Abstract
Once generated, neurons are thought to permanently exit the cell cycle and become irreversibly differentiated. However, neither the precise point at which this post-mitotic state is attained nor the extent of its irreversibility is clearly defined. Here we report that newly born neurons from the upper layers of the mouse cortex, despite initiating axon and dendrite elongation, continue to drive gene expression from the neural progenitor tubulin α1 promoter (Tα1p). These observations suggest an ambiguous post-mitotic neuronal state. Whole transcriptome analysis of sorted upper cortical neurons further revealed that neurons continue to express genes related to cell cycle progression long after mitotic exit until at least post-natal day 3 (P3). These genes are however down-regulated thereafter, associated with a concomitant up-regulation of tumor suppressors at P5. Interestingly, newly born neurons located in the cortical plate (CP) at embryonic day 18-19 (E18-E19) and P3 challenged with calcium influx are found in S/G2/M phases of the cell cycle, and still able to undergo division at E18-E19 but not at P3. At P5 however, calcium influx becomes neurotoxic and leads instead to neuronal loss. Our data delineate an unexpected flexibility of cell cycle control in early born neurons, and describe how neurons transit to a post-mitotic state.
Keywords: post-mitotic neuronal state, cell cycle, ionomycin, calcium influx, RNA-seq, channel rhodopsin
Introduction
A longstanding orthodoxy in neurobiology is that once formed, neurons never divide. Indeed, neurons are the quintessential 'post-mitotic' cell. Attempts to induce neurons to proliferate by either expressing oncogenes or by inactivating tumor suppressors are well documented in the literature, but have generally resulted in neuronal death instead1,2,3,4. In addition, aberrant cell cycle re-entry precedes neuronal loss under various neurotoxic conditions, and activation of cell cycle genes by neurons is elevated in certain neurodegenerative disorders such as Alzheimer's disease5,6. Together, these observations reinforce the notion that cell division is essentially incompatible with the survival of mature neurons. However, precisely when and how during its genesis a neuron attains this irreversible post-mitotic state is still poorly understood. The cell cycle machinery has a role during neuronal development. For instance, genes that regulate phases of the cell cycle also modulate neuronal migration, axon formation, and dendrite growth and branching7,8. However, comparatively little is known about the timing that defines the terminally differentiated state in neurons, or in other words, when a neuron truly becomes a post-mitotic cell. It has been proposed that during the acquisition of morphological features that define a neuron (i.e. axon and dendrites formation), proteins associated with cell cycle regulation participate in these morphological changes as part of the developmental machinery distinct from the cell cycle control8. Cell cycle exit and terminal differentiation generally are coordinated, although in some context they are separable, suggesting that cell cycle exit and differentiation could be different events9. For instance, in Drosophila it was shown that neurons from retina could continue cycling and undergo mitosis while maintaining characteristics of terminal differentiation, such as axon formation10. It is also possible, however, that newly born neurons are in an intermediate state between progenitors and neurons and that their post-mitotic nature is still not fully defined.
During neurogenesis in the cerebral cortex, neural progenitors divide to produce neurons in the ventricular zone (VZ) and sub-ventricular zone, which undergo further differentiation and migrate to their final destinations in the cortical plate in a systematic fashion11. A tight coordination between these events is mediated by calcium signaling, which modulates various aspects of neurogenesis, including neural induction, migration, formation of neural circuits and neurotransmission12,13,14,15,16. Importantly, calcium oscillations also affect the G1/S transition of the cell cycle and thereby regulate progenitor proliferation and differentiation12,17,18. Based on these findings and with the aim of further investigating the functions of calcium signaling in early-born neurons, we used in situ live imaging, and directly characterized the effects of perturbing calcium dynamics in the developing neocortex. Here, we show unexpected cell cycle control flexibility in early born migrating neurons in the CP, which might depend on calcium buffering. These results suggest that neurons transit to a post-mitotic stage gradually.
Results
Newly born neurons from upper cortical layers gradually attain a morphological and molecular differentiated state
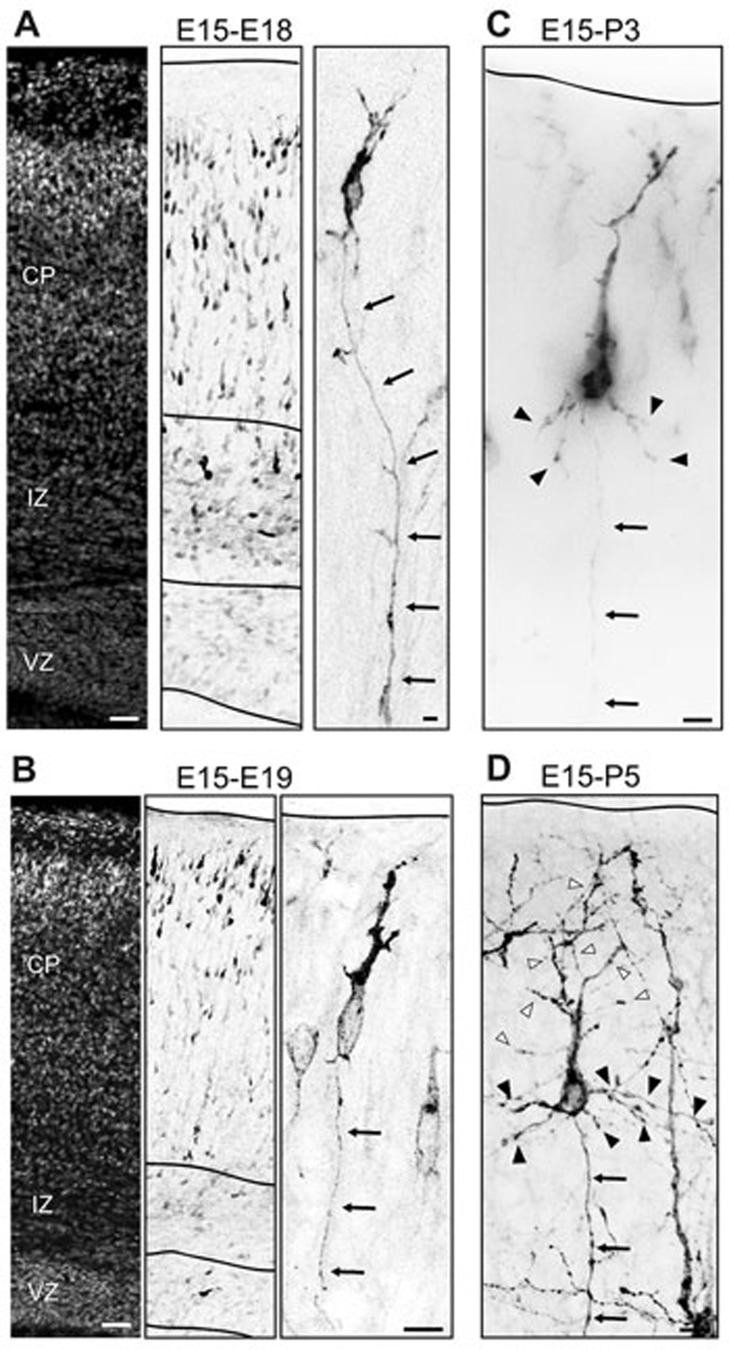
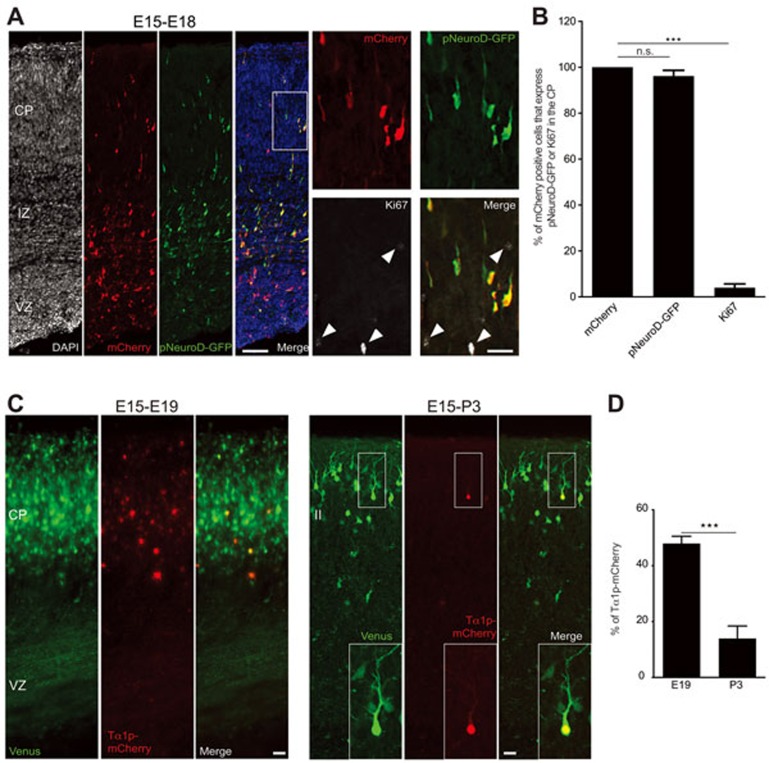
Neurons differentiating in the developing cortex undergo complex morphological changes11 that ultimately lead to brain wiring. To determine when during these morphological changes neurons establish their post-mitotic identity, recently generated neurons were labeled with membrane bound GFP (F-FGP) in utero at E15. These cells were migrating into the CP of the developing mouse cortex at E18-E19 and have initiated axon extension (Figure 1A and 1B11,19,20,21,22), suggesting that the neuronal identity may be established during the migrating phase in the CP. Accordingly, electroporated cells in the CP, which are migrating to their final positions, have voltage-dependent currents characteristic of young neurons11. Moreover, neuronal and glial progenitors are restricted to the VZ and the intermediate zone (IZ) and absent in the CP of the mouse-developing cortex23, before differentiated astrocytes proliferate locally in postnatal upper layers of the cortex24. At E18, E15-in utero electroporated cells in the CP are scattered and migrating to their final positions (Figure 1A). At E19, there is reduced migration of those transfected cells and they are arranged as a layer within the upper CP (Figure 1B). Eventually, transfected cells develop basal dendrites at P3 (Figure 1C) and their apical dendrite, or initially leading process, is remodeled and sculpted to a more complex structure (P5, Figure 1D). These results show that the morphological changes cells undergo within the CP/upper layer of the developing cortex (E18-P5) are associated with a well-defined neuronal identity. Furthermore, co-electroporation at E15 of the pCAG-mCherry construct with a vector expressing GFP under the control of the neuronal pNeuroD promoter25 revealed that at E18 the majority of mCherry-transfected cells in the CP co-express pNeuroD-GFP and do not express the cell cycle marker Ki67; thus, supporting their neuronal status (Figure 2A and 2B; GFP/mCherry = 96.21% ± 2.48%; Ki67/mCherry = 3.98% ± 1.67%; n = 590 cells from three brains from three litters; P < 0.0001 by one-way analysis of variance (ANOVA), post hoc Dunnett test ***P < 0.001; values are mean ± SEM). However, at E19 and P3 pCAG-EGFP-transfected cells in the CP/upper layer still drive mCherry expression under the Tα1 promoter, which is active in neuronal progenitors26,27,28 (Figure 2C). Expression of Tα1p-mCherry was not detected at P5 (data not shown) and at P3 the number of GFP/Tα1p-mCherry expressing cells decreased considerably compared with the number of GFP/ Tα1p-mCherry labeled cells detected at E19 (Figure 2D; E19 = 47.92% ± 2.617%; n = 3879 cells from three brains from three litters; P3 = 13.94% ± 4.480%; n = 2887 cells from three brains from three litters; ***P = 0.0002 by t-test; values are mean ± SEM). Accordingly, it was reported that Tα1p expression in vivo is maintained in developing neurons until early postnatal life, subsequent to which its expression decreased coincidently with neuronal maturation29. Altogether, these results suggest that even though E15-transfected cells at E18-P3 displayed morphological features of developing neurons, they might retain molecular signatures of the neurogenic stage, indicating that E18-P3 neurons bear a mixed, progenitor-committed neuron, identity. This scenario suggests that attainment of the final post-mitotic neuronal identity is a gradual process.
Figure 1.
Neurons in the CP and layer II initiated gradually neuronal morphological differentiation with the formation of axon and dendrites. (A) At E18, E15-in utero electroporated cells in the CP are scattered migrating to their final positions and have already initiated axon extension (arrows at the inset). (B) At E19, migration is reduced in transfected cells which are arranged as a layer within the upper CP with single axon (arrows at the inset). (C, D) At P3, transfected cells develop basal dendrites (dendrites, black arrowheads; axon, arrows) and at P5 their apical dendrite, or initially leading process, is remodeled and sculpted to a more complex structure (apical dendrite, white arrowheads; basal dendrites, black arrowheads; axon, arrows). Scale bar: 200 μm (left panels from A, B) and 10 μm (inset from A-D).
Figure 2.
Cells in the CP and layer II that drive gene expression from the neuronal promoter pNeuroD and the neurogenic intermediate neuronal progenitor promoter Tα1p. (A) Cells in the CP co-express pNeuroD-GFP/mCherry, and are not immunoreactive for Ki67 (white arrowheads, inset from the left panel), a marker of cycling cells. (B) Quantification of experiment from A (P < 0.0001 by one-way ANOVA, post hoc Dunnett test ***P < 0.001; values are mean ± SEM) (C) At E19 and P3, Tα1p-mCherry is expressed in Venus transfected cells from the CP and layer II, respectively. (D) Quantification of mCherry/Venus-transfected cells in the CP and layer II at E19 and P3, respectively (***P = 0.0002 by t-test). Scale bar: 200 μm (A) and 20 μm (inset from A, C).
To test this hypothesis we performed unbiased genome-wide transcriptional profiling of upper layer neurons at P3 and P5, the time window in which we detected the Tα1 promoter to be active or silent. We electroporated cortices with an enhanced YFP (eYFP)-encoding plasmid driven by a neuronal promoter (pNeuroD)25 at E15 and isolated eYFP-positive upper layer (layer II) neurons by fluorescence activated cell sorting (FACS) of dissociated mouse cortices at either P3 or P5 (Supplementary Information, Figure S1). We then extracted whole RNA from neurons sorted in this manner and performed unbiased genome-wide transcriptional profiling using Illumina next-generation sequencing (RNA-seq)30. Analysis of sequencing data using the Cufflinks platform31,32 revealed a total of 322 genes that were differentially expressed between P3 and P5. Of these, 253 genes were up-regulated in P5 relative to P3, whereas 69 genes were down-regulated (Supplementary Information, Figure S2; GEO accession number: GSE61845; and Supplementary information, Table S1 for complete list of genes analyzed). We confirmed a number of these gene expression changes through analysis using quantitative reverse transcription-PCR (qRT-PCR) (Supplementary Information, Figure S3A and Supplementary information, Table S2).
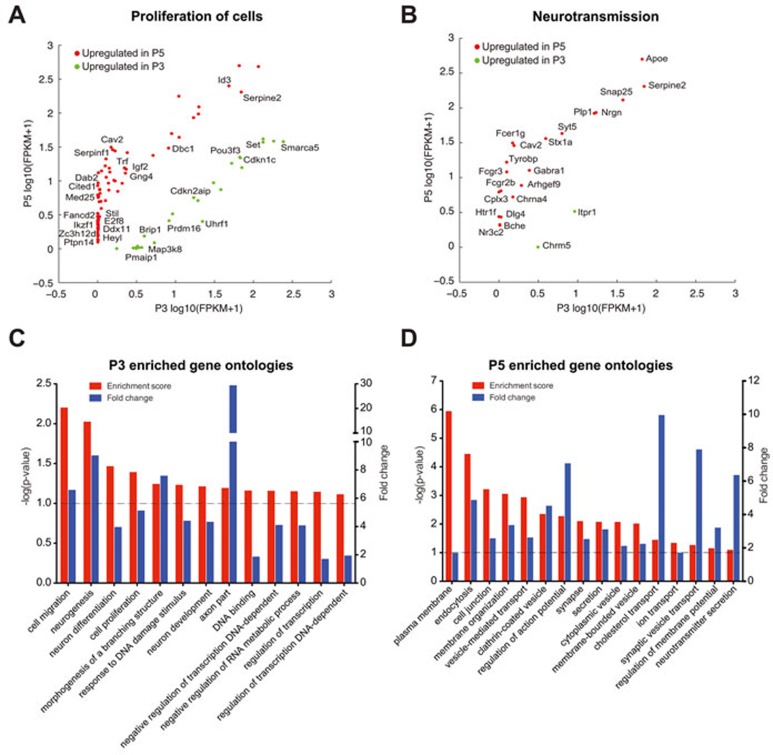
To gain a global perspective of the biological pathways that could be affected by the observed transcriptional changes between P3 and P5, we subjected our list of differentially expressed genes to Ingenuity Pathway Analysis (IPA) (Ingenuity Systems; www.ingenuity.com) and to an analysis of overrepresented gene ontology (GO) terms using DAVID. Surprisingly, networks and bio functions associated with cell proliferation featured prominently with the IPA analysis (networks: (a) cellular function and maintenance, cellular growth and proliferation, cell cycle; (b) cell cycle, nervous system development and function, cellular growth and proliferation; Supplementary Information, Figure S3B; bio-functions: proliferation of cells; P = 5.84 × 10−5; Figure 3A and Supplementary information, Table S3 for a complete list of genes of this bio function) suggesting that the initial neuronal post-mitotic stage could either be more plastic than originally thought or that neurons express a mixed molecular signature between cycling and post-mitotic cells. Specifically, genes that were expressed in P3 but were down-regulated in P5 included those such as UHRF1, BRIP1, and FANCB. Uhrf1 facilitates progression into S phase by repressing the activities of p21 and the retinoblastoma protein (pRb)33,34. Additionally, Uhrf1 is important for DNA replication itself35 and its overexpression can trigger S-phase re-entry in terminally differentiated and quiescent cells36. In the developing cortex, Uhrf1 is expressed in neuronal stem cells37. Interestingly, it was reported that a transcriptional hallmark of adult neuronal stem cells is the increased mRNA expression level of neurogenic regulators, such as UHRF138 and that these genes could preferentially regulate neurogenesis38. Moreover, it was shown that UHRF1 is expressed in early born upper developing cortical neurons39. Brip1 and Fancb are known to participate in the repair of replication-associated DNA damage40,41. Thus, upper cortical neurons at P3 continue to express gene products that could potentially support DNA replication.
Figure 3.
Transcriptional landscape of cortical layer II neurons at P3 and P5 suggests a not well-defined post-mitotic identity at P3 compared to P5. (A, B) Scatter plots showing genes up-regulated at P3 and P5 that are represented in proliferation of cells, and neurotransmission bio-functions. (C, D) Summary of selected categories of over-represented GO terms at P3 and P5 as calculated by DAVID. The GO categories are ranked by their associated −log(P-value). Horizontal bar (threshold) indicates P < 0.1.
In contrast, genes that were upregulated at P5 consisted mainly of important negative modulators of cell proliferation and tumor suppressors, such as ZC3H12D, E2F8, RPRM, DAB2, DBC1, and IKZF1 (Figure 3A and Supplementary Information, Figure S3B)42,43,44,45,46,47. Interestingly, Zc3h12d inhibits the progression of cells from G1 to S phase by suppressing the phosphorylation (i.e., inactivation) of pRb48. Rb-mediated silencing of cell cycle genes is thought to be critical for maintaining the post-mitotic state of various cell types, including neurons and cardiomyocytes49,50,51,52. The up-regulation of Zc3h12d and the concomitant down-regulation of Uhrf1 suggest that an elevation in pRb activity could be particularly important for the creation of a non-permissive state for cell cycle in P5 neurons. Accordingly, differentiated neurons in the retina or brain have been observed to dedifferentiate to give rise to tumors upon reduced Rb family function or induction by oncogenes, respectively53,54. Concomitant with the up-regulation of these cell cycle inhibitors, we found that a number of genes implicated in neurotransmission and synaptic functions were also substantially up regulated in P5 neurons (STX1A, SYT5, SYT10, DLG4, NGRN, SNAP25, C1QA, and C1QB; Figure 3B). Finally, GO analysis supported this general view with categories related to cell proliferation (neurogenesis, P-value 0.0094; cell proliferation, P-value 0.0405; response to DNA damage stimulus, P-value 0.0585), cell migration, axon part, and neuron development among others over-represented at P3 (Figure 3C and Supplementary information, Table S4), while categories related with synapse and membrane dynamics are enriched at P5 (Figure 3D and Supplementary information, Table S4). Together, these results suggest that substantial changes to the transcriptional landscape at P5 could underlie imposition of a post-mitotic state concomitant with neuronal specialization. In contrast, at P3 upper layer neurons seem to have a mixed identity between developing neurons and cells that still retain the potential to re-enter the cell cycle.
Newly born neurons express mAG-hGem upon calcium influx
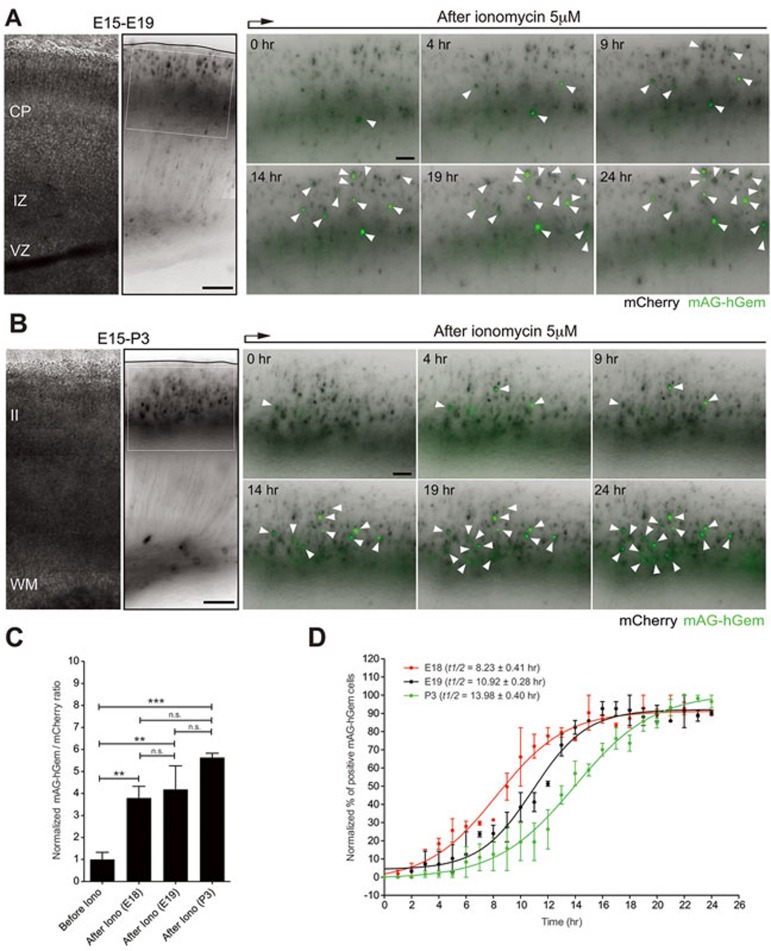
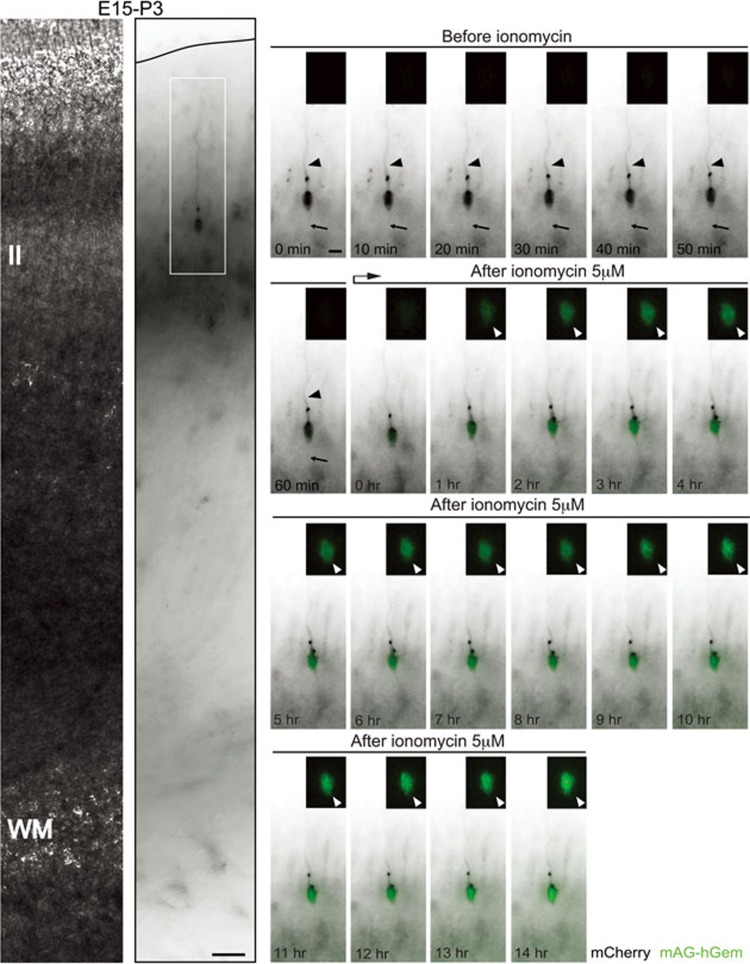
Based on our observations, we wondered whether the expression of proliferation genes is sufficient to induce cell division even when morphological differentiation has occurred. Calcium oscillations affect the G1/S transition of the cell cycle and thereby regulate progenitor proliferation and differentiation12,17,18. In addition, calcium transients regulate neuronal migration55. For instance, granule cells exhibit distinct frequencies of the transient calcium elevations as they migrate in different cortical layers, with less frequent transients when approaching their final destination and complete the migration only after the loss of calcium elevations55. Therefore, we used in situ live imaging to determine whether perturbing calcium influx is a sufficient stimulus to evoke proliferation in cortical neurons during the E18-P5 period. Specifically, we asked whether calcium influx in newly born neurons might evoke features of proliferating cells. We tested different concentrations of the calcium ionophore ionomycin (5 μM, 10 μM, and 50 μM) and found that 5 μM of ionomycin was a concentration at which pCAG-mCherry-transfected neurons in the CP survived the 12 to 24 h time-lapse imaging session and do not express caspase-3 (Supplementary Information, Figure S4). To visualize cell cycle re-entry we utilized monomeric Azami-Green 1 (mAG), a genetically encoded fluorescent probe, fused to a truncated version of the human DNA replication licensing factor Geminin (mAG-hGem, amino acids 1-110). mAG-hGem selectively accumulates in the nucleus of transfected cells specifically in the S/G2/M phases of the cell cycle56. Importantly, non-cycling cells that are quiescent or post-mitotic do not show any signal with this construct56. Therefore the expression of mAG-hGem can be used to indicate a cell that has entered the cell division cycle. We introduced pCAG-mAG-hGem and pCAG-mCherry plasmids into the cortex of E15 mouse embryos via in utero electroporation. Time-lapse imaging analysis was then performed in acute coronal cortical slices at E18-P5 before and after the addition of the calcium ionophore ionomycin (5μM). This paradigm allowed for the assessment of mAG-hGem expression at various ages by neurons that share the same birth date. Neurons in ionomycin-treated slices collected at P5 initiated the nuclear expression of mAG-hGem but then lost both mCherry and mAG-hGem fluorescence in a manner indicating cell death (Supplementary Information, Figure S5A and S5B). Concomitantly, we observed a substantial increase in pyknotic nuclei (nuclei displaying increased chromatin condensation) among mCherry-transfected neurons following ionomycin treatment at P5 relative to either untreated or similarly treated slices at P3 (Supplementary Information, Figure S6; Control (P3) = 7.43% ± 0.80%, n = 120 cells from three slices from three brains from three litters; Ionomycin (P3) = 9.86% ± 1.48%, n = 90 cells from three slices from three brains from three litters; Ionomycin (P5) = 87.93% ± 6.03%, n =150 cells from three slices from three brains from three litters; ****P< 0.0001 by one-way ANOVA, post hoc Dunnett test ***P< 0.001; values are mean ± SEM). Importantly, upon ionomycin treatment neurons in slices collected at E18-P3 expressed nuclear mAG-hGem and survived the 14-24 h imaging session. 24 h ionomycin treatment induced nuclear mAG-hGem expression in similar percentages of mCherry-electroporated neurons from the CP at E18-E19 and layer II at P3 (Figure 4A-4C; E18 = 28.04% ± 3.94%, n = 389 cells from 3 slices from three brains from three litters; E19 = 30.90% ± 7.94%, n = 471 cells from 3 slices from three brains from three litters; P3 = 41.58% ± 1.48%, n = 297 cells from 3 slices from three brains from three litters; P = 0.2307 by one-way ANOVA; values are mean ± SEM). However, the nuclear mAG-hGem expression was faster in neurons from the CP (E18-E19) compared to treated-layer II neurons at P3 (E18 t1/2 = 8.23 ± 0.41 h; E19 t1/2 = 10.92 ± 0.28 h; P3 t1/2 = 13.98 ± 0.40 h; ****P < 0.0001 by one-way ANOVA, post hoc Tukey test: E18 vs. E19 = ns; E18 vs. P3 = ***P < 0.001; E19 vs. P3 = **P < 0.01; values are mean ± SEM; Figure 4D). These results suggest that final commitment or post-mitotic state is established in a gradual process in upper layer neurons following a precise temporal sequence: it is weakly defined during neuronal migration (E18-E19), gets stronger once migration is completed (P3), and reach the irreversible stage at P5. This could be reflected by the increased time to express mAG-hGem at P3 compared to E18-E19 and the observed neurotoxicity at P5 upon calcium influx.
Figure 4.
Calcium influx induces faster nuclear mAG-hGem expression in neurons from the CP compared to neurons from layer II. (A) mCherry/mAG-hGem-transfected cells located in the CP at E19 express the S/G2/M marker mAG-hGem (white arrowheads) upon ionomycin treatment (5 μM). Left panels: cortical slice used for time-lapse at E19 (in utero electroporated at E15). Right panels: Time-lapse sequence of CP-neurons upon ionomycin treatment (inset from left panel). (B) mCherry/mAG-hGem-transfected cells located in layer II at P3 express the S/G2/M marker mAG-hGem (white arrowheads) upon ionomycin treatment (5 μM). Left panels: cortical slice used for time-lapse at P3 (in utero electroporated at E15). Right panels: Time-lapse sequence of layer II-neurons upon ionomycin treatment (inset from left panel). (C) 24 h ionomycin treatment induced similarly nuclear mAG-hGem expression in mCherry-electroporated neurons from the CP at E18-E19 and layer II at P3 (****P < 0.0001 by one-way ANOVA, post hoc Dunnett test; ***P < 0.001, **P < 0.01; values are mean ± SEM). (D) Nuclear mAG-hGem expression in neurons from the CP at E18-E19 is faster compared to layer II neurons at P3 (one-way ANOVA, post hoc Tukey test; E18 vs E19, not significant; E18 vs P3, ***P < 0.001; E19 vs P3, **P < 0.01; values are mean ± SEM). Scale bar: 150 μm (left panels) and 100 μm (right panel).
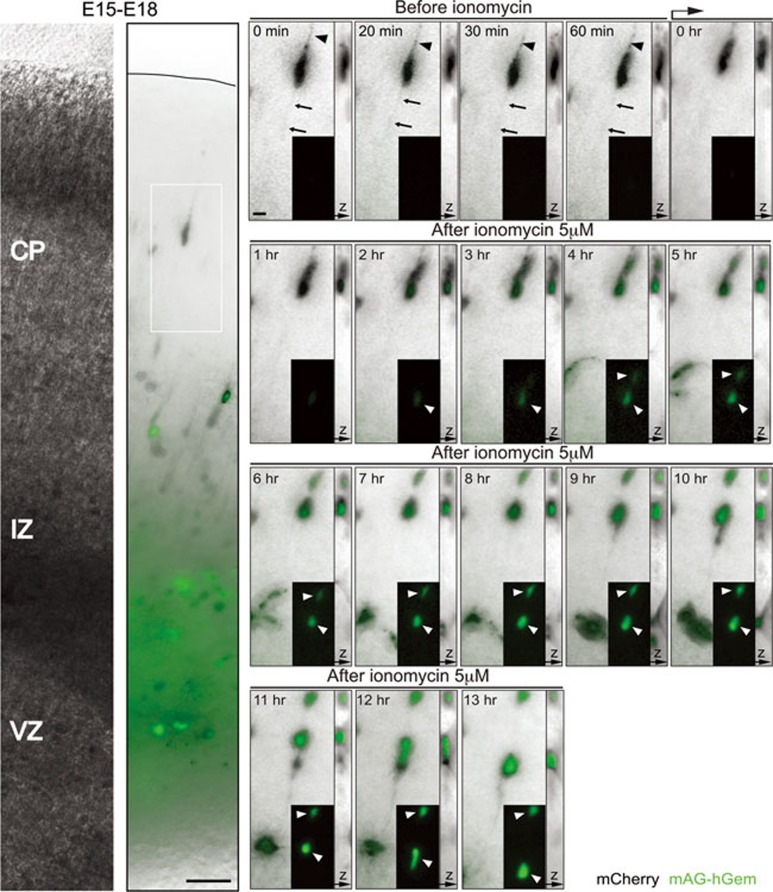
Although calcium influx induced the nuclear expression of mAG-hGem in upper cortical neurons at P3, which already express the neuronal marker NeuN (Supplementary Information, Figure S7A), we were not able to detect cell divisions after nuclear mAG-hGem expression during the 14-24 h imaging session (Figure 4B and Figure 5). In contrast, ionomycin treatment caused a significant increase of the neuronal progenitor marker Tbr2 immunoreactivity among mCherry-positive cells in layer II, compared to untreated controls (82% ± 3%, n = 233 cells from 3 slices vs. 35% ± 7%, n = 362 cells from 4 slices, respectively; P = 0.0003 by t-test; Supplementary Information, Figure S7D and S7E). We therefore decided to explore whether successful neuronal divisions could be induced at earlier cortical developmental stages. To this end, we analyzed E15-transfected cells in the CP of E18-19 cortices. Close analysis of imaged cells in the CP at E18-19 revealed that ionomycin treatment induced migrating neurons with a leading process and a trailing process or axon (labeled with membrane-GFP, F-GFP), to cease migration and undergo a transformation from the bipolar morphology that is typical of migrating neurons to a multipolar morphology (66% ± 3%, n = 163 cells from 4 slices from four brains from three litters; Supplementary Information, Figure S8). More surprisingly however, a subset of pCAG-mAG-hGem and pCAG-mCherry electroporated cells in the CP at E18 and E19 underwent cell division (13% of 100 imaged cells, four slices from four brains from four litters; Figure 6 and Supplementary Information, Figure S9). These results show that neurons in the developing CP can express mAG-hGem and undergo division following calcium influx.
Figure 5.
Calcium influx induces mAG-hGem expression but not cell division in developing neurons from layer II at P3. Neuron with apical dendrite and axon (arrowhead and arrow respectively, in the time-lapse sequence) in layer II from P3 coronal brain slice (left panels, in utero electroporated at E15 with mCherry and mAG-hGem plasmids) express the S/G2/M marker mAG-hGem (white arrowheads, insets from time-lapse sequence) upon ionomycin treatment (5 μM). Scale bar: 50 μm (left panel) and 10 μm (time-lapse sequence).
Figure 6.
Calcium influx induces mAG-hGem expression and cell division in developing neurons from the CP. Neuron with a leading process and trailing process or axon (arrowhead and arrow respectively, in the time-lapse sequence) in the CP from E18 coronal brain slice (left panels, in utero electroporated at E15 with mCherry and mAG-hGem plasmids) express the S/G2/M marker mAG-hGem (white arrowheads, insets from time-lapse sequence) and divide upon ionomycin treatment (5 μM). Scale bar: 50 μm (left panel) and 10 μm (time-lapse sequence).
We next explored whether the morphological changes associated with calcium influx are cell autonomous. Because bath application of ionomycin affects the whole tissue, we instead electroporated a plasmid expressing the light-activated ion channel channelrhodopsin-2 (pCAG-mCherry-ChR2) into cortices of E15 embryos and again prepared brain slices for time-lapse imaging at E18. Induction of depolarization using a short light pulse (30-40 s; 473 nm) caused cells in the CP to stop migrating, retract their leading and trailing processes, and produce instead multiple small neurites that extend and retract actively (Supplementary Information, Figure S10A). Furthermore, similar to ionomycin treatment, a subset of light-activated mCherry-ChR2 expressing cells also underwent cell division (Supplementary Information, Figure S10B), suggesting that cell division decision is in fact cell autonomous. In contrast, control mCherry-electroporated cells in the CP assumed neither a multipolar morphology, nor underwent cell division following the administration of a light pulse (Supplementary Information, Figure S10C).
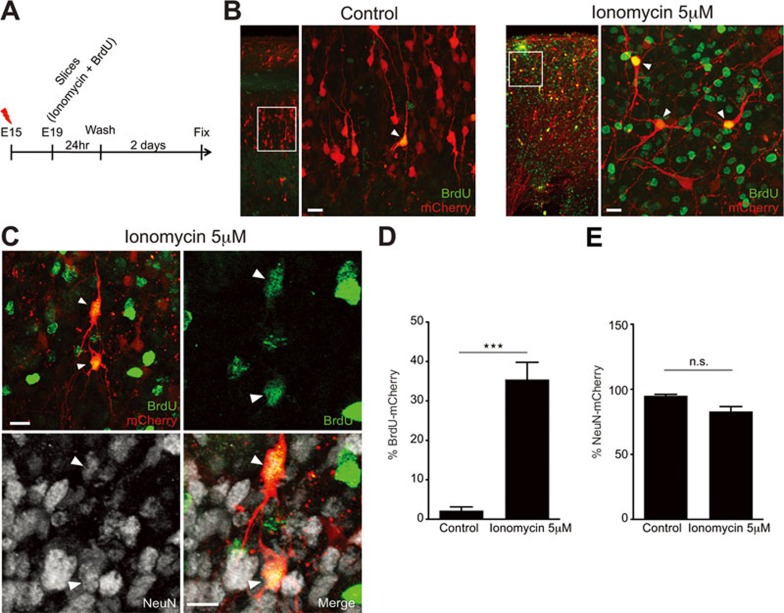
Based on our findings showing neuronal divisions in the CP at E18-E19, we attempted to characterize the fate of the daughter cells that are produced from such division events. Accordingly, we prepared acute coronal slices at E19 from mCherry-electroporated brains at E15, and then incubated the slices with ionomycin together with the thymidine analogue, 5-bromodeoxyuridine (BrdU; 5 μM), an indicator of DNA replication, for 24 h followed by washout and fresh media exchange. Forty-eight hours later, a duration when neuronal progenitors have been shown to clonally expand11, we fixed the slices and evaluated BrdU incorporation into the dividing cells using immunohistochemistry (Figure 7A). As expected, mCherry-positive cells in untreated control slices were mainly negative for BrdU (Figure 7B), whereas a substantial number of ionomycin-treated mCherry-positive cells incorporated BrdU (Figure 7B and 7D; Control = 2.17% ± 0.96%, n = 144 cells from three slices from three brains from three litters; Ionomycin = 35.44% ± 4.36%, n = 194 cells from seven slices from three brains from three litters; ***P = 0.0003 by t-test; values are mean ± SEM). To test whether these cells express neuronal markers, we assessed immunoreactivity for the neuron-specific protein, NeuN. A substantial fraction of the BrdU-positive cells also expressed NeuN (Figure 7C), and there were no significant differences in the total number of NeuN-positive cells among mCherry-transfected cells between ionomycin-treated slices and untreated controls (Figure 7E; Control = 94.95% ± 1.17%, n = 117 cells from three slices from three brains from three litters; Ionomycin = 82.91% ± 3.79%, n = 205 cells from four slices from three brains from three litters; P = 0.0814 by t-test; values are mean ± SEM). Together, these results suggest that cortical neurons that divide upon calcium influx exhibit a limited proliferative potential akin to intermediate progenitors and may exclusively produce neurons when they divide.
Figure 7.
Calcium influx in developing neurons induces a limited proliferative potential. (A) Diagram depicting the steps followed during the experiments. (B) Control neurons in the CP are negative for BrdU and display a radial organization. Slices treated with ionomycin (5 μM) and washout for 2-3 days, contain mCherry-transfected cells in the CP positive for BrdU without a radial organization (white arrowheads, inset from the left panel). (C) Slices treated with ionomycin (5 μM) and washout for 2-3 days contain mCherry-transfected cells in the CP, positive for BrdU and NeuN (white arrowheads, inset from the left panel) that lost radial dendritic organization. (D) Quantification of mCherry-transfected cells in the CP positive for BrdU (***P = 0.0003 by t-test; values are mean ± SEM). (E) Quantification of mCherry-transfected cells in the CP positive for NeuN (P = 0.0814 by t-test; values are mean ± SEM). Scale bar: 10 μm.
Discussion
Recently it was demonstrated that early neurons from upper cortical layers could be instructed to reprogram their identity from one neuronal subtype to another57,58. This plasticity was observed during a short window of development and was lost gradually, thus suggesting not only that the acquisition of neuronal identity is a gradual process, but also that the neuronal post-mitotic state might not be acquired right after mitosis. It is generally believed that cell division is incompatible with differentiation. However, the exact boundary between these biological processes is not well defined. Despite the fact our calcium ionopohore treatment bypasses aspects of normal cell cycle control, our work uncovers a hitherto unheard of flexibility in the development of cortical neurons, wherein newly born neurons retain the potential to divide. We think our results contribute to the advance of the field of neuronal development and should raise intriguing questions such as: what are the physiological cues that promote mitosis in the developing cortex and why newly born neurons no longer respond? Are the mitotic signals lost or are the cells unable to respond? Indeed, our work suggests that changes in calcium buffering alter cell division activity during neuronal migration in the developing cortex. Thus we could speculate that migrating neurons might have the capacity to divide but they are not exposed to the physiological cues that change intracellular calcium levels and promote mitosis. Furthermore, our data suggest that during the formation of axons and dendrites, neurons do not have a terminally defined post-mitotic state. In fact, the post-mitotic or terminally differentiated state might be established through a gradual process that starts with neuronal migration and becomes well defined only when neuronal connectivity is achieved.
Supporting this view, our results show that cortical upper layer neurons might attain an irreversible post-mitotic status, concomitant with a neurotoxic effect of calcium influx and up-regulation of tumor suppressors and genes associated with synapse function, only until P5. On the other hand, at earlier time points (E18-P3) calcium influx in upper layer neurons induced the accumulation of mAG-hGem without inducing neurotoxicity. Importantly, upon calcium influx, the nuclear accumulation of mAG-hGem is faster at E18-E19 compared to P3, and cell division occurs at E18-E19 but not at P3. Therefore, the molecular signature at E18-E19 might be more associated with proliferation despite the fact that these cells have already initiated axon extension. Accordingly, the neuronal progenitor Tα1 promoter is more active at E19 compared to P3, suggesting that upper layer neurons exit the proliferation state gradually. Similar observations have also recently been made for cardiomyocytes in both zebrafish and mice, where proliferation of cardiomyctes was induced in response to partial surgical resection of the heart59,60. Interestingly, in the mouse heart, this regenerative potential is also transient and is lost by P760. However, the question remains: what determines the point of no return during neuronal differentiation? We found up-regulation of negative regulators of cell proliferation and tumor suppressors at P5, such as ZC3H12D, E2F8, RPRM, DAB2, DBC1, and IKZF142,43,44,45,46,47. These genes might determine the establishment of a post-mitotic state. For instance, Zc3h12d inhibits the progression of cells from G1 to S phase by suppressing the phosphorylation and inactivation of pRb48. Rb-mediated silencing of cell cycle genes is critical for maintaining the post-mitotic state of various cell types, including neurons in the developing cortex and cardiomyocytes49,50. Accordingly, differentiated neurons in the retina or brain could dedifferentiate to give rise to tumors upon reduced Rb family function or induction by oncogenes, respectively53,54. Additionally, alterations in centrosome integrity could contribute to the neuronal post-mitotic state as it was described for cardiomyocytes61. Interestingly, loss of centrosome integrity during neuronal development was reported in cultured neurons62. Therefore, it is plausible that in the developing cortex, once neuronal migration is completed, neurons transit to a post-mitotic stage due to the loss of centrosome integrity that might occur during dendrite outgrowth. Altogether, our results not only suggest that the post-mitotic neuronal cell identity is achieved gradually but also shed light on potential regulatory programs that function later on to maintain this terminally differentiated state63.
Materials and Methods
Fluorescent protein constructs
Venus (pCAGIG-GFP), pNeuroD-GFP, and mCherry (pCAGIG) were kindly provided by Z. Xie (Boston University). The GAP43-GFP (F-GFP; pCAGIG) construct was a gift from A. Gartner (the University of Leuven, Belgium). The mCherry-ChR2 (pCAGIG) was a gift from M. Carlen (Karolinska Institutet). mAG-hGem (pCAGIG)56.
In utero electroporation
The Institutional Animal Care and Use Committee of the Massachusetts Institute of Technology approved all experiments. Pregnant Swiss Webster mice were anesthetized by intraperitoneal injections of ketamine 1%/xylazine 2 mg/ml (0.01 μl/g body weight), uterine horns were exposed and plasmids mixed with Fast Green (Sigma) were microinjected into the lateral ventricles of embryos. Five current pulses (50 ms pulse/950 ms interval; 35-36 V) were delivered across the head of the embryos.
Organotypic slice cultures
Mouse embryos were electroporated at E15 and acute coronal brain slices (240 μm) were prepared at E18, E19, P3, and P5 as previously described19. Slices were transferred onto slice culture inserts (Millicell) in cell culture dishes (35 mm × 10 mm, Corning) with Neurobasal medium (Gibco) containing: B27 (1%), glutamine (1%), penicillin/streptomycin (1%), horse serum (5%), and N2 (1%). Slices were used for imaging (1-2 h after slicing) or for pharmacological treatments (incubated at 37 °C in 5% CO2, for 1-2 days). We selected slices from at least three different brains harvested at different days. We did time-lapse analysis in brain slices before and after the treatment. Samples were not included in the analysis if brain slices were not healthy and maintained their shape for the duration of the experiment. Brain slices were not considered in the experiment if they detached from the agarose.
Time-lapse imaging
F-GFP- and mCherry-positive cells were imaged on an inverted Nikon microscope (TE 2000-S) with a 20× objective (NA 0.45). During the time-lapse imaging, slices were kept in an acrylic chamber at 37 °C in 5% CO2. We captured time-lapse images with a CoolSNAP EZ camera (Roper Scientific) using NIS-Elements software (Nikon).
Immunofluorescence
Cortical sections Brains were removed and kept overnight in 4% FA and thereafter transferred to 30% sucrose/PBS (4 °C, overnight). Brains were embedded in OCT compound and sectioned in a cryostat. The 20-30 μm cryosections were incubated overnight at room temperature with 1° antibodies.
Organotypical slice sections Slices were fixed by immersion in 4% FA (30 min). Slices were removed from the slice culture inserts and floating sections were blocked at room temperature for 1 h with 2% goat serum and 0.2% Triton X-100 in PBS. Primary antibodies were applied for 24-48 h at room temperature, followed by thorough washing in 1 × PBS.
Antibodies
The following antibodies were used: rabbit anti-Ki67 (SP6, NeoMarkers, 1:3 000), mouse anti-BrdU (M0744, Clone Bu20a, DakoCytomation, 1:500), rabbit anti-NeuN (ABN78, Millipore, 1:1 000), rabbit anti-cleaved caspase-3 (9661, Cell Signaling, 1:500), rabbit anti-Pax6 (PRB-278P, Covance, 1:200), rabbit ant-Sox2 (AB5603, Millipore, 1:200), rabbit anti-Tbr2 (ab23345, abcam, 1:200). Nuclei were visualized with Hoechst (Invitrogen). Alexa-conjugated secondary antibodies (Jackson Immuno., 1:1 000) were applied for 1-2 h at 20 °C.
Confocal imaging
Images were taken with a Zeiss LSM 510 confocal microscope. Z-series images were collected with 1 μm steps. To perform 3D reconstructions on stacks of images of transfected cells, only Z sections in the same focal plane as GFP were used for analysis and for producing figures. 3D reconstructions and Z-stack analysis were created and performed with ImageJ software.
Image processing
Adjustments of brightness and contrast were performed on images. Epifluorescent images of several focal planes from the time-lapse analysis were assembled using AxioVision software (Zeiss). Some time-lapse sequences were processed using the Gaussian filter to reduce noise.
FACS
Cortices from P3 and P5 pups electroporated at E15 were dissected. Cortices from each litter were pooled and cells were enzymatically dissociated and re-suspended in PBS. GFP positive cells were isolated using the FACS Aria 1 (BD Bioscience) system. Gating for GFP fluorescent cells was set using non-transfected cortical cells. Cells were directly sorted into RLT (QIAGEN) buffer to enhance RNA quality and efficiency.
RNA preparation, high-throughput sequencing, and bioinformatic analyses
Total RNA from GFP positive sorted cells from P3 and P5 cortices of at least 6 brains for P3 and at least 6 brains for P5 from three different litters (2-3 brains per litter) was prepared using QIAGEN kit according to manufacturer's instructions. Total RNA was qualified using an Agilent 2100 Bioanalyzer and prepared by for sequencing using Illumina RNA-sequencing kit following manufacturer's instructions. High-throughput sequencing was done on an Illumina HiSeq 2000 platform. Sequence reads were aligned to mouse mm9 genome with Bowtie64. Reads with mapping quality less than 30 were filtered out with samtools65. Differential gene expression (a gene was considered differentially expressed if P value < 0.1), pathway, and GO analysis were then generated through the use of Cuffdiff, IPA32 and DAVID66, respectively. The P-value is reported by Cuffdiff software and is not multiple-comparison adjusted. The cutoff 0.1 was chosen to select around 200 differentially expressed genes, which is a reasonable number for the following validation and functional analysis approaches, such as DAVID and IPA (32 and Trapnell C, personal communication).
qRT-PCR analysis
RNA was extracted (QIAGEN) from GFP positive sorted cells from transfected cortices of at least 6 brains. Total RNA was reverse-transcribed (Invitrogen) and quantitatively amplified on a thermal cycler (Bio-Rad) using SYBR green (Bio-Rad) and gene-specific primers (Supplementary information, Table S2). The comparative Ct method67 was used to examine differences in gene expression.
Statistical analysis
Compiled data are expressed as mean ± SEM. We used the two-tailed Student's t-test and one-way ANOVA for statistical analysis. Fisher's exact test was used for the transcriptome analysis.
Author Contributions
This study was designated, directed and coordinated by FCdA. RM, DR, OD, KM, MR, BS, and AM provided help with the experiments. JM did the bioinformatics analysis. JG did the qRT-PCR analysis. LHT provided conceptual and technical guidance for the project. The manuscript was written by FCdA and RM, and commented on by all authors.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank CG Dotti, P Soba, K Duncan, and E Kramer for critical reading of the manuscript and providing suggestions. Deutsche Forschungsgemeinschaft (DFG) Grant (FOR 2419; CA1495/1-1), ERA-NET Neuron Grant (Bundesministerium für Bildung und Forschung, BMBF, 01EW1410 ZMNH AN B1), Landesforschungsförderung Hamburg (Z-AN LF), and University Medical Center Hamburg-Eppendorf (UKE) provided support to FCdA.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
FACS of GFP positive cells from transfected cortices.
Heat maps showing the up-regulated genes at P3 and P5.
Quantitative RT-PCR of genes up regulated at P3 and P5 corroborate the RNA-seq data.
Ionomycin has a dose-response effect in developing cortical neurons.
Calcium influx induces cell death in developing neurons from layer II at P5.
Ionomycin treatment induces pyknotic nuclei in upper layer neurons at P5.
Newly born neurons express Tbr2 upon calcium influx.
Calcium influx induces the formation of multipolar cells in the CP.
Calcium influx induces mAG-hGem expression and cell division in developing neurons from the CP.
Cell morphological changes in the CP upon depolarization are cell-autonomous.
Primer sequences used for exon amplification
References
- Feddersen RM, Ehlenfeldt R, Yunis WS, Clark HB, Orr HT. Disrupted cerebellar cortical development and progressive degeneration of Purkinje cells in SV40 T antigen transgenic mice. Neuron 1992; 9:955–966. [DOI] [PubMed] [Google Scholar]
- al-Ubaidi MR, Hollyfield JG, Overbeek PA, Baehr W. Photoreceptor degeneration induced by the expression of simian virus 40 large tumor antigen in the retina of transgenic mice. Proc Natl Acad Sci USA 1992; 89:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, et al. Requirement for a functional Rb-1 gene in murine development. Nature 1992; 359:328–330. [DOI] [PubMed] [Google Scholar]
- Herrup K, Neve R, Ackerman SL, Copani A. Divide and die: cell cycle events as triggers of nerve cell death. J Neurosci 2004; 24:9232–9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci 2007; 8:368–378. [DOI] [PubMed] [Google Scholar]
- Kim D, Tsai LH. Linking cell cycle reentry and DNA damage in neurodegeneration. Ann NY Acad Sci 2009; 1170:674–679. [DOI] [PubMed] [Google Scholar]
- Frank CL, Tsai LH. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron 2009; 62:312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K. Post-mitotic role of the cell cycle machinery. Curr Opin Cell Biol 2013; 25:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta LA, Edgar BA. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol 2007; 19:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta LA, Katzaroff AJ, Perez CL, de la Cruz A, Edgar BA. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev Cell 2007; 12:631–643. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 2004; 7:136–144. [DOI] [PubMed] [Google Scholar]
- Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron 2004; 43:647–661. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Selective role of N-type calcium channels in neuronal migration. Science 1992; 257:806–809. [DOI] [PubMed] [Google Scholar]
- Yuste R, Peinado A, Katz LC. Neuronal domains in developing neocortex. Science 1992; 257:665–669. [DOI] [PubMed] [Google Scholar]
- Llinas R, Sugimori M, Silver RB. Microdomains of high calcium concentration in a presynaptic terminal. Science 1992; 256:677–679. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Calcium signalling remodelling and disease. Biochem Soc Trans 2012; 40:297–309. [DOI] [PubMed] [Google Scholar]
- Kapur N, Mignery GA, Banach K. Cell cycle-dependent calcium oscillations in mouse embryonic stem cells. Am J Physiol Cell Physiol 2007; 292:C1510–1518. [DOI] [PubMed] [Google Scholar]
- Lin JH, Takano T, Arcuino G, et al. Purinergic signaling regulates neural progenitor cell expansion and neurogenesis. Dev Biol 2007; 302:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda FC, Meletis K, Ge X, Rei D, Tsai LH. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci 2010; 30:10391–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A, Sato T, Ando R, Noguchi N, Masaoka M, Miyata T. Dynamics of centrosome translocation and microtubule organization in neocortical neurons during distinct modes of polarization. Cereb Cortex 2014; 24:1301–1310. [DOI] [PubMed] [Google Scholar]
- Namba T, Kibe Y, Funahashi Y, et al. Pioneering axons regulate neuronal polarization in the developing cerebral cortex. Neuron 2014; 81:814–829. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Yamauchi K. Excitatory cortical neurons with multipolar shape establish neuronal polarity by forming a tangentially oriented axon in the intermediate zone. Cereb Cortex 2013; 23:105–113. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci 2005; 25:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature 2012; 484:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, Anton ES. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron 2007; 54:429–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci 2006; 26:1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 2007; 449:351–355. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Yamamoto A, Kawaguchi A, et al. Direct isolation of committed neuronal progenitor cells from transgenic mice coexpressing spectrally distinct fluorescent proteins regulated by stage-specific neural promoters. J Neurosci Res 2001; 65:220–227. [DOI] [PubMed] [Google Scholar]
- Gloster A, Wu W, Speelman A, et al. The T alpha 1 alpha-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci 1994; 14:7319–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc M, Mousli M, Hopfner R, et al. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene 2005; 24:7337–7345. [DOI] [PubMed] [Google Scholar]
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 2009; 37:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EM, Bonsu NM, Price RJ, Lindsay HD. Depletion of Uhrf1 inhibits chromosomal DNA replication in Xenopus egg extracts. Nucleic Acids Res 2013; 41:7725–7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace IM, Latella L, Papait R, et al. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J Cell Biol 2002; 157:909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murao N, Matsuda T, Noguchi H, Koseki H, Namihira M, Nakashima K. Characterization of Np95 expression in mouse brain from embryo to adult: A novel marker for proliferating neural stem/precursor cells. Neurogenesis 2014; 1:e976026. Doi: 10.4161/23262133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R, Tripathi P, Ninkovic J, et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 2010; 7:744–758. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Goff LA, Brettler AC, et al. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 2015; 85:275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 2001; 105:149–160. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 2008; 9:297–308. [DOI] [PubMed] [Google Scholar]
- Minagawa K, Yamamoto K, Nishikawa S, et al. Deregulation of a possible tumour suppressor gene, ZC3H12D, by translocation of IGK@ in transformed follicular lymphoma with t(2;6)(p12;q25). Br J Haematol 2007; 139:161–163. [DOI] [PubMed] [Google Scholar]
- Mok SC, Chan WY, Wong KK, et al. DOC-2, a candidate tumor suppressor gene in human epithelial ovarian cancer. Oncogene 1998; 16:2381–2387. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Gill JH, Pitt E, Kennedy W, Knowles MA. Negative regulation of G(1)/S transition by the candidate bladder tumour suppressor gene DBCCR1. Oncogene 2001; 20:2956–2964. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453:110–114. [DOI] [PubMed] [Google Scholar]
- Ohki R, Nemoto J, Murasawa H, et al. Reprimo, a new candidate mediator of the p53-mediated cell cycle arrest at the G2 phase. J Biol Chem 2000; 275:22627–22630. [DOI] [PubMed] [Google Scholar]
- Christensen J, Cloos P, Toftegaard U, et al. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res 2005; 33:5458–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa K, Katayama Y, Nishikawa S, et al. Inhibition of G(1) to S phase progression by a novel zinc finger protein P58(TFL) at P-bodies. Mol Cancer Res 2009; 7:880–889. [DOI] [PubMed] [Google Scholar]
- Sdek P, Zhao P, Wang Y, et al. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J Cell Biol 2011; 194:407–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa M, Okada K, Nakajima K, Ajioka I. Cortical excitatory neurons become protected from cell division during neurogenesis in an Rb family-dependent manner. Development 2013; 140:2310–2320. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell 2005; 8:541–551. [DOI] [PubMed] [Google Scholar]
- Sage C, Huang M, Karimi K, et al. Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science 2005; 307:1114–1118. [DOI] [PubMed] [Google Scholar]
- Ajioka I, Martins RA, Bayazitov IT, et al. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 2007; 131:378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 2012; 338:1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada T, Komuro H. Completion of neuronal migration regulated by loss of Ca(2+) transients. Proc Natl Acad Sci USA 2004; 101:8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132:487–498. [DOI] [PubMed] [Google Scholar]
- Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat Cell Biol 2013; 15:214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rossa A, Bellone C, Golding B, et al. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat Neurosci 2013; 16:193–200. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010; 464:606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science 2011; 331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebrowski DC, Vergarajauregui S, Wu CC, et al. Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. elife 2015; 4:e05563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiess M, Maghelli N, Kapitein LC, et al. Axon extension occurs independently of centrosomal microtubule nucleation. Science 2010; 327:704–707. [DOI] [PubMed] [Google Scholar]
- Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nat Neurosci 2014; 17:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FACS of GFP positive cells from transfected cortices.
Heat maps showing the up-regulated genes at P3 and P5.
Quantitative RT-PCR of genes up regulated at P3 and P5 corroborate the RNA-seq data.
Ionomycin has a dose-response effect in developing cortical neurons.
Calcium influx induces cell death in developing neurons from layer II at P5.
Ionomycin treatment induces pyknotic nuclei in upper layer neurons at P5.
Newly born neurons express Tbr2 upon calcium influx.
Calcium influx induces the formation of multipolar cells in the CP.
Calcium influx induces mAG-hGem expression and cell division in developing neurons from the CP.
Cell morphological changes in the CP upon depolarization are cell-autonomous.
Primer sequences used for exon amplification