Abstract
The biological processes governing brain development and maturation depend on complex patterns of gene and protein expression, which can be influenced by many factors. One of the most overlooked is the long noncoding class of RNAs (lncRNAs), which are known to play important regulatory roles in an array of biological processes. Little is known about the distribution of lncRNAs in the sensory systems of the brain, and how lncRNAs interact with other mechanisms to guide the development of these systems. In this study, we profiled lncRNA expression in the mouse auditory forebrain during postnatal development at time points before and after the onset of hearing (P7, P14, P21, adult). First, we generated lncRNA profiles of the primary auditory cortex (A1) and medial geniculate body (MG) at each age. Then, we determined the differential patterns of expression by brain region and age. These analyses revealed that the lncRNA expression profile was distinct between both brain regions and between each postnatal age, indicating spatial and temporal specificity during maturation of the auditory forebrain. Next, we explored potential interactions between functionally-related lncRNAs, protein coding RNAs (pcRNAs), and associated proteins. The maturational trajectories (P7 to adult) of many lncRNA – pcRNA pairs were highly correlated, and predictive analyses revealed that lncRNA-protein interactions tended to be strong. A user-friendly database was constructed to facilitate inspection of the expression levels and maturational trajectories for any lncRNA or pcRNA in the database. Overall, this study provides an in-depth summary of lncRNA expression in the developing auditory forebrain and a broad-based foundation for future exploration of lncRNA function during brain development.
Keywords: lncRNA, non-coding RNA, sequencing, transcriptome, brain, synapse, plasticity, development, maturation, cortex, thalamus, neurotransmission, neuromodulation, sensory
1. INTRODUCTION
The biological processes governing the development and maturation of the brain depend on a complex network of gene and protein expression, which can be influenced by many factors. These expression patterns are incompletely understood and represent many interesting avenues of study in terms of temporospatial expression and regulation.
Especially in the sensory systems of the brain, the onset of sensory experience represents a dramatic shift in activity that could affect the mechanisms guiding their development and maturation. In altricial animals, for example, the marked increase of neuronal activity in the central auditory pathways upon opening of the ear canals in early postnatal life stimulates maturation of the brain’s auditory circuitry. This window provides an opportunity to study the impact of hearing onset on neurophysiological response properties (Bao, 2015; Barkat et al., 2011; Chang et al., 2003; Chun et al., 2013; de Villers-Sidani and Merzenich, 2011; Froemke and Jones, 2011b; Hensch, 2005; Kral, 2013; Oswald and Reyes, 2011; Sanes and Bao, 2009; Sanes and Woolley, 2011; Yang et al., 2012) and critical periods for sound processing (Bao et al., 2001; Brown and Kaczmarek, 2011; Dorrn et al., 2010; Edeline et al., 2011; Froemke et al., 2013; Froemke and Jones, 2011a; Hurley and Sullivan, 2012; Kilgard and Merzenich, 1998; Metherate and Hsieh, 2003; O'Neil et al., 2011; Schachtele et al., 2011; Sun et al., 2010; Sutor and Hagerty, 2005; Venkataraman and Bartlett, 2013). Undoubtedly, these maturational events are supported by alterations in neuronal circuitry at the cellular and molecular levels, including changes in gene and protein expression; however, documentation of these properties is incomplete. To advance inquiry along these lines, we recently sequenced the transcriptome in divisions of the auditory forebrain of C57bl/6J mice from postnatal day 7 (P7) through adulthood, which spans the onset of hearing (~P11-P13) in this species (Hackett et al., 2015). In addition to generating a database of the entire transcriptome, approximately 5,000 protein-coding RNAs (pcRNAs) were profiled in detail. Maturational changes in expression were observed in scores of gene families with important roles in brain structure and function.
In addition to pcRNAs, non-coding RNA (ncRNA) expression is also essential to develop a complete understanding of the genomic landscape during brain development. Among the many ncRNA subtypes that could be explored, interest in long non-coding RNAs (lncRNAs) has increased considerably as awareness of their functional importance has grown. There are roughly 10,000 lncRNAs in mammalian genomes (Cabili et al., 2011; Harrow et al., 2012; Ilott and Ponting, 2013; Rinn and Chang, 2012). Traditionally believed to be non-functional, lncRNAs have recently been shown to possess functional roles(Dinger et al., 2009; Mercer et al., 2009), including roles in high-order chromosomal dynamics (Amaral and Mattick, 2008), embryonic stem cell differentiation (Dinger et al., 2008), telomere biology (Schoeftner and Blasco, 2008), subcellular structural organization (Mercer et al., 2008), and breast cancer (Bhan et al., 2014; Bhan et al., 2013). LncRNAs are usually defined as non-coding RNA with length more than 200 base pairs (Mercer et al., 2009; Perkel, 2013). Structurally, lncRNAs and mRNAs are very similar, as both can exhibit poly-adenylation (poly(A)). The number of definable lncRNAs varies by study. An early study in 2007 estimated that there are four times more lncRNAs than pcRNAs (Kapranov et al., 2007). Another study claims to have identified 35,000 lncRNAs (Carninci et al., 2005), and many of them have characteristics similar to mRNA, such as 5’ capping, splicing, and poly-adenylation, with the exception of open reading frames. In the latest effort to quantify human lncRNA, the Encyclopedia of DNA Elements (ENCODE) (Djebali et al., 2012) project identified 13,333 lncRNAs and further categorized them into four sub-classes: 1) antisense, 2) large intergenic non-coding RNAs (lincRNA), 3) sense intronic, and 4) processed transcripts. Compared to pcRNAs, lncRNAs tend to have much lower expression levels, often due to cell-type specific expression (Cabili et al., 2011; Guttman et al., 2010; Liu et al., 2016), but transcript abundance is not known to be related to function (Ulitsky and Bartel, 2013).
As a group, lncRNAs are relatively highly expressed the adult and developing brain (Derrien et al., 2012; Lin et al., 2011; Lipovich et al., 2012; Mercer et al., 2008; Ng et al., 2012; Smalheiser et al., 2008; Washietl et al., 2014). The functions of most are unknown, but many are now known to have regulatory influence over the expression of other genes and proteins (Carninci et al., 2005; Carrieri et al., 2012; Guttman et al., 2011; Guttman and Rinn, 2012; Halley et al., 2014; Katayama et al., 2005; Khalil et al., 2009; Kornienko et al., 2013; Kurokawa, 2011; Magistri et al., 2012; Mattick, 2007; Meng et al., 2012; Onoguchi et al., 2012; St Laurent and Wahlestedt, 2007; Tsai et al., 2010; Vance et al., 2014; Wu et al., 2013; Zhang et al., 2012; Zhao et al., 2013). With respect to nervous system development, lncRNAs may also influence maturational processes such as neurogenesis, synaptogenesis, cell migration, cell type specification, neurite outgrowth, and synaptic plasticity (Aprea et al., 2013; Berghoff et al., 2013; Bernard et al., 2010; Bond et al., 2009; Feng et al., 2006; Kraus et al., 2013; Lin et al., 2014a; Ling et al., 2011; Lipovich et al., 2012; Liu et al., 2016; Modarresi et al., 2012; Ng et al., 2012; Onoguchi et al., 2012; Tarabykin et al., 2001; Ulitsky et al., 2011; Vance et al., 2014).
As observed for protein coding genes, lncRNA expression patterns in the brain tend to be spatially and temporally restricted. Spatially, expression levels may vary substantially between major brain regions (e.g., hippocampus versus cerebral cortex) (Amaral et al., 2009; Kadakkuzha et al., 2015; Ling et al., 2009; Ling et al., 2011; Lv et al., 2013; Mercer et al., 2008; Ponjavic et al., 2009; Sauvageau et al., 2013; Spigoni et al., 2010; Ziats and Rennert, 2013), and also by subdivision or compartment within a region (e.g., cortical layer) (Aprea et al., 2013; Belgard et al., 2011; Kadakkuzha et al., 2015; Mercer et al., 2008; Sasaki et al., 2008; Sauvageau et al., 2013; Spigoni et al., 2010). Spatial specificity is also apparent at the cellular level, where expression may be restricted to subpopulations of neurons, glia, or even subcellular compartments (e.g., nuclei, cytoplasm)(Aprea et al., 2013; Kadakkuzha et al., 2015; Korneev et al., 2008; Liu et al., 2016; Mercer et al., 2008; Mercer et al., 2010; Pollard et al., 2006; Sasaki et al., 2008; Sauvageau et al., 2013; Sone et al., 2007; Tochitani and Hayashizaki, 2008). Temporally, lncRNA expression in a given locus (e.g., region, subregion, cell type) often changes over the long course of nervous system development, most notably between key developmental stages or significant events (e.g., the onset of sensory experience) (Amaral et al., 2009; Aprea et al., 2013; Lin et al., 2011; Ling et al., 2009; Ling et al., 2011; Lipovich et al., 2012; Liu et al., 2016; Mercer et al., 2010; Ponjavic et al., 2009; Spigoni et al., 2010; Tarabykin et al., 2001). Accordingly, the roles played by lncRNAs in brain development may well depend on the precise timing and location of a given event. A number of outstanding reviews of this rapidly growing literature are available (Aprea and Calegari, 2015; Clark and Blackshaw, 2014; Geisler and Coller, 2013; Guttman and Rinn, 2012; Knauss and Sun, 2013; Mattick, 2007; Mehler and Mattick, 2007; Ng et al., 2013; Qureshi et al., 2010; Qureshi and Mehler, 2012; St Laurent and Wahlestedt, 2007; Wu et al., 2013).
The specificity in temporal and spatial expression patterns among lncRNAs suggest that profiles differ between brain region and cellular subtype, as well as developmental stage. Informed by knowledge of those patterns, subsequent studies may be implemented to identify regulatory relationships, interactions, and functional pathways greater specificity.
In the present study, we used high throughput sequencing of total RNA (RNAseq) to profile lncRNA expression patterns in two different divisions of the auditory forebrain at key postnatal ages relative to the onset of hearing. RNAseq has been traditionally used as a replacement for microarray technology to profile pcRNAs (Asmann et al., 2009; Cloonan et al., 2008; Guo et al., 2013b; Marioni et al., 2008; Wang et al., 2009c). While the majority of studies focus solely on pcRNAs, high throughput sequencing allows us to perform advanced data mining (Han et al., 2014; Samuels et al., 2013; Vickers et al., 2015; Ye et al., 2014). One of the primary minable yet underutilized products of RNAseq data is lncRNA. Based on previous findings (Guo, 2015), total RNAseq produces data better suited for studying lncRNA than RNAseq data produced from a poly(A) RNA library. Taking advantage of the unique properties of total RNAseq, we profiled lncRNA expression in the mouse auditory forebrain at four postnatal time points (postnatal days P7, P14, P21, and adult), spanning the period before and after the onset of hearing (i.e., ~P11). The primary goal was to characterize the development of lncRNA over time by employing differential expression analyses of the transcriptome between time points and brain regions. The findings augment our previous characterization of pcRNA in the auditory forebrain acquired from the same animal subjects (Hackett et al., 2015).
2. MATERIALS AND METHODS
2.1 Tissue acquisition
All procedures were approved by the Animal Care and Use Committee at Massachusetts Eye and Ear Infirmary and adhered to the guidelines established by the National Institutes of Health for the care and use of laboratory animals. The morning that a new litter of pups was first observed was designated P0. Brains were collected from 24 adult (8 – 10 weeks) and juvenile (P7, P14, and P21) male and female C57BL/6J mice (Jackson Labs 000664) (N = 6 per age, equal numbers of males and females, total = 24). Animals were euthanized intraperitoneally with a lethal dose of ketamine and xylazine (200/50 mg/kg, respectively). Brains were removed immediately, flash frozen on dry ice, and stored at −80° C.
2.2 Sample acquisition
Frozen brains from 6 animals in each age group (3 male, 3 female) were sectioned in the coronal plane (rostral to caudal) on a sliding microtome and viewed through a surgical microscope. As areas targeted for sampling became visible (A1, primary auditory cortex; MG, medial geniculate body), they were extracted using a sterile tissue punch or curette of a size appropriate to the brain region. A1 samples were obtained using a 0.5 mm diameter punch, with the ventral edge beginning approximately 1 mm dorsal to the rhinal fissure. MG samples were harvested with a curette after using a micro-dissecting scalpel to circumscribe its perimeter. Auditory cortex samples were centered on A1 but potentially included some tissue in the adjacent auditory field dorsal to A1. For the MG, the microdissection procedure was intended to exclude the lateral geniculate nucleus (LGN) and adjoining nuclei dorsal, medial, and ventral to the MG. The extreme rostral and caudal poles of the MG were largely excluded from these samples. Punches from homologous areas of both hemispheres were combined in a sterile tube containing 400 ul of Trizol, homogenized for 45 seconds using a mechanized sterile pestle, flash frozen on dry ice, then stored at −80° C.
2.3 RNA extraction and sequencing
For each Trizol lysate, 100μl of Reagent Grade Chloroform (Fisher Scientific, S25248) was added. The samples were centrifuged for 3 minutes on a desktop centrifuge to fractionate the aqueous and organic layers. After centrifugation, the resulting aqueous layer was carefully removed and transferred to 2.0ml Sarstedt tubes (Sarstedt, 72.694), which were run on the QIAsymphony using the QIAsymphony RNA Kit (Qiagen, 931636) and protocol RNA_CT_400_V7, which incorporates DNAse treatment. Prior to each run, the desk was uv-irradiated using the programmed cycle. The resulting RNA was eluted to 100μl of RNase free water and stored at −80°C in 2.0ml Sarstedt tubes until use. Samples were initially quantitated using a Qubit RNA assay. Additional analyses of purity and the quantitation of total RNA were performed using a NanoDrop spectrophotometer (Thermo Scientific) and Agilent RNA 6000 Pico chip (Agilent) using the protocol, reagents, chips, and ladder provided in the kit. RNA Quality control data for the 48 samples sequenced are contained in Supplementary Table S1.
RNAseq was performed by the Vanderbilt Technologies for Advanced Genomics core (VANTAGE). Total RNA was isolated with the Aurum Total RNA Mini Kit. All samples were quantified on the QuBit RNA assay. RNA quality was verified using an Agilent Bioanalyzer. RNAseq data was obtained by first using the Ribo-Zero Magnetic Gold Kit (Human/Mouse/Rat) (Epicentre) to perform ribosomal reduction on 1μg total RNA following the manufacturer’s protocol. After ribosomal RNA (rRNA) depletion, samples were then purified using the Agencourt RNAClean XP Kit (Beckman Coulter) according to Epicentre protocol specifications. After purification, samples were eluted in 11μl RNase-free water. Next, 1μl ribosomal depleted samples were run on the Agilent RNA 6000 Pico Chip to confirm rRNA removal. After confirmation of rRNA removal, 8.5μl rRNA-depleted samples were put into the Illumina TruSeq Stranded RNA Sample Preparation kit (Illumina) for library preparation. Libraries were multiplexed six per lane and sequenced on the HiSeq 2500 to obtain at least 30 million paired end (2x50 bp) reads per sample.
The complete set of raw sequencing files is available from the National Center for Biotechnology Information (NCBI) database under accession number SRP053237 (http://www.ncbi.nlm.nih.gov/projects/geo/). All other supporting data are included in the Supplementary files.
2.4 RNAseq data processing
The RNAseq data went through multiple stages of thorough quality control as recommended by Guo et al (Guo et al., 2013c). Raw data and alignment quality control were performed using QC3 (Guo et al., 2014a), and gene quantification quality control was conducted using MultiRankSeq (Guo et al., 2014b). Raw data were aligned with TopHat2 (Kim et al., 2013) against the mm10 mouse reference genome, and read counts per gene were obtained using HTSeq (Anders et al., 2014). Normalized read counts (used in all plots) were obtained by normalizing each gene’s read count against the sample’s total read count, then multiplied by a constant (1 × 106). pcRNA and lncRNA were annotated using references file MM10 v38.82 downloaded from Ensembl. Hierarchical clustering analysis and heatmaps were produced using the Heatmap3 (Zhao et al., 2014) package from R. For all samples, quality control data are contained in Table S2.
Differential expression analyses between all postnatal ages and brain regions were performed using MultiRankSeq (Guo et al., 2014b) with three methods for RNAseq analysis: DESeq (Anders and Huber, 2010); edgeR (Robinson et al., 2010); baySeq (Hardcastle and Kelly, 2010). These three methods were chosen based on results of several previous studies in which multiple RNAseq differential analysis methods were compared for accuracy and sensitivity of read count-based data (Dillies et al., 2013; Guo et al., 2013a; Kvam et al., 2012; Robles et al., 2012; Soneson and Delorenzi, 2013). In analyses of the same dataset, the methods typically differ in numbers of differentially expressed genes identified in a comparison of any two samples and also in the direction of expression (up- or down-regulation). False discovery rate (FDR < 0.05) was used to correct multiple testing. The differential expression datasets associated with each pairwise comparison (4 ages × 2 brain areas) are contained in supplementary Tables S5 – S10. Trend analysis of lncRNA expression across the four age points (P7 → P14 → P21 → Adult) was conducted using the Mann-Kendall trend test (Hirsch et al., 1982).
Potential interactions between lncRNAs and pcRNAs were identified using Spearman correlation analysis. To evaluate lncRNA coding potential, we employed the Coding-Potential Assessment Tool (CPAT) (Wang et al., 2013b) (Table S11). BEDTools (Quinlan and Hall, 2010) was used to extract the genomic sequences of lncRNA as input for CPAT. We also performed network analysis using Cytoscape (Saito et al., 2012) and function analysis using WebGestalt (Wang et al., 2013a) based on the correlation results (Table S12). To ensure high correlations were not due to static low expression values across all samples, we filtered out the lowest 25% of all RNAs based on standard deviation. For a subset of genes, lncPro (Lu et al., 2013) was applied to obtain interaction scores between lncRNAs and selected protein targets.
2.5 Database and Look-Up tool for generating lncRNA maturational profiles
Table S4 contains the raw read counts, differential analyses, and pcRNA correlations for all lncRNAs. Tables S5 – 10 contain the differential expression analyses for comparisons of postnatal age and brain region. To facilitate screening and extraction of maturational profiles from the database, a Look-Up tool was developed (Table S13). The tool automatically plots the maturational profiles and correlation matrices for any single lncRNA gene or list of genes (up to 25 at a time) by brain region. It also generates a listing of the normalized counts for all samples by age and brain region for custom applications.
3. RESULTS
3.1 Data quality
RNAseq data were obtained from 48 samples and quality controlled. Sample information (sample ID, brain region, age, sex, and quality assessments) is contained in Table S1. On average, each sample was sequenced with 33.8 million reads (range: 27.6-45.1 million). Sample 10 failed sequencing with less than half million reads produced, and thus was removed from subsequent analyses. No other quality issue was observed. The raw data statistics are contained in Table S2. Alignment quality control was conducted, revealing an average of 77.19% of all reads (range: 51.86% – 83.01%) were aligned to coding RNA regions (Table S3). The complete raw read count information can be found in Table S4.
3.2 Cluster analysis
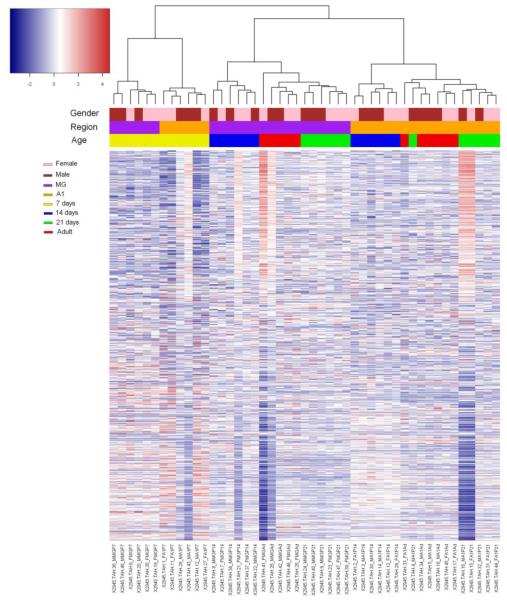
To examine differences in lncRNA expression associated with age and brain region, we first performed an unsupervised cluster analysis using Heatmap3 (Zhao et al., 2014). This analysis revealed that lncRNA expression patterns are distinctively associated with postnatal age and brain region (Fig. 1). Of special interest were the stronger differences by brain region in lncRNA expression patterns for older mice. That is, P7 samples were clustered together first, then clustered by brain regions. P14 to adult samples were separated into two large clusters by brain region and then by age within each regional cluster. This suggests that lncRNA expression in the A1 and MG regions was relatively similar for younger mice before hearing onset. Then, with maturation, the lncRNA expression patterns became more regionally distinct. Gender, on the other hand, had no significant role in lncRNA expression in A1 or MG regions. The unsupervised cluster analysis showed that by using lncRNA expression information alone, we can distinguish brain regions and postnatal age.
Fig. 1. Grand summary of lncRNA expression in MG and A1 from P7 to adult.
Top: Unsupervised hierarchical clustering of samples by sex, brain region, and age. Bottom: Heatmap summarizing total lncRNA expression for each sample, arranged in columns by cluster. Each bar represents one gene. Color code denotes expression level.
3.3 Differential expression analyses
Differential expression analyses were carried out by comparing A1 and MG at different ages and collectively. Because we used three RNAseq expression analysis packages, differential expression for a gene was considered to be significant if all three methods identified it as significant. Summaries of various comparisons are contained in Tables 1 and 2. The detailed results (including fold change and raw and adjusted p-values of all genes) of the comparisons can be found in Tables S5-S10. The results reveal several trends. First, in comparing brain regions, there were substantial regional differences in expression at all ages. The total numbers were fairly stable from P7 to P14, decreased by nearly 60% from P14 to P21, and then by an additional 53% from P21 to adulthood (Table 1). Similarly, comparing successive ages within each region, the numbers of differentially expressed genes were greatest in the P7-P14 interval as compared to all other intervals (Table 2, top). These results indicate that regional differences in lncRNA expression are greatest during the earlier stages of postnatal development, and before and shortly after the onset of hearing. Second, the total numbers of differentially expressed genes from P7 to adult were 70% greater in A1 (N=554) as compared to MG (N=388) (Table 2, top). This suggests a greater degree of genomic modification in A1 from P7 to maturity. Finally, of the differentially expressed genes in one age interval (e.g., P7-P14), a minority of the same genes was also differentially expressed at another age interval (e.g., P14-P21, P21-adult) (Table 2, middle and bottom). Only a handful of the same genes that were differentially expressed between P7 and P14 exhibited significantly changed expression from P21 to adult (A1, N=0; MG, N=3). This indicates that lncRNA genes whose expression levels change with age tend to be different between age intervals.
Table 1.
Differential expression of lncRNAs between A1 and MG by age.
| A1 vs MG | Differentially Expressed lncRNA |
|---|---|
| P7 | 375 |
| P14 | 430 |
| P21 | 180 |
| Adult | 85 |
The total numbers of lncRNAs that were differentially expressed between A1 and MG are listed by postnatal age.
Table 2.
Differential expression of lncRNAs between age groups by brain region.
| Age | P14 | A1 P21 |
Adult | P14 | MG P21 |
Adult |
|---|---|---|---|---|---|---|
| P7 | 215 | 134 | 554 | 206 | 493 | 388 |
| P14 | --- | 1 | 196 | --- | 55 | 67 |
| P21 | --- | --- | 0 | --- | --- | 9 |
| Age | P7-P21 | P7-Adult | A1 P14-P21 |
P14-Adult | P21-Adult |
|---|---|---|---|---|---|
| P7-P14 | 69 | 169 | 1 | 48 | 0 |
| P7-P21 | --- | 115 | 1 | 32 | 0 |
| P7-Adult | --- | --- | 1 | 136 | 0 |
| P14-P21 | --- | --- | --- | 1 | 0 |
| P14-Adult | --- | --- | --- | --- | 0 |
| Age | P7-P21 | P7-Adult | MG P14-P21 |
P14-Adult | P21-Adult |
|---|---|---|---|---|---|
| P7-P14 | 182 | 156 | 21 | 17 | 3 |
| P7-P21 | --- | 298 | 51 | 46 | 8 |
| P7-Adult | --- | --- | 45 | 58 | 4 |
| P14-P21 | --- | --- | --- | 27 | 3 |
| P14-Adult | --- | --- | --- | --- | 4 |
In each panel, the number of differentially expressed lncRNAs are given for each comparison within the brain region indicated (A1 or MG). Top, comparisons of each age with all other ages. Middle (A1) and bottom (MG), interactions between all possible age group comparisons by brain region. Totals reflect the numbers of differentially expressed genes from each age group comparison (e.g., P7-P14) that were also differentially expressed in all others (e.g., P7-P21, P7-Adult, etc.).
Table 3 lists the top 50 up- and down-regulated genes in A1 and MG, ranked by log2 fold change (FC) in expression from P7 to adult. A minority of these genes were up-regulated (N=11) or down-regulated (N=9) in both regions. This indicates that the lncRNA genes with the greatest changes in expression during maturation tended to be preferentially expressed in A1 or MG. Only two genes (GM26924, GM15564) were up-regulated in one region (MG) and down-regulated in another (A1). These findings are evidence of strong regional specificity in lncRNA expression during maturation.
Table 3.
Top 50 up- and down-regulated genes in A1 and MG.
| MG | A1 | ||||||
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | ||||
| lncRNA | FC | lncRNA | FC | lncRNA | FC | lncRNA | FC |
| Gm13905 | 6.99 | H19 | −6.79 | Gm16291 | 4.81 | Rp24-86o15.2 | −5.60 |
| 1500015l24rik | 5.98 | Gm4926 | −5.46 | Gm26747 | 4.45 | A430048g15rik | −5.58 |
| A730090h04rik | 4.42 | Gm19980 | −5.02 | Gm12576 | 4.17 | Rp23-442i7.1 | −5.54 |
| Gm16336 | 4.30 | Ai314831 | −4.42 | Gm13905 | 4.15 | Gm14273 | −4.43 |
| 1810009n23rik | 4.28 | Gm11738 | −4.33 | Gm13373 | 3.92 | Rp23-302f9.2 | −4.39 |
| 6720416l17rik | 4.27 | E130006d01rik | −4.25 | 1500015l24rik | 3.86 | Rp23-302f9.4 | −4.19 |
| Gm12530 | 4.10 | Gm11375 | −4.23 | Gm26684 | 3.86 | Rp24-465o15.1 | −3.98 |
| 1700042g15rik | 4.01 | Gm12408 | −4.20 | 7530420f21rik | 3.74 | Nespos | −3.77 |
| Gm13293 | 3.97 | D830044d21rik | −4.11 | Gm13558 | 3.69 | Gm26576 | −3.64 |
| Gm26512 | 3.54 | Gm14696 | −4.05 | A330074k22rik | 3.58 | Gm13398 | −3.60 |
| Gm13558 | 3.53 | Rp24-68f22.7 | −3.84 | Gm14532 | 3.52 | 2310002f09rik | −3.53 |
| Gm26924 | 3.40 | Gm14697 | −3.80 | Gm14285 | 3.48 | 1500016l03rik | −3.49 |
| 2210411m09rik | 3.33 | Rp23-45g16.5 | −3.79 | 4930588k23rik | 3.46 | Gm26771 | −3.42 |
| Gm13943 | 3.33 | D430036j16rik | −3.67 | Gm12300 | 3.38 | Gm15564 | −3.37 |
| Gm12541 | 3.16 | Gm12371 | −3.59 | Gm12446 | 3.32 | 4930412b13rik | −3.36 |
| Gm16291 | 2.99 | 5730457n03rik | −3.56 | Gm12907 | 3.31 | Gm4926 | −3.19 |
| Gm16302 | 2.99 | Rp23-442i7.1 | −3.56 | 0610043k17rik | 3.30 | Gm26924 | −3.17 |
| Gm11521 | 2.77 | 4930556l07rik | −3.48 | Nphs1os | 3.25 | Gm20667 | −3.02 |
| Gm11840 | 2.76 | Gm13481 | −3.47 | Gm13322 | 3.23 | Gm11267 | −3.02 |
| Gm13403 | 2.73 | Gm14339 | −3.46 | Gm13833 | 3.18 | Rp23-302f9.1 | −3.01 |
| Gm15564 | 2.62 | Gm27040 | -3.42 | 1810009n23rik | 3.17 | Gm11820 | −3.01 |
| Gm13833 | 2.62 | Gm20618 | −3.40 | 4933428c19rik | 3.16 | E130006d01rik | −2.97 |
| Gm16013 | 2.62 | Gm12923 | −3.39 | Gm13834 | 3.14 | H19 | −2.93 |
| Gm15864 | 2.53 | Gm11419 | −3.38 | Gm15523 | 3.13 | Gm14100 | −2.88 |
| A330074k22rik | 2.47 | 4930453o03rik | −3.35 | 9330179d12rik | 3.11 | Gm11837 | −2.83 |
| Gm10804 | 2.45 | Gm4473 | −3.32 | Gm14011 | 3.11 | 4732490b19rik | −2.74 |
| Gm12027 | 2.44 | Gm26626 | −3.30 | Aw495222 | 3.08 | A530058n18rik | −2.65 |
| Rp24-530j21.3 | 2.43 | Gm15295 | −3.30 | 4930563e18rik | 3.03 | Gm11738 | −2.64 |
| Gm1667 | 2.41 | 8430436n08rik | −3.29 | Gm20478 | 3.01 | Gm16863 | −2.64 |
| Gm11213 | 2.40 | Gm11754 | −3.26 | Gm15856 | 2.94 | Gm26654 | −2.60 |
| Gm5535 | 2.36 | Gm2990 | −3.25 | Gm13629 | 2.92 | G630018n14rik | −2.60 |
| B230344g16rik | 2.33 | Rp24-465o15.1 | −3.25 | Dio3os | 2.92 | Gm4425 | −2.57 |
| Rp24-480m3.2 | 2.31 | 4930515l19rik | −3.24 | A630023p12rik | 2.90 | B930086a06rik | −2.54 |
| Gm17180 | 2.27 | Gm26654 | −3.23 | 4930596m17rik | 2.82 | Gm14224 | −2.53 |
| Gm14424 | 2.27 | 2010300f17rik | −3.23 | Gm14029 | 2.78 | Gm12167 | −2.53 |
| Gm17179 | 2.21 | C130021i20rik | −3.22 | 4930486i03rik | 2.77 | Gm13849 | −2.53 |
| Gm12576 | 2.20 | 3110099e03rik | −3.20 | Bc039966 | 2.76 | 9930014a18rik | −2.52 |
| Gm11634 | 2.20 | 9130024f11rik | −3.20 | Gm11681 | 2.74 | Gm13404 | −2.52 |
| Gm13629 | 2.19 | 2310065f04rik | −3.18 | Gm15411 | 2.73 | Gm7518 | −2.51 |
| Gm12756 | 2.18 | 7330404k18rik | −3.18 | Tmem61 | 2.72 | Gm16292 | −2.49 |
| 9530072k05rik | 2.14 | 1700028i16rik | −3.17 | Gm27019 | 2.72 | Gm16876 | −2.48 |
| 5133400j02rik | 2.14 | Gm14225 | −3.16 | Gm13643 | 2.67 | Rp23-45g16.5 | −2.45 |
| Gm16535 | 2.13 | D030055h07rik | −3.13 | Bc051537 | 2.65 | Gm17750 | −2.42 |
| A330023f24rik | 2.12 | Gm15202 | −3.12 | Gm12536 | 2.60 | Gm26640 | −2.42 |
| Gm13807 | 2.11 | Gm14967 | −3.12 | Gm14317 | 2.60 | Gm2990 | −2.40 |
| Gm11681 | 2.09 | 4930558j18rik | −3.11 | Gm14397 | 2.55 | Gm11986 | −2.40 |
| Gm13362 | 2.08 | 5530401n12rik | −3.11 | Gm26795 | 2.55 | Gm13066 | −2.35 |
| Gm14426 | 2.07 | Gm15222 | −3.09 | A330048o09rik | 2.54 | Gm26569 | −2.33 |
| Gm12536 | 2.04 | Gm15326 | −3.09 | Gm12828 | 2.53 | Gm12108 | −2.30 |
| 9530085p06rik | 2.03 | Gm11376 | −3.04 | 0610040f04rik | 2.52 | Mir17hg | −2.28 |
The top 50 up-regulated and down-regulated genes in MG and A1 were ranked based on average log2 fold change (FC) magnitude comparing P7 to adult (FC = average of DESeq2, EdgeR, BaySeq). Genes that were up- or down-regulated in both regions are in bold text.
Table 4 lists the 50 genes at each age that were more highly expressed in MG than A1 (left) or A1 than MG (right). Several trends were noted. First, a relatively large number of genes exhibited regional dominance at only one age (MG, N=47; A1, N=56). This implies that many genes are differentially regulated in A1 or MG at a particular age. Second, a minority of genes was regionally dominant in A1 or MG at all ages (MG, N=18; A1, N=11) or three out of four ages (MG, N=3; A1, N=11). These genes have strong regional specificity, regardless of age. Third, only one gene (1700080N15RIK) was regionally dominant in A1 at one age and MG at another. This lncRNA was more highly expressed in MG in adults, but in A1 at P7. The rarity of genes with such patterns is further evidence of strong regional specificity among the majority of lncRNA genes.
Table 4.
Top 50 differentially expressed genes in MG and A1.
| MG > A1 | A1 > MG | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| P7 | P14 | P21 | Adult | P7 | P14 | P21 | Adult |
| Gm2373 | 2810011l19rik | Gm13544 | Gm13544 | A830009l08rik | Gm12371 | 9130024f11rik | Gm12371 |
| Gm13112 | Gm13112 | 2810011l19rik | 1700081h22rik | 9930014a18rik | Gm13389 | A830009l08rik | A830009l08rik |
| Rmst | A830019p07rik | 4930448n21rik | B230323a14rik | Dlx1os | A830009l08rik | Gm12371 | 9130024f11rik |
| 2610028e06rik | Gm13544 | Gm13112 | Gm12828 | Gm13389 | Gm15810 | Gm14317 | Dlx6os1 |
| 2810011l19rik | B230323a14rik | Six3os1 | 4930448n21rik | Gm12027 | 9130024f11rik | Gm15810 | Gm15810 |
| F730016j06rik | Gm26512 | B230323a14rik | 2810011l19rik | Dlx6os1 | Gm11766 | Gm13601 | Gm11766 |
| Gm12828 | Gm12828 | Gm12828 | Gm17634 | Gm26580 | Gm26803 | 1700047f07rik | C130071c03rik |
| A330074k22rik | Gm2373 | Gm2373 | A330074k22rik | 9130024f11rik | Gm17322 | Gm12300 | Dlx1os |
| Gm15663 | 4930448n21rik | Gm16168 | 9330185c12rik | 6530403h02rik | Gm15581 | A330009n23rik | Gm13601 |
| Rp23-131o4.2 | 1500016l03rik | 1500016l03rik | Gm12750 | Gm26803 | A830036e02rik | Dlx6os1 | A330009n23rik |
| 4933436c20rik | Rmst | Gm10863 | Gm2373 | Gm15581 | 2610017i09rik | A630023p12rik | Gm26805 |
| 4930455g09rik | Gm17634 | 9330185c12rik | Gm16154 | Gm26801 | Gm27002 | Gm26911 | Gm16295 |
| Gm12827 | Rp23-448c3.1 | Gm12750 | 1500016l03rik | Gm26871 | 4930467d21rik | 5830416p10rik | 5330416c01rik |
| B230323a14rik | Gm13629 | Rp23-448c3.1 | 4930579o11rik | Gm15810 | 9230115e21rik | 5031425f14rik | A830036e02rik |
| Gm15893 | Gm12750 | Gm14210 | Gm13905 | A830036e02rik | Gm15813 | Gm26565 | Gm15813 |
| Gm26641 | Gm3510 | 1700081h22rik | Rp23-448c3.1 | Gm11766 | 9930014a18rik | Gm15813 | Gm26803 |
| 1500016l03rik | 1700081h22rik | 4930579o11rik | Rmst | Rp24-86o15.2 | Gm15338 | Gm13659 | 4930447m23rik |
| Gm26777 | Gm26560 | Rp23-302f9.2 | Six3os1 | Gm15813 | 5830416p10rik | Gm20603 | 2610017i09rik |
| Gm17634 | Gm26641 | Rmst | Gm13112 | Gm26565 | Gm26578 | Gm26580 | Gm14317 |
| Gm26597 | Six3os1 | Gm16154 | Gm16168 | 4930467d21rik | A330009n23rik | 4930467d21rik | 1700047f07rik |
| A330033j07rik | 1700040n02rik | 4933436c20rik | Gm26512 | A430048g15rik | Gm26911 | Gm11767 | Gm17202 |
| 4930593c16rik | F730016j06rik | Gm4221 | Gm15511 | Gm20705 | Gm14317 | Gm15581 | Gm11767 |
| Bb031773 | Gm17080 | 1700040n02rik | Gm17566 | 4833422c13rik | Gm13601 | G630016g05rik | Gm13659 |
| Gm14259 | 9330185c12rik | Gm17634 | Gm13629 | 4930545l23rik | Gm20603 | 2700033n17rik | Gm14290 |
| Gm26782 | Gm15511 | Gm17566 | Sox2ot | 2310075c17rik | A630023p12rik | Gm13389 | Ai115009 |
| Gm15417 | 4930593c16rik | Gm17080 | Gm17080 | Gm20687 | Gm26580 | Gm15411 | Gm13389 |
| Gm11454 | 4930579o11rik | A830019p07rik | Gm16336 | 4930419g24rik | Gm14290 | Gm26924 | 1700001l05rik |
| Gm12750 | 4933436c20rik | Gm13629 | Neat1 | Gm17750 | Gm1976 | Gm17202 | Gm17322 |
| Gm13629 | Gm15417 | A330074k22rik | 1700040n02rik | Gm13749 | Gm16295 | Gm16339 | Gm26578 |
| Gm20478 | Gm26782 | Sox2ot | Gm12843 | Gm17089 | Gm11767 | 2310065f04rik | Gm15411 |
| Rp24-113d21.1 | 4921504a21rik | A730090h04rik | Gm12827 | Gm26794 | C130071c03rik | Gm12907 | Ai854517 |
| Rp23-392m3.2 | Gm17566 | Gm26747 | 4933436c20rik | Gm15829 | Rp23-230f2.8 | Gm16295 | 2900079g21rik |
| 1700125g02rik | Sox2ot | Gm20635 | A330033j07rik | 2610017i09rik | Dlx1os | 3110099e03rik | D030055h07rik |
| Gm26560 | Gm12843 | Gm26512 | 4930552p12rik | 9230115e21rik | Gm13749 | 4930545l23rik | Gm20603 |
| 4732491k20rik | A330074k22rik | Gm12843 | Rp23-302f9.2 | Gm12360 | 6530403h02rik | 6530403h02rik | Gm26871 |
| 4932441j04rik | Gm26694 | 5330434g04rik | Gm13293 | Gm13963 | 4933413l06rik | 1700108n11rik | 7530420f21rik |
| 1700040n02rik | 0610007n19rik | Gm15511 | Gm26742 | Gm17088 | Gm26871 | 2610017i09rik | Gm14291 |
| 5330434g04rik | Gm11454 | Gm17552 | Gm26674 | 4930578m01rik | Gm26565 | A830036e02rik | Gm12300 |
| A230006k03rik | Gm16154 | Gm14662 | Gm26641 | Gm15513 | 4930538e20rik | Rp23-230f2.8 | 2700033n17rik |
| Gm15511 | Rp23-302f9.2 | Rp23-302f9.1 | 4930593c16rik | Gm17116 | Gm13963 | 4930556i23rik | Bb114351 |
| 6430710c18rik | Bc039966 | Gm11454 | Gm2366 | 1700080n15rik | Gm12063 | Dlx1os | 5830416p10rik |
| Gm12843 | Gm14259 | Gm10687 | Bc039966 | 5830416p10rik | 2310075c17rik | Gm13749 | Gm1976 |
| 1700071m16rik | Gm12406 | Gm26641 | Gm26888 | Gm17608 | Gm17202 | Gm26795 | Gm13704 |
| Rp24-351j24.2 | Gm26674 | Gm2366 | A430010j10rik | Gm11767 | 1110002j07rik | Gm26805 | Gm2415 |
| Six3os1 | Rp23-131o4.2 | 4930593c16rik | Gm14210 | Gm13584 | Gm26794 | Gm15564 | Gm26911 |
| Gm15345 | Gm16168 | 4921504a21rik | Gm15417 | Gm13398 | 4930545l23rik | Gm16229 | Gm27002 |
| A730036i17rik | Gm10863 | Gm14259 | Gm13189 | Gm26569 | B230216n24rik | Gm17322 | G630016g05rik |
| Rp24-312b12.1 | Gm10516 | 2700069i18rik | Rp23-302f9.4 | 4930533b01rik | Dlx6os1 | Gm12295 | C530050e15rik |
| Gm20686 | Rp23-302f9.4 | Gm13293 | 1700080n15rik | 9430029a11rik | 5031415h12rik | Gm27002 | 4930556i23rik |
| C130021i20rik | Gm26780 | Bc039966 | 1700042g15rik | C130071c03rik | 4930556i23rik | Gm2415 | 2810029c07rik |
The top 50 differentially expressed lncRNA genes between MG and A1 are listed by postnatal age. Left columns, genes with significantly higher expression levels in MG as compared to A1. Right columns, genes with higher expression levels in A1 as compared to MG. Significance (p < 0.05) and ranking determined by differential expression analyses.
3.4 Expression trend analysis
Expression trend analyses were carried out to identify genes with different expression growth patterns with age. We focused on three lncRNA expression patterns: monotonically increasing, monotonically decreasing and static. A gene was monotonically increasing if its expression continuously increased at each time point and the change between P7 and adult was statistically significant. Monotonically decreasing genes were defined in the same fashion, but had decreased expression at each time point. A gene was static if the absolute fold change of the expression value between any two time points was less than 1.5. LncRNA that had other patterns of expression between P7 and adult (e.g., increasing, then decreasing) were categorized as “other.” The numbers of significantly monotonically decreasing, increasing and static genes in A1, MG and A1+MG are given in Table 5, along with the numbers of genes with a different trajectory (Other). In A1, about 7% lncRNAs had an increasing pattern, 10% had a decreasing trajectory, 32% had static profiles, and 51% had trajectories classified as “other.” For MG, about 5% of lncRNAs had an increasing pattern, 13% had a decreasing pattern, 32% were static, and 50% had “other” trajectories. A minority of the genes with increasing (N=29) or decreasing (N=96) profiles were common to both A1 and MG (A1 | MG). In contrast, many of the lncRNAs with static or other profiles were common to both regions. These findings indicate that expression levels for the majority of lncRNAs were changing during maturation, and also indicate strong regional specificity for genes with monotonically increasing or decreasing trajectories.
Table 5.
Monotonic lncRNA expression patterns (all genes).
| Direction | A1 | MG | A1 | MG |
|---|---|---|---|
| Increasing | 216 | 143 | 29 |
| Decreasing | 325 | 427 | 96 |
| Static | 1044 | 1034 | 1157 |
| Other | 1628 | 1609 | 1931 |
The total numbers of genes with maturational trajectories categorized as monotonically increasing, monotonically decreasing, static, or other (from P7 to adult) are tallied for A1 and MG. The numbers of genes that were common to both A1 and MG are tallied in the third column (A1 | MG).
3.5 Analysis of protein coding potential
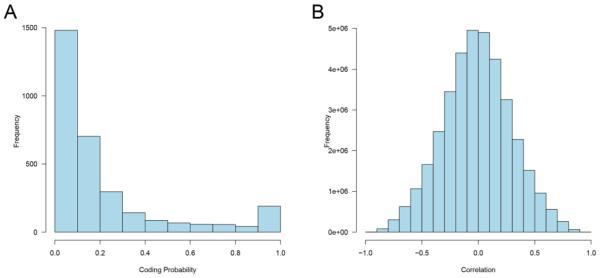
We evaluated all lncRNAs’ protein coding potential using CPAT (Wang et al., 2013b) (Table S11). CPAT summarizes each lncRNA’s coding potential with a coding probability. The majority of the lncRNAs have near zero coding probability, but a relatively small group has high coding probabilities (Figure 2A). Of these, 190 lncRNA genes had a coding probability greater than 0.90 (see Table S11), many of which are currently listed as provisional protein-coding genes in the NCBI drosophila gene database (source: flybase.org).
Fig. 2. Protein coding probabilities and correlations.
A: Histogram of the coding probabilities for all lncRNAs that were expressed in mouse brain. B: Histogram of Spearman’s correlation coefficient between all lncRNA – pcRNA pairs. LncRNAs and pcRNAs with nominal expression levels were filtered out to avoid artificially high correlations.
3.6 Analyses of potential interactions with pcRNAs and proteins
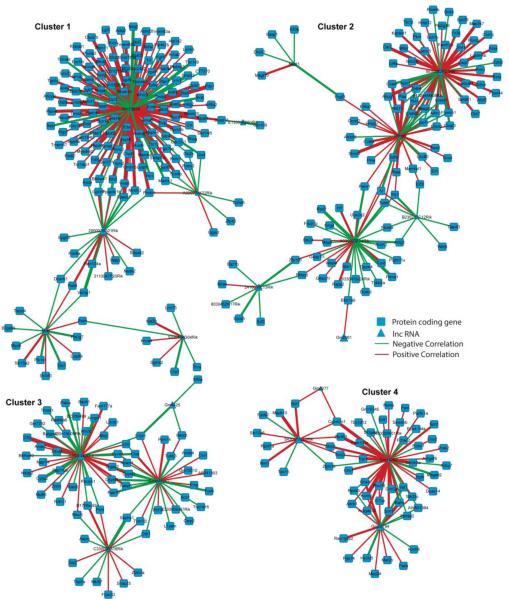
To learn more about potential interactions involving lncRNAs, we computed Spearman’s correlations (expression levels by age) between all possible lncRNA and pcRNA pairs and identified an abundance of highly correlated pairs. There are over 600 million possible lncRNA – pcRNA pairs, and the Spearman’s correlations followed a normal distribution (Figure 2B). Only pairs with the highest correlations were selected for further functional analysis (Table S12). Using Spearman correlation |r| > 0.95 as the cutoff, there were 321 positively correlated pairs and 221 negatively correlated pairs. Putting these pairs into Cytoscape, we identified four major regulation network clusters in the mouse auditory forebrain (Fig. 3). These clusters were mainly associated with four sets of lncRNAs; Cluster 1: Gm13629, A330074k22rik, 2900079g21rik, Miat, 5330434g04rik; Cluster 2: 9530082p21rik, Snhg6, Neat1, B230217c12rik, A330023f24rik, 2410018l13rik; Cluster 3: Rp23-44217.1, Ccdc41os1, C330006a16rik, Gm4425; Cluster 4: Gm14290, Gm26794, 5430417l22rik.
Fig. 3. Cluster network clusters of correlated pairs.
Cluster network built in Cytoscape from the correlation results in Fig. 2B. Four major network clusters were visible.
Additional functional analysis was carried out for the pcRNA within each cluster using WebGestalt (Wang et al., 2013a) (Table 6). Functional categories differed between the four clusters. These functions included general cellular processes (e.g., intracellular signal transduction) as well as those specifically related to brain development (e.g., neuron projection development), suggesting a wide range of potential regulatory effects by lncRNA. Clusters 1 and 3, for example, contained numerous pcRNAs that are essential for normal brain structure and function in the adult and developing brain.
Table 6.
Functional gene ontology categories of pcRNA genes in Fig. 3.
| Cluster | Sub Category | Genes | p |
|---|---|---|---|
| 1 | Intracellular Signal Transduction | 27 | 0.0015 |
| 1 | Cellular Protein Modification Process | 31 | 0.0021 |
| 1 | Regulation Of Ion Transmembrane Transport | 9 | 0.0021 |
| 1 | Phosphate-Containing Compound Metabolic Process | 33 | 0.0030 |
| 1 | Plasma Membrane | 49 | 0.0000 |
| 1 | Cytoplasm | 81 | 0.0001 |
| 1 | Golgi Apparatus | 19 | 0.0004 |
| 2 | Cellular Component Organization | 33 | 0.0002 |
| 2 | Organic Substance Metabolic Process | 55 | 0.0004 |
| 2 | Phosphate-Containing Compound Metabolic Process | 25 | 0.0016 |
| 2 | Primary Metabolic Process | 51 | 0.0019 |
| 2 | Cytoplasm | 57 | 0.0000 |
| 2 | Intracellular Organelle | 60 | 0.0002 |
| 2 | Membrane-Bounded Organelle | 54 | 0.0005 |
| 3 | Neuron Projection Development | 11 | 0.0001 |
| 3 | Cell Projection Part | 9 | 0.0008 |
| 3 | Neuron Projection | 10 | 0.0008 |
| 3 | Cytoskeleton | 15 | 0.0008 |
| 4 | Rho Protein Signal Transduction | 6 | 0.0012 |
| 4 | Cellular Protein Modification Process | 15 | 0.0023 |
| 4 | Regulation Of Nucleotide Metabolic Process | 6 | 0.0023 |
| 4 | Neuronal Cell Body | 9 | 0.0000 |
| 4 | Postsynaptic Membrane | 6 | 0.0001 |
| 4 | Postsynaptic Density | 5 | 0.0001 |
For each cluster, the functional subcategories with significant enrichment are listed, along with the number of genes included in each.
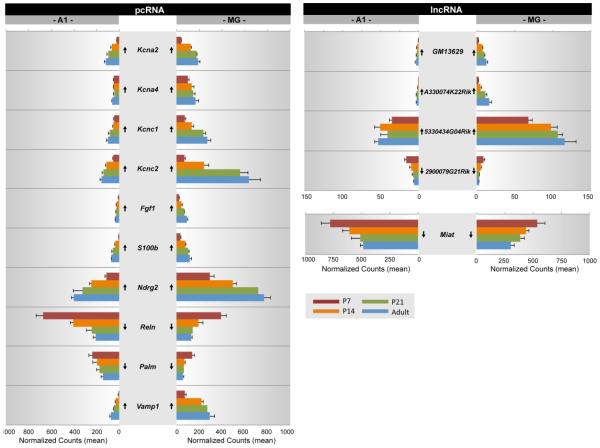
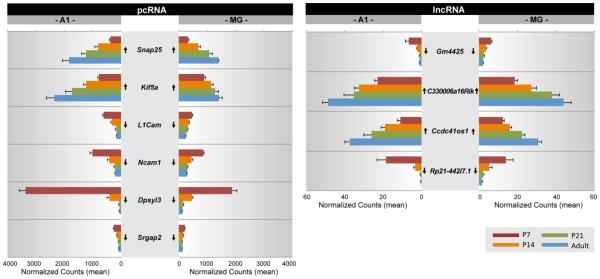
To highlight potential interactions in these clusters, several protein coding transcripts with known involvement in brain development or brain activity were selected as exemplars from Clusters 1 and 3. In Figs. 4 and 5, the expression levels of genes in both Clusters were plotted as a function of postnatal age. Arrows indicate whether the maturational trajectories from P7 to adult were significantly increased or decreased (p < 0.05)(from Tables S9-S10). A number of general observations were observed. First, the expression levels of pcRNAs and lncRNAs (mean normalized read counts) varied within and sometimes between regions. As a group, lncRNAs tended to have lower overall expression levels, which is typical of lncRNAs (Ulitsky and Bartel, 2013). A notable exception was Miat, which had very high expression in both A1 and MG, even higher than many pcRNAs. Second, most genes exhibited some degree of regional asymmetry (A1 vs MG) in expression. Among those with the most pronounced asymmetries in Figs. 4 and 5 were Dpsyl3, Kcnc2, Reln and 5330434G04Rik. Third, all genes were significantly up- or down-regulated between P7 and adult. Generally, the increase (or decrease) tended to be steady over the age range, although relatively large shifts in expression were sometimes observed between P7 and P14 (immediately before and after hearing onset) in one or both regions (e.g., pcRNAs: Ncam1, Dpsyl3, Kcna2, Reln, Vamp1; lncRNAs: Gm4425, Rp21-442l7.1). Fourth, maturational trajectories were the same in A1 and MG (up or down) for all genes included in Figs. 4 and 5. However, the trajectories of many genes differ between brain regions (Hackett et al., 2015), indicating that regional asymmetries in expression level and maturational trajectory are common. Fifth, within a Cluster, some pcRNAs were correlated with more than one lncRNA, yet the maturational trajectory of a given pcRNA was not always predictive of its correlations. For example, in Cluster 1, Kcnc1 was positively correlated with both Gm13629 and A330074k22Rik, Vamp1 was negatively correlated with Miat and 2900079g21Rik, while Palm was positively correlated with 5330434g0Rik and negatively correlated with Miat. Similarly, in Cluster 3, Ncam1, Dpys13, and Srgap2 were positively correlated with Rp23-442l7.1 (1110015O18Rik) and negatively correlated with Ccdc41os1.
Fig. 4. Gene expression profiles of lncRNA and pcRNA pairs from Cluster 1 (see Fig. 3).
For each gene, mean normalized counts are plotted by postnatal age (P7, P14, P21, Adult) and brain region (A1, MG). Expression trajectory is indicated by arrows (up, down). Arrows indicate significant up- or down-regulation from P7-Adult was significant (p < 0.05).
Fig. 5. Gene expression profiles of lncRNA and pcRNA pairs from Cluster 3 (see Fig. 3).
For each gene, mean normalized counts are plotted by postnatal age (P7, P14, P21, Adult) and brain region (A1, MG). Expression trajectory is indicated by arrows (up, down). Arrows indicate significant up- or down-regulation from P7-Adult was significant (p < 0.05).
Tables 7 and 8 contain the Spearman correlations (r) between each pcRNA – lncRNA pair, along with the lncRNA-protein interaction scores (is) computed by the lncPro analysis. Several observations were notable. First, while the cutoff for the clustering of pcRNA – lncRNA pairs in Fig. 3 was set at |r| = 0.95, most pcRNAs also had significant correlations with the other lncRNAs in its Cluster. Second, the majority of pcRNA – lncRNA pairs with high correlations also had moderately-high to high predicted protein interaction scores, although some exceptions were also noted. For example, the pcRNA Kcnc1 (voltage-gated potassium channel Kv3.1) was linked with two lncRNAs in Cluster 1 (Gm13629, A330074k22rik). The correlations with Kcnc1 were high at r = 0.95, while the predicted protein interaction scores were moderate for Gm13629 and high for A330074k22rik (Table 7). Similarly, Vamp1 (vesicle-associated membrane protein 1) was highly correlated with 5330434g0Rik, but the protein interaction score was rather low (i.s. = 46.82). Conversely, some pairs with relatively low correlations (e.g., Kcnc2 – Miat) had high interaction scores. Kcnc2 (voltage-gated potassium channel Kv3.2) was modestly correlated with Miat (r = 0.50), but their interaction score was high (i.s. = 90.72). Finally, both positive and negative correlations were observed for some pairs with high protein interaction scores (e.g., Reln, Kif5a), suggesting potential interactions between pairs with opposing maturational trajectories.
Table 7.
lncRNA – pcRNA correlations and interactions.
| Gm13629 | A330074k22rik | 5330434g04rik | Miat | 2900079g21rik | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 2 | 8 | X | 5 | 9 | |||||||
| Chr | r | Is | r | Is | r | Is | r | is | r | is | |
| Kcna2 | 3 | 0.95 | 62.56 | 0.89 | 82.90 | 0.76 | 90.00 | −0.50 | 79.34 | −0.91 | 57.68 |
| Kcna4 | 11 | 0.82 | 77.79 | 0.83 | 78.01 | 0.95 | 82.14 | −0.92 | 71.89 | −0.87 | 57.75 |
| Kcnc1 | 7 | 0.95 | 61.31 | 0.95 | 93.38 | 0.79 | 67.07 | −0.84 | 93.18 | −0.92 | 59.12 |
| Kcnc2 | 12 | 0.96 | 62.94 | 0.91 | 87.39 | 0.80 | 78.21 | −0.58 | 90.72 | −0.89 | 59.78 |
| Fgf1 | 18 | 0.97 | 83.01 | 0.91 | 94.01 | 0.82 | 74.64 | −0.83 | 94.85 | −0.92 | 84.06 |
| S100b | 10 | 0.97 | 97.00 | 0.91 | 90.92 | 0.76 | 60.67 | −0.85 | 77.89 | −0.93 | 70.64 |
| Ndrg2 | 14 | 0.96 | 90.87 | 0.94 | 77.77 | 0.84 | 78.19 | −0.85 | 75.79 | −0.93 | 49.37 |
| Reln | 5 | -0.96 | 90.22 | −0.94 | 97.32 | −0.82 | 96.03 | 0.88 | 96.44 | 0.94 | 80.74 |
| Palm | 10 | −0.83 | 96.29 | −0.70 | 77.58 | −0.95 | 77.19 | 0.95 | 76.46 | 0.88 | 78.21 |
| Vamp1 | 6 | 0.94 | 57.77 | 0.93 | 77.77 | −0.88 | 46.82 | −0.95 | 77.36 | −0.97 | 57.19 |
Correlations and predicted protein interaction scores between pcRNA and lncRNA genes involved in neurotransmission and plasticity in the brain (selected from Fig. 3, Cluster 1). Spearman correlations (r) and lncPro interaction scores (is; score range 1 - 100) are listed for each pcRNA-lncRNA pair. Bold type denotes pcRNA – lncRNA pairs illustrated in Cluster 1 with correlations of at least |0.95|. Raw data in Table S12.
Table 8.
lncRNA – pcRNA correlations and interactions.
| Gm4425 | C330006A16Rik | Ccdc41os1 | RP23-442I7.1* | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 12 | 2 | 10 | 3 | ||||||
| Chr | r | is | r | is | r | is | r | is | |
| Kif5a | 10 | −0.95 | 95.05 | 0.87 | 93.73 | 0.90 | 98.81 | −0.94 | 98.89 |
| Snap25 | 2 | −0.90 | 79.13 | 0.95 | 82.73 | 0.94 | 97.14 | −0.93 | 97.60 |
| L1cam | X | 0.93 | 60.77 | −0.87 | 72.71 | −0.95 | 79.01 | 0.93 | 79.12 |
| Ncam1 | 9 | 0.92 | 77.44 | −0.86 | 63.34 | −0.95 | 78.49 | 0.95 | 78.65 |
| Dpys13 | 18 | 0.94 | 59.98 | −0.91 | 75.32 | −0.96 | 64.70 | 0.96 | 95.66 |
| Srgap2 | 1 | 0.92 | 78.96 | −0.90 | 76.22 | −0.95 | 98.14 | 0.96 | 98.12 |
Correlations and predicted protein interaction scores between pcRNA and lncRNA genes involved in neurotransmission and plasticity in the brain (selected from Fig. 3, Cluster 3). Spearman correlations (r) and lncPro interaction scores (is; score range 1 - 100) are listed for each pcRNA-lncRNA pair.
alternative name: 1110015O18Rik (ENSMUSG00000098659). Bold type denotes pcRNA – lncRNA pairs illustrated in Cluster 3 with correlations of at least |0.95|. Raw data is in Table S12.
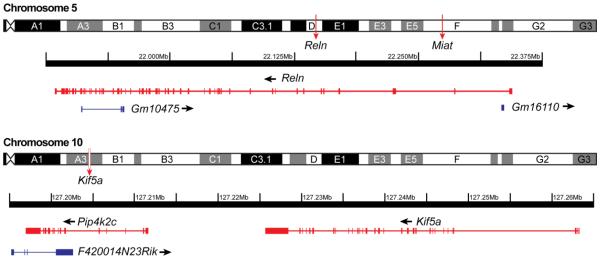
To further explore potential lncRNA – pcRNA interactions, we inspected the genomic loci of two highly-correlated pairs from Tables 7 and 8 with sequences located on the same chromosome, as sequences with overlapping or nearby domains tend to have higher correlations and greater interaction potential (see Discussion). Their genomic loci are illustrated in Fig. 6.
Fig. 6. Genomic loci of three pcRNAs involved in brain development and nearby lncRNAs.
A: Chromosome 10 loci of pcRNAs Pip4k2c and Kif5a, and overlap with lncRNA F420014N23Rik. The distant Miat locus on chromosome 10 is indicated, but the sequence is not illustrated. B: Chromosome 5 loci of the pcRNA Reln, overlapped by lncRNAs Gm10475 and Gm16110. Arrows indicate the direction of transcription.
From chromosome 5, the pcRNA Reln (Reelin) and lncRNA Miat had a strong correlation (r = 0.88) and protein interaction score (i.s. = 96.44). Miat is located nearly 90Mb downstream of Reln in a region occupied by several lincRNAs and other non-coding genes, and are transcribed in the same direction. Its distant location from Reln on chromosome 5 is indicated in Fig. 6, but the sequence was not illustrated. By comparison, two antisense lncRNAs (Gm16110, Gm10475) overlapped regions of the Reln sequence. Although, their correlations fell below the |r| = 0.80 cutoff, all three genes had significant downward trajectories in both A1 and MG (see Tables S9, S10).
On chromosome 10, the pcRNA Kif5a (Kinesin heavy chain isoform 5a) is located just downstream of the lincRNA F420014N23Rik and transcribed in opposite directions. Kif5a was strongly upregulated from P7 to adult in A1 and MG. F420014N23Rik expression was upregulated in A1 and static in MG. Of additional interest was that the F420014N23Rik locus overlaps another pcRNA, Pip4k2c (Phosphatidylinositol 4-phosphate 5-kinase), also strongly upregulated in both brain regions, which is in a family of proteins with roles in brain development and vesicular transport. Thus, potential interactions may be possible with one or both pcRNAs.
Overall, these results suggest that lncRNAs that are highly correlated with a given pcRNA may be more likely to interact with the associated protein, but the predictive value of correlated expression is uncertain. Functional studies will be needed in the future to further characterize potential relationships.
4. DISCUSSION
In the present study, we set out to achieve two goals. The first was to generate complete lncRNA transcriptome profiles of A1 and MG during postnatal development before and after the onset of hearing. As a foundation for future study, our second goal was to create a user-friendly searchable database and Look-Up Tool to facilitate the examination and exploration of trends in expression by brain region and postnatal age. Overall, the differential analyses of global lncRNA expression revealed significant differences between brain regions and changes in both regions with postnatal age. Based on lncRNA expression profiles alone, we could distinguish between brain regions and ages. Analyses of potential interactions between lncRNA and pcRNAs or proteins revealed how maturational changes may be manifested within functional categories.
4.1 Regional and temporal specificity in lncRNA expression
The expression of lncRNAs was regionally-specific. Globally, expression profiles in A1 were distinct from MG at every postnatal age examined. A relative minority was strongly up- or down-regulated in both regions. This is consistent with the expression patterns of pcRNA genes in the auditory forebrain (Hackett et al., 2015), and with studies of lncRNAs in other brain regions (Belgard et al., 2011; Liu et al., 2016). Indeed, numerous studies have found that lncRNA expression patterns vary by brain region (Amaral et al., 2009; Kadakkuzha et al., 2015; Ling et al., 2009; Ling et al., 2011; Lv et al., 2013; Mercer et al., 2008; Ponjavic et al., 2009; Sauvageau et al., 2013; Spigoni et al., 2010; Ziats and Rennert, 2013). In addition, the distributions of many genes are specific to restricted loci, such as cortical layer, cell type, or subcellular compartment (Aprea et al., 2013; Belgard et al., 2011; Kadakkuzha et al., 2015; Korneev et al., 2008; Liu et al., 2016; Mercer et al., 2008; Mercer et al., 2010; Pollard et al., 2006; Sasaki et al., 2008; Sauvageau et al., 2013; Sone et al., 2007; Spigoni et al., 2010; Tochitani and Hayashizaki, 2008). Thus, the functional roles of many pcRNAs and lncRNAs in the developing and mature brain are spatially-specific.
In addition to spatial specificity, several studies have reported that lncRNA expression is often temporally specific (Amaral et al., 2009; Aprea et al., 2013; Lin et al., 2011; Ling et al., 2009; Ling et al., 2011; Lipovich et al., 2012; Liu et al., 2016; Mercer et al., 2010; Ponjavic et al., 2009; Spigoni et al., 2010; Tarabykin et al., 2001). These findings are consistent with the maturational trends that we observed in the auditory forebrain. Within each brain region (A1, MG), lncRNA expression profiles were distinct at each postnatal age. In both regions, the majority of lncRNAs had maturational trajectories that reflected a change in expression level between at least two consecutive age groups. Only about one-third of lncRNAs had static profiles from P7 through adult. Although speculative, it may be hypothesized that genes with static profiles are more likely to play roles in housekeeping or other maintenance, while those with changing levels are involved in the regulation of key maturational events. Moreover, the strong correlations and predicted interactions between certain pairs of lncRNA and pcRNAs are suggestive of potential interactions among RNA subtypes within various functional pathways. As most of these pathways have not been characterized, there is much room for discovery.
4.2 Functional roles of lncRNAs in auditory forebrain maturation: where to begin?
The function of most lncRNAs is unknown. Consequently, network analyses (e.g., functional gene ontology analysis) are of limited value at this time. Fortunately, interest in this subject is growing, and loss and gain of function experiments are being use to reveal the identity and putative functions of many lncRNAs, including those active in the developing brain. Several have regulatory influence over the expression of other genes and proteins (Carninci et al., 2005; Carrieri et al., 2012; Guttman et al., 2011; Guttman and Rinn, 2012; Halley et al., 2014; Katayama et al., 2005; Khalil et al., 2009; Kornienko et al., 2013; Kurokawa, 2011; Magistri et al., 2012; Mattick, 2007; Meng et al., 2012; Onoguchi et al., 2012; St Laurent and Wahlestedt, 2007; Tsai et al., 2010; Vance et al., 2014; Wu et al., 2013; Zhang et al., 2012; Zhao et al., 2013). Through these interactions, lncRNAs can influence a variety of maturational processes such as neurogenesis, synaptogenesis, cell migration, cell type specification, neurite outgrowth, and synaptic plasticity (Aprea et al., 2013; Berghoff et al., 2013; Bernard et al., 2010; Bond et al., 2009; Feng et al., 2006; Kraus et al., 2013; Lin et al., 2014a; Ling et al., 2011; Lipovich et al., 2012; Liu et al., 2016; Modarresi et al., 2012; Ng et al., 2012; Onoguchi et al., 2012; Tarabykin et al., 2001; Ulitsky et al., 2011; Vance et al., 2014). For example, the growth factors Bdnf (Brain-derived neurotrophic factor) and Fgf2 (Fibroblast growth factor 2) are intensively-studied genes with key roles in brain development and plasticity. Both are regulated by their antisense sequences (Bdnf-AS, Fgf2-AS) in a manner that impacts neuronal proliferation, neurite outgrowth, and maturation (MacFarlane et al., 2010; Modarresi et al., 2012). Similarly, the functions of other lncRNAs appear to be linked to neuronal activity, as suggested by altered expression with changes in activity (Barry et al., 2014; Kim et al., 2010; Lipovich et al., 2012).
Although functional characterization was beyond the scope of the present study, we made efforts to identify lncRNAs with the potential to interact with selected pcRNAs or their proteins in the auditory forebrain. One approach was to select lncRNA-pcRNA pairs with high correlations in the P7-adult expression trajectory. From these, several genes with known involvement in brain development or plasticity were selected for predictive analysis of lncRNA – protein interactions. This tact was explored because the interactions of lncRNAs are not necessarily restricted to mRNA, as lncRNAs may also interact with proteins (Hacisuleyman et al., 2014) and can regulate post-transcriptional processes (Yoon et al., 2013). We found that most of the highly correlated genes also had strong predicted interaction scores, based on sequence matching and secondary structure (Lu et al., 2013). Naturally, predicted interactions require experimental validation. However, prior data suggest that the screening of correlated lncRNA-pcRNA pairs for potential interactions is an efficient way to identify genes that are promising for direct functional study.
For example, among the most highly expressed and upregulated lncRNAs in both auditory forebrain regions was the lincRNA, Malat1 (Metastasis associated lung adenocarcinoma transcript 1). Malat1 is strongly expressed in the brain, mainly by neurons (Bernard et al., 2010), and localized to nuclear speckles (Clemson et al., 2009; Hutchinson et al., 2007). In cultured hippocampal neurons, Bernard et al (2010) found that Malat1 expression levels increased steadily from P0 through P28, consistent with our findings in A1 and MG. They also showed that overexpression of Malat1 increased presynaptic bouton density on dendrites, while knock-down reduced synaptic density. Accordingly, gene ontology categories linked to synaptic and dendritic formation were enriched after Malat1 overexpression, although not all genes in these categories were affected. Further, cells transfected with Malat1 oligonucleotides exhibited reduced expression of the postsynaptic proteins neuroligin 1 (Nlgn1) and synaptic cell adhesion molecule 1 (Cadm1), while the Eph receptor B2 (Ephb2) and neuronal pentraxin 2 (Nptx2 or Narp) were relatively unchanged. This suggests that Malat1 regulates expression of a subset of genes involved in synapse formation in cultured hippocampal neurons. In the auditory forebrain, where Malat1 was strongly upregulated from P7 to adult, we found that Nlgn1, Cadm1 and Ephb2 were significantly downregulated in one or both auditory regions, whereas Nptx2 was upregulated in A1 and static in MG (Hackett et al., 2015). The differences between hippocampus and auditory forebrain suggest that regulation of the same genes by Malat1 can vary by gene and brain region. Accordingly, the potential for interactions between lncRNAs and pcRNAs to be regionally and temporally specific suggests that the interactions identified in one brain region or cell population may not apply to others.
An additional approach that has been used to identify potential interactions between lncRNAs and pcRNAs involves inspection of the genomic loci. Generally, lncRNA expression tends to be positively correlated with the expression of neighboring or overlapping sequences (Cabili et al., 2011; Dinger et al., 2008; Mercer et al., 2008; Ponjavic et al., 2009; Ulitsky et al., 2011). And, genes involved in the same biological pathways tend to have higher correlations when located in nearby genomic domains (Al-Shahrour et al., 2010). Although our inspection of such relationships was limited to a handful of genes, we found that some correlated pairs had overlapping or nearby genomic loci, while others were located at distant loci or on other chromosomes. Thus, in selecting lncRNA – pcRNA pairs that may interact for functional studies, it may be prudent to begin with pairs that have closely-associated loci, such as natural antisense transcripts (Carrieri et al., 2012; Katayama et al., 2005; Smalheiser et al., 2008). The relationship between Bdnf and Bdnf-As mentioned above is a frequently-cited example. A second example provides additional context for discussion of such relationships. The expression of two lncRNAs (Dlx1os, Dlx6os1) are in flux during the generation and migration of GABAergic interneurons in early development (Mercer et al., 2010), where they appear to exert transcriptional control over interneuron specification in the hippocampus (Berghoff et al., 2013; Bond et al., 2009; Feng et al., 2006; Kraus et al., 2013). These studies indicated that loss of Dlx1os (aka Dlx1-AS), which partially overlaps the associated transcription factor, Dlx1, in the antisense direction (Dinger et al., 2008), resulted in increased Dlx1 transcript expression and increased interneuron number. Similarly, loss of Dlx6os1 (aka Evf2) resulted in increased expression of the transcription factor, Dlx6, but decreased interneuron number. Thus, loss of the antisense transcripts led to increased expression of the associated transcription factors, but disparate effects of interneuron number. In the auditory forebrain, two observations were notable with respect to the expression patterns of these genes. First, Dlx1os and Dlx6os1 expression was restricted to A1 (absent in MG), suggesting regional specificity in the auditory forebrain. Referring to in situ hybridization assays in the Allen Brain Atlas (http://mouse.brain-map.org), we noted that expression of Dlx1 and Dlx6 overlapped their antisense lncRNA counterparts in A1 and most other cortical areas, where signals are concentrated in subpopulations of putative GABAergic neurons in layers 1 – 3. The absence of expression in MG probably corresponds with the absence of GABAergic neurons in that structure (Hackett et al., 2016). Thus, these transcription factors and associated antisense lncRNAs have overlapping anatomical distributions in the auditory forebrain that are also restricted to a particular neuronal subclass in cortex. Second, the expression of Dlx1 and Dlx1os decreased significantly from P7 to adult in A1, mainly between P7 and P14. Dlx6 and Dlx6os1 were expressed at very low levels, but with downward trends that did not reach significance. Thus, we observed tandem decreases in expression between these pcRNA-lncRNA pairs in A1 during maturation, while the loss of the same antisense lncRNAs led to increased pcRNA expression in developing hippocampus.
The reasons for these regional and temporal differences are not known, but could be informative with respect to the varied regulatory roles of lncRNAs in the adult and developing brain. However, based on the examples discussed above, we conclude that while prediction of pcRNA-lncRNA interactions based on proximity or correlated expression levels may be useful, those relationships could also be inaccurate and misleading. Carefully designed studies are needed to explore and better understand these relationships.
4.3 Species differences in lncRNA expression
In addition to cellular and regional differences in gene expression, species differences in pcRNA and protein expression evident in the central auditory pathway (Bush and Hyson, 2008; Wang et al., 2009a; Wang et al., 2009b), and are widespread throughout the brain (Bernard et al., 2012; Lin et al., 2014b; Mashiko et al., 2012; Nehme et al., 2012; Shukla et al., 2014; Van der Zee and Keijser, 2011; Watakabe et al., 2009; Zeng et al., 2012). Although not as extensively explored so far, species differences in lncRNA expression are notable, with several genes identified as rodent or primate-specific (e.g., Bdnf-As, Fmr4, Har1f, Bace1-as, Disc2, Scaant1)(Brandon et al., 2009; Faghihi et al., 2008; Khalil et al., 2009; Lipovich et al., 2012; Millar et al., 2004; Modarresi et al., 2012; Modarresi et al., 2011; Pollard et al., 2006; Pruunsild et al., 2007; Sopher et al., 2011; Tay et al., 2009; Washietl et al., 2014).
As an example, the pcRNA, brain-derived neurotrophic factor (Bdnf), has comparable expression patterns in mice, nonhuman primates, and humans, but the antisense lncRNA, Bdnf-As, is only expressed in primates. Thus, regulation of Bdnf by Bdnf-As is unique to primates (Lipovich et al., 2012), and therefore its regulation would appear to involve other mechanisms in mice or other species.
The proximity and genomic loci of interacting gene pairs may also be species dependent. One of the genes found by Bernard et al (2010) to be regulated by Malat1 in cultured hippocampal neurons, Cadm1, is located downstream of Malat1 on human chromosome 11, whereas in mice, Malat1 is located on chromosome 19 and Cadm1 is on chromosome 9. Similarly, Malat1 (aka Neat2) is just downstream of the lncRNA, Neat1, on mouse chromosome 19. Malat1 and Neat1 were strongly upregulated in A1 and MG. Moreover, Neat1 has been linked to promotion of differentiation and maturation of neurons and oligodendrocytes (Ip and Nakagawa, 2012; Mercer et al., 2010), and is associated with neuroprotection in Huntington’s disease (Sunwoo et al., 2016). Interestingly, the genomic loci of Malat1 and Neat1 are flanked by several pcRNAs with no known functional roles in brain development or maturation (e.g., Frmd8, Slc25a45, Dpf2, Tigd3, Pola2, Capn1, Scyl1, Ltbp3, Ehbp1l1, Map3k11, Sipa1, Kat5). In fact, very few genes located on chromosome 19 are currently associated with these functions. One of these, Flrt1, was upregulated in A1 and MG from P7 through adult, is in a family of genes with varied roles in brain development (Haines et al., 2006; Wheldon et al., 2010; Yamagishi et al., 2011), but there is no known association with either Malat1 or Neat1.
Given these observations, we would argue that documentation of species differences is absolutely essential to make informed conclusions and predictions about the roles of particular genes, and we must be vigilant to consider such differences in the interpretation and application of profiling data.
4.4 Applications of the lncRNA database
RNAseq is a powerful tool for mRNA profiling and transcriptome analyses with broad potential applicability in neurobiology (Han et al., 2014). Relatively small amounts of starting material (< 10 ng) are sufficient to conduct whole transcriptome sequencing of discrete brain areas or cell populations. The reduction in sequencing costs, development of bioinformatics tools, and availability of genomic libraries further add to the attractiveness of this approach (McGettigan, 2013; Sengupta et al., 2011). The dataset generated by this study comprises an extensive lncRNA reference library that indexes the expression of all known lncRNAs in A1 and MG from P7 to adult. In addition to information about these structures during postnatal development, the dataset is also a rich source of information about mature animals. We envision several potential applications of this lncRNA dataset by those interested in the structure and function of the auditory forebrain.
4.4.1 Reference database and guide for functional studies of the auditory forebrain
At present, none of the known lncRNAs have demonstrated functional roles in the auditory system. A major goal of this study was to establish a database of lncRNA expression in the auditory forebrain that would provide a broad foundation to guide focused studies of lncRNA function and regulation of mechanisms that govern the maturation of auditory processing in the forebrain. By identifying the most highly expressed lncRNAs in each brain region, and those that were strongly up- and down-regulated with age, numerous lncRNAs in A1 and MG are candidates for additional study. Then, as illustrated for a subset of genes, examination of lncRNA – pcRNA correlations, shared genomic loci, and predictive analyses of protein interactions, genes with the highest potential for interaction may be identified. Thus, a potentially powerful application of this database is as a screening tool to explore novel roles and interactions. A second, and related, application is to provide a baseline for experimental studies of hearing (e.g., altered sound exposure during development, hearing loss, aging, other pathology)(Clarkson et al., 2012; Holt et al., 2005; Sharma et al., 2009; Sun et al., 2008) [109, 110, 35, 36]. Transcriptomic analyses of global or targeted gene expression are powerful means to identify genes that are changing the most (or the least). The Look-Up Tool (Table S13) provides a convenient and user-friendly means to view expression levels and trends of any lncRNA and pcRNA by brain region and postnatal age.
4.4.2 Detailed anatomical profiling of lncRNA expression
In addition to regional differences in global lncRNA expression, insight into potential functional interactions may be gained by inspection of detailed anatomical distributions of lncRNA transcripts. Most lncRNA are expressed in the nucleus, but many have cytoplasmic expression, signaling potential regulation of gene expression in the cytoplasm (Batista and Chang, 2013). While only partially known, many lncRNA – pcRNA pairs in the developing brain may be co-localized within the same cells (Ponjavic et al., 2009). In addition, it is likely that lncRNAs are differentially expressed in distinct classes of neurons (e.g., glutamatergic, GABAergic, other) and glia (e.g., astrocytes, oligodendrocytes, microglia) (Cahoy et al., 2008; Zeisel et al., 2015; Zhang et al., 2014). Thus, for lncRNAs that are candidates for functional studies, it would be prudent to conduct assays designed to reveal the cellular and subcellular localization of their transcripts. To some extent, resources such as the Allen Brain Atlas can be consulted to identify the regional and subcellular distributions of transcripts for some lncRNAs. Clearly, the inclusion of lncRNAs within such anatomical resources would be helpful for identification of genes expressed in target brain areas, and for validation of other detection tools (e.g., qRT-PCR, RNAseq).
5. Conclusions
The lncRNA transcriptome of the mouse auditory forebrain was profiled at four postnatal ages before and after the onset of hearing. Globally, lncRNA expression was significantly different between brain regions (A1, MG) and at each postnatal age (P7, P14, P21, Adult). These patterns match trends observed for pcRNA in a prior study (Hackett et al., 2015), indicating that both RNA classes have spatial and temporal specificity in the developing (maturing) auditory forebrain. The results also identify lncRNAs with high expression levels and those with strong up- or down-regulation from P7 to Adult in both regions. Their expression levels are highly correlated with numerous pcRNAs, suggesting potential interactions. Additional analyses of selected highly-correlated pairs revealed that predicted interactions with associated proteins was often, but not always, strong, suggesting that the correlations may serve as an initial screen for potential interactions. Further study of those interactions may lead to new insights into the regulatory relationships between lncRNAs, pcRNAs, and proteins. A user-friendly database and Look-Up Tool were provided as supplementary files to facilitate inspection of the expression levels and maturational trajectories for any lncRNA or pcRNA in the database.
Supplementary Material
Highlights.
lncRNA expression was profiled in 2 auditory forebrain regions during maturation
Expression profiles differed between brain regions at each age (P7-P14-P21-Adult)
For each brain region (A1, MG), expression profiles differed between each age group
Expression trajectories (P7 to adult) correlated with subsets of protein coding genes
Database and look-up tools were created to simplify inspection of the entire dataset
ACKNOWLEDGEMENTS
YG was supported by Cancer Center Support Grant P30 CA68485. T.A.H was supported by NIH/NIDCD grant K18 DC012527 and Vanderbilt Kennedy Center infrastructure grant P30-HD015052-33. We would like to thank Page Hoskins for editorial support.
Abbreviations
- A1
Primary auditory cortex, area 1
- lncRNA
Long non-coding RNA
- MG
Medial geniculate body, thalamus
- pcRNA
Protein-coding RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Al-Shahrour F, Minguez P, Marques-Bonet T, Gazave E, Navarro A, Dopazo J. Selection upon genome architecture: conservation of functional neighborhoods with changing genes. PLoS Comput Biol. 2010;6:e1000953. doi: 10.1371/journal.pcbi.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS. Noncoding RNA in development. Mammalian genome : official journal of the International Mammalian Genome Society. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea J, Calegari F. Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. EMBO J. 2015;34:2865–2884. doi: 10.15252/embj.201592655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D, Lesche M, Dahl A, Groszer M, Hiller M, Calegari F. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32:3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, Kocher JP. 3' tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genomics. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. Perceptual learning in the developing auditory cortex. Eur J Neurosci. 2015;41:718–724. doi: 10.1111/ejn.12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Chan VT, Merzenich MM. Cortical remodelling induced by activity of ventral tegmental dopamine neurons. Nature. 2001;412:79–83. doi: 10.1038/35083586. [DOI] [PubMed] [Google Scholar]
- Barkat TR, Polley DB, Hensch TK. A critical period for auditory thalamocortical connectivity. Nat Neurosci. 2011;14:1189–1194. doi: 10.1038/nn.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ, Mattick JS. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MM, Serikawa K, Lemon T, Morgan R, Copeland C, Smith K, Cullen V, Davis-Turak J, Lee CK, Sunkin SM, Loboda AP, Levine DM, Stone DJ, Hawrylycz MJ, Roberts CJ, Jones AR, Geschwind DH, Lein ES. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Millar JK, Korth C, Sive H, Singh KK, Sawa A. Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci. 2009;29:12768–12775. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Kaczmarek LK. Potassium channel modulation and auditory processing. Hear Res. 2011;279:32–42. doi: 10.1016/j.heares.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush AL, Hyson RL. Effects of lithium and deafferentation on expression of glycogen synthase kinase-3beta, NFkappaB, beta-catenin and pCreb in the chick cochlear nucleus. Brain Res. 2008;1203:18–25. doi: 10.1016/j.brainres.2008.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. Journal of Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chang EH, Kotak VC, Sanes DH. Long-term depression of synaptic inhibition is expressed postsynaptically in the developing auditory system. Journal of Neurophysiology. 2003;90:1479–1488. doi: 10.1152/jn.00386.2003. [DOI] [PubMed] [Google Scholar]
- Chun S, Bayazitov IT, Blundon JA, Zakharenko SS. Thalamocortical long-term potentiation becomes gated after the early critical period in the auditory cortex. J Neurosci. 2013;33:7345–7357. doi: 10.1523/JNEUROSCI.4500-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Frontiers in Genetics. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson C, Herrero-Turrion MJ, Merchan MA. Cortical Auditory Deafferentation Induces Long-Term Plasticity in the Inferior Colliculus of Adult Rats: Microarray and qPCR Analysis. Front Neural Circuits. 2012;6:86. doi: 10.3389/fncir.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, Robertson AJ, Perkins AC, Bruce SJ, Lee CC, Ranade SS, Peckham HE, Manning JM, McKernan KJ, Grimmond SM. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- de Villers-Sidani E, Merzenich MM. Lifelong plasticity in the rat auditory cortex: basic mechanisms and role of sensory experience. Prog Brain Res. 2011;191:119–131. doi: 10.1016/B978-0-444-53752-2.00009-6. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan XA, Ruan YJ, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloe D, Le Gall C, Schaeffer B, Le Crom S, Guedj M, Jaffrezic F, French StatOmique C. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrn AL, Yuan K, Barker AJ, Schreiner CE, Froemke RC. Developmental sensory experience balances cortical excitation and inhibition. Nature. 2010;465:932–936. doi: 10.1038/nature09119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeline JM, Manunta Y, Hennevin E. Induction of selective plasticity in the frequency tuning of auditory cortex and auditory thalamus neurons by locus coeruleus stimulation. Hear Res. 2011;274:75–84. doi: 10.1016/j.heares.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froemke RC, Carcea I, Barker AJ, Yuan K, Seybold BA, Martins AR, Zaika N, Bernstein H, Wachs M, Levis PA, Polley DB, Merzenich MM, Schreiner CE. Long-term modification of cortical synapses improves sensory perception. Nat Neurosci. 2013;16:79–88. doi: 10.1038/nn.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froemke RC, Jones BJ. Development of auditory cortical synaptic receptive fields. Neurosci Biobehav R. 2011a;35:2105–2113. doi: 10.1016/j.neubiorev.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froemke RC, Jones BJ. Development of auditory cortical synaptic receptive fields. Neurosci Biobehav Rev. 2011b;35:2105–2113. doi: 10.1016/j.neubiorev.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li CI, Ye F, Shyr Y. Evaluation of read count based RNAseq analysis methods. BMC Genomics. 2013a;14(Suppl 8):S2. doi: 10.1186/1471-2164-14-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Sheng Q, Li J, Ye F, Samuels DC, Shyr Y. Large Scale Comparison of Gene Expression Levels by Microarrays and RNAseq Using TCGA Data. PLoS One. 2013b;8:e71462. doi: 10.1371/journal.pone.0071462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform. 2013c doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhao S, Sheng Q, Ye F, Li J, Lehmann B, Pietenpol J, Samuels DC, Shyr Y. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 2014a;103:323–328. doi: 10.1016/j.ygeno.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. Biomed Res Int. 2014b;2014:248090. doi: 10.1155/2014/248090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhao S, Sheng Q, Guo M, Lehmann BD, Pietenpol J, Samuels DC, Shyr Y. RNAseq by Total RNA Library Identifies Additional RNAs Compared to Poly(A) RNA library. Biomed Res Int. 2015 doi: 10.1155/2015/862130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett TA, Clause AR, Takahata T, Hackett NJ, Polley DB. Differential maturation of vesicular glutamate and GABA transporter expression in the mouse auditory forebrain during the first weeks of hearing. Brain Struct Funct. 2016;221:2619–2673. doi: 10.1007/s00429-015-1062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett TA, Guo Y, Clause A, Hackett NJ, Garbett K, Zhang P, Polley DB, Mirnics K. Transcriptional maturation of the mouse auditory forebrain. BMC Genomics. 2015;16:606. doi: 10.1186/s12864-015-1709-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines BP, Wheldon LM, Summerbell D, Heath JK, Rigby PW. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol. 2006;297:14–25. doi: 10.1016/j.ydbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Halley P, Kadakkuzha BM, Faghihi MA, Magistri M, Zeier Z, Khorkova O, Coito C, Hsiao J, Lawrence M, Wahlestedt C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Vickers KC, Samuels DC, Guo Y. Alternative applications for distinct RNA sequencing strategies. Brief Bioinform. 2014 doi: 10.1093/bib/bbu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- Hirsch RM, Slack JR, Smith RA. Techniques of Trend Analysis for Monthly Water-Quality Data. Water Resources Research. 1982;18:107–121. [Google Scholar]
- Holt AG, Asako M, Lomax CA, MacDonald JW, Tong L, Lomax MI, Altschuler RA. Deafness-related plasticity in the inferior colliculus: gene expression profiling following removal of peripheral activity. J Neurochem. 2005;93:1069–1086. doi: 10.1111/j.1471-4159.2005.03090.x. [DOI] [PubMed] [Google Scholar]
- Hurley LM, Sullivan MR. From behavioral context to receptors: serotonergic modulatory pathways in the IC. Front Neural Circuits. 2012;6:58. doi: 10.3389/fncir.2012.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott NE, Ponting CP. Predicting long non-coding RNAs using RNA sequencing. Methods. 2013;63:50–59. doi: 10.1016/j.ymeth.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ. 2012;54:44–54. doi: 10.1111/j.1440-169X.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- Kadakkuzha BM, Liu XA, McCrate J, Shankar G, Rizzo V, Afinogenova A, Young B, Fallahi M, Carvalloza AC, Raveendra B, Puthanveettil SV. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front Cell Neurosci. 2015;9:63. doi: 10.3389/fncel.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Group, R.G.E.R. Genome Science, G. Consortium, F. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgard MP, Merzenich MM. Plasticity of temporal information processing in the primary auditory cortex. Nat Neurosci. 1998;1:727–731. doi: 10.1038/3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–U165. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev SA, Korneeva EI, Lagarkova MA, Kiselev SL, Critchley G, O'Shea M. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA. 2008;14:2030–2037. doi: 10.1261/rna.1084308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral A. Auditory critical periods: a review from system's perspective. Neuroscience. 2013;247:117–133. doi: 10.1016/j.neuroscience.2013.05.021. [DOI] [PubMed] [Google Scholar]
- Kraus P, Sivakamasundari V, Lim SL, Xing X, Lipovich L, Lufkin T. Making sense of Dlx1 antisense RNA. Dev Biol. 2013;376:224–235. doi: 10.1016/j.ydbio.2013.01.035. [DOI] [PubMed] [Google Scholar]
- Kurokawa R. Long noncoding RNA as a regulator for transcription. Prog Mol Subcell Biol. 2011;51:29–41. doi: 10.1007/978-3-642-16502-3_2. [DOI] [PubMed] [Google Scholar]
- Kvam VM, Liu P, Si Y. A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am J Bot. 2012;99:248–256. doi: 10.3732/ajb.1100340. [DOI] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, Lachman HM. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, Head SR, Duester G, Dong PD, Rana TM. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014a;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, Gingeras TR, Ecker JR, Snyder MP. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A. 2014b;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KH, Hewitt CA, Beissbarth T, Hyde L, Banerjee K, Cheah PS, Cannon PZ, Hahn CN, Thomas PQ, Smyth GK, Tan SS, Thomas T, Scott HS. Molecular networks involved in mouse cerebral corticogenesis and spatio-temporal regulation of Sox4 and Sox11 novel antisense transcripts revealed by transcriptome profiling. Genome Biol. 2009;10:R104. doi: 10.1186/gb-2009-10-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KH, Hewitt CA, Beissbarth T, Hyde L, Cheah PS, Smyth GK, Tan SS, Hahn CN, Thomas T, Thomas PQ, Scott HS. Spatiotemporal regulation of multiple overlapping sense and novel natural antisense transcripts at the Nrgn and Camk2n1 gene loci during mouse cerebral corticogenesis. Cereb Cortex. 2011;21:683–697. doi: 10.1093/cercor/bhq141. [DOI] [PubMed] [Google Scholar]
- Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, Loeb JA. Activity-Dependent Human Brain Coding/Noncoding Gene Regulatory Networks. Genetics. 2012;192:1133. doi: 10.1534/genetics.112.145128. −+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, Lim DA. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QS, Ren SJ, Lu M, Zhang Y, Zhu DH, Zhang XG, Li TT. Computational prediction of associations between long non-coding RNAs and proteins. Bmc Genomics. 2013:14. doi: 10.1186/1471-2164-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Cui W, Liu H, He H, Xiu Y, Guo J, Liu H, Liu Q, Zeng T, Chen Y, Zhang Y, Wu Q. Identification and characterization of long non-coding RNAs related to mouse embryonic brain development from available transcriptomic data. PLoS One. 2013;8:e71152. doi: 10.1371/journal.pone.0071152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane LA, Gu Y, Casson AG, Murphy PR. Regulation of fibroblast growth factor-2 by an endogenous antisense RNA and by argonaute-2. Mol Endocrinol. 2010;24:800–812. doi: 10.1210/me.2009-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko H, Yoshida AC, Kikuchi SS, Niimi K, Takahashi E, Aruga J, Okano H, Shimogori T. Comparative anatomy of marmoset and mouse cortex from genomic expression. J Neurosci. 2012;32:5039–5053. doi: 10.1523/JNEUROSCI.4788-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17:4–11. doi: 10.1016/j.cbpa.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet. 2012;21:3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metherate R, Hsieh CY. Regulation of glutamate synapses by nicotinic acetylcholine receptors in auditory cortex. Neurobiol Learn Mem. 2003;80:285–290. doi: 10.1016/s1074-7427(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Millar JK, James R, Brandon NJ, Thomson PA. DISC1 and DISC2: discovering and dissecting molecular mechanisms underlying psychiatric illness. Ann Med. 2004;36:367–378. doi: 10.1080/07853890410033603. [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature biotechnology. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Patel NS, Sahagan BG, Wahlestedt C, Lopez-Toledano MA. Knockdown of BACE1-AS Nonprotein-Coding Transcript Modulates Beta-Amyloid-Related Hippocampal Neurogenesis. Int J Alzheimers Dis. 2011;2011:929042. doi: 10.4061/2011/929042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme B, Henry M, Mouginot D, Drolet G. The Expression Pattern of the Na(+) Sensor, Na(X) in the Hydromineral Homeostatic Network: A Comparative Study between the Rat and Mouse. Front Neuroanat. 2012;6:26. doi: 10.3389/fnana.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- O'Neil JN, Connelly CJ, Limb CJ, Ryugo DK. Synaptic morphology and the influence of auditory experience. Hear Res. 2011;279:118–130. doi: 10.1016/j.heares.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci U S A. 2012;109:16939–16944. doi: 10.1073/pnas.1202956109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald AM, Reyes AD. Development of inhibitory timescales in auditory cortex. Cereb Cortex. 2011;21:1351–1361. doi: 10.1093/cercor/bhq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel JM. Visiting "noncodarnia". Biotechniques. 2013;54(301):303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr., Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles JA, Qureshi SE, Stephen SJ, Wilson SR, Burden CJ, Taylor JM. Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing. BMC genomics. 2012;13:484. doi: 10.1186/1471-2164-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DC, Han L, Li J, Quanghu S, Clark TA, Shyr Y, Guo Y. Finding the lost treasures in exome sequencing data. Trends Genet. 2013 doi: 10.1016/j.tig.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes DH, Bao S. Tuning up the developing auditory CNS. Curr Opin Neurobiol. 2009;19:188–199. doi: 10.1016/j.conb.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes DH, Woolley SM. A behavioral framework to guide research on central auditory development and plasticity. Neuron. 2011;72:912–929. doi: 10.1016/j.neuron.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Tabata H, Tachikawa K, Nakajima K. The cortical subventricular zone-specific molecule Svet1 is part of the nuclear RNA coded by the putative netrin receptor gene Unc5d and is expressed in multipolar migrating cells. Mol Cell Neurosci. 2008;38:474–483. doi: 10.1016/j.mcn.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D'Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtele SJ, Losh J, Dailey ME, Green SH. Spine formation and maturation in the developing rat auditory cortex. J Comp Neurol. 2011;519:3327–3345. doi: 10.1002/cne.22728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Bolin JM, Ruotti V, Nguyen BK, Thomson JA, Elwell AL, Stewart R. Single read and paired end mRNA-Seq Illumina libraries from 10 nanograms total RNA. J Vis Exp. 2011:e3340. doi: 10.3791/3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Nag TC, Wadhwa S, Roy TS. Temporal distribution of mRNA expression levels of various genes in the developing human inferior colliculus. Neurosci Lett. 2009;461:229–234. doi: 10.1016/j.neulet.2009.06.049. [DOI] [PubMed] [Google Scholar]
- Shukla R, Watakabe A, Yamamori T. mRNA expression profile of serotonin receptor subtypes and distribution of serotonergic terminations in marmoset brain. Front Neural Circuits. 2014;8:52. doi: 10.3389/fncir.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Torvik VI, Mise N, Ikeda R, Abe K. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci Res. 2008;62:236–239. doi: 10.1016/j.neures.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics. 2013;14:91. doi: 10.1186/1471-2105-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, Hurley JB, Thienes CP, Gaasterland T, Filippova GN, La Spada AR. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigoni G, Gedressi C, Mallamaci A. Regulation of Emx2 expression by antisense transcripts in murine cortico-cerebral precursors. PLoS One. 2010;5:e8658. doi: 10.1371/journal.pone.0008658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Sun W, Zhang L, Lu J, Yang G, Laundrie E, Salvi R. Noise exposure-induced enhancement of auditory cortex response and changes in gene expression. Neuroscience. 2008;156:374–380. doi: 10.1016/j.neuroscience.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YJ, Wu GK, Liu BH, Li P, Zhou M, Xiao Z, Tao HW, Zhang LI. Fine-tuning of pre-balanced excitation and inhibition during auditory cortical development. Nature. 2010;465:927–931. doi: 10.1038/nature09079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo JS, Lee ST, Im W, Lee M, Byun JI, Jung KH, Park KI, Jung KY, Lee SK, Chu K, Kim M. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington's Disease. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9928-9. [DOI] [PubMed] [Google Scholar]
- Sutor B, Hagerty T. Involvement of gap junctions in the development of the neocortex. Biochim Biophys Acta. 2005;1719:59–68. doi: 10.1016/j.bbamem.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Tay SK, Blythe J, Lipovich L. Global discovery of primate-specific genes in the human genome. Proc Natl Acad Sci U S A. 2009;106:12019–12024. doi: 10.1073/pnas.0904569106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Hayashizaki Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008;372:691–696. doi: 10.1016/j.bbrc.2008.05.127. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Zee EA, Keijser JN. Localization of pre- and postsynaptic cholinergic markers in rodent forebrain: a brief history and comparison of rat and mouse. Behav Brain Res. 2011;221:356–366. doi: 10.1016/j.bbr.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman Y, Bartlett EL. Postnatal development of synaptic properties of the GABAergic projection from the inferior colliculus to the auditory thalamus. J Neurophysiol. 2013;109:2866–2882. doi: 10.1152/jn.00021.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Roteta LA, Hucheson-Dilks H, Han L, Guo Y. Mining diverse small RNA species in the deep transcriptome. Trends Biochem Sci. 2015;40:4–7. doi: 10.1016/j.tibs.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brozoski TJ, Turner JG, Ling L, Parrish JL, Hughes LF, Caspary DM. Plasticity at glycinergic synapses in dorsal cochlear nucleus of rats with behavioral evidence of tinnitus. Neuroscience. 2009a;164:747–759. doi: 10.1016/j.neuroscience.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Turner JG, Ling L, Parrish JL, Hughes LF, Caspary DM. Age-related changes in glycine receptor subunit composition and binding in dorsal cochlear nucleus. Neuroscience. 2009b;160:227–239. doi: 10.1016/j.neuroscience.2009.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic acids research. 2013a;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013b;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009c;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Sadakane O, Shimegi S, Takahata T, Higo N, Tochitani S, Hashikawa T, Naito T, Osaki H, Sakamoto H, Okamoto M, Ishikawa A, Hara S, Akasaki T, Sato H, Yamamori T. Enriched expression of serotonin 1B and 2A receptor genes in macaque visual cortex and their bidirectional modulatory effects on neuronal responses. Cereb Cortex. 2009;19:1915–1928. doi: 10.1093/cercor/bhn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheldon LM, Haines BP, Rajappa R, Mason I, Rigby PW, Heath JK. Critical role of FLRT1 phosphorylation in the interdependent regulation of FLRT1 function and FGF receptor signalling. PLoS One. 2010;5:e10264. doi: 10.1371/journal.pone.0010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi S, Hampel F, Hata K, Del Toro D, Schwark M, Kvachnina E, Bastmeyer M, Yamashita T, Tarabykin V, Klein R, Egea J. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. EMBO J. 2011;30:2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EJ, Lin EW, Hensch TK. Critical period for acoustic preference in mice. Proc Natl Acad Sci U S A 109 Suppl. 2012;2:17213–17220. doi: 10.1073/pnas.1200705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Samuels DC, Clark T, Guo Y. High-throughput sequencing in mitochondrial DNA research. Mitochondrion. 2014 doi: 10.1016/j.mito.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He LQ, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, Smith KA, Ebbert AJ, Swanson B, Kuan L, Page DT, Overly CC, Lein ES, Hawrylycz MJ, Hof PR, Hyde TM, Kleinman JE, Jones AR. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao Y, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector D. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen KN, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng SY, Liddelow SA, Zhang CL, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. Journal of Neuroscience. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Guo Y, Sheng Q, Shyr Y. Advanced heat map and clustering analysis using heatmap3. Biomed Res Int. 2014;2014:986048. doi: 10.1155/2014/986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, Gao YJ, Hoffman PN, Cui H, Li M, Dong X, Tao YX. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J Mol Neurosci. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.