Abstract
The existing biobanks of remnant tissue from clinically indicated kidney biopsies are attractive potential reservoirs for quantification of clinically relevant human tissue proteins by quantitative proteomics. However, a significant caveat of this strategy is that the tissues are often preserved in optimal cutting temperature (OCT) medium. Although OCT is an effective method of preserving the morphologic and immunohistological characteristics of tissues for later study, it significantly impacts efforts to quantify protein expression by liquid chromatography–tandem mass spectrometry methods. We report here a simple, reproducible, and cost-effective procedure to extract proteins from OCT-embedded tissue samples. Briefly, the excess frozen OCT medium was scraped before thawing from the tissue specimens stored at –80°C for ∼3 months. The tissue samples were homogenized and diethyl ether/methanol extraction was performed to remove the remaining OCT medium. The recovered protein was denatured, reduced, and alkylated. The second step of protein extraction and desalting was performed by chloroform/methanol/water extraction of denatured proteins. The resultant protein pellet was trypsin-digested and the marker proteins of various kidney cellular compartments were quantified by targeted selective reaction monitoring proteomics. Upon comparison of peptide signals from OCT-embedded tissue and flash-frozen tissue from the same donors, both individual protein quantities, and their interindividual variabilities, were similar. Therefore, the approach reported here can be applied to clinical reservoirs of OCT-preserved kidney tissue to be used for quantitative proteomics studies of clinically relevant proteins expressed in different parts of the kidney (including drug transporters and metabolizing enzymes).
Introduction
Twenty-six million Americans adults have chronic kidney disease (CKD) (http://www.niddk.nih.gov/health-information/health-statistics/Pages/kidney-disease-statistics-united-states.aspx), and people with CKD are at higher risk for cardiovascular and all-cause mortality (Tonelli et al., 2006). However, development of new diagnostic and therapeutic tools for CKD is hampered by our incomplete understanding of its underlying pathophysiology. In this direction, targeted mass spectrometry is a powerful tool, enabling simultaneous quantification of several prespecified proteins in biologic samples (Hood et al., 2012). Determining the abundance of protein components or targets of a given pathway using quantitative proteomics can generate a snapshot of the pathway status and activity within these samples. More recently, kidney tissue has gained notice as a more informative biologic sample for proteomics studies aiming to understand CKD pathophysiology. However, although kidneys play a critical role in the elimination of many drugs (Feng et al., 2012; Yacovino and Aleksunes, 2012; Moss et al., 2014), we have a limited quantitative understanding of the protein expression of drug transporters and metabolizing enzymes in the kidney tissue. Further, these data are scarcely available from CKD patients. Quantitative characterization of the transporters and enzymes in healthy as well as diseased tissues is indispensable for building pharmacokinetic and pharmacodynamic models to predict drug disposition and response. As such, both targeted and untargeted, or “shotgun,” proteomics methods have been applied to blood and urine in attempts to gain insight into mechanisms underlying CKD (Rossing et al., 2008; Filip et al., 2014). However, lack of power of such studies owing to the limitation of sample size often leads to inconclusive outcomes.
To overcome the sample-size problem, an attractive potential reservoir for such tissue is the existing biobanks of remnant tissue from clinically indicated kidney biopsies. Unfortunately, a significant caveat of this strategy is that these clinically stored tissues are often preserved in the optimal cutting temperature (OCT) medium, which interferes with mass spectrometry signal. On the other hand, mass spectrometric analyses perform best on protein extracted from flash-frozen tissue. Here, we describe optimization and evaluation of a method for membrane or structural protein extraction and targeted liquid chromatography–tandem mass spectrometry (LC-MS/MS), selective reaction monitoring (SRM) on OCT-embedded kidney tissue and assess the method’s performance compared with the gold-standard samples preserved by flash freezing.
Materials and Methods
Chemicals.
The ProteoExtract Native Membrane Protein Extraction Kit was procured from Calbiochem/MerckMillipore (Darmstadt, Germany). The protein quantification bicinchoninic acid (BCA) assay kit, sequencing-grade trypsin, iodoacetamide, and dithiothreitol were purchased from Pierce Biotechnology (Rockford, IL). Synthetic heavy peptides for all the surrogate peptides (Supplemental Table 1S) were obtained as internal standards from Thermo Fisher Scientific (Rockford, IL). Synthetic light peptides for the renal transporters OAT1, OAT3, OCT2, and P-glycoprotein (P-gp) were obtained as calibrators from New England Peptides (Boston, MA). Chloroform, ethyl ether, high-performance liquid chromatography (HPLC)-grade acetonitrile/methanol, and formic acid were purchased from Fischer Scientific (Fair Lawn, NJ). Ammonium bicarbonate (98% purity) and sodium deoxycholate (DOC, 98% purity) were obtained from Thermo Fisher Scientific (Rockford, IL) and MP Biomedicals (Santa Ana, CA), respectively.
Sample Procurement, OCT Removal, and Trypsin Digestion.
Histologically normal sections of kidney tissue from healthy donors (n = 5, 3 male, 2 female, 53–67 years old) undergoing clinically indicated partial nephrectomy (e.g., removal of a kidney mass) were obtained after informed consent (Proteogenex, Culver City, CA). Specimens weighed 450–700 mg and were divided into two equal sections, with one section immediately frozen in liquid nitrogen and stored at –80°C. The second half was embedded in the OCT medium using a protocol followed by the University of Washington Department of Pathology; samples were subsequently transferred to –80°C. After 3 months of storage at –80°C, excess OCT around the tissue block was removed with a scalpel without compromising the embedded tissue. Two milliliters of extraction buffer I and 10 μl of protease inhibitor (Calbiochem ProteoExtract Native Protein Extraction Kit) were added to both sets of tissue samples. The samples were homogenized until completely suspended and then incubated on ice for 20 minutes on a rocker table to prevent the formation of cell clumps. The samples were then sonicated for 60 seconds and centrifuged at 16,000g, 4°C, for 15 minutes. The supernatant (the cytosolic fraction) was removed without disturbing the pellet and transferred to a new tube and set aside. One milliliter of extraction buffer II plus 5 μl of protease inhibitor was added to the pellet, which was then resuspended by pipetting up and down ten times or until the pellet was completely broken apart. The sample was incubated on ice for 30 minutes on a rocker table to prevent cell clumps from forming, then centrifuged at 16,000g, 4°C, for 15 minutes. The supernatant (membrane fraction) was removed without disturbing the pellet and transferred completely to a new tube. Total protein was quantified using BCA assay kit (Pierce Biotechnology) and the sample was diluted to 2 mg/ml before analysis.
To remove OCT contamination a previously developed method was modified (Weston and Hummon, 2013). First, 800 μl of diethyl ether and 200 μl of methanol were added to 50 μl of diluted protein sample (100 μg total protein). The sample was vortexed for 5 minutes and then centrifuged at 10,000g for 10 minutes. The upper and lower liquid layers were removed carefully so as not to disturb the interfacial pellet, and air dried. The pellet was resuspended in 80 μl of ammonium bicarbonate, 20 μl of DOC (3%), and 15 μl of 250 mM dithiothreitol and incubated for 5 minutes at 95°C. iodoacetamide (IAA) (18 μl of 500 mM) was added to the samples, and then they were incubated in the dark for 20 minutes. Beyond the previously reported method, a second cleaning step was added to decrease ion suppression in liquid chromatography–tandem mass spectrometry (LC-MS/MS) and to remove any contamination that could affect trypsinization. Five-hundred microliters of methanol, 100 μl of chloroform, and 400 μl of water were added and the samples were vortexed for 1 minute. The samples were centrifuged at 6000g, 4°C, for 5 minutes. The upper and lower liquid layers were removed without disturbing the interfacial layer. The pellet was washed with 1 ml of methanol followed by centrifugation at 6000g for 5 minutes. Then the methanol was completely removed and the sample was air dried. The pellet was completely resuspended in 20 μl of sodium DOC (3%) and 40 μl of 100 mM ammonium bicarbonate. Twenty microliters of 0.16 μg/μl trypsin was added to the sample, which was then incubated for 18 hours at 37°C, with gentle shaking to assist trypsin digestion. The reaction was quenched by adding 30 μl of heavy peptide cocktail in 80% acetonitrile containing 0.1% formic acid. The sample was centrifuged at 3000g for 2 minutes and 100 μl of the supernatant was transferred into LC-MS vials for analysis. The absolute quantification of OAT1, OAT3, OCT2, and P-gp was performed by using light surrogate peptides as calibrators per published protocol (Prasad et al., 2016).
LC-MS/MS Analysis.
LC-MS/MS was performed to quantify 23 structural proteins expressed in different sections of the nephron, the functional unit of the kidney (Table 1). The surrogate peptides of the markers of various cellular compartments of kidney were quantified using triple-quadrupole LC-MS instruments [Xevo TQ-S coupled to ACQUITY UPLC (Waters, Milford, MA)] in positive electrospray ionization mode. Approximately 10 μg of the trypsin digest (5 μl) was injected onto the column (Waters 2.1 μm, C18 100A; 150 × 2.1 mm; Phenomenex, Torrance, CA) and eluted at 0.3 ml/min. A mobile phase consisting of water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B) was employed. A flow rate of 0.3 ml/min was used in a gradient manner (Supplemental Table 1S). MS/MS analysis was performed by monitoring the surrogate peptides and the internal standards using instrument parameters provided in Supplemental Table 1S.
TABLE 1 .
Proteins and peptides quantified in the kidney tissue
| Protein | Tissue Localizationa | Detected Peptide |
|---|---|---|
| Nephrin | Podo | ELVLVTGPSDNQAK |
| Podocalyxin-like | Podo | LGDQGPPEEAEDR |
| Podocin | Podo | LPAGLQHSLAVEAEAQR and SLTEILLER |
| CD34 | Mes, Glom endo | LGILDFTEQDVASHQSYSQK and SWSPTGER |
| Aldolase B | PT | ELSEIAQSIVANGK |
| Arginosuccinate synthase 1 | PT | APNTPDILEIEFK |
| Cathepsin H | PT | VNHAVLAVGYGEK |
| Dicarbonyl/L-xylulose reductase | PT | TQADLDSLVR |
| Dipeptidase 1 | PT | VASLIGVEGGHSIDSSLGVLR |
| OAT1 | PT | TSLAVLGK |
| OAT3 | PT | TVLAVFGK |
| OCT2 | PT | SPGVAELSLR and LNPSFLDLVR |
| SERPINA1 | PT | DTEEEDFHVDQVTTVK |
| SGLT2 | PT | LEDISEDPSWAR |
| P-gp (MDR1) | PT, Mes | NTTGALTTR |
| Tamm-Horsfall protein | TAL | DWVSVVTPAR |
| Golgi membrane protein 1 | DTINLLDQR | |
| Aquaporin 2 | CD | QSVELHSPQSLPR and SLAPAVVTGK |
| Carbonic anhydrase 2 | DCT, CD | YDPSLKPLSVSYDQATSLR |
| Tight junction protein 1 (ZO1) | Podo, TAL, DCT, CD | DNPHFQSGETSIVISDVLK |
| Plakoglobin | PT, TAL, DCT, CD | LLNQPNQWPLVK |
| Claudin 5 | Endo | EFYDPSVPVSQK and VYDSVLALSTEVQAAR |
| Intercellular adhesion molecule 2 | Endo | ILLDEQAQWK |
Podo, glomerular podocytes; Mes, glomerular mesangial cells; PT, proximal tubule; TAL, thick ascending limb of the loop of Henle; DCT, distal convoluted tubule; CD, collecting duct; Endo, endothelial cells
Data Analysis.
LC-MS/MS data were processed by integrating the peak areas generated from the reconstructed ion chromatograms for the surrogate peptides and the internal standards using MassLynx 4.1 (Waters, Hertfordshire, UK). The peak response for two to three transitions from each peptide was averaged for quantification of samples or standards. Paired Student’s t test was used to compare peptide recovery between flash-frozen and OCT-embedded samples.
Results
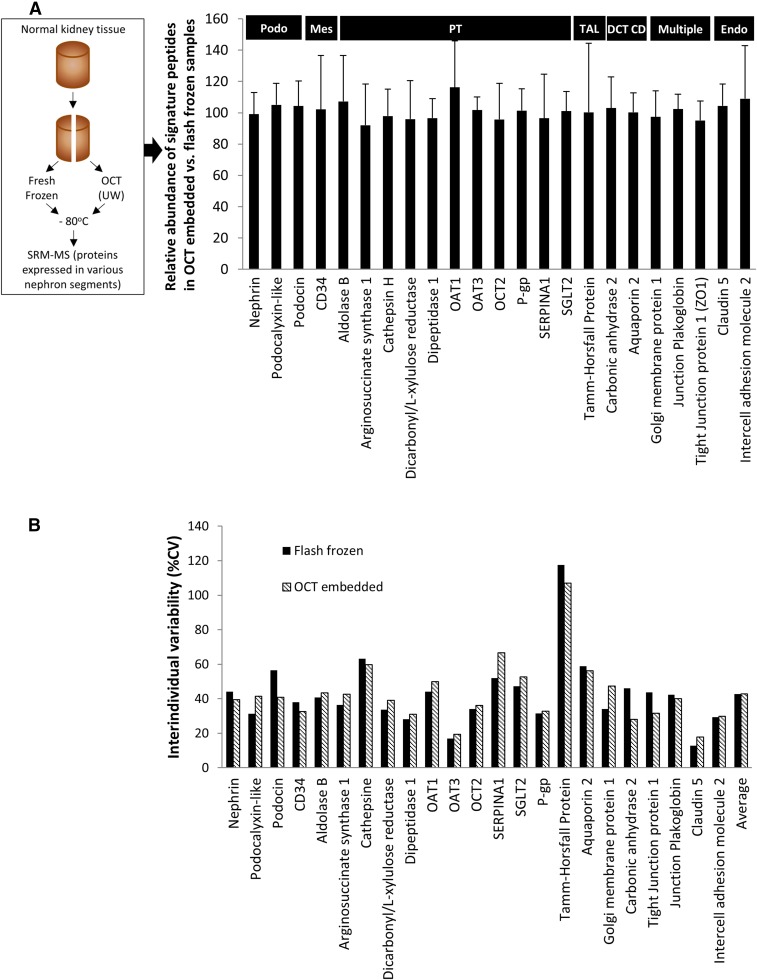
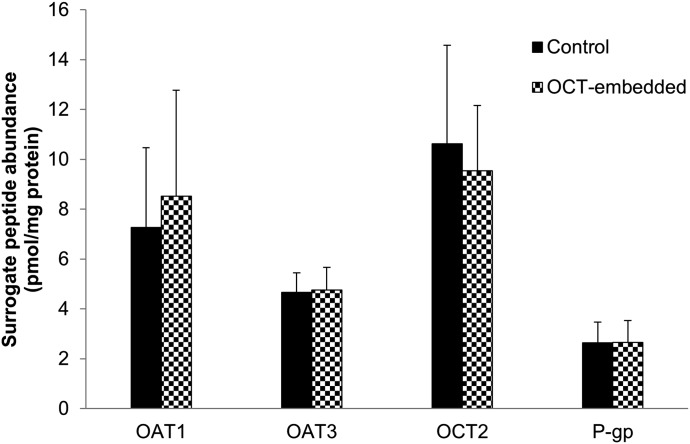
The results of LC-MS/MS quantification of 23 structural proteins expressed in different sections of the nephron are presented as %expression of each protein in the OCT-embedded samples compared with the paired flash-frozen control sample (Fig. 1A). When comparing peptide signal from OCT-embedded tissue to flash frozen tissue, we observed a cumulative peptide signal recovery of 101.1 ± 5.3%. Individual protein quantities in OCT were 92.1–116.2% of that in flash-frozen tissue (Fig. 1A). Interindividual variability of all the targeted proteins was also comparable in OCT-embedded versus flash-frozen tissue (42.7% and 42.9%, respectively; Fig. 1B). At absolute level, OAT1, OAT3, OCT2, and P-gp expression (pmol/mg total protein) was 7.26 ± 3.21, 4.67 ± 0.79, 10.62 ± 3.96, and 2.64 ± 0.83 (control) versus 8.53 ± 4.25, 4.76 ± 0.92, 9.54 ± 2.63, and 2.66 ± 0.87 (OCT-embedded), respectively (Fig. 2). The latter indicates 100% recovery of the proteins from the OCT-embedded kidney samples.
Fig. 1.
Expression levels of selected proteins, shown as percent expression detected in OCT-embedded sample versus expression detected in flash frozen samples (A). The protein signals in OCT-embedded samples were measured at 101.1 ± 5.3% of the signal in flash frozen samples, indicating good recovery of proteins. Interindividual variability (%CV) of tissue protein expression determined in flash-frozen and OCT-embedded samples (B)
Fig. 2.
Absolute protein abundance of OAT1, OAT3, OCT2, and P-gp in control (flash-frozen, solid black) and OCT-embedded (black and white pattern) human kidney samples (n = 5). The protein abundance is expressed as pmol/mg total membrane protein (mean ± standard deviation).
Discussion
The method reported here yields comparable protein quantification by targeted mass spectrometry from OCT-embedded tissue versus the gold standard of flash-frozen tissue and enables the use of clinical stores of OCT-preserved kidney tissue for quantitative proteomics studies. Methods have been reported for protein extraction and mass spectrometry from formalin-fixed and paraffin-embedded (FFPE) tissue; however, a limited number of reports exist on mass spectrometry proteomic analysis of OCT embedded tissue samples (Azimzadeh et al., 2010; Tian et al., 2011; Quesada-Calvo et al., 2015; Zhang et al., 2015). These published reports are mainly discovery proteomics data, which focuses on qualitative appearance of peptide signals rather than absolute peptide quantitation.
OCT and FFPE media are not ideal matrices for mass spectrometry quantification. OCT is mainly composed of polyvinyl alcohol (PVA), polyethylene glycol (PEG), and nonreactive ingredients, which serve as cryopreservative medium and support matrix for tissue sectioning. Unless completely removed, PVA and PEG can suppress ionization of the targeted peptides (Schwartz et al., 2003). Starting from recently developed methods for OCT removal (Tian et al., 2011; Johnson and White, 2014; Zhang et al., 2015), we optimized a simple and cost-effective method to extract proteins from OCT-embedded kidney tissue samples for quantitative mass spectrometry applications. This method can be applied to clinical reservoirs of OCT-preserved kidney tissue for use in quantitative proteomics studies of clinically relevant proteins expressed in different parts of the kidney (including drug transporters and metabolizing enzymes). Differential interindividual variability of proteins was observed, indicating unique regulation (e.g., mediated by individual genetic and epigenetic factors) of these proteins. The highest variability was observed for Tamm-Horsfall protein, consistent with reported values (Fu et al., 2016). However, as the main aim of this report was methodological, unique interindividual variability has no effect on our conclusions. Quantitative characterization of the clinically relevant proteins, including transporters and enzymes, in healthy as well as diseased kidney tissues is indispensable for building better pharmacokinetic and pharmacodynamic models to predict drug disposition and response.
Abbreviations
- CKD
chronic kidney disease
- DOC
sodium deoxycholate
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- OCT
optimal cutting temperature
- OCT2
organic cation transporter 2
- P-gp
P-glycoprotein
Authorship Contributions
Participated in research design: Vrana, Goodling, Afkarian, Prasad.
Conducted experiments: Vrana, Prasad.
Performed data analysis: Vrana, Afkarian, Prasad.
Wrote or contributed to the writing of the manuscript: Vrana, Goodling, Afkarian, Prasad.
Footnotes
Dr. Afkarian’s effort was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Disease [K23DK089017 and R01DK104706] and a Norman S. Coplon Extramural Grant from Satellite Healthcare. Dr. Prasad’s effort was funded by Department of Pharmaceutics, University of Washington, Seattle, WA.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aires I, Santos AR, Bogarin R, Genc G, Pratas J, Ozkaya O, Carvalho F, Rueff J, Nolasco F, Calado J. (2013) Disruption of urate transport in familial renal glucosuria and report on SGLT2 expression in normal and patholocal kidney. Port J Nephrol Hypert 27:261–267. [Google Scholar]
- Azimzadeh O, Barjaktarovic Z, Aubele M, Calzada-Wack J, Sarioglu H, Atkinson MJ, Tapio S. (2010) Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J Proteome Res 9:4710–4720. [DOI] [PubMed] [Google Scholar]
- Ernest S, Rajaraman S, Megyesi J, Bello-Reuss EN. (1997) Expression of MDR1 (multidrug resistance) gene and its protein in normal human kidney. Nephron 77:284–289. [DOI] [PubMed] [Google Scholar]
- Feng B, El-Kattan AF, Radi ZA.(2012) Renal transporters in drug disposition, drug-drug interactions, and nephrotoxicity . Curr Protoc Toxicol Chapter 23:Unit 23.3:1–15. [DOI] [PubMed] [Google Scholar]
- Filip S, Pontillo C, Peter Schanstra J, Vlahou A, Mischak H, Klein J. (2014) Urinary proteomics and molecular determinants of chronic kidney disease: possible link to proteases. Expert Rev Proteomics 11:535–548. [DOI] [PubMed] [Google Scholar]
- Fu Q, Grote E, Zhu J, Jelinek C, Köttgen A, Coresh J, Van Eyk JE. (2016) An empirical approach to signature peptide choice for selected reaction monitoring: quantification of uromodulin in urine. Clin Chem 62:198–207. [DOI] [PubMed] [Google Scholar]
- Fujinaka H, Iida T, Yamamoto T. (2014) The proteome-transcriptome-combined database of specific nephron segment proteins for novel urinary biomarker discovery. J Clinic Nephrol Res 1:1014–1019. [Google Scholar]
- Higgins JP, Wang L, Kambham N, Montgomery K, Mason V, Vogelmann SU, Lemley KV, Brown PO, Brooks JD, van de Rijn M. (2004) Gene expression in the normal adult human kidney assessed by complementary DNA microarray. Mol Biol Cell 15:649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood LE, Omenn GS, Moritz RL, Aebersold R, Yamamoto KR, Amos M, Hunter-Cevera J, Locascio L, Workshop Participants (2012) New and improved proteomics technologies for understanding complex biological systems: addressing a grand challenge in the life sciences. Proteomics 12:2773–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H, White FM. (2014) Quantitative analysis of signaling networks across differentially embedded tumors highlights interpatient heterogeneity in human glioblastoma. J Proteome Res 13:4581–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusniemi AM, Kestilä M, Patrakka J, Lahdenkari AT, Ruotsalainen V, Holmberg C, Karikoski R, Salonen R, Tryggvason K, Jalanko H. (2004) Tissue expression of nephrin in human and pig. Pediatr Res 55:774–781. [DOI] [PubMed] [Google Scholar]
- Kwon O, Myers BD, Sibley R, Dafoe D, Alfrey E, Nelson WJ. (1998) Distribution of cell membrane-associated proteins along the human nephron. J Histochem Cytochem 46:1423–1434. [DOI] [PubMed] [Google Scholar]
- Miyanaka K, Gotoh T, Nagasaki A, Takeya M, Ozaki M, Iwase K, Takiguchi M, Iyama KI, Tomita K, Mori M. (1998) Immunohistochemical localization of arginase II and other enzymes of arginine metabolism in rat kidney and liver. Histochem J 30:741–751. [DOI] [PubMed] [Google Scholar]
- Moss DM, Neary M, Owen A. (2014) The role of drug transporters in the kidney: lessons from tenofovir. Front Pharmacol 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Nakao Y, Masuda S, Katsura T, Kamba T, Ogawa O, Inui K. (2013) Precise comparison of protein localization among OCT, OAT, and MATE in human kidney. J Pharm Sci 102:3302–3308. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Sakurai Y, Saito H, Masuda S, Urakami Y, Goto M, Fukatsu A, Ogawa O, Inui K. (2002) Gene expression levels and immunolocalization of organic ion transporters in the human kidney. J Am Soc Nephrol 13:866–874. [DOI] [PubMed] [Google Scholar]
- Prasad B, Gaedigk A, Vrana M, Gaedigk R, Leeder JS, Salphati L, Chu X, Xiao G, Hop CE, Evers R, et al. (2016) Ontogeny of hepatic drug transporters as quantified by LC-MS/MS proteomics. Clin Pharmacol Ther DOI:10.1002/cpt.409 (published ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-Calvo F, Bertrand V, Longuespée R, Delga A, Mazzucchelli G, Smargiasso N, Baiwir D, Delvenne P, Malaise M, De Pauw-Gillet MC, et al. (2015) Comparison of two FFPE preparation methods using label-free shotgun proteomics: Application to tissues of diverticulitis patients. J Proteomics 112:250–261. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Lamas M, Martin D, del Carmen Namorado M, Islas S, Luna J, Tauc M, González-Mariscal L. (2002) The renal segmental distribution of claudins changes with development. Kidney Int 62:476–487. [DOI] [PubMed] [Google Scholar]
- Rossing K, Mischak H, Dakna M, Zürbig P, Novak J, Julian BA, Good DM, Coon JJ, Tarnow L, Rossing P, PREDICTIONS Network (2008) Urinary proteomics in diabetes and CKD. J Am Soc Nephrol 19:1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SA, Reyzer ML, Caprioli RM. (2003) Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom 38:699–708. [DOI] [PubMed] [Google Scholar]
- Tian Y, Bova GS, Zhang H. (2011) Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal Chem 83:7013–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli M, Wiebe N, Culleton B, House A, Rabbat C, Fok M, McAlister F, Garg AX. (2006) Chronic kidney disease and mortality risk: a systematic review. J Am Soc Nephrol 17:2034–2047. [DOI] [PubMed] [Google Scholar]
- Weston LA, Hummon AB. (2013) Comparative LC-MS/MS analysis of optimal cutting temperature (OCT) compound removal for the study of mammalian proteomes. Analyst (Lond) 138:6380–6384. [DOI] [PubMed] [Google Scholar]
- Yacovino LL, Aleksunes LM. (2012) Endocrine and metabolic regulation of renal drug transporters. J Biochem Mol Toxicol 26:407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sakashita S, Taylor P, Tsao MS, Moran MF. (2015) Comprehensive proteome analysis of fresh frozen and optimal cutting temperature (OCT) embedded primary non-small cell lung carcinoma by LC-MS/MS. Methods 81:50–55. [DOI] [PubMed] [Google Scholar]