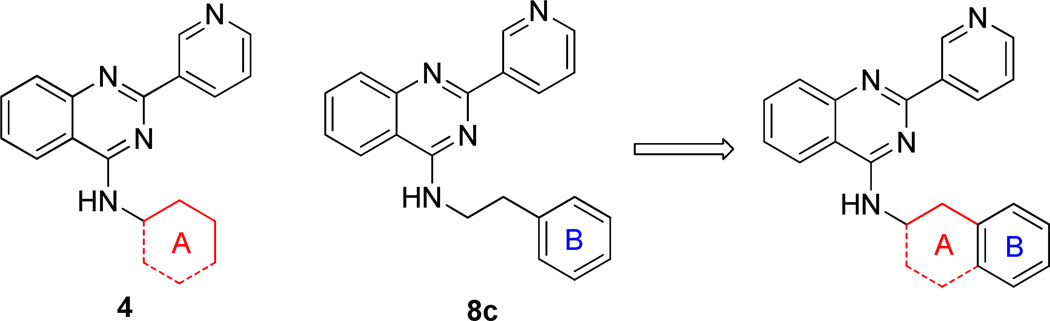
To understand the nature of the binding site, a phenyl ring with different length linkers was introduced into the molecules (8a-e, Table 3). Compound 8a, by replacement of the cyclohexyl ring in 4 with a phenyl ring, lost activity. Insertion of a 1–4 carbon linker between the phenyl and quinazoline rings gave 8b-8e. Interestingly, 8c, with a phenylethyl group, was slightly more potent than 4. Extension of the linker did not benefit activity, suggesting that a two carbon length linker between the phenyl ring and secondary amino group of 8c may allow optimal binding of the phenyl group. Substitution of the phenyl group in 8 with a 2- or 3-pyridine ring (8f or 8g) sharply diminished potency, indicating a repulsive effect of the pyridine nitrogen atom.
Table 3.
Structure and inhibitory activity of 2-(pyridin-3-yl)quinazoline derivatives with aromatic ringsa
 | |||||
|---|---|---|---|---|---|
| Comp. | R | IC50 (µM) | Comp. | R | IC50 (µM) |
| 8a | 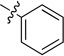 |
inactive | 8e | 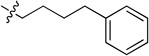 |
1.28 ± 0.13 |
| 8b | 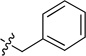 |
5.77 ± 0.82 | 8f | 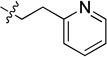 |
1.40 ± 0.12 |
| 8c | 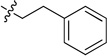 |
0.097 ± 0.009 | 8g | 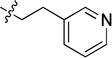 |
4.29 ± 0.57 |
| 8d | 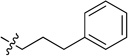 |
1.53 ± 0.22 | |||
Experiments were performed in triplicate, and the mean ± SD is shown.
To enhance the binding affinity of the compounds with GCase, a new series of compounds was designed to integrate both the hydrophobic interaction of the A ring and the π-π interaction of the B ring by fusing the cyclohexyl ring (4) with a phenyl ring (Figure 2 and Table 4). These derivatives (9a-c) exhibited single digit nanomolar inhibitory activity against GCase. Stereochemistry did not seem to be important (9a and 9b). The compound with an indane ring (9c) gave comparable activity to that of the tetralin ring (9a). Attachment of the quinazoline ring to the tetralin ring (9d) and indane ring (9e) at position 1 instead of position 2 (9a and 9c) dramatically increased the IC50 values to the low micromolar range, indicating the importance of the orientation of this substituent for binding activity of these inhibitors. The introduction of an oxygen atom to give a chromane (9f) did not significantly affect the potency. These results suggest important hydrophobic and π-π interactions in the binding of this series of compounds to GCase.
Figure 2.
Rational design of a new series of potent quinazoline inhibitors
Table 4.
Structure and inhibitory activity of 2-(pyridin-3-yl)quinazoline derivatives with fused ringsa
 | |||||
|---|---|---|---|---|---|
| Comp. | R | IC50 (nM) | Comp. | R | IC50 (nM) |
| 9a | 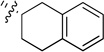 |
8.7 ± 1.1 | 9d | 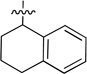 |
872 ± 71 |
| 9b | 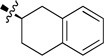 |
9.9 ± 1.3 | 9e | 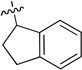 |
821 ± 73 |
| 9c | 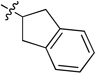 |
8.3 ± 1.0 | 9f | 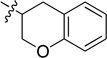 |
35 ± 3 |
Experiments were performed in triplicate, and the mean ± SD is shown.
Finally, we examined the substituent effect on the pyridine ring. A methyl group was introduced at different positions of the 3-pyridinyl ring of 9c to give 11a-c (Table 5). Methylation of the pyridine ring decreased the potency. Whereas the compound with a methyl group at position 4 of the pyridinyl ring (11a) showed only moderate potency, the other two compounds (11b and 11c) with a methyl group at positions 5- and 6 were much more potent, comparable to that when a 2-furanyl group replaced the pyridine ring, but still not as potent as 9c. The replacement of the 3-pyridinyl ring in 9c with either phenyl or 3-thienyl groups, however, gave compounds that retained the same potency as 9c, suggesting that more groups could be introduced at position 2 of the quinazoline ring.
Table 5.
Structure and inhibitory activity of 4-(2,3-dihydro-1H-2-indenamino)quinazoline derivatives with aromatic ringsa
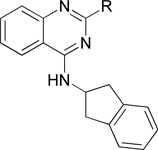 | |||||
|---|---|---|---|---|---|
| Comp. | R | IC50 (nM) | Comp. | R | IC50 (nM) |
| 11a | 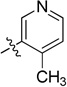 |
122 ± 12 | 11e | 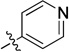 |
168 ± 18 |
| 11b | 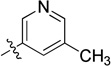 |
12.5 ± 1.7 | 11f | 8.2 ± 1.0 | |
| 11c | 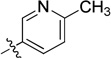 |
29.4 ± 3.6 | 11g | 5.2 ± 0.6 | |
| 11d | 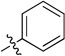 |
6.5 ± 0.7 | 11h | 21.1 ± 2.4 | |
Experiments were performed in triplicate, and the mean ± SD is shown.
We also evaluated the activity of these selected compounds at various pH values (Table 6). Interestingly, compared to the inhibitory activity at pH 5.9, the activity of 9a, 9b, and 9c with a 3-pyridinyl ring decreased by 3 fold at pH 5.0, while the activity of 11d, 11f, and 11g only dropped slightly at both pH conditions. Although the cause for the change in IC50 values is not clear, an explanation may involve the protonation state of the compounds or the variability of the enzyme efficacy at different pH values.
Table 6.
GCase inhibitory activity of 9a, 9b, 9c, 11d, 11g, and 11f at pH 5.0, pH 5.9, and pH 7.0a
| Comp | Structure | IC50 (nM) |
||
|---|---|---|---|---|
| pH 5.0 | pH 5.9 | pH 7.0 | ||
| 9a | 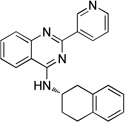 |
22.4 ± 3.4 | 8.7 ± 1.1 | 16.9 ± 2.7 |
| 9b | 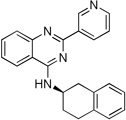 |
30.1 ± 4.5 | 9.9 ± 1.3 | 18.8 ± 2.5 |
| 9c | 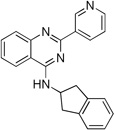 |
25.9 ± 3.9 | 8.3 ± 1.0 | 12.7 ± 1.4 |
| 11d | 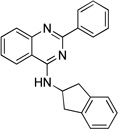 |
10.5 ± 1.5 | 6.5 ± 0.7 | 9.4 ± 1.4 |
| 11f | 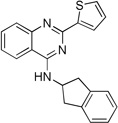 |
13.9 ± 1.9 | 8.2 ± 1.0 | 12.9 ± 2.0 |
| 11g | 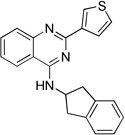 |
8.9 ± 1.3 | 5.2 ± 0.6 | 6.6 ± 1.0 |
Experiments were performed in triplicate, and the mean ± SD is shown.
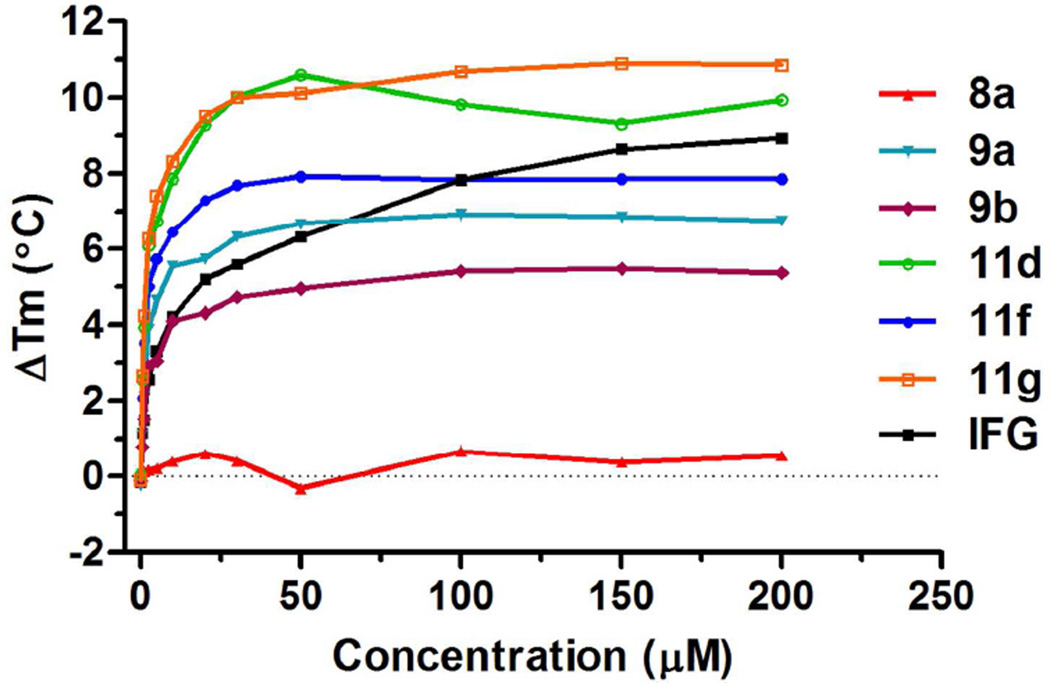
Compounds shown to act as pharmacological chaperones for GCase (or other lysosomal enzymes) also stabilize the enzyme against thermal denaturation. A fluorescent thermal shift assay was developed to evaluate the binding affinity of ligands with protein30. To evaluate their abilities to stabilize GCase, the most potent compounds, 9a, 9b, 11d, 11g and 11f were accessed in a wild-type GCase fluorescent thermal shift assay with a negative control (8a) and a positive control (IFG) at pH 5.0. The selected compounds increased the GCase melting point in a dose-dependent manner (Figure 3), while inactive compound 8a did not change the melting points significantly. Most compounds exhibited greater ability to stabilize GCase than IFG at lower concentrations, and 11g showed the maximum thermal shift up to around 11 °C. It should be noted that the increment in melting point has a direct correlation with the compound’s binding affinity31. The maximum thermal shift of our compounds corresponded to their inhibitory activity at pH 5.0, suggesting a close correlation of the binding affinity and the inhibitory activity for these compounds.
Figure 3.
Fluorescent thermal shift analysis of selected compounds. Compound 9a, 9b, 11d, 11f, 11g, and IFG showed their ability to stabilize wild-type GCase in a dose-dependent manner. Data represent the results of three independent experiments performed with three replicates per sample.
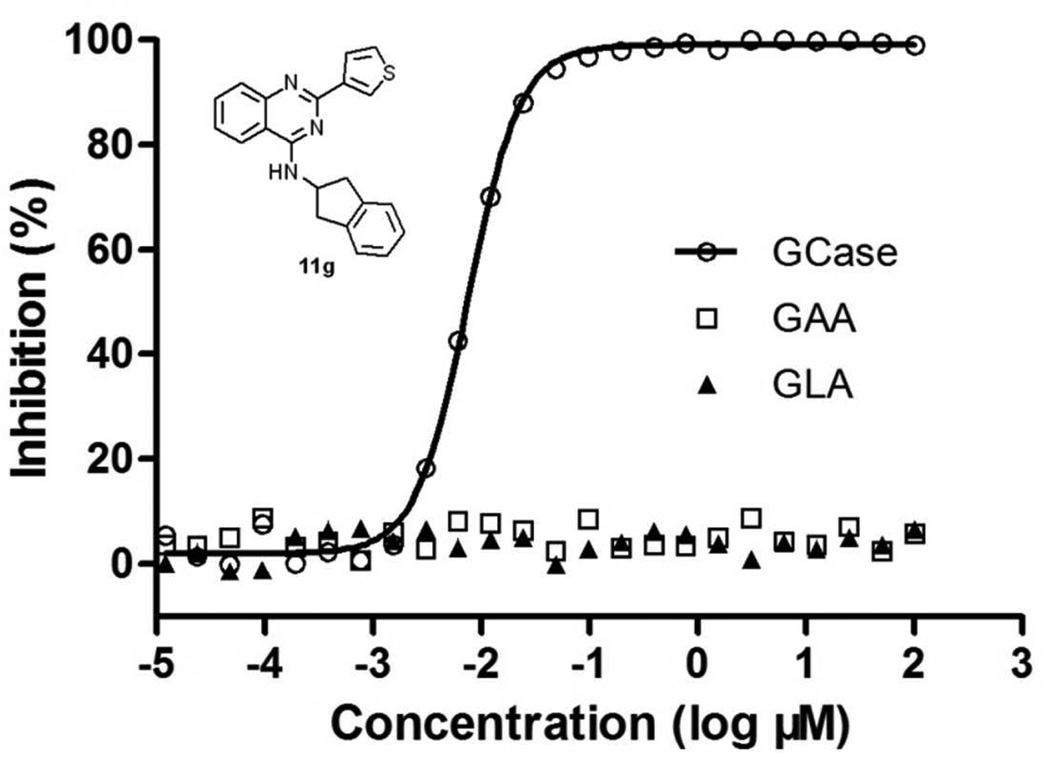
The selected compounds (9a, 9b, 11d, 11g, and 11f) were further evaluated against two other lysosomal hydrolases, acid α-glucosidase (GAA) and α-galactosidase A (GLA). The activities of tested enzymes were not significantly changed by compound treatments up to 100 µM (representative results shown for 11g in Figure 4).
Figure 4.
Selectivity of inhibitor 11g with related hydrolases. 11g was tested on GCase, acid α-glucosidase (GAA), and α-galactosidase A (GLA). Data represent the results of three independent experiments performed with three replicates per sample.
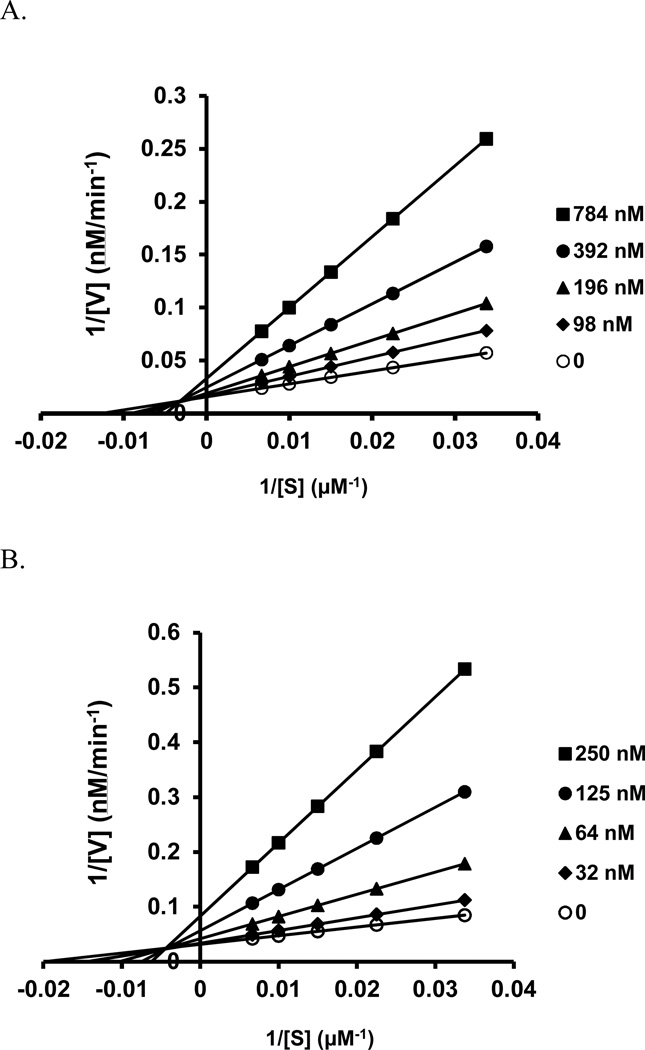
We further tested compounds 4 and 11g by measuring GCase activity at various substrate concentrations (30–150 µM) and in the absence or presence of increasing concentrations of GCase inhibitors. Similarly to reported non-iminosugar inhibitors20, both of our inhibitors exhibited linear mixed inhibition, with an increase in Km and decrease in Vmax values upon increasing inhibitor concentrations (Figure 5A and 5B)
Figure 5.
Lineweaver–Burk plots of the enzyme kinetics of GCase inhibitors (A) 4 and (B) 11g. Each inhibitor was tested in triplicate in two independent assays at the concentrations shown in the legend box of each graph, with (○) indicating the absence of inhibitor. (A) Compounds 4 and (B) Compound 11g showed an increase in Km and a decrease in Vmax, indicating linear mixed inhibition.
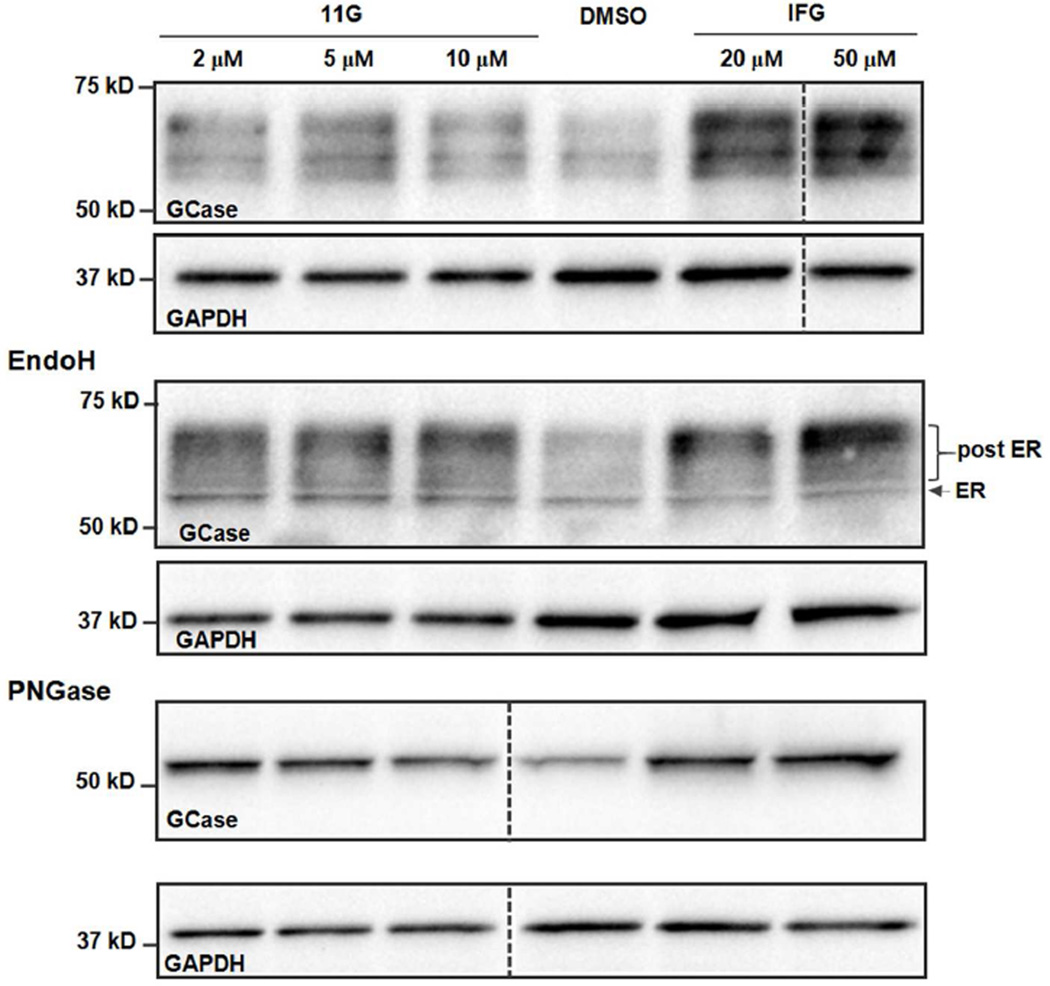
Finally, we tested 11g in patient-derived N370S fibroblasts. The fibroblasts were treated with 11g and IFG for 3 days, and levels of GCase protein were determined by Western blot. We found that 11g treatments (2, 5, and 10 µM) significantly increased the concentration of GCase, while IFG showed a similar effect only at the higher concentrations (20 and 50 µM) (Figure 6, top). We also used endoH and PNGase F to digest the cell lysates to determine whether 11g treatment affected ER maturation. The Western blot (Figure 6, middle) showed that most of the GCase bands were resistant to endoH digestion, indicating that the major GCase signal after treatment is a post-ER form. PNGase F digestion gave a single band with increased GCase concentration after 11g treatment (Figure 6, bottom). In addition to this finding, we also found that a 3-day treatment of compound 11g (2 µM) increased 50% of GCase activity in a separate enzyme activity assay, suggesting that 11g increased both GCase levels and the activity of patient-derived N370S fibroblasts.
Figure 6.
Modulator 11g increases GCase protein levels in patient-derived Gaucher’s disease fibroblasts. Top: Western blot of N370S fibroblast treated with 11g or IFG for 3 days. Middle: Western blot of endoH digestion of N370S fibroblast after a 3-day treatment of 11g, IFG, or DMSO; endoH resistance of proteins indicates their post-ER localization; Bottom: Western blot of PNGase F digestion of N370S fibroblast after a 3-day treatment of 11g, IFG, or DMSO. The GAPDH signal was used for the loading control, n = 3.
CONCLUSIONS
In this paper, we describe the design and SAR of a series of quinazoline GCase inhibitors having single digit nanomolar potency. The SAR suggested that a hydrophobic interaction and a π-π interaction may be involved in compound binding to GCase. These quinazoline derivatives also stabilized GCase, as indicated by thermal shift assays, and exhibited high selectivity against other lysosomal hydrolases. Furthermore, the most potent compound (11g) increased the mature post-ER form of GCase and the enzyme activity in patient-derived N370S fibroblasts. It will be of interest to further test these compounds in other biological assays and models of Gaucher’s and Parkinson’s disease.
EXPERIMENTAL SECTION
Materials and Methods
Chemistry
Commercially available reagents and solvents were used without further purification. All reactions were monitored by thin-layer chromatography (TLC) using 0.25 mm Silicycle extra hard 250 µM TLC plates (60 F254). Purification of reaction products was carried out by flash chromatography using an Agilent 971-FP flash purification system with Silicycle silica gel columns. The yields are not optimized. The purity of all compounds was over 95% as analyzed with an Agilent 1260 Infinity HPLC system and an Agilent Poroshell 120 EC-C18 (4.6 × 50 mm, 2.7 µm) reverse phase column, detecting with UV absorbance (254 nm). 1H NMR and 13C NMR spectra were obtained using a Bruker Avance III 500 MHz system (500 MHz for 1H NMR and 125 MHz for 13C NMR) spectrometer. Chemical shifts are reported relative to chloroform (δ = 7.26 for 1H NMR and δ = 77.16 for 13C NMR spectra) or dimethyl sulfoxide (δ = 2.50 for 1H and δ = 39.52 for 13C NMR spectra). Data are reported as br = broad, s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet. Mass spectra were obtained using a Bruker AmaZon SL system. High resolution mass spectra (HRMS) were performed using an Agilent 6210A LC-TOF instrument with a dual spray ESI source, with a high resolution Time of Flight (TOF) Mass analyzer and collecting in a 2GigHz detector mode, coupled with an Agilent 1200 HPLC.
Preparation of 4-chloro-2-(pyridin-3-yl)quinazoline (5)25
To a solution of 2-aminobenzonitrile (5.90 g, 50 mmol) in sulfolane (20 mL) was added nicotinoyl chloride hydrochloride (12.0 g, 67.4 mmol), and the mixture was stirred at 100°C for 16 h. PCl5 (18.2 g, 87.5 mmol) was added in one portion and stirred at 100 °C for 10 h. The mixture was cooled to room temperature, and carefully poured into 400 mL of saturated sodium bicarbonate solution cooling in an ice bath. The solid was filtered, washed with water, dried, and purified by flash chromatography to give 5 as a pale-yellow solid (5.50 g, 46%); mp 160–163 °C. 1H NMR (400 MHz, CDCl3) δ 9.76 (d, J = 1.3 Hz, 1H), 8.82 (dt, J = 8.0, 1.9 Hz, 1H), 8.73 (dd, J = 4.7, 1.4 Hz, 1H), 8.26 (dd, J = 8.4, 0.8 Hz, 1H), 8.10 (d, J = 8.4 Hz, 1H), 7.95 (ddd, J = 8.4, 7.0, 1.4 Hz, 1H), 7.69 (ddd, J = 8.2, 7.0, 1.1 Hz, 1H), 7.44 (dd, J = 7.5, 4.8 Hz, 1H). 13C NMR: (100 MHz, CDCl3) δ 162.9, 158.3, 151.8, 151.7, 150.3, 136.1, 135.2, 132.4, 129.1, 128.9, 126.0, 123.5, 122.8. ESI-MS m/z: 242 (M+H)+.
General Procedure for Compound 4, 6a-6i, 7a-7i, 8a-8g and 9a-9f
A mixture of 4-chloro-2-(pyridin-3-yl)quinazoline 5 (72 mg, 0.3 mmol), amine (0.3 mmol), and potassium carbonate (69 mg, 0.3 mmol) in DMF (3 mL) was stirred at room temperature or 60 °C overnight. Water (20 mL) was added, and the formed solid was filtered, washed with water, and dried in vacuo to give product. The products were usually pure (>95% purity). Those products without sufficient purity were purified by flash chromatography.
Analytical Data for Compounds
N-Cyclohexyl-2-(pyridin-3-yl)quinazolin-4-amine (4)
Off-white solid (64 mg, 70%); mp 108–111 °C. 1H NMR (500 MHz, CDCl3) δ 9.74 (s, 1H), 8.84 (d, J = 6.6 Hz, 1H), 8.70 (s, 1H), 7.95 (d, J = 7.1 Hz, 1H), 7.74 (t, J = 7.7 Hz, 2H), 7.49 – 7.35 (m, 2H), 5.77 (s, 1H), 4.48 – 4.32 (m, 1H), 2.28 – 2.20 (m, 2H), 1.93 – 1.79 (m, 2H), 1.79 – 1.67 (m, 1H), 1.61 – 1.46 (m, 2H), 1.46 – 1.23 (m, 3H). 13C NMR (125 MHz, CDCl3) δ 158.9, 158.8, 150.7, 150.5, 150.3, 135.7, 134.6, 132.7, 129.0, 125.8, 123.2, 120.5, 113.9, 50.1, 33.0, 25.9, 25.1. HRMS (ESI): calcd for C19H21N4 [M+H]+, 305.1761; found, 305.1764.
N-(4-Methylcyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (cis/trans = 3/2) (6a)
White solid (54 mg, 43%); mp 130–134 °C.
Cis isomer: 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.80 (d, J = 7.8 Hz, 1H), 8.69 (s, 1H), 7.92 (d, J = 8.3 Hz, 1H), 7.77 – 7.71 (m, 2H), 7.48 – 7.39 (m, 2H), 5.87 (d, J = 5.8 Hz, 1H), 4.65 – 4.58 (m, 1H), 2.02–1.94 (m, 2H), 1.86–1.81 (m, 2H), 1.76 – 1.62 (m, 3H), 1.36 – 1.21 (m, 2H), 1.00 (d, J = 6.5 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.8, 150.8, 150.4, 150.3, 135.8, 134.5, 132.8, 128.8, 125.9, 123.3, 120.5, 113.9, 47.2, 30.8, 30.4, 29.2, 21.3.
Trans isomer: 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.80 (d, J = 7.8 Hz, 1H), 8.69 (s, 1H), 7.92 (d, J = 8.3 Hz, 1H), 7.77 – 7.70 (m, 2H), 7.48 – 7.39 (m, 2H), 5.63 (d, J = 5.3 Hz, 1H), 4.38 – 4.26 (m, 1H), 2.28 (d, J = 9.9 Hz, 2H), 1.86–1.81 (m, 2H), 1.54 – 1.40 (m, 1H), 1.40 – 1.30 (m, 2H), 1.30 – 1.14 (m, 2H), 0.98 (d, J = 6.5 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 159.1, 158.8, 150.8, 150.5, 150.4, 135.8, 134.5, 132.8, 129.0, 125.9, 123.3, 120.7, 114.0, 50.5, 34.1, 33.1, 32.3, 22.4. HRMS (ESI): calcd for C20H23N4 [M+H]+, 319.1917; found, 319.1921.
N-(4-Ethylcyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (cis/trans = 3/2) (6b)
Pale-yellow solid (38 mg, 30%); mp 62–65 °C.
Cis isomer: 1H NMR (500 MHz, CDCl3) δ 9.74 (s, 1H), 8.81 (d, J = 7.7 Hz, 1H), 8.68 (s, 1H), 7.92 (d, J = 8.2 Hz, 1H), 7.77 – 7.71 (m, 1H), 7.48 – 7.39 (m, 2H), 5.85 (d, J = 5.8 Hz, 1H), 4.67 – 4.61 (m, 1H), 2.02–1.70 (m, 5H), 1.38 – 1.15 (m, 4H), 0.93 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.8, 150.8, 150.4, 135.8, 134.5, 132.8, 128.9, 125.9, 123.3, 120.5, 113.9, 47.5, 39.0, 30.8, 29.7, 29.3, 28.1, 11.8.
Trans isomer: 1H NMR (500 MHz, CDCl3) δ 9.74 (s, 1H), 8.81 (d, J = 7.7 Hz, 1H), 8.68 (s, 1H), 7.92 (d, J = 8.2 Hz, 1H), 7.77 – 7.71 (m, 1H), 7.48 – 7.39 (m, 2H), 5.63 (brs, 1H), 4.38 – 4.30 (m, 1H), 2.28 (d, J = 9.9 Hz, 2H), 1.99–1.70 (m, 2H), 1.54 – 1.40 (m, 1H), 1.40 – 1.30 (m, 2H), 1.30 – 1.15 (m, 2H), 0.93 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 159.1, 158.8, 150.8, 150.4, 135.8, 134.5, 132.8, 129.0, 125.9, 123.3, 120.7, 114.0, 50.8, 37.7, 33.1, 31.7, 28.3, 11.8. HRMS (ESI): calcd for C21H25N4 [M+H]+, 333.2074; found, 333.2080.
Trans-N-(4-Methylcyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (6c)
White solid (52 mg, 55%); mp 173–175 °C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.80 (d, J = 7.8 Hz, 1H), 8.69 (s, 1H), 7.92 (d, J = 8.3 Hz, 1H), 7.77 – 7.71 (m, 1H), 7.70 (d, J = 8.2 Hz, 1H), 7.53 – 7.33 (m, 2H), 5.63 (d, J = 5.3 Hz, 1H), 4.38 – 4.26 (m, 1H), 2.28 (d, J = 9.9 Hz, 2H), 1.84 (d, J = 12.6 Hz, 2H), 1.54 – 1.40 (m, 1H), 1.40 – 1.30 (m, 2H), 1.30 – 1.14 (m, 2H), 0.98 (d, J = 6.5 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.8, 150.8, 150.5, 150.4, 135.7, 134.6, 132.7, 129.0, 125.7, 123.2, 120.5, 113.9, 50.4, 34.1, 33.1, 32.3, 22.3. HRMS (ESI): calcd for C20H23N4 [M+H]+, 319.1917; found, 319.1921.
N-(4-Methoxycyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (6d)
White solid (50 mg, 50%); mp 163–165 °C. 1H NMR (500 MHz, CDCl3) δ 9.73 (s, 1H), 8.81 (d, J = 7.7 Hz, 1H), 8.70 (d, J = 3.4 Hz, 1H), 7.95 (d, J = 8.2 Hz, 1H), 7.81 – 7.57 (m, 2H), 7.52 – 7.32 (m, 2H), 5.69 (d, J = 0.8 Hz, 1H), 4.52 – 4.27 (m, 1H), 3.40 (s, 3H), 3.31 – 3.13 (m, 1H), 2.35 (d, J = 12.5 Hz, 2H), 2.18 (d, J = 11.3 Hz, 2H), 1.61 – 1.22 (m, 4H). 13C NMR (125 MHz, CDCl3) δ 159.1, 158.7, 150.8, 150.3, 135.8, 134.3, 132.8, 128.9, 125.9, 123.3, 120.6, 113.8, 78.4, 56.0, 49.7, 30.6, 30.4. HRMS (ESI): calcd for C20H23N4O [M+H]+, 335.1866; found, 335.1866.
N-(4,4-Dimethylcyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (6e)
White solid (65 mg, 65%); mp 171–172 °C. 1H NMR (500 MHz, CDCl3) δ 9.74 (s, 1H), 8.80 (d, J = 7.9 Hz, 1H), 8.69 (d, J = 3.7 Hz, 1H), 7.90 (s, 1H), 7.79 – 7.63 (m, 2H), 7.45 (s, 1H), 7.41 (d, J = 3.1 Hz, 1H), 5.66 (d, J = 5.5 Hz, 1H), 4.43 – 4.25 (m, 1H), 2.16 – 2.00 (m, 2H), 1.64 – 1.38 (m, 6H), 1.01 (d, J = 3.0 Hz, 6H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.8, 150.8, 150.4, 135.7, 134.5, 132.7, 129.0, 125.8, 123.2, 120.6, 113.9, 50.3, 37.9, 31.8, 29.9, 28.7, 25.2. HRMS (ESI): calcd for C21H25N4 [M+H]+, 333.2074; found, 333.2076.
N-(4,4-Difluorocyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (6f)
White solid (52 mg, 51%); mp 196–197 °C. 1H NMR (500 MHz, CDCl3) δ 9.72 (s, 1H), 8.80 (d, J = 7.8 Hz, 1H), 8.70 (d, J = 2.6 Hz, 1H), 7.95 (d, J = 8.3 Hz, 1H), 7.80 – 7.75 (m, 1H), 7.73 (d, J = 8.1 Hz, 1H), 7.47 (t, J = 7.5 Hz, 1H), 7.43 (dd, J = 7.7, 4.8 Hz, 1H), 5.70 (d, J = 6.2 Hz, 1H), 4.60 – 4.45 (m, 1H), 2.32 (d, J = 12.6 Hz, 2H), 2.28 – 2.12 (m, 2H), 2.12 – 1.93 (m, 2H), 1.85 – 1.70 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 159.1, 158.5, 150.8, 150.3, 150.1, 135.8, 134.3, 133.1, 129.0, 126.2, 123.6 (q, JC-F = 241 Hz), 123.4, 120.5, 113.8, 48.1, 32.5 (t, JC-F = 24.7 Hz), 28.6 (d, JC-F = 9.2 Hz). HRMS (ESI): calcd for C19H19F2N4 [M+H]+, 341.1572; found, 341.1577.
N-(4-Dimethylaminocyclohexyl)-2-(pyridin-3-yl)quinazolin-4-amine (6g)
Pale-yellow solid (63 mg, 60%); mp 231–235°C. 1H NMR (500 MHz, CDCl3) δ 9.70 (s, 1H), 8.76 (d, J = 7.8 Hz, 1H), 8.65 (d, J = 3.6 Hz, 1H), 7.87 (d, J = 8.2 Hz, 1H), 7.78 (d, J = 8.1 Hz, 1H), 7.75 – 7.66 (m, 1H), 7.42 (t, J = 7.5 Hz, 1H), 7.37 (dd, J = 7.5, 4.9 Hz, 1H), 6.02 (d, J = 6.7 Hz, 1H), 4.64 (s, 1H), 2.38 (s, 6H), 2.30 (s, 1H), 2.05 (s, 2H), 1.83 (s, 6H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.6, 150.6, 150.3, 150.2, 135.7, 134.6, 132.7, 128.8, 125.9, 123.2, 120.8, 114.0, 62.2, 46.0, 42.5, 27.9, 25.6. HRMS (ESI): calcd for C21H26N5 [M+H]+, 348.2183; found, 348.2184.
N-(Tetrahydro-2H-pyran-3-yl)-2-(pyridin-3-yl) quinazolin-4-amine (6h)
White solid (48 mg, 53%); mp 112–113 °C. 1H NMR (500 MHz, CDCl3) δ 9.72 (s, 1H), 8.80 (dt, J = 7.9, 1.9 Hz, 1H), 8.68 (d, J = 3.7 Hz, 1H), 7.93 (d, J = 8.3 Hz, 1H), 7.80 (d, J = 8.2 Hz, 1H), 7.78 – 7.73 (m, 1H), 7.50 – 7.44 (m, 1H), 7.41 (dd, J = 7.8, 4.8 Hz, 1H), 6.18 (d, J = 7.1 Hz, 1H), 4.68 – 4.59 (m, 1H), 4.01 (dd, J = 11.5, 2.7 Hz, 1H), 3.89 – 3.80 (m, 1H), 3.80 – 3.66 (m, 2H), 2.08 – 2.00 (m, 3H), 1.95 – 1.83 (m, 1H), 1.73 – 1.61 (m, 1H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.6, 150.7, 150.3, 150.2, 135.9, 134.4, 133.0, 128.9, 126.1, 123.3, 120.7, 113.9, 71.5, 68.6, 46.5, 27.8, 23.0. HRMS (ESI): calcd for C18H19N4O [M+H]+, 307.1553; found, 307.1556.
N-(1-Methylpiperidin-4-yl)-2-(pyridin-3-yl)quinazolin-4-amine (6i)
Pale-yellow solid (34 mg, 35%); mp 132–133 °C. 1H NMR (500 MHz, CDCl3) δ 9.72 (s, 1H), 8.78 (d, J = 7.5 Hz, 1H), 8.68 (d, J = 3.4 Hz, 1H), 7.90 (d, J = 8.1 Hz, 1H), 7.73 (t, J = 7.4 Hz, 1H), 7.49 – 7.34 (m, 3H), 5.73 (d, J = 6.1 Hz, 1H), 4.48 – 4.30 (m, 1H), 2.91 (d, J = 9.1 Hz, 2H), 2.36 (s, 3H), 2.32 – 2.17 (m, 4H), 1.72 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.7, 150.8, 150.4, 150.3, 135.6, 134.5, 132.8, 129.0, 125.9, 123.2, 120.7, 113.9, 54.7, 48.0, 46.3, 32.1. HRMS (ESI): calcd for C19H22N5 [M+H]+, 320.1870; found, 320.1872.
N-(1-Isopropylpiperidin-4-yl)-2-(pyridin-3-yl)quinazolin-4-amine (6j)
Yellow solid (49 mg, 47%); mp 191–192 °C. 1H NMR (500 MHz, CDCl3) δ 9.73 (d, J = 1.2 Hz, 1H), 8.79 (dt, J = 7.8, 1.5 Hz, 1H), 8.68 (dd, J = 4.6, 1.2 Hz, 1H), 7.91 (d, J = 8.3 Hz, 1H), 7.77 – 7.73 (m, 1H), 7.71 (d, J = 8.2 Hz, 1H), 7.49 – 7.43 (m, 1H), 7.41 (dd, J = 7.8, 4.8 Hz, 1H), 5.68 (d, J = 7.1 Hz, 1H), 4.49 – 4.35 (m, 1H), 2.99 (d, J = 11.1 Hz, 2H), 2.90 – 2.79 (m, 1H), 2.47 (t, J = 11.6 Hz, 2H), 2.28 (d, J = 11.5 Hz, 2H), 1.80 – 1.66 (m, 2H), 1.13 (d, J = 6.4 Hz, 6H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.7, 150.8, 150.5, 150.4, 135.7, 134.5, 132.8, 129.0, 125.9, 123.3, 120.6, 113.9, 54.9, 48.7, 47.8, 32.3, 18.4. HRMS (ESI): calcd for C21H26N5 [M+H]+, 348.2183; found, 348.2184.
N-(1-Benzylpiperidin-4-yl)-2-(pyridin-3-yl)quinazolin-4-amine (6k)
Yellow solid (67 mg, 57%); mp 180–182 °C. 1H NMR (500 MHz, CDCl3) δ 9.72 (d, J = 1.6 Hz, 1H), 8.78 (dt, J = 7.9, 1.9 Hz, 1H), 8.69 (dd, J = 4.8, 1.6 Hz, 1H), 7.91 (d, J = 8.2 Hz, 1H), 7.77 – 7.72 (m, 1H), 7.71 (d, J = 8.1 Hz, 1H), 7.50 – 7.43 (m, 1H), 7.41 (dd, J = 7.8, 5.0 Hz, 1H), 7.39 – 7.27 (m, 5H), 5.66 (d, J = 7.1 Hz, 1H), 4.49 – 4.37 (m, 1H), 3.62 (s, 2H), 2.98 (d, J = 10.6 Hz, 2H), 2.38 – 2.29 (m, 2H), 2.24 (d, J = 11.4 Hz, 2H), 1.81 – 1.69 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.7, 150.8, 150.5, 150.3, 135.6, 134.5, 132.8, 129.4, 129.0, 128.4, 127.4, 125.9, 123.2, 120.6, 113.9, 63.2, 52.5, 48.4, 32.0. HRMS (ESI): calcd for C25H26N5 [M+H]+, 396.2183; found, 396.2184.
N-Cyclooctyl-2-(pyridin-3-yl)quinazolin-4-amine (7a)
White solid (65 mg, 66%); mp 140–142 °C. 1H NMR (500 MHz, CDCl3) δ 9.75 (d, J = 1.4 Hz, 1H), 8.82 (d, J = 7.9 Hz, 1H), 8.69 (dd, J = 4.7, 1.4 Hz, 1H), 7.92 (d, J = 8.3 Hz, 1H), 7.76 – 7.72 (m, 1H), 7.70 (d, J = 8.1 Hz, 1H), 7.51 – 7.35 (m, 2H), 5.73 (d, J = 6.2 Hz, 1H), 4.73 – 4.56 (m, 1H), 2.18 – 2.00 (m, 2H), 2.00 – 1.43 (m, 12H). 13C NMR (125 MHz, CDCl3) δ 158.8, 158.6, 150.7, 150.3, 150.2, 135.8, 134.5, 132.7, 128.9, 125.8, 123.3, 120.5, 114.0, 51.0, 32.7, 27.2, 26.1, 24.2. HRMS (ESI): calcd for C21H25N4 [M+H]+, 333.2074; found, 333.2076.
N-Cycloheptyl-2-(pyridin-3-yl)quinazolin-4-amine (7b)
Off-white solid (70 mg, 74%); mp 158–159°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.82 (d, J = 7.9 Hz, 1H), 8.68 (s, 1H), 7.93 (d, J = 8.3 Hz, 1H), 7.78 – 7.65 (m, 2H), 7.50 – 7.37 (m, 2H), 5.76 (d, J = 5.6 Hz, 1H), 4.64 – 4.46 (m, 1H), 2.25 – 2.12 (m, 2H), 1.83 – 1.52 (m, 10H). 13C NMR (125 MHz, CDCl3) δ 158.7, 158.6, 150.7, 150.3, 150.2, 135.8, 134.5, 132.7, 128.8, 125.9, 123.3, 120.6, 113.9, 52.3, 34.9, 28.3, 24.6. HRMS (ESI): calcd for C20H23N4 [M+H]+, 319.1917; found, 319.1918.
N-Cyclopentyl-2-(pyridin-3-yl)quinazolin-4-amine (7c)
Off-white solid (52 mg, 60%); mp 147–148°C. 1H NMR (500 MHz, CDCl3) δ 9.76 (d, J = 1.5 Hz, 1H), 8.81 (dt, J = 7.9, 1.9 Hz, 1H), 8.68 (dd, J = 4.8, 1.7 Hz, 1H), 7.91 (d, J = 8.2 Hz, 1H), 7.76 – 7.71 (m, 1H), 7.70 (d, J = 8.2 Hz, 1H), 7.46 – 7.42 (m, 1H), 7.40 (dd, J = 7.9, 4.8 Hz, 1H), 5.73 (d, J = 6.1 Hz, 1H), 4.80 – 4.72 (m, 1H), 2.34 – 2.25 (m, 2H), 1.86 – 1.71 (m, 4H), 1.67 – 1.59 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 159.4, 158.8, 150.7, 150.4, 150.3, 135.7, 134.6, 132.7, 129.0, 125.8, 123.2, 120.6, 113.9, 53.2, 33.4, 24.1. HRMS (ESI): calcd for C18H19N4 [M+H]+, 291.1604; found, 291.1604.
N-Cyclobutyl-2-(pyridin-3-yl)quinazolin-4-amine (7d)
Off-white solid (47 mg, 57%); mp 173–174°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.87 – 8.79 (m, 1H), 8.69 (d, J = 4.2 Hz, 1H), 7.93 (dd, J = 8.0, 4.1 Hz, 1H), 7.74 (t, J = 8.2 Hz, 2H), 7.47 – 7.43 (m, 1H), 7.43 – 7.39 (m, 1H), 6.03 (s, 1H), 4.96 – 4.97 (m, 1H), 2.66 – 2.54 (m, 2H), 2.16 – 2.03 (m, 2H), 1.96 – 1.85 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 158.8, 150.7, 150.5, 150.4, 135.7, 134.5, 132.8, 129.0, 125.9, 123.2, 120.6, 113.7, 46.8, 31.4, 15.6. HRMS (ESI): calcd for C17H17N4 [M+H]+, 277.1448; found, 277.1449.
N-Cyclopropyl-2-(pyridin-3-yl)quinazolin-4-amine (7e)
Pale-yellow solid (30 mg, 38%); mp 180–181°C. 1H NMR (500 MHz, CDCl3) δ 9.81 (s, 1H), 8.89 (d, J = 7.9 Hz, 1H), 8.70 (d, J = 3.7 Hz, 1H), 7.96 (d, J = 8.3 Hz, 1H), 7.77 – 7.73 (m, 1H), 7.71 (d, J = 8.2 Hz, 1H), 7.47 – 7.36 (m, 2H), 6.14 (s, 1H), 3.17 (td, J = 6.9, 3.1 Hz, 1H), 1.07 – 0.95 (m, 2H), 0.79 – 0.67 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 160.9, 158.9, 150.8, 150.5, 150.4, 135.8, 134.5, 132.8, 129.0, 126.0, 123.2, 120.6, 113.9, 24.5, 7.5. HRMS (ESI): calcd for C16H15N4 [M+H]+, 263.1291; found, 263.1291.
N-(Adamantan-1-yl)-2-(pyridin-3-yl)quinazolin-4-amine (7f)
White solid (37 mg, 35%); mp 114–116°C. 1H NMR (500 MHz, CDCl3) δ 9.73 (d, J = 1.6 Hz, 1H), 8.83 (d, J = 7.8 Hz, 1H), 8.69 (dd, J = 4.7, 1.5 Hz, 1H), 7.92 (d, J = 8.1 Hz, 1H), 7.80 – 7.69 (m, 1H), 7.66 (d, J = 8.1 Hz, 1H), 7.51 – 7.32 (m, 2H), 5.58 (s, 1H), 2.38 (d, J = 2.3 Hz, 6H), 2.25 – 2.18 (m, 3H), 1.88 – 1.74 (m, 6H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.1, 150.7, 150.3, 135.8, 134.5, 132.6, 128.9, 125.9, 123.3, 120.6, 114.2, 53.6, 41.7, 36.7, 29.7. HRMS (ESI): calcd for C23H25N4 [M+H]+, 357.2074; found, 357.2074.
N-(Bicyclo[2.2.1]heptan-2-yl)-2-(pyridin-3-yl)quinazolin-4-amine (7g)
White solid (47 mg, 50%); mp 145–146°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.82 (dt, J = 7.9, 1.8 Hz, 1H), 8.68 (d, J = 3.9 Hz, 1H), 7.91 (d, J = 8.3 Hz, 1H), 7.75 – 7.71 (m, 1H), 7.69 (d, J = 8.2 Hz, 1H), 7.49 – 7.35 (m, 2H), 5.64 (d, J = 5.5 Hz, 1H), 4.27 – 4.16 (m, 1H), 2.51 (d, J = 4.1 Hz, 1H), 2.39 (s, 1H), 2.05 (ddd, J = 13.2, 7.9, 2.2 Hz, 1H), 1.73 – 1.61 (m, 1H), 1.61 – 1.53 (m, 1H), 1.51 (d, J = 10.1 Hz, 1H), 1.48 – 1.36 (m, 2H), 1.34 – 1.20 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 158.9, 158.7, 150.7, 150.3, 150.2, 135.8, 134.5, 132.7, 128.9, 125.8, 123.3, 120.5, 113.9, 55.1, 42.1, 41.0, 36.0, 35.9, 28.3, 26.7. HRMS (ESI): calcd for C20H21N4 [M+H]+, 317.1761; found, 317.1763.
N-(Cyclohexylmethyl)-2-(pyridin-3-yl)quinazolin-4-amine (7h)
Pale-yellow solid (67 mg, 71%); mp 121–123°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.81 (d, J = 7.8 Hz, 1H), 8.68 (d, J = 3.8 Hz, 1H), 7.91 (d, J = 8.2 Hz, 1H), 7.79 – 7.65 (m, 2H), 7.50 – 7.34 (m, 2H), 5.98 (s, 1H), 3.65 (t, J = 6.1 Hz, 2H), 1.87 (d, J = 12.6 Hz, 2H), 1.82 – 1.61 (m, 4H), 1.32 – 1.18 (m, 3H), 1.14 – 1.04 (m, 1H). 13C NMR (125 MHz, CDCl3) δ 159.9, 158.8, 150.7, 150.3, 135.7, 134.6, 132.7, 128.9, 125.8, 123.2, 120.5, 114.0, 47.6, 38.0, 31.3, 26.5, 26.0. HRMS (ESI): calcd for C20H23N4 [M+H]+, 319.1917; found, 319.1920.
N-(2-Cyclohexylethyl)-2-(pyridin-3-yl)quinazolin-4-amine (7i)
White solid (48 mg, 48%); mp 153–154°C. 1H NMR (500 MHz, CDCl3) δ 9.76 (s, 1H), 8.86 – 8.78 (m, 1H), 8.69 (d, J = 4.1 Hz, 1H), 7.92 (dd, J = 7.9, 5.4 Hz, 1H), 7.79 – 7.67 (m, 2H), 7.48 – 7.37 (m, 2H), 5.84 (s, 1H), 3.86 – 3.75 (m, 2H), 1.83 (d, J = 12.3 Hz, 2H), 1.76 – 1.60 (m, 5H), 1.50 – 1.38 (m, 1H), 1.33 – 1.12 (m, 3H), 1.08 – 0.96 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 159.7, 158.8, 150.7, 150.4, 150.3, 135.8, 135.7, 134.5, 132.8, 132.7, 128.9, 128.9, 125.9, 123.2, 120.6, 114.0, 39.4, 37.0, 35.7, 33.4, 26.6, 26.3. HRMS (ESI): calcd for C21H25N4 [M+H]+, 333.2074; found, 333.2074.
N-Phenyl-2-(pyridin-3-yl)quinazolin-4-amine (8a)
Off-white solid (55 mg, 62%); mp 179–180°C. 1H NMR (500 MHz, CDCl3) δ 9.73 (d, J = 1.8 Hz, 1H), 8.82 (dt, J = 7.9, 1.9 Hz, 1H), 8.70 (dd, J = 4.8, 1.6 Hz, 1H), 8.01 (d, J = 8.3 Hz, 1H), 7.95 (d, J = 8.1 Hz, 1H), 7.86 (d, J = 7.6 Hz, 2H), 7.84 – 7.80 (m, 1H), 7.69 (brs, 1H), 7.61 – 7.52 (m, 1H), 7.50 – 7.45 (m, 2H), 7.43 (dd, J = 7.9, 4.8 Hz, 1H), 7.21 (t, J = 7.4 Hz, 1H). 13C NMR (125 MHz, CDCl3) δ 158.4, 157.6, 150.8, 150.7, 150.2, 138.4, 136.1, 134.2, 133.3, 129.3, 129.2, 126.8, 124.6, 123.4, 121.6, 120.5, 114.1. HRMS (ESI): calcd for C19H15N4 [M+H]+, 299.1291; found, 299.1292.
N-Benzyl-2-(pyridin-3-yl)quinazolin-4-amine (8b)
Off-white solid (65 mg, 70%); mp 140–141°C. 1H NMR (500 MHz, CDCl3) δ 9.56 (s, 1H), 8.61 (d, J = 7.9 Hz, 1H), 8.47 (d, J = 3.6 Hz, 1H), 7.74 (d, J = 8.3 Hz, 1H), 7.61 (d, J = 8.2 Hz, 1H), 7.58 – 7.44 (m, 1H), 7.28 – 7.18 (m, 4H), 7.18 – 7.13 (m, 2H), 7.13 – 7.07 (m, 1H), 6.39 (s, 1H), 4.78 (d, J = 5.4 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.6, 158.7, 150.6, 150.4, 150.2, 138.5, 135.8, 134.5, 132.8, 128.8, 128.0, 127.7, 126.0, 123.2, 120.9, 113.9, 45.4. HRMS (ESI): calcd for C20H17N4 [M+H]+, 313.1448; found, 313.1453.
N-Phenethyl-2-(pyridin-3-yl)quinazolin-4-amine (8c)
White solid (43 mg, 44%); mp 140–141°C. 1H NMR (500 MHz, CDCl3) δ 9.79 (s, 1H), 8.85 (d, J = 7.1 Hz, 1H), 8.70 (s, 1H), 7.94 (d, J = 8.4 Hz, 1H), 7.78 – 7.69 (m, 1H), 7.60 (d, J = 8.0 Hz, 1H), 7.46 – 7.39 (m, 2H), 7.38 – 7.31 (m, 2H), 7.30 – 7.25 (m, 3H), 5.98 (s, 1H), 4.11 – 3.99 (m, 2H), 3.11 (t, J = 6.9 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.6, 158.7, 150.8, 150.3, 150.2, 139.0, 135.8, 134.4, 132.9, 129.0, 128.9, 126.8, 126.0, 123.3, 120.5, 113.9, 42.6, 35.4. HRMS (ESI): calcd for C21H19N4 [M+H]+, 327.1604; found, 327.1603.
N-(3-Phenylpropyl)-2-(pyridin-3-yl)quinazolin-4-amine (8d)
Off-white sticky oil (60 mg, 59%). 1H NMR (500 MHz, CDCl3) δ 9.74 (s, 1H), 8.76 (dt, J = 7.9, 1.8 Hz, 1H), 8.68 (d, J = 3.6 Hz, 1H), 7.90 (d, J = 8.3 Hz, 1H), 7.78 – 7.66 (m, 1H), 7.53 (d, J = 7.8 Hz, 1H), 7.45 – 7.34 (m, 2H), 7.33 – 7.27 (m, 2H), 7.25 – 7.19 (m, 3H), 5.94 (s, 1H), 3.89 – 3.75 (m, 2H), 2.81 (t, J = 7.4 Hz, 2H), 2.13 (p, J = 7.2 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.7, 158.7, 150.7, 150.3, 150.2, 141.6, 135.8, 134.5, 132.7, 128.8, 128.7, 128.5, 126.2, 125.8, 123.2, 120.6, 113.9, 41.2, 33.7, 30.7. HRMS (ESI): calcd for C22H21N4 [M+H]+, 341.1761; found, 341.1758.
N-(4-Phenylbutyl)-2-(pyridin-3-yl)quinazolin-4-amine (8e)
Pale-yellow solid (73 mg, 69%); mp 108–109°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.81 (d, J = 7.9 Hz, 1H), 8.69 (d, J = 3.7 Hz, 1H), 7.93 (d, J = 8.3 Hz, 1H), 7.77 – 7.71 (m, 1H), 7.70 (d, J = 8.1 Hz, 1H), 7.51 – 7.35 (m, 2H), 7.28 (d, J = 7.7 Hz, 2H), 7.23 – 7.10 (m, 3H), 5.90 (s, 1H), 3.89 – 3.70 (m, 2H), 2.72 (t, J = 6.7 Hz, 2H), 1.90 – 1.75 (m, 4H). 13C NMR (125 MHz, CDCl3) δ 159.7, 158.7, 150.8, 150.3, 150.1, 142.1, 135.8, 134.4, 132.8, 128.8, 128.5, 128.5, 126.0, 125.9, 123.3, 120.6, 113.9, 41.4, 35.7, 29.0, 28.9. HRMS (ESI): calcd for C23H23N4 [M+H]+, 355.1917; found, 355.1919.
N-(2-(Pyridin-2-yl)ethyl)-2-(pyridin-3-yl)quinazolin-4-amine (8f)
White solid (57 mg, 58%); mp 138–139°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (d, J = 1.2 Hz, 1H), 8.82 (dt, J = 7.9, 1.9 Hz, 1H), 8.70 – 8.66 (m, 1H), 8.51 (s, 1H), 8.47 (d, J = 4.1 Hz, 1H), 7.93 (d, J = 8.2 Hz, 1H), 7.77 – 7.72 (m, 1H), 7.68 (d, J = 8.1 Hz, 1H), 7.59 (d, J = 7.8 Hz, 1H), 7.46 – 7.38 (m, 2H), 7.24 (dd, J = 7.7, 4.9 Hz, 1H), 6.28 (t, J = 5.3 Hz, 1H), 4.10 – 3.96 (m, 2H), 3.13 (t, J = 7.0 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.7, 158.6, 150.7, 150.2, 150.1, 148.1, 136.6, 135.8, 134.7, 134.4, 133.0, 128.9, 126.2, 123.8, 123.4, 120.7, 113.9, 42.4, 32.6. HRMS (ESI): calcd for C20H18N5 [M+H]+, 328.1557; found, 328.1561.
N-(2-(Pyridin-3-yl)ethyl)-2-(pyridin-3-yl)quinazolin-4-amine (8g)
Off-white solid (67 mg, 68%); mp 102–104°C. 1H NMR (500 MHz, CDCl3) δ 9.77 (d, J = 1.4 Hz, 1H), 8.83 (dt, J = 7.9, 1.9 Hz, 1H), 8.68 (dd, J = 4.8, 1.7 Hz, 1H), 7.91 (d, J = 8.0 Hz, 1H), 7.73 (ddd, J = 8.3, 7.0, 1.3 Hz, 1H), 7.57 (d, J = 7.8 Hz, 1H), 7.46 – 7.37 (m, 2H), 7.07 (d, J = 8.3 Hz, 2H), 6.72 – 6.62 (m, 2H), 5.96 – 5.87 (m, 1H), 4.03 – 3.94 (m, 2H), 2.98 (t, J = 6.9 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.6, 158.8, 150.7, 150.3, 145.1, 135.8, 134.5, 132.8, 129.8, 128.9, 128.8, 125.9, 123.3, 120.6, 115.6, 114.0, 42.8, 34.5. HRMS (ESI): calcd for C20H18N5 [M+H]+, 328.1557; found, 328.1560.
(S)-2-(Pyridin-3-yl)-N-(1,2,3,4-tetrahydronaphthalen-2-yl)quinazolin-4-amine (9a)
Off-white solid (74 mg, 70%); mp 150–152°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.82 (d, J = 7.9 Hz, 1H), 8.68 (d, J = 3.7 Hz, 1H), 7.95 (d, J = 8.3 Hz, 1H), 7.75 (t, J = 7.7 Hz, 1H), 7.70 (d, J = 8.1 Hz, 1H), 7.47 – 7.42 (m, 1H), 7.41 (dd, J = 7.9, 4.8 Hz, 1H), 7.24 – 7.11 (m, 4H), 5.87 (d, J = 5.2 Hz, 1H), 5.00 – 4.87 (m, 1H), 3.43 (dd, J = 16.2, 5.0 Hz, 1H), 3.12 – 2.88 (m, 3H), 2.40 – 2.30 (m, 1H), 2.13 – 2.01 (m, 1H). 13C NMR (125 MHz, CDCl3) δ 159.2, 158.7, 150.8, 150.2, 135.8, 135.7, 134.3, 134.1, 132.9, 129.7, 129.1, 128.9, 126.4, 126.2, 126.0, 123.3, 120.7, 113.9, 47.1, 35.8, 28.5, 27.5. HRMS (ESI): calcd for C23H21N4 [M+H]+, 353.1761; found, 328.1762.
(R)-2-(Pyridin-3-yl)-N-(1,2,3,4-tetrahydronaphthalen-2-yl)quinazolin-4-amine (9b)
Off-white solid (65 mg, 62%); mp 153–156°C. 1H NMR (500 MHz, CDCl3) δ 9.75 (s, 1H), 8.82 (d, J = 7.9 Hz, 1H), 8.68 (d, J = 3.7 Hz, 1H), 7.95 (d, J = 8.3 Hz, 1H), 7.75 (t, J = 7.7 Hz, 1H), 7.70 (d, J = 8.1 Hz, 1H), 7.47 – 7.42 (m, 1H), 7.41 (dd, J = 7.9, 4.8 Hz, 1H), 7.24 – 7.11 (m, 4H), 5.87 (d, J = 5.2 Hz, 1H), 5.00 – 4.87 (m, 1H), 3.43 (dd, J = 16.2, 5.0 Hz, 1H), 3.12 – 2.88 (m, 3H), 2.40 – 2.30 (m, 1H), 2.13 – 2.01 (m, 1H). 13C NMR (125 MHz, CDCl3) δ 159.2, 158.7, 150.8, 150.2, 135.8, 135.7, 134.3, 134.1, 132.9, 129.7, 129.1, 128.9, 126.4, 126.2, 126.0, 123.3, 120.7, 113.9, 47.1, 35.8, 28.5, 27.5. HRMS (ESI): calcd for C23H21N4 [M+H]+, 353.1761; found, 328.1764.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(pyridin-3-yl)quinazolin-4-amine (9c)
Grey solid (75 mg, 74%); mp 167–168°C. 1H NMR (500 MHz, CDCl3) δ 9.78 (d, J = 1.7 Hz, 1H), 8.84 (dd, J = 7.9, 1.7 Hz, 1H), 8.69 (d, J = 4.7 Hz, 1H), 7.93 (d, J = 8.3 Hz, 1H), 7.77 – 7.70 (m, 1H), 7.68 (d, J = 8.1 Hz, 1H), 7.47 – 7.37 (m, 2H), 7.32 – 7.27 (m, 2H), 7.25 – 7.20 (m, 2H), 6.04 (s, 1H), 5.37 – 5.29 (m, 1H), 3.60 (dd, J = 16.2, 7.2 Hz, 2H), 3.08 (dd, J = 16.2, 4.8 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.4, 158.6, 150.7, 150.3, 141.1, 135.8, 134.4, 132.9, 128.9, 127.0, 126.0, 125.1, 123.3, 120.7, 113.9, 52.6, 40.3. HRMS (ESI): calcd for C24H21NO [M+H]+, 339.1618; found, 339.1612.
N-(1,2,3,4-Tetrahydronaphthalen-1-yl)-2-(pyridin-3-yl) quinazolin-4-amine (9d)
Pale-yellow sticky oil (35 mg, 33%). 1H NMR (500 MHz, CDCl3) δ 9.78 (d, J = 0.9 Hz, 1H), 8.89 (d, J = 7.8 Hz, 1H), 8.69 (d, J = 3.5 Hz, 1H), 8.00 (d, J = 7.8 Hz, 1H), 7.83 – 7.73 (m, 1H), 7.68 (d, J = 8.0 Hz, 1H), 7.51 – 7.41 (m, 2H), 7.39 (d, J = 7.6 Hz, 1H), 7.25 – 7.22 (m, 1H), 7.22 – 7.15 (m, 2H), 6.02 (s, 1H), 5.94 – 5.86 (m, 1H), 3.00 – 2.80 (m, 2H), 2.34 – 2.23 (m, 1H), 2.20 – 2.12 (m, 1H), 2.00 – 1.93 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.7, 150.6, 150.2, 138.1, 136.8, 136.1, 133.0, 129.5, 129.1, 128.8, 127.7, 126.6, 126.1, 123.4, 120.7, 113.8, 49.2, 29.6, 29.5, 20.2. HRMS (ESI): calcd for C23H21N4 [M+H]+, 353.1761; found, 353.1767.
N-(2,3-Dihydro-1H-inden-1-yl)-2-(pyridin-3-yl)quinazolin-4-amine (9e)
Brown solid (57 mg, 56%); mp 128–130°C. 1H NMR (500 MHz, CDCl3) δ 9.78 (d, J = 1.5 Hz, 1H), 8.84 (dt, J = 7.9, 1.9 Hz, 1H), 8.67 (dd, J = 4.8, 1.6 Hz, 1H), 7.95 (d, J = 8.4 Hz, 1H), 7.80 – 7.73 (m, 1H), 7.70 (d, J = 8.0 Hz, 1H), 7.50 – 7.37 (m, 3H), 7.35 (d, J = 7.5 Hz, 1H), 7.33 – 7.28 (m, 1H), 7.26 – 7.20 (m, 2H), 6.17 (q, J = 7.4 Hz, 1H), 6.02 (d, J = 7.5 Hz, 1H), 3.12 (ddd, J = 15.8, 8.7, 4.0 Hz, 1H), 3.04 (dt, J = 16.0, 8.0 Hz, 1H), 2.92 – 2.83 (m, 1H), 2.16 – 2.01 (m, 1H). 13C NMR (125 MHz, CDCl3) δ 159.5, 158.8, 150.8, 150.5, 150.3, 143.9, 143.4, 135.8, 134.5, 132.9, 129.0, 128.4, 127.0, 126.0, 125.2, 124.3, 123.3, 120.7, 113.8, 56.5, 34.1, 30.5. HRMS (ESI): calcd for C22H19N4 [M+H]+, 339.1604; found, 339.1610.
N-(Chroman-3-yl)-2-(pyridin-3-yl)quinazolin-4-amine (9f)
White solid (67 mg, 63%); mp 145–148°C. 1H NMR (500 MHz, CDCl3) δ 9.74 (s, 1H), 8.80 (dt, J = 7.9, 1.9 Hz, 1H), 8.68 (d, J = 3.6 Hz, 1H), 7.90 (d, J = 7.9 Hz, 1H), 7.72 (ddd, J = 8.3, 7.0, 1.3 Hz, 1H), 7.63 (d, J = 7.7 Hz, 1H), 7.45 – 7.34 (m, 2H), 7.19 – 7.12 (m, 1H), 7.07 (d, J = 6.6 Hz, 1H), 6.95 – 6.88 (m, 2H), 6.09 (d, J = 7.3 Hz, 1H), 5.11 (ddt, J = 7.3, 3.6, 1.7 Hz, 1H), 4.43 (ddd, J = 11.0, 3.8, 2.2 Hz, 1H), 4.35 (dd, J = 11.0, 1.7 Hz, 1H), 3.34 (dd, J = 16.8, 5.2 Hz, 1H), 3.10 (d, J = 16.8 Hz, 1H). 13C NMR (125 MHz, CDCl3) δ 159.0, 158.4, 153.9, 150.7, 150.3, 150.1, 135.6, 134.2, 132.9, 130.6, 128.9, 127.8, 126.0, 123.2, 121.5, 120.7, 119.5, 117.0, 113.7, 68.0, 44.0, 30.4. HRMS (ESI): calcd for C22H19N4O [M+H]+, 355.1553; found, 355.1551.
Preparation of 2-chloro-N-(2,3-dihydro-1H-inden-2-yl)quinazolin-4-amine (10)
A mixture of 2,4-dichloroquinazoline (398 mg, 2.0 mmol), 2,3-dihydro-1H-inden-2-amine (266 mg, 2.0 mmol), and potassium carbonate (276 mg, 2.0 mmol) in DMF (5 mL) was stirred at room temperature for 5 h. Water (20 mL) was added, and the formed solid was filtered, washed with water, and solid was dried to give 10 as an off-white solid (390 mg, 66 %); mp 239–240°C.
1H NMR (500 MHz, CDCl3) δ 7.76 (d, J = 8.2 Hz, 1H), 7.74 – 7.69 (m, 1H), 7.60 (d, J = 8.2 Hz, 1H), 7.41 (t, J = 7.4 Hz, 1H), 7.30 – 7.26 (m, 2H), 7.24 – 7.19 (m, 2H), 6.08 (d, J = 6.7 Hz, 1H), 5.27 – 5.15 (m, 1H), 3.53 (dd, J = 16.2, 7.0 Hz, 2H), 3.00 (dd, J = 16.2, 4.0 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 160.6, 157.8, 151.0, 140.7, 133.6, 128.0, 127.1, 126.2, 125.1, 120.8, 113.3, 52.6, 40.2. MS (ESI) m/z [M + H]+: calcd, 296.09; found, 296.13.
General Procedure for 11a-11h
A mixture of 2-chloro-N-(2,3-dihydro-1H-inden-2-yl)quinazolin-4-amine 10 (148 mg, 0.5 mmol), boronic acid (0.5 mmol), Pd(PPh3)4 (58 mg, 0.05 mmol), potassium carbonate (276 mg, 2.0 mmol) in dioxane (10 mL) and water (1.5 mL) was heated at 85 °C under an argon atmosphere for 16 h. Water (5 mL) was added, and the mixture was extracted with EtOAc (25 mL × 3). The combined organic phase was washed with brine (15 mL), dried (Na2SO4), filtered, evaporated, and purified by flash chromatography to give product.
Analytical Data for Compounds 11a-11h
N-(2,3-Dihydro-1H-inden-2-yl)-2-(4-methylpyridin-3-yl)quinazolin-4-amine (11a)
White solid (125 mg, 71%); mp 170–172°C. 1H NMR (500 MHz, CDCl3) δ 9.16 (s, 1H), 8.50 (d, J = 5.0 Hz, 1H), 7.91 (d, J = 7.9 Hz, 1H), 7.75 (ddd, J = 8.3, 7.0, 1.2 Hz, 1H), 7.67 (d, J = 7.8 Hz, 1H), 7.49 – 7.42 (m, 1H), 7.29 (dd, J = 5.3, 3.4 Hz, 2H), 7.25 – 7.19 (m, 3H), 5.94 (d, J = 7.0 Hz, 1H), 5.26 (tdd, J = 7.1, 4.4, 2.6 Hz, 1H), 3.54 (dd, J = 16.2, 7.1 Hz, 2H), 3.04 (dd, J = 16.2, 4.4 Hz, 2H), 2.72 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 161.1, 159.1, 151.3, 150.1, 149.2, 146.6, 140.9, 135.4, 132.7, 128.8, 126.8, 125.9, 124.9, 120.7, 113.2, 52.3, 40.2, 21.0. HRMS (ESI): calcd for C23H21N4 [M+H]+, 353.1761; found, 353.1765.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(5-methylpyridin-3-yl)quinazolin-4-amine (11b)
Yellow solid (134 mg, 76%); mp 200–201°C. 1H NMR (500 MHz, CDCl3) δ 9.59 (d, J = 1.6 Hz, 1H), 8.63 (s, 1H), 8.53 (d, J = 1.6 Hz, 1H), 7.92 (d, J = 8.0 Hz, 1H), 7.77 – 7.71 (m, 1H), 7.66 (d, J = 7.8 Hz, 1H), 7.45 – 7.38 (m, 1H), 7.30 (dd, J = 5.3, 3.4 Hz, 2H), 7.23 (dd, J = 5.5, 3.2 Hz, 2H), 5.95 (d, J = 6.8 Hz, 1H), 5.40 – 5.29 (m, 1H), 3.61 (dd, J = 16.2, 7.2 Hz, 2H), 3.08 (dd, J = 16.2, 4.8 Hz, 2H), 2.45 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 159.3, 158.8, 151.2, 150.3, 147.6, 141.1, 136.0, 134.0, 132.7, 132.6, 128.7, 126.8, 125.7, 124.9, 120.8, 113.9, 52.4, 40.1, 18.5. HRMS (ESI): calcd for C23H21N4 [M+H]+, 353.1761; found, 353.1757.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(6-methylpyridin-3-yl)quinazolin-4-amine (11c)
Yellow solid (129 mg, 73%); mp 175–176°C. 1H NMR (500 MHz, CDCl3) δ 9.66 (d, J = 1.9 Hz, 1H), 8.69 (dd, J = 8.1, 2.2 Hz, 1H), 7.89 (d, J = 8.0 Hz, 1H), 7.73 – 7.68 (m, 1H), 7.63 (d, J = 7.9 Hz, 1H), 7.41 – 7.36 (m, 1H), 7.29 – 7.25 (m, 2H), 7.23 – 7.19 (m, 3H), 5.91 (d, J = 6.6 Hz, 1H), 5.33 – 5.23 (m, 1H), 3.58 (dd, J = 16.2, 7.2 Hz, 2H), 3.06 (dd, J = 16.1, 4.8 Hz, 2H), 2.62 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 159.8, 159.2, 158.8, 150.4, 149.7, 141.0, 136.0, 132.6, 131.6, 128.8, 126.9, 125.6, 124.9, 122.7, 120.5, 113.7, 52.5, 40.2, 24.5. HRMS (ESI): calcd for C23H21N4 [M+H]+, 353.1761; found, 353.1767.
N-(2,3-Dihydro-1H-inden-2-yl)-2-phenylquinazolin-4-amine (11d)
White solid (84 mg, 50%); mp 221–223°C. 1H NMR (500 MHz, CDCl3) δ 8.64 – 8.56 (m, 2H), 7.93 (d, J = 8.3 Hz, 1H), 7.72 (ddd, J = 8.3, 7.0, 1.3 Hz, 1H), 7.64 (d, J = 7.8 Hz, 1H), 7.53 – 7.46 (m, 3H), 7.39 (ddd, J = 8.1, 7.0, 1.1 Hz, 1H), 7.31 (dd, J = 5.3, 3.3 Hz, 2H), 7.24 (dd, J = 5.5, 3.2 Hz, 2H), 5.86 (d, J = 6.6 Hz, 1H), 5.38 (tdd, J = 7.1, 5.1, 2.1 Hz, 1H), 3.61 (dd, J = 16.1, 7.2 Hz, 2H), 3.08 (dd, J = 16.1, 5.0 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 160.4, 159.2, 150.5, 141.1, 138.9, 132.5, 130.1, 128.9, 128.4, 128.2, 126.8, 125.3, 125.0, 120.4, 113.6, 52.4, 40.2. HRMS (ESI): calcd for C23H20N3 [M+H]+, 338.1652; found, 338.1647.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(pyridin-4-yl)quinazolin-4-amine (11e)
Brown solid (59 mg, 35%); mp 246–247°C. 1H NMR (500 MHz, CDCl3) δ 8.74 (d, J = 5.5 Hz, 2H), 8.40 (d, J = 5.9 Hz, 2H), 7.93 (d, J = 8.2 Hz, 1H), 7.78 – 7.70 (m, 1H), 7.67 (d, J = 8.1 Hz, 1H), 7.48 – 7.39 (m, 1H), 7.29 (dd, J = 5.2, 3.4 Hz, 2H), 7.22 (dd, J = 5.4, 3.3 Hz, 2H), 5.99 (d, J = 6.7 Hz, 1H), 5.37 – 5.29 (m, 1H), 3.59 (dd, J = 16.2, 7.2 Hz, 2H), 3.07 (dd, J = 16.1, 4.8 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.4, 158.4, 150.2, 150.2, 146.4, 141.0, 132.8, 129.2, 126.9, 126.3, 125.0, 122.3, 120.6, 114.1, 77.2, 77.0, 76.7, 52.5, 40.1. HRMS (ESI): calcd for C22H19N4 [M+H]+, 339.1604; found, 339.1608.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(thiophen-2-yl)quinazolin-4-amine (11f)
Off-white solid (68 mg, 40%); mp 243–246°C. 1H NMR (500 MHz, CDCl3) δ 8.08 (d, J = 3.1 Hz, 1H), 7.85 (d, J = 8.4 Hz, 1H), 7.69 (t, J = 7.6 Hz, 1H), 7.61 (d, J = 8.1 Hz, 1H), 7.45 (d, J = 4.9 Hz, 1H), 7.38 – 7.33 (m, 1H), 7.32 – 7.27 (m, 2H), 7.23 (dd, J = 5.3, 3.3 Hz, 2H), 7.18 – 7.13 (m, 1H), 5.88 (d, J = 6.3 Hz, 1H), 5.31 – 5.24 (m, 1H), 3.60 (dd, J = 16.2, 7.2 Hz, 2H), 3.06 (dd, J = 16.2, 5.0 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.0, 157.3, 150.4, 145.2, 141.1, 132.6, 129.0, 128.5, 128.2, 127.9, 126.8, 125.1, 124.9, 120.5, 113.5, 52.5, 40.1. HRMS (ESI): calcd for C21H18N3S [M+H]+, 344.1216; found, 344.1218.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(thiophen-3-yl)quinazolin-4-amine (11g)
Yellow solid (75 mg, 44%); mp 232–233°C. 1H NMR (500 MHz, CDCl3) δ 8.34 (d, J = 3.0 Hz, 1H), 8.02 (dd, J = 5.0, 0.9 Hz, 1H), 7.87 (d, J = 8.3 Hz, 1H), 7.74 – 7.65 (m, 1H), 7.62 (d, J = 8.0 Hz, 1H), 7.42 – 7.34 (m, 2H), 7.30 (dd, J = 5.2, 3.4 Hz, 2H), 7.25 – 7.21 (m, 2H), 5.85 (d, J = 6.5 Hz, 1H), 5.36 – 5.25 (m, 1H), 3.58 (dd, J = 16.2, 7.2 Hz, 2H), 3.07 (dd, J = 16.1, 5.0 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.1, 157.8, 150.5, 143.1, 141.1, 132.5, 128.6, 127.9, 127.4, 126.8, 125.4, 125.1, 124.9, 120.5, 113.5, 52.3, 40.2. HRMS (ESI): calcd for C21H18N3S [M+H]+, 344.1216; found, 344.1215.
N-(2,3-Dihydro-1H-inden-2-yl)-2-(furan-2-yl)quinazolin-4-amine (11h)
Pale-yellow solid (80 mg, 49%); mp 231–232°C.1H NMR (500 MHz, CDCl3) δ 7.96 (d, J = 8.4 Hz, 1H), 7.73 – 7.67 (m, 1H), 7.67 – 7.61 (m, 2H), 7.41 – 7.32 (m, 2H), 7.32 – 7.26 (m, 2H), 7.25 – 7.21 (m, 2H), 6.57 (dd, J = 3.3, 1.7 Hz, 1H), 5.98 (d, J = 6.8 Hz, 1H), 5.34 – 5.20 (m, 1H), 3.57 (dd, J = 16.1, 7.2 Hz, 2H), 3.05 (dd, J = 16.1, 5.0 Hz, 2H). 13C NMR (125 MHz, CDCl3) δ 159.1, 153.7, 153.4, 150.1, 144.6, 141.0, 132.7, 128.8, 126.8, 125.3, 124.9, 120.5, 113.7, 113.1, 111.8, 52.3, 40.1. HRMS (ESI): calcd for C21H18N3O [M+H]+, 328.1444; found, 328.1448.
Enzymatic Assays
4-Methylumbelliferyl β-D-glucopyranoside (4MU-β-Glc), 4- methylumbelliferyl α-D-glucopyranoside,4-methylumbelliferyl α-D- galactopyranoside, and buffer components were purchased from Sigma-Aldrich (St. Louis, MO). The recombinant wild-type GCase enzyme velaglucerase alfa (Vpriv®, Shire Human Genetic Therapies, Inc.), acid α-glucosidase enzyme alglucosidase alfa (Lumizyme®, Genzyme Corporation), α-galactosidase A enzyme agalsidase beta (Fabrazyme®, Genzyme Corporation) were used in activity assays. The GCase activity assay buffer was composed of 50 mM citric acid, 176 mM K2HPO4, and 0.01% Tween-20 at pH5.0, pH 5.9 and pH7.0. A solution of 1 M sodium hydroxide and 1 M glycine (pH 10) was used as the stop solution for all three enzyme activity assays.
GCase Enzyme Activity Assay
The compounds in DMSO solution (0.5 µL/well) were transferred to a black 96-well plate (the final titration started from 100 µM, a 12 or 24-point 2-fold dilution series). Enzyme solution (33.5 µL, 7.5 nM final concentration, in pH 5.9 buffer) was transferred to the wells. After 5 min of incubation at room temperature, the enzyme reaction was initiated by the addition of blue substrate (4MU-β-Glc) (33 µL/well). The final concentration of the blue substrate was 1.5 mM. The blue substrate reaction was terminated by the addition of 33 µL/well stop solution (1 M NaOH and 1 M glycine mixture, pH 10) after 30 min of incubation at 37 °C. The fluorescence was then measured in a Biotek Synergy H1 multi-mode plate reader with Ex = 365 nm and Em = 440 nm. The selected compounds were further assayed under pH 5.0 and pH 7.0 to evaluate their selectivity under various pH conditions.
Enzyme Kinetic Assay20
The substrate resorufin β-D-glucopyranoside was diluted to five concentrations, ranging from 30 to 150 µM. Seven concentrations of inhibitors (between 0.5- and 5-fold of IC50 value) and a DMSO control were added to the enzyme solution. The final enzyme concentration was 10 nM to give a linear reaction over 10 min. Enzyme kinetics were measured by the addition of 66 µL of substrate to a 96-well assay plate, followed by 33 µL of enzyme solution (with or without inhibitor) using a dispense module on a Biotek Synergy H1 multi-mode plate reader. The increase in product fluorescence was measured at 1 min intervals for 10 min in the plate reader. The rate of product formation was calculated by converting the fluorescence units to nanomoles of product per minute using a standard curve of the free fluorophore, resorufin.
Enzyme Selectivity Assays
The acid α-glucosidase and α-galactosidase A enzyme activity assay methods were similar to the GCase enzyme activity assay above with slight modifications. The buffer for the two enzyme assays consisted of 50 mM citric acid, 176 mM K2HPO4, and 0.01% Tween-20 at pH 4.8. The final enzyme concentrations for acid α-glucosidase and α-galactosidase A were 8 and 1 nM, respectively. The substrate concentrations for these related enzymes were at 0.16 and 0.4 mM, respectively.
Fluorescence Thermal Shift Analysis32
A robotic pipeline in the High Throughput Analysis Laboratory (HTAL) was used for protein ligand screening by fluorescence thermal shift (FTS) analysis. The pipeline used a Mosquito robot (TTP Labtech) for protein dispensing and an Echo 550 (Labcyte) to add compounds. Thermal scanning coupled with fluorescence detection was performed on a real-time PCR machine CFX384 (Bio-Rad Laboratories). The assay was run in 384-well PCR plates, using 10 µL citric acid/ K2HPO4 buffer (50 mM citric acid, 150 mM K2HPO4, pH 5.0) per well. The assay concentration for protein was 1 µM and that for Sypro Orange (Invitrogen) was 5X. Protein was premixed with Sypro Orange and dispensed to a plate first, and compounds were added. Final concentrations of compounds ranged from 0.5 to 200 µM. Then the plate was sealed with an optical seal, shaken, and centrifuged. The thermal scan was from 10 to 95 °C with a temperature ramp rate of 1.5 °C/min. The fluorescence was recorded every 10 sec. Data analysis and report generation were performed using the in-house software excelFTS. The Tm of wild-type GCase was found to follow a logarithmic dose-dependent trend when denaturation was performed in the presence of isofagomine or selected compounds.
N370S Cell Culture and Compound Treatment
The N370S fibroblast cell line was obtained from Coriell, GM00372, cultured in DMEM medium (Life Tech) including 1% v/v L-glutamine 200 mM (Life Tech), 1% v/v pen strep (Life Tech), 10% FBS (Life Tech) at 37 °C and 5% CO2 and treated with different compounds at indicated concentrations. The same volume of DMSO (0.1% v/v) was used as a control. After a 3-day treatment, cells were washed with inhibitor free media three times and followed by 1% Triton X-100 lysis buffer to lyse cells. Protein concentrations were measured with a Bradford kit (Thermo), and the GCase activity was determined at pH 5.5.
Deglycosylation of Proteins/Molecular Shift Assay 33
To study the subcellular localization and transport of the various GC mutants (ER and post-ER localization), EndoH and PNGaseF digestions were performed. For both reactions 20 µg of protein was used, and the experimental procedure was performed according to the manufacturer’s handbook (New England Biolabs). A positive digestion resulted in a shift in molecular size after the protein was subjected to SDS/PAGE. Anti-hGC (From Johannes Aerts, Leiden University, Leiden, The Netherlands) was used to detect the different forms of GC.
Western Blot
Proteins were denatured in 20% SDS sample buffer at 100 °C for 10 min; 10% Bis-Tris gel (Life Tech) was used for gels; Trans-Blot Turbo PVDF kit (Bio-Rad) was used for membrane transfer. GCase antibody (Sigma-Aldrich) and GAPDH primary antibodies (EMD Millipore) were incubated with the membranes overnight, then the incubated membranes were treated with the secondary antibody (Peroxidase-AffiniPure Goat Anti-Rabbit/Mouse IgG (H+L), Jackson Immunoresearch Lab) for 30 min. Chemidoc MP system (Bio-Rad) was used to scan the membranes and analyze the imaging.
Supplementary Material
Acknowledgments
This work was supported by R01NS076054 (to D.K.). J Z. is supported by a fellowship from Lysosomal Therapeutics Inc. (Boston). This work made use of the IMSERC at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF NNCI-1542205), the State of Illinois, and the International Institute for Nanotechnology (IIN). We would like to thank H. Goudarzi and S. Shafaie in IMSERC at Northwestern University for their assistance with HRMS experiments and C. H. Luan in Northwestern University’s High Throughput Analysis Laboratory for his assistance with thermal denaturation experiments.
ABBREVIATIONS USED
- GD
Gaucher’s disease
- GCase
β-glucocerebrosidase
- ER
endoplasmic reticulum
- ERT’
enzyme replacement therapy
- SRT
substrate reduction therapy
- PC
pharmacological chaperone
- IFG
Isofagomine
- PD
Parkinson’s Disease
- DLB
dementia with Lewy bodies
- SAR
structure activity relationship
- GAA
acid α-glucosidase
- GLA
α-galactosidase A
Footnotes
The authors declare no competing financial interest.
Supporting Information Available: 1H and 13C NMR spectra of 4, 6c, 7a, 7b, 8c, 9a, 9b, 9c, 11b, 11d, 11f, 11g, and 11h (PDF)
Molecular formula strings (CSV)
REFERENCES
- 1.Grabowski GA. Phenotype, diagnosis, and treatment of Gaucher’s disease. Lancet. 2008;372:1263–1271. doi: 10.1016/S0140-6736(08)61522-6. [DOI] [PubMed] [Google Scholar]
- 2.Bennett LL, Mohan D. Gaucher disease and its treatment options. Ann. Pharmacother. 2013;47:1182–1193. doi: 10.1177/1060028013500469. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski GA, Zimran A, Ida H. Gaucher disease types 1 and 3: Phenotypic characterization of large populations from the ICGG Gaucher Registry. Am. J. Hematol. 2015;(90 Suppl 1):S12–S18. doi: 10.1002/ajh.24063. [DOI] [PubMed] [Google Scholar]
- 4.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum. Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 5.Sawkar AR, Cheng WC, Beutler E, Wong CH, Balch WE, Kelly JW. Chemical chaperones increase the cellular activity of N370S beta -glucosidase: a therapeutic strategy for Gaucher disease. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15428–15433. doi: 10.1073/pnas.192582899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tekoah Y, Tzaban S, Kizhner T, Hainrichson M, Gantman A, Golembo M, Aviezer D, Shaaltiel Y. Glycosylation and functionality of recombinant beta-glucocerebrosidase from various production systems. Biosci. Rep. 2013;33:e00071. doi: 10.1042/BSR20130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N. Engl. J. Med. 2004;351:1972–1977. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 8.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, De Marco EV, Durr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen GJ, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan EK, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu YR, Zabetian CP, Zhao Y, Ziegler SG. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schapira AHV, Olanow CW, Greenamyre JT, Bezard E. Slowing of neurodegeneration in Parkinson’s disease and Huntington’s disease: future therapeutic perspectives. Lancet. 2014;384:545–555. doi: 10.1016/S0140-6736(14)61010-2. [DOI] [PubMed] [Google Scholar]
- 10.Sidransky E, Lopez G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012;11:986–998. doi: 10.1016/S1474-4422(12)70190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin MK, Farrer MJ. Genetics and genomics of Parkinson’s disease. Genome Med. 2014;6:48. doi: 10.1186/gm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sardi SP, Clarke J, Viel C, Chan M, Tamsett TJ, Treleaven CM, Bu J, Sweet L, Passini MA, Dodge JC, Yu WH, Sidman RL, Cheng SH, Shihabuddin LS. Augmenting CNS glucocerebrosidase activity as a therapeutic strategy for parkinsonism and other Gaucher-related synucleinopathies. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3537–3542. doi: 10.1073/pnas.1220464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sybertz E, Krainc D. Development of targeted therapies for Parkinson’s disease and related synucleinopathies. J. Lipid Res. 2014;55:1996–2003. doi: 10.1194/jlr.R047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang GN, Reinkensmeier G, Zhang SW, Zhou J, Zhang LR, Zhang LH, Butters TD, Ye XS. Rational design and synthesis of highly potent pharmacological chaperones for treatment of N370S mutant Gaucher disease. J. Med. Chem. 2009;52:3146–3149. doi: 10.1021/jm801506m. [DOI] [PubMed] [Google Scholar]
- 16.Trapero A, Gonzalez-Bulnes P, Butters TD, Llebaria A. Potent aminocyclitol glucocerebrosidase inhibitors are subnanomolar pharmacological chaperones for treating gaucher disease. J. Med. Chem. 2012;55:4479–4488. doi: 10.1021/jm300342q. [DOI] [PubMed] [Google Scholar]
- 17.Sawkar AR, Schmitz M, Zimmer KP, Reczek D, Edmunds T, Balch WE, Kelly JW. Chemical chaperones and permissive temperatures alter localization of Gaucher disease associated glucocerebrosidase variants. ACS Chem. Biol. 2006;1:235–251. doi: 10.1021/cb600187q. [DOI] [PubMed] [Google Scholar]
- 18.Steet RA, Chung S, Wustman B, Powe A, Do H, Kornfeld SA. The iminosugar isofagomine increases the activity of N370S mutant acid beta-glucosidase in Gaucher fibroblasts by several mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13813–13818. doi: 10.1073/pnas.0605928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butters TD, Dwek RA, Platt FM. Imino sugar inhibitors for treating the lysosomal glycosphingolipidoses. Glycobiology. 2005;15:43R–52R. doi: 10.1093/glycob/cwi076. [DOI] [PubMed] [Google Scholar]
- 20.Zheng W, Padia J, Urban DJ, Jadhav A, Goker-Alpan O, Simeonov A, Goldin E, Auld D, LaMarca ME, Inglese J, Austin CP, Sidransky E. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13192–13197. doi: 10.1073/pnas.0705637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marugan JJ, Zheng W, Motabar O, Southall N, Goldin E, Westbroek W, Stubblefield BK, Sidransky E, Aungst RA, Lea WA, Simeonov A, Leister W, Austin CP. Evaluation of quinazoline analogues as glucocerebrosidase inhibitors with chaperone activity. J. Med. Chem. 2011;54:1033–1058. doi: 10.1021/jm1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marugan JJ, Huang W, Motabar O, Zheng W, Xiao J, Patnaik S, Southall N, Westbroek W, Lea WA, Simeonov A, Goldin E, Debernardi MA, Sidransky E. Non-iminosugar glucocerebrosidase small molecule chaperones. MedChemComm. 2012;3:56–60. doi: 10.1039/C1MD00200G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tropak MB, Kornhaber GJ, Rigat BA, Maegawa GH, Buttner JD, Blanchard JE, Murphy C, Tuske SJ, Coales SJ, Hamuro Y, Brown ED, Mahuran DJ. Identification of pharmacological chaperones for Gaucher disease and characterization of their effects on beta-glucocerebrosidase by hydrogen/deuterium exchange mass spectrometry. ChemBioChem. 2008;9:2650–2662. doi: 10.1002/cbic.200800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W, Zheng W, Urban DJ, Inglese J, Sidransky E, Austin CP, Thomas CJ. N4-phenyl modifications of N2-(2-hydroxyl)ethyl-6-(pyrrolidin-1-yl)-1,3,5-triazine-2,4-diamines enhance glucocerebrosidase inhibition by small molecules with potential as chemical chaperones for Gaucher disease. Bioorg. Med. Chem. Lett. 2007;17:5783–5789. doi: 10.1016/j.bmcl.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storz T, Heid R, Zeldis J, Hoagland SM, Rapisardi V, Hollywood S, Morton G. Convenient and practical one-pot synthesis of 4-chloropyrimidines via a novel chloroimidate annulation. Org. Process Res. Dev. 2011;15:918–924. [Google Scholar]
- 26.Urban DJ, Zheng W, Goker-Alpan O, Jadhav A, Lamarca ME, Inglese J, Sidransky E, Austin CP. Optimization and validation of two miniaturized glucocerebrosidase enzyme assays for high throughput screening. Comb. Chem. High Throughput Screening. 2008;11:817–824. doi: 10.2174/138620708786734244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilar-Moncayo M, Garcia-Moreno MI, Trapero A, Egido-Gabas M, Llebaria A, Fernandez JMG, Mellet CO. Bicyclic (galacto)nojirimycin analogues as glycosidase inhibitors: Effect of structural modifications in their pharmacological chaperone potential towards beta-glucocerebrosidase. Org. Biomol. Chem. 2011;9:3698–3713. doi: 10.1039/c1ob05234a. [DOI] [PubMed] [Google Scholar]
- 28.Trapero A, Alfonso I, Butters TD, Llebaria A. Polyhydroxylated bicyclic isoureas and guanidines are potent glucocerebrosidase inhibitors and nanomolar enzyme activity enhancers in Gaucher cells. J. Am. Chem. Soc. 2011;133:5474–5484. doi: 10.1021/ja111480z. [DOI] [PubMed] [Google Scholar]
- 29.Berger Z, Perkins S, Ambroise C, Oborski C, Calabrese M, Noell S, Riddell D, Hirst WD. Tool compounds robustly increase turnover of an artificial substrate by glucocerebrosidase in human brain lysates. PLoS One. 2015;10:e0119141. doi: 10.1371/journal.pone.0119141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 32.Filippova EV, Luan CH, Dunne SF, Kiryukhina O, Minasov G, Shuvalova L, Anderson WF. Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila. J. Struct. Funct. Genomics. 2014;15:33–40. doi: 10.1007/s10969-014-9176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zunke F, Andresen L, Wesseler S, Groth J, Arnold P, Rothaug M, Mazzulli JR, Krainc D, Blanz J, Saftig P, Schwake M. Characterization of the complex formed by beta-glucocerebrosidase and the lysosomal integral membrane protein type-2. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3791–3796. doi: 10.1073/pnas.1514005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.