Abstract
Caveolae are the primary route for internalization and transendothelial transport of macromolecules, such as insulin and albumin. Caveolae-mediated endocytosis is activated by Src-dependent caveolin-1 (Cav-1) phosphorylation and subsequent recruitment of dynamin-2 and filamin A (FilA), which facilitate vesicle fission and trafficking, respectively. Here, we tested the role of RalA and phospholipase D (PLD) signaling in the regulation of caveolae-mediated endocytosis and trafficking. The addition of albumin to human lung microvascular endothelial cells induced the activation of RalA within minutes, and siRNA-mediated down-regulation of RalA abolished fluorescent BSA uptake. Co-immunoprecipitation studies revealed that albumin induced the association between RalA, Cav-1, and FilA; however, RalA knockdown with siRNA did not affect FilA recruitment to Cav-1, suggesting that RalA was not required for FilA and Cav-1 complex formation. Rather, RalA probably facilitates caveolae-mediated endocytosis by activating downstream effectors. PLD2 was shown to be activated by RalA, and inhibition of PLD2 abolished Alexa-488-BSA uptake, indicating that phosphatidic acid (PA) generated by PLD2 may facilitate caveolae-mediated endocytosis. Furthermore, using a PA biosensor, GFP-PASS, we observed that BSA induced an increase in PA co-localization with Cav-1-RFP, which could be blocked by a dominant negative PLD2 mutant. Total internal reflection fluorescence microscopy studies of Cav-1-RFP also showed that fusion of caveolae with the basal plasma membrane was dependent on PLD2 activity. Thus, our results suggest that the small GTPase RalA plays an important role in promoting invagination and trafficking of caveolae, not by potentiating the association between Cav-1 and FilA but by stimulating PLD2-mediated generation of phosphatidic acid.
Keywords: caveolin, endocytosis, filamin, Ras protein, trafficking
Introduction
Caveolae-mediated transcellular transport of macromolecules through endothelial cells (ECs)2 lining blood vessels is a highly selective and regulated process (1). Although the signaling mechanisms regulating the invagination and internalization (endocytosis) of caveolae have not been fully characterized, it was shown that Src-dependent phosphorylation of the large GTPase dynamin-2 (2, 3) and its recruitment to the neck of caveolae (4), phosphorylation of caveolin-1 Tyr-14 (5) and subsequent recruitment of actin cross-linking protein filamin A (FilA) to caveolae (6), and dynamic actin remodeling (7) are some of the general requirements (8).
In addition to the large GTPase dynamin-2, which is required for fission of caveolae from the plasma membrane, several small GTPases detected in caveolin-enriched membrane fractions have been proposed to participate in caveolae-mediated transport. For example, Cdc42, a small GTPase of the Rho family, was detected in caveolae (9), where it is thought to control caveolae-mediated endocytosis by regulating actin polymerization and interactions between the actin cytoskeleton and intersectin, a scaffolding protein required for efficient fission and internalization of caveolae (10–12). Another small GTPase, Rab5, which is known to participate in endocytosis by regulating vesicle docking and fusion, directly binds to caveolin-1, and this interaction increases Rab5 activity (13). Moreover, Rab5 was shown to control the targeting of caveolae to early endosomes and to be essential for cholera toxin B accumulation in Golgi vesicles (14), suggesting that Rab5 facilitates intracellular trafficking of caveolae. There is also evidence that other small GTPases, such as Ras (15), RhoA (9), and Rac1 (16), localize to caveolin-enriched membrane microdomains; however, their function in caveolae-mediated endocytosis and trafficking has not yet been established.
Here, we hypothesized that yet another small GTPase, RalA, a member of the Ras superfamily of GTPases, also participates in the regulation of caveolae internalization and trafficking. RalA was previously shown to regulate receptor endocytosis, trafficking, and exocytosis as well as actin cytoskeletal dynamics (17, 18). It was demonstrated that agonist-dependent endocytosis of EGF, insulin, transferrin, and activin type II receptors depends on RalA-mediated activation of its effector RalBP1 (19). Activated RalBP1 interacts with the clathrin assembly protein complex, which includes a number of endocytic proteins, such as Eps15, epsin, and Rab11-FIP2, as well as clathrin adaptor protein AP-2 (20–22). Thus, RalBP1 activation is essential for assembly of the clathrin coat and clathrin-mediated endocytosis of various receptors.
We have previously shown that actin-binding protein FilA is recruited to caveolae following activation of Src kinase and Src-dependent phosphorylation of caveolin-1 and that FilA is required for endocytosis of albumin and trafficking of caveolae (6). Another group demonstrated that FilA interacts with active RalA (17). Therefore, in this work, we sought to determine the functional significance of RalA recruitment and association with FilA and Cav-1 in human lung microvascular endothelial cells (HLMVECs). We observed RalA activation and recruitment to EC caveolae upon stimulation of albumin, perhaps the best studied macromolecular cargo of endothelial caveolae (24), and demonstrated that FilA is not required for RalA activation by albumin, but rather that it is essential for RalA association with caveolae. Furthermore, a significant decrease in caveolae-mediated internalization of albumin was detected in cells treated with RalA siRNA, suggesting that RalA activation is critical for efficient caveolae-mediated transport. Also, we observed that PLD2-mediated production of PA downstream of RalA was involved in the regulation of caveolae fusion and fission events, arguably by promoting changes in membrane curvature and actin cytoskeleton reorganization.
Materials and Methods
Cell Culture
HLMVECs (Lonza, Walkersville, MD) were cultured on dishes coated with 0.2% gelatin in EBM-2-MV growth medium (Lonza) supplemented with 10% FBS, l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin.
Reagents
All reagents were obtained from Sigma-Aldrich unless stated otherwise. Filamin A mAb was obtained from Chemicon (Temecula, CA). Caveolin-1 polyclonal antibody was from BD Biosciences. Monoclonal antibodies for β-actin and RalA were from Sigma-Aldrich (Franklin Lakes, NJ). Normal mouse IgG was obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX). Antibody for PLD1 was purchased from Cell Signaling Technology (Danvers, MA), and the antibody for PLD2 was provided by Drs. Nozawa and Banno (Gifu International Institute of Biotechnology, Gifu, Japan). DAPI, Alexa-488 BSA, and all fluorescently labeled secondary antibodies were purchased from Molecular Probes, Inc. (Eugene, OR). HRP-conjugated goat-anti-mouse and goat-anti-rabbit secondary antibodies were from KPL (Gaithersburg, MD). n-Octyl glucoside was purchased from RPI (Mt. Prospect, IL). PLD1 inhibitor VU0359595 was from Cayman Chemical (Ann Arbor, MI), and PLD2 inhibitor VU 0364739 was from Tocris Bioscience (Bristol, UK).
Transfection and Infection of ECs
GFP-tagged human RalA as well as constitutively active (RalAG23V) and dominant negative (RalAS28N) mutants were kindly provided by Dr. Ferguson (John P. Robarts Research Institute, London, Canada). RalA siRNA (ON-TARGET plus SMART pool) was from Dharmacon (Pittsburgh, PA), which included four on-target sequences: oligonucleotide 1, GGACUACGCUGCAAUUAGA; oligonucleotide 2, CAAAUAAGCCCAAGGGUCA; oligonucleotide 3, GAGGAAGUCCAGAUCGAUA; and oligonucleotide 4, GAAAUUCGAGCGAGAAAGA.
Full-length Homo sapiens caveolin-1 was used as a template to generate C-terminal RFP-tagged caveolin-1 (Cav-1-RFP). HLMVECs were transfected with Cav-1 or RalA constructs alone or in combination with control, FilA siRNA, or RalA siRNA by nucleofection (Amaxa Inc., Gaithersburg, MD) according to the manufacturer's instructions. Cells were used for experiments 48–72 h after transfection.
GFP-PASS lentiviral construct was added to HLMVECs transfected with Cav-1-RFP for 48 h. Cells were then starved and stimulated with BSA (30 mg/ml). Adenoviral constructs, vector control, and dominant negative mutants of hPLD1 K898R and mPLD2 K758R were generated at the University of Iowa Gene Transfer Vector Core (Iowa City, IA). Adenoviral constructs (5 plaque-forming units/cell) of vector control, hPLD1 K898R, or mPLD2 K758R mutant were added to HLMVECs grown to ∼80% confluence in EBM-2-MV growth medium (Lonza) supplemented with 10% FBS. After overnight culture, the virus-containing medium was replaced with fresh complete medium and treated with BSA (30 mg/ml).
RalA Activation Assay
The RalA activation assay kit was purchased from Upstate (Temecula, CA) and used according to the manufacturer's instructions. Briefly, HLMVECs were serum-deprived for 5 h and then incubated with 30 mg/ml BSA at 37 °C for 5, 10, or 30 min. Cells were washed with ice-cold TBS and lysed in a buffer containing 50 mm Tris-HCl, pH 7.5, 0.2 m NaCl, 1% Nonidet P-40, 10 mm MgCl2, 0.5 mm dithiothreitol, 1 mm PMSF, and protein inhibitor mixture. Lysates were precleared with glutathione-agarose and incubated with agarose-conjugated RalBP1 for 30 min at 4 °C. The amount of precipitated active RalA was estimated by densitometry analysis from Western blots. The time course for RalA activation was calculated from three independent experiments.
Cell Fractionation by Density Gradient Centrifugation
Fractionation was conducted as described previously (25) with slight modifications. Briefly, two confluent 100-mm plates were washed and scraped into basal buffer (20 mm Tris-HCl, pH 7.8, 250 mm sucrose) supplemented with 1 mm CaCl2 and 1 mm MgCl2. Cells were centrifuged at 1,000 × g for 10 min, and the cell pellet was suspended in 1 ml of basal buffer containing 1 mm CaCl2, 1 mm MgCl2, and protease inhibitor mixture (Sigma-Aldrich). Cells were lysed by 40 strokes with a Dounce homogenizer followed by passage 10 times through a 27-gauge needle. Lysates were centrifuged at 10,000 × g for 10 min to remove unbroken cells and large cell fragments. Supernatants were collected and mixed with an equal volume of 50% OptiPrep and overlaid with 20, 15, 10, 5, and 0% OptiPrep gradient in basal buffer containing 1 mm sodium orthovanadate. Gradients were centrifuged at 52,000 × g in SW55Ti rotor for 10 h at 4 °C. Twelve fractions were collected starting from the top of the gradient, and equal volume samples of each fraction were analyzed by SDS-PAGE.
Immunoprecipitation and Western Blotting Analysis
For stimulation studies, cells were starved for 3 h and then treated with BSA (30 mg/ml for 30 min). For Western blotting, cells were lysed on ice for 30 min in lysis buffer containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 60 mm n-octyl glucoside, 1% Triton X-100, 1 mm PMSF, 1 mm sodium orthovanadate, and protease inhibitor mixture. All insoluble material was removed by centrifugation (100,000 × g for 45 min). For immunoprecipitations, lysates were incubated with M-280 Dynabeads coated with sheep anti-mouse IgG (Dynal Biotech, LLC) preconjugated with monoclonal anti-filamin A or anti-FLAG antibodies for 1 h at 4 °C. Proteins were resolved by SDS-PAGE and processed with an ECL Super Signal kit (Pierce), and then relative band intensities (densitometry) from scanned blots were determined using ImageJ software (National Institutes of Health).
Fluorescent Albumin Uptake, Immunostaining, and Confocal Microscopy
For uptake experiments, GFP construct, RalA cDNA constructs, and RalA siRNA were transfected into HLMVECs seeded on collagen-coated glass coverslips. Forty-eight h after transfection, cells were serum-deprived for 2–3 h in basal medium and then treated with 0.1% 1-butanol, 2-butanol, or 200 nm PLD inhibitors for 1 h. After treatment, cells were incubated with Alexa-488-BSA or Alexa-555-BSA (50 μg/ml) in basal medium containing 0.1 mg/ml unlabeled BSA for 30 min at 37 °C and washed with acid wash buffer (pH 2.5) followed by Hanks' balanced salt solution to remove surface-bound BSA, fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and stained with the nuclear marker DAPI (1 μg/ml). Uptake was quantified for 50 cells/sample. Each experiment was repeated three times. Alexa-488-BSA uptake was estimated from thresholded images using the ImageJ Particle Analysis module.
For immunostaining, non-transfected cells or HLMVECs transfected with empty GFP vector (EV), RalA constructs, or RalA siRNA were fixed with 4% paraformaldehyde, permeabilized, and incubated with caveolin-1 or RalA antibodies (1 μg/ml). Non-confocal DAPI images were acquired using mercury lamp excitation and a UV filter set. Confocal microscopy was performed using a Zeiss LSM 510 META microscope (Carl Zeiss MicroImaging, Inc.) with 488- and 543-nm excitation laser lines and pinhole set to achieve 1 Airy unit.
Total Internal Reflective Fluorescence (TIRF) Microscopy
HLMVECs transfected with Cav-1-RFP were infected with EV, hPLD1 K898R, or mPLD2 K758R mutants for 36 h and then starved for 3 h before stimulation with BSA (30 mg/ml). In some experiments, 200 nm PLD inhibitor was added into the system 1 h before BSA stimulation. Live cell TIRF images were acquired using a Zeiss Axio Observer Z1 microscope (Carl Zeiss MicroImaging, Inc.) with 561-nm excitation and a ×100/1.46 numeric aperture α-Plan-Fluor objective. Experiments were conducted at 37 °C in 5% CO2 in a Pecon XL TIRF S incubator chamber.
PLD Activation Assay
HLMVECs were labeled with [32P]orthophosphate (5 μCi/ml) in phosphate-free medium with 2% fetal bovine serum for 18–24 h (26, 27). Cells were then washed in minimal essential medium and treated with thrombin (0.05 units/ml) for 30 min in the presence of 0.1% 1-butanol. Treatment was terminated by the addition of 1 ml of methanol-concentrated HCl (100:1, v/v). Cellular lipids were extracted, and [32P]PBt, an indicator of PLD activation in vivo (28), which was formed as a result of PLD-mediated trans-phosphatidylation, was separated by TLC in 1% potassium oxalate-impregnated silica gel H plates as described (28). Briefly, the upper phase of ethyl acetate/2,2,4-trimethylpentane/glacial acetic acid/water (65:10:15:50, v/v/v/v) was used as the developing solvent system. Unlabeled PBt was introduced as a carrier, the lipids were separated by TLC and visualized by autoradiography, and radioactivity of PBt was quantified by liquid scintillation counting. Data generated were expressed as a percentage of control.
Statistical Analysis
Statistical comparisons were performed with GraphPad Prism5 using ANOVA with significance level set at p < 0.05; p < 0.01 and p < 0.001 were considered highly significant.
Results
RalA Is Activated by Albumin in HLMVECs
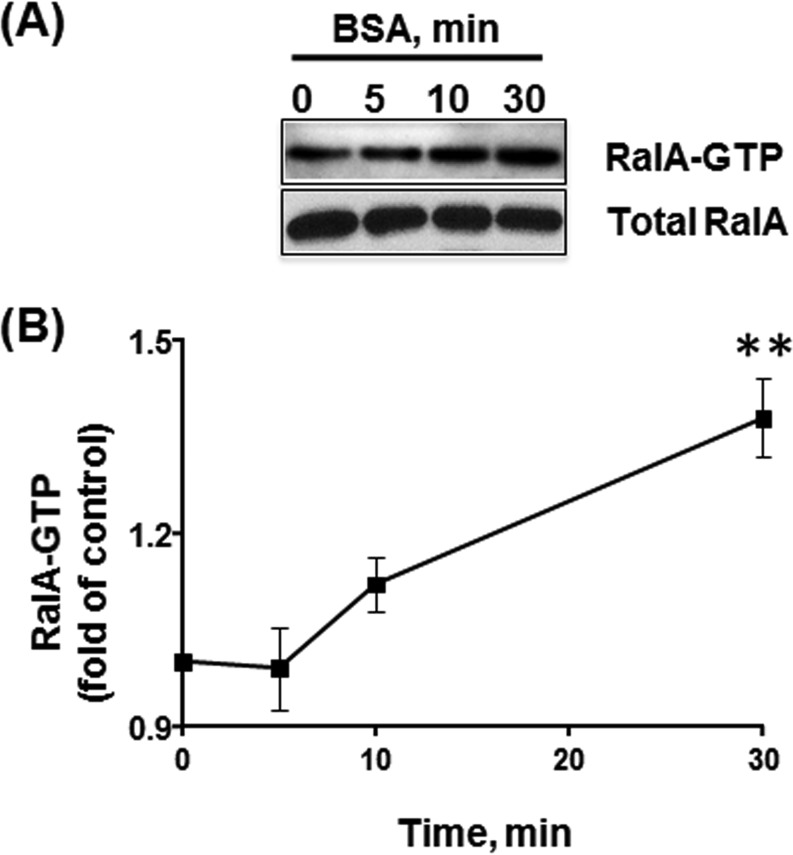
To test whether RalA is activated upon the addition of albumin to stimulate caveolae-mediated endocytosis, we assessed RalA-GTP/RalBP1 binding. Starved HLMVECs were stimulated with 30 mg/ml BSA for 5, 10, or 30 min. Clarified lysates were incubated with a RalBP1 fragment, and the amount of co-precipitated GTP-bound RalA was analyzed by Western blotting. An increase in RalA activity was detected after 10 min and remained elevated through 30 min of BSA stimulation (Fig. 1A). Densitometry analysis of Western blots revealed a 35% enhancement in the amount of GTP-bound RalA in the presence of BSA (Fig. 1B), suggesting that RalA is activated during caveolae-mediated endocytosis of albumin.
FIGURE 1.
RalA activation induced by BSA. A, serum-deprived HLMVECs were stimulated with 30 mg/ml BSA for 5, 10, and 30 min, and RalA activity was assessed by RalBP1 pull-down assay. The amount of active GTP-bound RalA co-precipitated with the RalA-binding domain of RalBP1 was analyzed by Western blotting. Note the increase in RalA activity following 30 min of BSA stimulation. B, graph represents the time course of RalA activation induced by BSA, and the data are expressed as a percentage of the RalA activity in unstimulated (control) cells (mean ± S.E., n = 6). BSA stimulation induced a 35% increase in RalA activity.
RalA Is Associated with Cargo-loaded Caveolae in Endothelial Cells
To investigate whether RalA is localized on caveolae, we examined RalA distribution in density gradient fractions of HLMVECs. The majority of endothelial RalA was localized to cholesterol-enriched fractions (fractions 2–4), which also contained Cav-1 and FilA (Fig. 2A). However, because centrifugation in the density gradient separates protein complexes and organelles based on the ratio of protein to lipid, RalA could be associated with lipid rafts and not necessarily caveolae. Co-immunoprecipitation analysis verified that in starved cells, RalA interacts with both Cav-1 and FilA (Fig. 2B), indicating that RalA may be associated with caveolae by directly binding to either FilA or Cav-1. Moreover, stimulation of caveolae-mediated endocytosis with 30 mg/ml BSA induced an increase in association between RalA, Cav-1, and FilA (Fig. 2B), implying that formation of the RalA·Cav-1·FilA complex occurs during caveolae-mediated endocytosis. To confirm that RalA associates with caveolae actively participating in albumin transport, we stimulated HLMVECs with Alexa-488-BSA and assessed RalA localization. RalA was concentrated at the plasma membrane and also in a vesicular pool that co-localized with the internalized BSA (Fig. 2C). Together with biochemical analysis of cholesterol-enriched membrane fractions and immunoprecipitation results, these data suggest that RalA is recruited to cargo-loaded caveolae.
FIGURE 2.
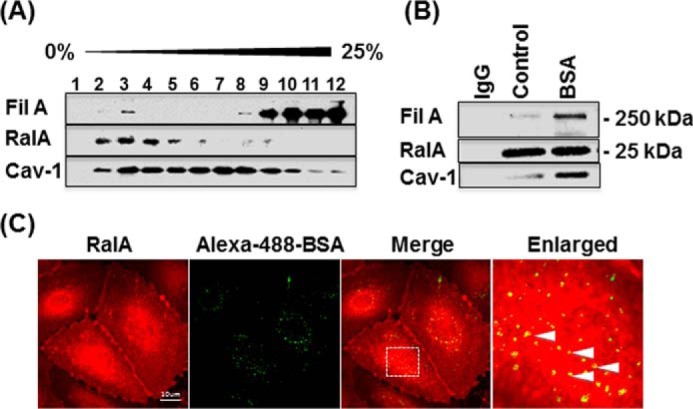
RalA is associated with caveolin-1 and filamin A during caveolae-mediated endocytosis. A, analysis of RalA distribution in sucrose density gradient fractions of HLMVECs revealed the majority of RalA in fractions 2–4 associated with buoyant fractions (fractions 2–4) that also contain Cav-1 and FilA. B, serum-deprived HLMVECs were stimulated with vehicle alone or 30 mg/ml BSA for 30 min, lysed, clarified, immunoprecipitated with anti-RalA antibody versus control IgG, and blotted for Cav-1 and FilA. Cav-1 and FilA interaction with RalA was dramatically increased upon stimulation with BSA, suggesting formation of the protein complex during caveolae-mediated endocytosis. C, serum-deprived HLMVECs were incubated with Alexa-488-BSA for 30 min, acid-washed, fixed, and stained for RalA. Co-localization of endogenous RalA with internalized BSA (see enlarged merged image) implies that RalA remains associated with internalized, BSA-loaded caveolae. Images are representative of three independent experiments.
Association between RalA and Cav-1 Is Nucleotide-dependent
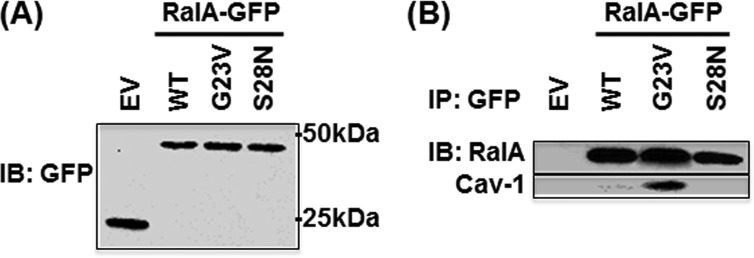
Increased RalA activity and association of RalA and Cav-1 in response to the addition of BSA raises the possibility that this interaction might be nucleotide-dependent. To test this hypothesis, we transfected HLMVECs with GFP-tagged WT RalA, constitutively active RalA mutant (RalA G23V), or dominant negative RalA mutant (RalA S28N) and assessed their interaction with Cav-1 by co-immunoprecipitation. Western blotting analysis confirmed equal expression of all RalA constructs (Fig. 3A), whereas immunoprecipitation of GFP-tagged WT or mutant RalA constructs revealed that Cav-1 primarily associated with constitutively active GTP-bound RalA G23V mutant (Fig. 3B). Neither overexpressed WT RalA nor dominant negative RalA S28N mutant was found in the complex with Cav-1, implying that RalA activation is essential for its recruitment to caveolae.
FIGURE 3.
Nucleotide-dependent interaction between RalA and caveolin-1. HLMVECs transfected with EV or GFP-tagged RalA constructs were lysed 48 h after transfection and blotted with anti-GFP antibody. A, expression level of WT, constitutively active (G23V), and dominant negative (S28N) GFP-tagged RalA analyzed by Western blotting (IB) shows equal expression. B, GFP-tagged proteins were immunoprecipitated (IP) with anti-GFP antibody and blotted for RalA and caveolin-1. Note that Cav-1 associated with GTP-bound constitutively active RalA mutant. Results are representative of two independent experiments.
Expression of Dominant Negative RalA Mutant Inhibits Caveolae-mediated Endocytosis of Albumin
Because RalA interacts with Cav-1 in a GTP-dependent manner, we tested whether RalA activation is required for caveolae-mediated endocytosis. HLMVECs transfected with GFP-tagged RalA mutants were incubated with Alexa-555-BSA for 30 min, acid-washed to remove surface-bound albumin, and fixed. As shown in Fig. 4A, cells expressing constitutively active RalAG23V-GFP mutant accumulated internalized BSA in perinuclear regions, whereas in cells expressing dominant negative RalAS28N-GFP mutant, BSA accumulation was reduced and appeared as small vesicles evenly distributed throughout the cytoplasm (Fig. 4A). Quantification of BSA uptake in GFP-positive cells revealed that dominant negative RalA mutant reduced caveolae-mediated endocytosis by ∼35% in comparison with cells transfected with empty vector or non-transfected (NTF) cells used as an internal control for each condition (Fig. 4B). Although overexpression of WT RalA or constitutively active RalA G23V mutant did not significantly increase BSA uptake, dominant negative RalA S28N mutant significantly reduced caveolae-mediated endocytosis, suggesting that endogenous RalA expression is not rate-limiting.
FIGURE 4.

Effect of RalA mutants on caveolae-mediated endocytosis. A, HLMVECs were transfected with EV, GFP-tagged RalA, constitutively active RalA (G23V), or dominant negative RalA (S28N) mutants. Forty-eight h after transfection, cells were starved, incubated with 0.1 mg/ml BSA + 50 μg/ml Alexa-555-BSA for 30 min, acid-washed, and fixed, and confocal images of GFP and Alexa-555 BSA fluorescence were collected. Alexa-555-BSA images of transfected and non-transfected (NTF) cells were analyzed using ImageJ software (note the image mask of transfected cells). B, quantification of Alexa-555-BSA uptake. Dominant negative RalA (S28N) mutant reduced BSA endocytosis by 35% in comparison with non-transfected cells and cells expressing EV, WT RalA, or constitutively active (G23V) RalA. Data are mean ± S.E. (error bars) (n = 5); **, p < 0.01 versus EV by ANOVA.
Knockdown of FilA Blocks Albumin-mediated Increase in Cav-1/RalA Association
To investigate the role of FilA in RalA activation and recruitment to caveolae following the addition of BSA, HLMVECs were transfected with control or FilA siRNA (6). Seventy-two h after transfection, cells were serum-deprived for 4 h, stimulated with 30 mg/ml BSA for 30 min, and lysed. RalA activation assay revealed that knockdown of FilA did not inhibit the increase in RalA GTP binding in response to BSA (Fig. 5A), implying that RalA binding to FilA is not required for RalA activation. However, when RalA was immunoprecipitated from cells transfected with FilA siRNA, we did not detect an increase in RalA/Cav-1 association in the presence of BSA (Fig. 5B). These data suggest that FilA may serve as a scaffold for RalA·Cav-1 complex formation and that FilA is required for recruitment of activated RalA to cargo-loaded caveolae.
FIGURE 5.
Effect of filamin A siRNA on RalA activation and interaction with caveolin-1 and effect of RalA siRNA on interaction of filamin A and caveolin-1. A, HLMVECs transfected with control or FilA siRNA were stimulated with 30 mg/ml BSA for 30 min and lysed. Lysates were incubated with the RalA binding domain of RalBP1, and the amount of RalA-GTP was estimated by Western blotting. Quantification of RalA-GTP is shown in the graph (n = 3/group; *, p < 0.05 versus control siRNA + BSA 0 min; #, p < 0.05 versus FilA siRNA + BSA 0 min by ANOVA). B, starved or BSA-stimulated cells transfected with control or FilA siRNA were lysed, and RalA was immunoprecipitated from the whole cell lysates. Quantification of co-immunoprecipitated Cav-1 is shown in the graph (n = 3/group; *, p < 0.05 versus control siRNA + BSA 0 min by ANOVA). C, Western blotting analysis of RalA and Cav-1 in HLMVECs treated 48 h with SMART POOL (SP) RalA siRNA or each of four oligonucleotides separately versus scrambled control siRNA (Cont). D, HLMVECs transfected with control or RalA siRNA were stimulated with 30 mg/ml BSA for 30 min and lysed, and then FilA was immunoprecipitated. Immunoblotting for caveolin-1 revealed that knocking down RalA did not have a significant effect on Cav-1/FilA association. Results shown in C and D are representative of three independent experiments. IP, immunoprecipitation; IB, immunoblotting. AU, arbitrary units.
Knockdown of RalA Blocks Albumin Endocytosis but Not the Interaction between Cav-1 and FilA
Having demonstrated that dominant negative RalA reduces the endocytosis of albumin, we next treated HLMVECs with RalA siRNA versus scrambled control siRNA for 48 h, serum-deprived the cells for 2–3 h, and either stimulated with 30 mg/ml BSA for 30 min and lysed or incubated with Alexa-488-BSA for 30 min, acid-washed, and fixed for confocal imaging. As shown in Fig. 5C, we tested the SMART POOL (SP) RalA siRNA mixture as well as each of the four oligonucleotides from the SMART POOL individually. RalA expression was reduced specifically with no effect on Cav-1 expression in HLMVECs. Because recruitment of FilA to caveolae is crucial for albumin endocytosis, we next tested whether RalA inhibition affects Cav-1 and FilA interaction. By immunoprecipitation and Western blotting, no difference was detected with regard to Cav-1 and FilA association in the RalA siRNA group (Fig. 5D). However, endocytosis of BSA was blocked by both the RalA SMART POOL siRNA and the four individual oligonucleotides (Fig. 6, A and B). In addition, by rescuing RalA expression with RalA-GFP in RalA-depleted cells (Fig. 6C), BSA uptake was restored (Fig. 6D). Thus, RalA plays a role in endocytosis of albumin, although not by promoting interaction between Cav-1 and Fil A as we had assumed.
FIGURE 6.

RalA siRNA and PLD inhibitors block caveolae-mediated endocytosis of BSA. A, starved HLMVECs treated with SMART POOL (SP) RalA siRNA or individually with each of the four oligonucleotides for 48 h were incubated with Alexa-488-BSA for 30 min, acid-washed, and fixed. Images of internalized fluorescent albumin were acquired by confocal microscopy. B, quantification of fluorescence intensity of Alexa-488-BSA demonstrates that RalA siRNA blocked BSA uptake (n = 10/group; ***, p < 0.001 versus siCont by ANOVA). C, HLMVECs treated with SMART POOL RalA siRNA for 48 h were transfected with RalA-GFP to rescue RalA expression as compared with GFP, which was transfected as a negative control. Cells were then lysed and examined by Western blotting (IB) to confirm RalA rescue. D, quantification of fluorescence intensity of Alexa-555-BSA uptake in RalA-depleted and -repleted HLMVECs (n = 10/group; ***, p < 0.001 by ANOVA). E, quantification of fluorescence intensity of Alexa-488-BSA demonstrates that both RalA knockdown by siRNA and 1-butanol dramatically inhibited BSA uptake (n = 6/group; ***, p < 0.001 versus control siRNA by ANOVA). F, HLMVECs treated with 200 nm VU0359595 (PLD1 inhibitor) or VU0364739 (PLD2 inhibitor) for 1 h were incubated with Alexa-488-BSA for 30 min. Quantification of Alexa-488-BSA fluorescence indicates that PLD2 inhibitor but not PLD1 inhibitor blocked albumin uptake (n = 7/group; ***, p < 0.001 versus control by ANOVA). Error bars, S.E.
1-Butanol, mPLD2 K758R Mutant, and PLD2 Inhibitor VU0364739 Block Uptake of Fluorescent Albumin
Our studies with RalA mutants and RalA siRNA suggested that RalA plays a role in promoting caveolae-mediated endocytosis; however, the mechanism was not yet clear. RalA was shown not to be required for the association of Cav-1 and FilA, and thus in an attempt to determine the specific function of RalA, we assessed whether it plays a role in modulating caveolae internalization by looking at downstream effector PLD, which was previously reported to facilitate clathrin-mediated receptor internalization (29). To test whether PLD regulates caveolae internalization, we first treated cells with 1-butanol, a primary alcohol known to serve as an acceptor of PA generated by PLD to form phosphatidylbutanol, as catalyzed by the transphosphatidylase activity of PLD. As shown in Fig. 6E, uptake of BSA in HLMVECs was inhibited by 1-butanol, whereas no change was observed in the presence of the inactive secondary alcohol, 2-butanol. Nevertheless, after RalA siRNA treatment, inhibition of BSA uptake in the presence of 1-butanol was not different from that observed in the presence of RalA siRNA alone (Fig. 6E). Although 1-butanol has been used to study the role of PLD-generated PA signaling, it is known to have nonspecific side effects. To rule out other possibilities that may affect BSA uptake, we employed specific PLD inhibitors. Because there are two PLD isoforms expressed in mammalian cells, VU0359595 (PLD1 inhibitor) and VU0364739 (PLD2 inhibitor) were added to HLMVECs before BSA stimulation. As shown in Fig. 6F, uptake of BSA was inhibited by PLD2 inhibitor, whereas no effect was observed in the presence of PLD1 inhibitor. These data suggest that RalA facilitates caveolae-mediated endocytosis by activating PLD2.
PLD2 Inhibitor and PLD2 Dominant Negative Mutant Reduce PA Generation on Caveolae and Trafficking of Cav-1-positive Vesicles
PLD activation is known to stimulate PA generation, which induces negative curvature of the plasma membrane during both endocytosis and exocytosis (30, 31). We thus next assessed the correlation between PA generation and Cav-1 trafficking by live cell imaging. To block PA generation, we expressed PLD1 and PLD2 dominant negative mutants (Fig. 7A) for which the efficiency of inhibition was examined before use. Due to the short half-life of PA, it is difficult to measure its production; however, activation of PLD can be measured by the addition of primary alcohol, such as 1-butanol, to the incubation system. In the presence of 1-butanol, the transphosphatidylase activity of PLD transfers the PA to 1-butanol instead of water to generate PBt, which serves as an indicator of PLD-mediated PA generation. As shown in Fig. 7B, PLD activity in both basal and thrombin-stimulated conditions was reduced only by the mPLD2 K758R mutant. To visualize Cav-1-positive vesicle trafficking, HLMVECs were first transfected with Cav-1-RFP and then 12 h later were infected with hPLD1 K898R or mPLD2 K758R mutants or empty adenoviral vector and then, after 24 h, starved for 2–3 h and stimulated with 30 mg/ml BSA. Accumulation of Cav-1-RFP-positive vesicles in the TIRF plane (abluminal surface), which includes the basal membrane of cultured HLMVECs, was enhanced by BSA. As predicted, the increase in the fluorescent signal associated with Cav-1-positive vesicles was blocked by the mPLD2 K758R mutant (Fig. 7, C and D). The same result was observed in the presence of PLD2 inhibitor VU0364739 (Fig. 7E).
FIGURE 7.
Cav-1-RFP-positive vesicle trafficking and fusion. A, Western blotting analysis of overexpressed hPLD1 K898R (PLD1 mutant) and mPLD2 K758R (PLD2 mutant) versus EV and control cells. B, HLMVECs were infected with EV, hPLD1 K898R, or mPLD2 K758R mutant in adenovirus, labeled with [32P]ATP, and stimulated with thrombin (0.005 units/ml). [32P]PBt formation as a result of PLD activation indicated that PLD2 mutant inhibited thrombin-induced PLD activity, whereas PLD1 mutant had no effect. n = 3/group; *, p < 0.05; **, p < 0.01 by ANOVA. C, HLMVECs transfected with Cav-1-RFP and infected with EV, hPLD1-K898R, or mPLD2 K758R mutant were serum-deprived for 2 h, treated with 30 mg/ml BSA, and then imaged by live cell TIRF microscopy every 5 min for 20 min. Note the time-dependent appearance of Cav-1-RFP in the TIRF plane (abluminal aspect of the cell). D, quantification of relative fluorescence intensity of Cav-1-RFP (n = 3 regions/group from three independent experiments; **, p < 0.01 versus control; ***, p < 0.001 versus EV control + BSA by ANOVA). E, HLMVECs transfected with Cav-1-RFP and after 24 h were serum-deprived for 2 h, treated with 200 nm VU0359595 (PLD1 inhibitor) or VU0364739 (PLD2 inhibitor) for 1 h, and then stimulated with 30 mg/ml BSA and imaged by live cell TIRF microscopy every 5 min for 20 min (n = 3 regions/group from three independent experiments; ***, p < 0.001 versus control without BSA; **, p < 0.01 versus control with BSA by ANOVA). Error bars, S.E. AU, arbitrary units.
We also measured the co-localization of Cav-1-RFP vesicles and GFP-PASS (phosphatidic acid biosensor with superior sensitivity), which monitors PA production. As shown in Fig. 8, A and B, PLD2 but not PLD1 mutant blocked PA generation, indicating that PLD2 mediates PA generation in Cav-1-enriched membrane microdomains. Taken together, these results indicate that activation of PLD2, a downstream effector of RalA, facilitates the generation of PA, which in turn enhances the fission and fusion of caveolin-enriched membrane, thereby promoting caveolae trafficking.
FIGURE 8.

Effect of PLD2 mutant on PA generation in caveolae. A, HLMVECs were co-transfected with GFP-PASS and Cav-1-RFP following infection with EV, hPLD1-K898R, or mPLD2 K758R mutant; serum-deprived for 2 h; treated with 30 mg/ml BSA for 30 min; and imaged by confocal microscopy. B, confocal images representative of three independent experiments were used to determine Pearson's correlation coefficient of co-localized GFP-PASS and Cav-1-RFP fluorescence. Results indicate that BSA increases and PLD2 mutant reduces co-localization of PA and Cav-1 (n = 10/group; ***, p < 0.001 versus EV + BSA by ANOVA). Error bars, S.E.
Discussion
In the present study, we tested the hypothesis that RalA is required for caveolae-mediated internalization of albumin, the primary macromolecular cargo of EC caveolae. We showed that the addition of BSA to ECs induced a 35% increase in RalA activity within 30 min. Co-immunoprecipitation as well as co-localization analysis of Cav-1 with constitutively active RalA mutant (RalA G23V), but not dominant negative mutant (RalA S28N), provided evidence that RalA interacts with Cav-1 in a GTP-dependent manner.
Quantification of albumin uptake in HLMVECs expressing RalA mutants revealed that dominant negative RalA reduced BSA internalization by ∼35%, whereas overexpression of wild type RalA or constitutively active RalA did not have a statistically significant effect on caveolae-mediated albumin uptake. Reduction in endocytosis by dominant negative RalA is probably due to its ability to compete with endogenous RalA and prevent the activation of downstream effectors that participate in caveolae-mediated endocytosis. The lack of effect of WT RalA or G23V constitutively active mutant on endocytosis suggests that RalA activity is not limiting in this model of caveolae-mediated endocytosis. In this context, it is possible that we did not detect an increase in albumin uptake in RalA G23V-expressing cells because RalA effectors may have been depleted by overexpression of constitutively active RalA. Further, it is also possible that the concentration of BSA used in these studies saturated albumin uptake via caveolae, and thus we were not able to detect an increase in cells transfected with additional WT or constitutively active RalA mutant. To specifically and directly address the role of RalA, endogenous RalA was depleted using siRNA, and in this system, significant inhibition of BSA uptake was observed.
Because we had previously shown that FilA is essential for caveolae-mediated endocytosis (6), we tested whether FilA is required for RalA activation and/or association with caveolae. We showed that RalA activation in response to the addition of albumin to serum-deprived cells was not affected by FilA siRNA transfection; however, knockdown of FilA prevented BSA-mediated increase in RalA/Cav-1 association. Thus, these data suggest that FilA is not required for RalA activation but rather that it is essential for targeting RalA to caveolae. Based on our earlier study indicating that the interaction of FilA and Cav-1 is crucial for caveolae-mediated endocytosis and trafficking away from the membrane, and also because RalA was shown to function downstream of Cdc42 to induce filopodia formation by recruiting FilA (17), we tested whether RalA promotes caveolae-mediated endocytosis by recruiting FilA to caveolae. Surprisingly, knockdown of RalA did not affect association of FilA with Cav-1, indicating that the role of RalA in caveolae-mediated endocytosis is dependent on other downstream effectors.
An important RalA effector is PLD. PA generated at the membrane or on vesicles is known to have a conical shape (32, 33). In addition, PA generated at the neck of vesicles promotes negative membrane curvature and facilitates vesicle fusion and fission (30). PA can also recruit and increase the activity of regulators of the actin cytoskeleton, such as phosphatidylinositol phosphate kinase (PIPKI), which generates another signaling lipid, phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) (34). Shen et al. (35) reported that EGF-induced receptor internalization and degradation were increased by overexpression of PLD1 or PLD2 and inhibited by catalytically inactive mutants of either PLD1 or PLD2 as well as the primary alcohol, 1-butanol, which is known to divert the PA generated by PLD to the primary alcohol, resulting in accumulation of PBt. Thus, in the presence of the primary alcohol 1-butanol, PA is unavailable for signaling or modulating PA-dependent targets in cells (36). Moreover, Shen et al. (35) also showed that RalA regulates EGFR endocytosis by activating PLD. In addition, Lee et al. (37) demonstrated that during endocytosis, PLD can function as a GTPase-activating protein, directly stimulating dynamin activation and receptor internalization. To determine whether RalA can activate PLD and thereby promote caveolae-mediated endocytosis, we treated HLMVECs with 1-butanol in the absence or presence of RalA siRNA and assessed fluorescent BSA uptake. The results of these studies revealed that the effects of maximum concentrations of RalA siRNA and 1-butanol were not additive, which suggests that RalA facilitates BSA endocytosis by activating PLD. To prove the validity of this result, we additionally treated cells with PLD inhibitors and observed inhibition of BSA uptake but only in the presence of PLD2 inhibitor. Hence, we conclude that RalA mediates its stimulatory effect on endocytosis by activating PLD, presumably PLD2.
Cav-1-dependent trafficking is a dynamic process that requires both fission of caveolae from the apical membrane during endocytosis and fusion of caveolae with the basal membrane during exocytosis. Our studies show that the addition of BSA to serum-deprived HLMVECs increased the number of Cav-1-RFP-positive vesicles within the TIRF plane containing the basal membrane ∼20-fold compared with cells treated with vehicle alone. Furthermore, in cells expressing mutant PLD2, we observed 40% fewer caveolae appearing within the TIRF plane, suggesting that PLD2 plays a critical role in vesicle fission and fusion. Similar results were obtained using PLD1 and PLD2 inhibitors. These results provide evidence that caveolae trafficking is dependent on PLD2/PA signaling in HLMVECs.
The PA biosensor, GFP-PASS (37), provided an additional approach, which was to monitor PA generation in live cells. We assessed co-localization of PA with Cav-1-RFP following infection with dominant negative PLD2 mutant. Results from this study indicate that PLD2 facilitates endocytosis by generating PA on or near caveolae.
The role of RalA in caveolae-mediated transport is probably more complicated than presented here. Pelkmans and Helenius (38) suggested that vesicle internalization requires local disassembly of the cortical actin cytoskeleton, followed by formation of actin tails on cargo-loaded caveolae to propel them through the cytoplasm. There is also evidence that PLD interacts directly with the actin cytoskeleton. Komati et al. (39) showed that PLD regulates myogenesis by inducing PA-dependent actin fiber formation. Roach et al. (34) showed that PA regulates actin cytoskeletal reorganization by controlling the localization and function of PIPKI, an enzyme that produces the key actin cytoskeleton-regulating lipid, PI(4,5)P2. In plant cells (40) PA was shown to be a regulator of pollen tube F-actin dynamics, whereas others found that 1-butanol blocks actin polymerization and motility of sperm, which can be rescued by the addition of PA (41). Hence, PA generated by PLD is not only involved in formation of negative membrane curvature, but it also participates in reorganization of the actin cytoskeleton and thus may facilitate vesicle movement away from the plasma membrane. In addition, another established regulator of PLD2, the small GTPase ADP-ribosylation factor-6 (ARF6), was shown to regulate vesicle trafficking and remodeling of the actin cytoskeleton (42, 43). Although we cannot exclude the possibility that ARF6 may also participate in albumin-activated PA production and caveolae trafficking, the potential link between ARF6 activation, PA production, actin dynamics, and FilA recruitment to caveolin-enriched membrane microdomains requires further investigation.
In addition to regulating albumin uptake and transcytosis, the Cav-1/FilA/RalA/PLD2/PA signaling pathway described herein may be crucial for other biological processes as well. For example, insulin-stimulated glucose uptake has been shown to be regulated by Cav-1 and RalA. Knockdown of Cav-1 inhibits the recruitment of glucose transporter 4 (Glut4) to the plasma membrane and thereby insulin-stimulated glucose transport (44). Furthermore, Anna et al. (45) detected increased levels of Glut4 in caveolae following insulin stimulation and found that Glut4 internalization was reduced by inhibitors of caveolae formation. Moreover, insulin induced recruitment of Glut4 to the plasma membrane was demonstrated to be dependent on RalA activation (46, 47), and downstream RalA-dependent activation of the exocyst complex is known to play a role in vesicle fusion (48–50). Although the underlying mechanism of insulin-stimulated glucose transport requires further investigation, results of the present study support the notion that the Cav-1/FilA/RalA/PLD2/PA signaling pathway may be involved. For example, upon insulin stimulation, recruitment of RalA to Glut4 storage vesicles via phosphorylated Cav-1/FilA interaction may activate PLD2-mediated PA generation and formation of the exocyst complex, leading to membrane fusion of Glut4 storage vesicles. Because a defect in Glut4 membrane translocation causes insulin resistance, stimulation of PA generation near or on caveolae could be of significant therapeutic benefit for patients with type II diabetes mellitus.
In summary, we show for the first time that RalA activation and formation of a Cav-1·FilA·RalA complex is necessary for efficient caveolae-mediated endocytosis and trafficking. Upon cargo loading, RalA is recruited by FilA to caveolae, where it accumulates in its activated form and stimulates PLD2-mediated generation of PA. We speculate that PA accumulation at the neck of cargo-loaded caveolae prompts the development of negative plasma membrane curvature. Finally, with the recruitment of dynamin-2 (4), vesicles are pinched off from the plasma membrane and internalized (Fig. 9). Likewise, RalA activation followed by PLD2-dependent PA formation is also required for caveolae fusion with the basal membrane during exocytosis, a process known to be dependent on yet another downstream RalA effector, the exocyst complex (23, 51, 52).
FIGURE 9.

Proposed model for role of RalA in caveolae-mediated endocytosis. In the presence of albumin, RalA is recruited to caveolae, where it associates with Cav-1 and FilA, ultimately becoming activated upon GTP binding. Activated RalA triggers downstream effector PLD2-mediated generation of PA, which promotes caveolae-mediated endocytosis and transcytosis.
Author Contributions
Y. J. and M. S. S. performed the experiments and analyzed data. Y. J. wrote the paper. R. D. M. and V. N. conceived the study, edited the paper, and supervised all of the work. P. T. T. provided assistance with microscopy. L. H. conducted PBt measurements. G. D. and Y. L. edited the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Ferguson (John P. Robarts Research Institute) for providing RalA constructs. We thank Maricela Castellon (Department of Anesthesiology, University of Illinois), Dr. Ke Ma (Research Resources Center Core Imaging Facility, University of Illinois), and Tiffany Sharma (Department of Pharmacology Molecular Core, University of Illinois) for excellent technical assistance.
This work was supported by National Institutes of Health Grants R01 HL071626 and HL125356 (to R. D. M.), P01 HL060678 Project 4 and Core D (to R. D. M.), P01 HL98050 (to V. N.), R01 HL119478 (to G. D.), and T32 HL007692 (to M. S. S.); National Natural Science Foundation of China Grants 11272083 and 31470906; and the China Scholarship Council (CSC). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EC
- endothelial cell
- PBt
- phosphatidylbutanol
- PIPKI
- phosphatidylinositol phosphate kinase
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- EV
- empty GFP vector
- HLMVEC
- human lung microvascular endothelial cell
- TIRF
- total internal reflective fluorescence
- PLD
- phospholipase D
- ANOVA
- analysis of variance
- PA
- phosphatidic acid.
References
- 1. Parton R. G., and del Pozo M. A. (2013) Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 14, 98–112 [DOI] [PubMed] [Google Scholar]
- 2. Ahn S., Maudsley S., Luttrell L. M., Lefkowitz R. J., and Daaka Y. (1999) Src-mediated tyrosine phosphorylation of dynamin is required for β2-adrenergic receptor internalization and mitogen-activated protein kinase signaling. J. Biol. Chem. 274, 1185–1188 [DOI] [PubMed] [Google Scholar]
- 3. Ahn S., Kim J., Lucaveche C. L., Reedy M. C., Luttrell L. M., Lefkowitz R. J., and Daaka Y. (2002) Src-dependent tyrosine phosphorylation regulates dynamin self-assembly and ligand-induced endocytosis of the epidermal growth factor receptor. J. Biol. Chem. 277, 26642–26651 [DOI] [PubMed] [Google Scholar]
- 4. Shajahan A. N., Timblin B. K., Sandoval R., Tiruppathi C., Malik A. B., and Minshall R. D. (2004) Role of Src-induced dynamin-2 phosphorylation in caveolae-mediated endocytosis in endothelial cells. J. Biol. Chem. 279, 20392–20400 [DOI] [PubMed] [Google Scholar]
- 5. Zimnicka A. M., Husain Y. S., Shajahan A. N., Sverdlov M., Chaga O., Chen Z., Toth P. T., Klomp J., Karginov A. V., Tiruppathi C., Malik A. B., and Minshall R. D. (2016) Src-dependent phosphorylation of caveolin-1 Tyr14 promotes swelling and release of caveolae. Mol. Biol. Cell 27, 2090–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sverdlov M., Shinin V., Place A. T., Castellon M., and Minshall R. D. (2009) Filamin A regulates caveolae internalization and trafficking in endothelial cells. Mol. Biol. Cell 20, 4531–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orth J. D., and McNiven M. A. (2003) Dynamin at the actin-membrane interface. Curr. Opin. Cell Biol. 15, 31–39 [DOI] [PubMed] [Google Scholar]
- 8. Sverdlov M., Shajahan A. N., and Minshall R. D. (2007) Tyrosine phosphorylation-dependence of caveolae-mediated endocytosis. J. Cell Mol. Med. 11, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gingras D., Gauthier F., Lamy S., Desrosiers R. R., and Béliveau R. (1998) Localization of RhoA GTPase to endothelial caveolae-enriched membrane domains. Biochem. Biophys. Res. Commun. 247, 888–893 [DOI] [PubMed] [Google Scholar]
- 10. Hussain N. K., Jenna S., Glogauer M., Quinn C. C., Wasiak S., Guipponi M., Antonarakis S. E., Kay B. K., Stossel T. P., Lamarche-Vane N., and McPherson P. S. (2001) Endocytic protein intersectin-1 regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3, 927–932 [DOI] [PubMed] [Google Scholar]
- 11. Predescu S. A., Predescu D. N., Timblin B. K., Stan R. V., and Malik A. B. (2003) Intersectin regulates fission and internalization of caveolae in endothelial cells. Mol. Biol. Cell 14, 4997–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein I. K., Predescu D. N., Sharma T., Knezevic I., Malik A. B., and Predescu S. (2009) Intersectin-2L regulates caveola endocytosis secondary to Cdc42-mediated actin polymerization. J. Biol. Chem. 284, 25953–25961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hagiwara M., Shirai Y., Nomura R., Sasaki M., Kobayashi K., Tadokoro T., and Yamamoto Y. (2009) Caveolin-1 activates Rab5 and enhances endocytosis through direct interaction. Biochem. Biophys. Res. Commun. 378, 73–78 [DOI] [PubMed] [Google Scholar]
- 14. Pelkmans L., Bürli T., Zerial M., and Helenius A. (2004) Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell 118, 767–780 [DOI] [PubMed] [Google Scholar]
- 15. Song K. S., Li S., Okamoto T., Quilliam L. A., Sargiacomo M., and Lisanti M. P. (1996) Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. J. Biol. Chem. 271, 9690–9697 [DOI] [PubMed] [Google Scholar]
- 16. Michaely P. A., Mineo C., Ying Y.-S., and Anderson R. G. W. (1999) Polarized distribution of endogenous Rac1 and RhoA at the cell surface. J. Biol. Chem. 274, 21430–21436 [DOI] [PubMed] [Google Scholar]
- 17. Ohta Y., Suzuki N., Nakamura S., Hartwig J. H., and Stossel T. P. (1999) The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. U.S.A. 96, 2122–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hazelett C. C., and Yeaman C. (2012) Sec5 and Exo84 mediate distinct aspects of RalA-dependent cell polarization. PLoS One 7, e39602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakashima S., Morinaka K., Koyama S., Ikeda M., Kishida M., Okawa K., Iwamatsu A., Kishida S., and Kikuchi A. (1999) Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 18, 3629–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaguchi A., Urano T., Goi T., and Feig L. A. (1997) An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J. Biol. Chem. 272, 31230–31234 [DOI] [PubMed] [Google Scholar]
- 21. Cullis D. N., Philip B., Baleja J. D., and Feig L. A. (2002) Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 277, 49158–49166 [DOI] [PubMed] [Google Scholar]
- 22. Miliaras N. B., and Wendland B. (2004) EH proteins: multivalent regulators of endocytosis (and other pathways). Cell Biochem. Biophys. 41, 295–318 [DOI] [PubMed] [Google Scholar]
- 23. Balasubramanian N., Meier J. A., Scott D. W., Norambuena A., White M. A., and Schwartz M. A. (2010) RalA-exocyst complex regulates integrin-dependent membrane raft exocytosis and growth signaling. Curr. Biol. 20, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dobrinskikh E., Okamura K., Kopp J. B., Doctor R. B., and Blaine J. (2014) Human podocytes perform polarized, caveolae-dependent albumin endocytosis. Am. J. Physiol. Renal Physiol. 306, F941–F951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macdonald J. L., and Pike L. J. (2005) A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 46, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 26. Parinandi N. L., Scribner W. M., Vepa S., Shi S., and Natarajan V. (1999) Phospholipase D activation in endothelial cells is redox sensitive. Antioxid. Redox Signal. 1, 193–210 [DOI] [PubMed] [Google Scholar]
- 27. Parinandi N. L., Roy S., Shi S., Cummings R. J., Morris A. J., Garcia J. G., and Natarajan V. (2001) Role of Src kinase in diperoxovanadate-mediated activation of phospholipase D in endothelial cells. Arch. Biochem. Biophys. 396, 231–243 [DOI] [PubMed] [Google Scholar]
- 28. Natarajan V., Vepa S., al-Hassani M., and Scribner W. M. (1997) The enhancement by wortmannin of protein kinase C-dependent activation of phospholipase D in vascular endothelial cells. Chem. Phys. Lipids 86, 65–74 [DOI] [PubMed] [Google Scholar]
- 29. Du G., Huang P., Liang B. T., and Frohman M. A. (2004) Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol. Biol. Cell 15, 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kooijman E. E., Chupin V., de Kruijff B., and Burger K. N. (2003) Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 4, 162–174 [DOI] [PubMed] [Google Scholar]
- 31. Testerink C., and Munnik T. (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361 [DOI] [PubMed] [Google Scholar]
- 32. Kooijman E. E., Chupin V., Fuller N. L., Kozlov M. M., de Kruijff B., Burger K. N., and Rand P. R. (2005) Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 44, 2097–2102 [DOI] [PubMed] [Google Scholar]
- 33. Roth M. G. (2008) Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 34. Roach A. N., Wang Z., Wu P., Zhang F., Chan R. B., Yonekubo Y., Di Paolo G., Gorfe A. A., and Du G. (2012) Phosphatidic acid regulation of PIPKI is critical for actin cytoskeletal reorganization. J. Lipid Res. 53, 2598–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen Y., Xu L., and Foster D. A. (2001) Role for phospholipase D in receptor-mediated endocytosis. Mol. Cell. Biol. 21, 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morris A. J., Frohman M. A., and Engebrecht J. (1997) Measurement of phospholipase D activity. Anal. Biochem. 252, 1–9 [DOI] [PubMed] [Google Scholar]
- 37. Zhang F., Wang Z., Lu M., Yonekubo Y., Liang X., Zhang Y., Wu P., Zhou Y., Grinstein S., Hancock J. F., and Du G. (2014) Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol. Cell. Biol. 34, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pelkmans L., and Helenius A. (2002) Endocytosis via caveolae. Traffic 3, 311–320 [DOI] [PubMed] [Google Scholar]
- 39. Komati H., Naro F., Mebarek S., De Arcangelis V., Adamo S., Lagarde M., Prigent A. F., and Némoz G. (2005) Phospholipase D is involved in myogenic differentiation through remodeling of actin cytoskeleton. Mol. Biol. Cell 16, 1232–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pleskot R., Potocký M., Pejchar P., Linek J., Bezvoda R., Martinec J., Valentová O., Novotná Z., and Zárský V. (2010) Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J. 62, 494–507 [DOI] [PubMed] [Google Scholar]
- 41. Itach S. B., Finklestein M., Etkovitz N., and Breitbart H. (2012) Hyper-activated motility in sperm capacitation is mediated by phospholipase D-dependent actin polymerization. Dev. Biol. 362, 154–161 [DOI] [PubMed] [Google Scholar]
- 42. Schafer D. A., D'Souza-Schorey C., and Cooper J. A. (2000) Actin assembly at membranes controlled by ARF6. Traffic 1, 892–903 [DOI] [PubMed] [Google Scholar]
- 43. Hiroyama M., and Exton J. H. (2005) Localization and regulation of phospholipase D2 by ARF6. J. Cell. Biochem. 95, 149–164 [DOI] [PubMed] [Google Scholar]
- 44. González-Muñoz E., López-Iglesias C., Calvo M., Palacín M., Zorzano A., and Camps M. (2009) Caveolin-1 loss of function accelerates glucose transporter 4 and insulin receptor degradation in 3T3-L1 adipocytes. Endocrinology 150, 3493–3502 [DOI] [PubMed] [Google Scholar]
- 45. Ros-Baro A., Lopez-Iglesias C., Peiro S., Bellido D., Palacin M., Zorzano A., and Camps M. (2001) Lipid rafts are required for GLUT4 internalization in adipose cells. Proc. Natl. Acad. Sci. U.S.A. 98, 12050–12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen X. W., Leto D., Chiang S. H., Wang Q., and Saltiel A. R. (2007) Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev. Cell 13, 391–404 [DOI] [PubMed] [Google Scholar]
- 47. Karunanithi S., Xiong T., Uhm M., Leto D., Sun J., Chen X. W., and Saltiel A. R. (2014) A Rab10:RalA G protein cascade regulates insulin-stimulated glucose uptake in adipocytes. Mol. Biol. Cell 25, 3059–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inoue M., Chang L., Hwang J., Chiang S. H., and Saltiel A. R. (2003) The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature 422, 629–633 [DOI] [PubMed] [Google Scholar]
- 49. Inoue M., Chiang S. H., Chang L., Chen X. W., and Saltiel A. R. (2006) Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol. Biol. Cell 17, 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen X. W., Leto D., Xiao J., Goss J., Wang Q., Shavit J. A., Xiong T., Yu G., Ginsburg D., Toomre D., Xu Z., and Saltiel A. R. (2011) Exocyst function is regulated by effector phosphorylation. Nat. Cell Biol. 13, 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lopez J. A., Kwan E. P., Xie L., He Y., James D. E., and Gaisano H. Y. (2008) The RalA GTPase is a central regulator of insulin exocytosis from pancreatic islet β cells. J. Biol. Chem. 283, 17939–17945 [DOI] [PubMed] [Google Scholar]
- 52. Rondaij M. G., Bierings R., van Agtmaal E. L., Gijzen K. A., Sellink E., Kragt A., Ferguson S. S. G., Mertens K., Hannah M. J., van Mourik J. A., Fernandez-Borja M., and Voorberg J. (2008) Guanine exchange factor RalGDS mediates exocytosis of Weibel-Palade bodies from endothelial cells. Blood 112, 56–63 [DOI] [PubMed] [Google Scholar]