Abstract
Structural characterization of amyloid rich in cross-β structures is crucial for unraveling molecular basis of protein misfolding and amyloid formation associated with a wide range of human disorders. Elucidation of the β-sheet structure in non-crystalline amyloid has, however, remained an enormous challenge. Here we report structural analyses of the β-sheet structure in full-length transthyretin amyloid using solid-state NMR spectroscopy. Magic-angle-spinning (MAS) solid-state NMR was employed to investigate native-like β-sheet structures in amyloid state using selective labeling schemes for more efficient solid-state NMR studies. Analyses of extensive long-range 13C-13C correlation MAS spectra obtained with selectively 13CO- and 13Cα-labeled TTR reveal that the two main β-structures in the native state, the CBEF and DAGH β-sheets, remain intact after amyloid formation. The tertiary structural information would be of great use for examining quaternary structure of TTR amyloid.
Keywords: Transthyretin, amyloid, solid-state NMR, cross-β structure, protein misfolding, magic-angle spinning
Graphical Abstract
Transthyretin (TTR) is one of more than 30 human proteins that undergo an aberrant conformational change and misassemble into β-structured amyloid implicated in numerous degenerative diseases such as Alzheimer’s and Parkinson’s diseases, and amyloidoses. 1–5 The 127-residue homo-tetrameric protein (55 kDa) circulating in blood transports thyroid hormone (T4) and retinal binding protein. TTR amyloid formation process involves rate-limiting tetramer dissociation, followed by monomer misfolding and aggregation, which are associated with numerous amyloidoses including senile systemic amyloidosis (SSA), familial amyloidotic polyneuropathy (FAP) and familial amyloidotic cardiomyopathy (FAC).6,7 Structural investigation of amyloid is essential to understanding molecular mechanism of the misfolding and amyloid formation, which will help to develop not only therapeutic agents that can target pathogenic amyloids, but also biomarkers that can detect early stages of amyloid diseases.
Amyloid formation of globular proteins requires a conformational change from natively folded states to partially or globally unfolded aggregation-prone intermediates that self-assemble into cross-β structured amyloid. Recent biophysical studies have also suggested that subtle structural changes of the natively folded proteins may expose aggregation-prone segments susceptible for intermolecular interactions, triggering amyloid formation.8–13 The aggregation-prone intermediates populated by the local structural changes of natively folded states may lead to amyloid with extensive native-like structural features. Elucidations of the native-like structural features in amyloid would, therefore, be of great importance in understanding the molecular mechanism of protein misfolding and amyloid formation.
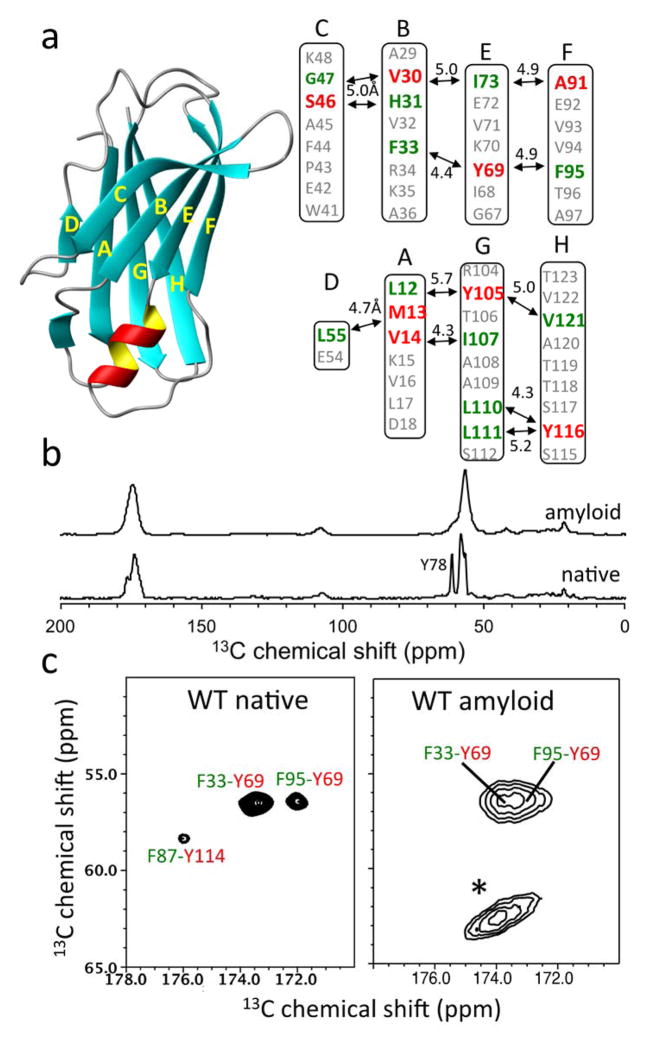
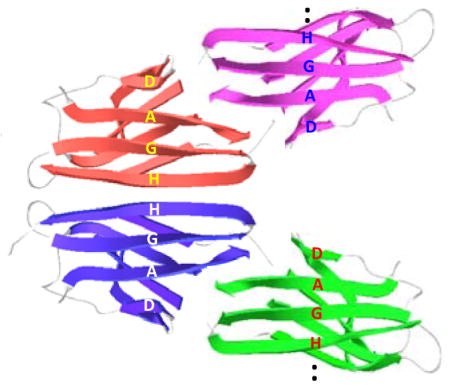
The native TTR monomer is rich in β-sheet conformations, adopting a β-sandwich consisting of two β-sheets comprising strands CBEF and DAGH (Figure 2a).14 Although extensive structural studies have been conducted to investigate β-sheet structures in TTR amyloid, structural features of TTR amyloid has been controversial. 6,15–18 It was originally proposed that the native BEF and AGH β-structures are retained in TTR amyloid, while strands C and D move away from the β-sheet structure. 6,15,17 Recent protease digestion studies, however, proposed a different structural model with non-native β-sheet conformations. 8 Our recent solution NMR experiments also suggested that the monomeric precursor state of TTR contains the native-like CBEF β-sheet structure, while DAGH β-sheet is destabilized.19,20 Here we employed solid-state NMR for structural studies of the β-sheet structures in TTR amyloid. Previous solid-state NMR structural studies of amyloid formed by a short peptide fragment TTR(105-115) provided valuable structural information of the amyloid state. 21 The full-length protein may, however, have different amyloidogenic properties and form distinct amyloid, requiring structural analyses of amyloid derived from full-length TTR. In this report, we made use of magic-angle spinning (MAS) solid-state NMR to scrutinize the extent of native-like structural features in full-length TTR amyloid, focusing on structural changes within the CBEF and DAGH β-sheets during amyloid formation.
Figure 2.
(a) Ribbon diagram representation of TTR monomer and labeling schemes for the solid-state NMR experiments. The 13CO (green) and 13Cα (red) carbons of the amino acids are labeled if the internuclear distance in the native TTR tetramer are between 4 and 6 Å. (b) 13C CPMAS spectra of the native and amyloid states of TTR prepared from 13CO-Phe and 13Cα-Tyr labeled TTR. (c) 2D PDSD spectra of the native and amyloid states of TTR with a contour level of 1.5 % with respect to the diagonal peak obtained using a mixing time of 500 ms at a 1H frequency of 830 MHz. * denotes spinning sidebands. For the solid-state NMR experiments, the native tetrameric TTR at pH 7.3 was precipitated using 90 % ammonium sulfate and dried under Ar gas. Both dried native and amyloid samples were rehydrated with 5 μL water.
Experimental Methods
Protein Expression
Wild type and mutant forms of TTR were expressed in BL21 (DE3) E. coli, and were purified as previously described. 6 Recombinant expression of the wild type and all of the mutant forms (V30M, F33V, L55P, Y69H, and Y116S) used in this study produced soluble tetrameric states of TTR at pH 7. Two different types of isotopically enriched TTR samples were prepared for solid-state NMR experiments. For the selectively 13CO- and 13Cα-labeled samples, M9 media supplemented with unlabeled amino acids (100 mg per liter culture) and 13C-labeled amino acids (50 mg/L) were used for protein expression. In our bacterial expression system, serine was synthesized from glycine, and thus 13Cα-Gly was used to label serine and both Cα and Cβ carbons were enriched in serine. In order to prepare protein samples with four uniformly 13C/15N labeled amino acids (Ala, Asp, Leu, and Val) while the rest of the amino acids unlabeled, 15N-NH4Cl and 13C-glucose were used in M9 media supplemented with the other sixteen unlabeled amino acids (100 mg/L).
Amyloid Formation
Amyloid samples were obtained by incubating the protein (0.2 mg/mL) in 200 mM acetate buffer (100 mM KCl, 1 mM EDTA, pH 4.4) for a period of 30 days at 37 °C. The insoluble amyloid was spun down and washed twice with deionized water to remove remaining soluble tetramers and soluble aggregates. The TTR amyloid was examined by transmission electron microscopy (TEM) and thioflavin T binding assay in our previous studies.20
CD Spectroscopy
The CD spectra were recorded by scanning from 260 nm to 190 nm on a Jasco J-810 spectropolarimeter (Easton, MD) using a 1 mm path length Suprasil quartz cell at pH 4.4 and 7.0. The protein samples (0.14 mg/ml) were pre-equilibrated at 20 °C for 5 min before acquisitions.
NMR Spectroscopy
For solid-state NMR experiments, the protein aggregates were dried using Ar gas at 25 °C and packed into a 3.2 mm rotor. A small amount of water (5 μL) was then added for rehydration. For the native state, tetrameric TTR at pH 7.3 was precipitated using 90% ammonium sulfate and dried under Ar gas. The dried native TTR was packed into the rotor and rehydrated with 5 μL water. The tetrameric TTR obtained from the ammonium sulfate precipitation was confirmed to maintain natively folded conformations based on the Cα chemical shifts in the solid-state NMR spectra that are identical to those in solution NMR spectra for the tetrameric TTR (BMRM 5507).
Solid-state NMR spectra were acquired using Bruker 800 and 830 MHz spectrometers equipped with a 3.2 mm CP-MAS probe at room temperature. The two-dimensional solid-state NMR spectra for the selectively labeled samples were mainly recorded using the Bruker DRX 830 MHz spectrometer with an ultra-narrow bore (31 mm) 19.6 T magnet. A Low-E MAS probe22 developed at the NHMFL with a Revolution NMR stator and 3.2 mm pencil rotors was used for the measurements.
A linear amplitude ramp on the 1H channel was used for the 1H/13C cross-polarization with a contact time of 1 ms. The 90° pulse-lengths for 1H and 13C were 3.0 μs and 5.0 μs, respectively. The two-pulse phase-modulated (TPPM) decoupling scheme was employed with a radio-frequency field strength of 80 kHz. Two-dimensional 13C-13C correlation NMR spectra were recorded using a proton-driven spin diffusion (PDSD) mixing scheme at spinning frequencies of 11 – 12 kHz. The spinning speed was set close to the Δωiso = 2ωr rotational resonance (RR) condition23,24 for efficient polarization transfer. Since the second spinning sideband from 13CO carbons is still observable under the second RR condition, the spinning speed was set slight below for each 13CO/13Cα spin pairs such that the sideband of the carbonyl carbon appears ~ 5 ppm away from the NMR resonance of the 13Cα carbon. For the 2D PDSD spectra, complex data points of 1024 × 362 and 1024 × 145 were collected for the native and amyloid states, respectively, with acquisition delay of 2 sec, and 48 – 64 FIDs were accumulated for each t1 data point.
Results
Native-like structural features in amyloidogenic TTR
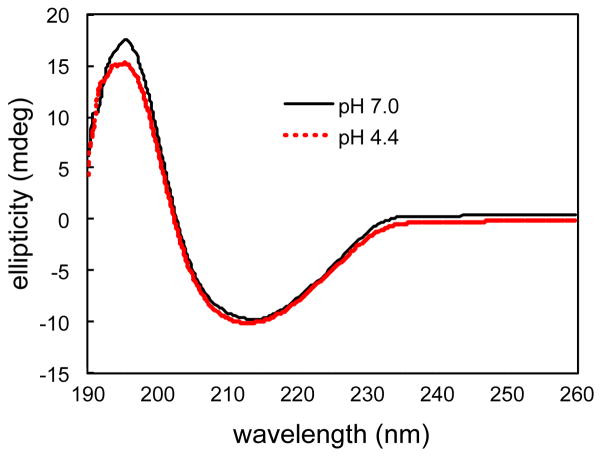
Our previous solution NMR studies showed that aggregation-prone state of TTR populated at mildly acidic pH of 4.4 contains extensive native-like β-sheet conformations. 19,20 In particular, the native CBEF β-sheet remains unchanged in the amyloidogenic intermediate state of TTR, while the other β-sheet (DAGH) undergoes conformational fluctuations in millisecond time scales. The structural feature of the aggregation-prone states of TTR was investigated with CD spectroscopy (Figure 1). The CD spectrum collected at the native condition (pH 7.0) displays a minimum at ~ 215 nm indicative of typical β-sheet conformations, clearly indicating that TTR adopts β-sheet conformations at the physiological pH. Notably the CD spectrum recorded at the amyloidogenic pH of 4.4 is almost identical to that observed at the native condition (pH 7.0). The signal intensity at ~196 nm slightly decreases at pH 4.4, suggesting that TTR becomes slightly more disordered presumably due to a local unfolding transition at the amyloidogenic pH.
Figure 1.
CD spectra of WT TTR (0.14 mg/ml) in 10 mM PBS (pH 7.0) and 10 mM sodium acetate (pH 4.4) buffer recorded at 20 °C.
Under the amyloidogenic condition of pH 4.4, WT TTR tetramers are dissociated to monomers and reach a dynamic equilibrium with the amyloidogenic TTR monomers. Previous ultracentrifugation experiments showed that approximately 25 % of the tetramers are observed at a protein concentration of 0.2 mg/ml. 6 Thus about 20 – 25 % of tetramers might be present under our experimental conditions (0.14 mg/ml), which indicates that the CD signals recorded at pH 4.4 mostly come from amyloidogenic TTR monomers. Previous biophysical studies also revealed that after the rate-determining step of tetramer dissociations to aggregation-prone monomers, TTR effectively forms amyloid via downhill mechanism in which major conformational changes are not required for amyloid formation. 25 These combined results suggest that TTR amyloid may retain the native-like β-sheet structures probed by our CD (Figure 1) and solution NMR experiments for the monomeric aggregation-prone state of TTR.
Labeling schemes for Solid-State NMR
We employed solid-state NMR to examine the native-like β-sheet structures observed in the amyloidogenic intermediate states. Magic-angle-spinning (MAS) solid-state NMR has provided valuable structural information on various amyloids, such as the location of β-strands within the amyloid. 26–37 Despite the great progress in solid-state NMR methodology, full structural characterization of the amyloid derived from this 127-residue protein remains an enormous challenge. Secondary structure information from chemical shift values obtained by sequential resonance assignments using various 3D MAS experiments would not be sufficient to identify arrangement of the β-strands into tertiary β-sheet structure.
In the current study, we utilized specific labeling schemes, which generate isolated 13CO-13Cα dipolar-coupled spin pairs in the native β-sheets (Figure 2a, Table 1, and Supplemental Figure S1), to specifically probe the native β-sheet structure in the amyloid state. For example, 13CO-Phe and 13Cα-Tyr labeled protein has two dipolar coupled 13CO-13Cα spin pairs at distances of 4–6 Å in the BEF strands. The dipolar-coupled spin pairs in the native tetramer can then be probed and compared to those in amyloid using solid-state NMR experiments that can detect spin-pairs with a separation of up to 6 Å. Since the crystal structure of tetrameric TTR has already been determined, the native state of TTR can be used as a standard sample for the solid-state NMR structural studies of amyloid state of TTR.
Table 1.
List of the labeling schemes used for the structural studies and single-point mutations for the resonance assignment.
| substructure | labeling scheme | TTR mutant samples for the resonance assignment |
|---|---|---|
| BEF | Phe-Tyr; Ile-Ala (Met) | Y69H, V30M |
| CB | His-Ser; Gly-Val | V30M |
| DA | Leu-Met | L55P |
| AGH | Tyr-Val; Ile-Val; Leu-Tyr | Y116S |
The 13C CPMAS spectrum of the F/Y labeled TTR confirms the enrichment of 13CO and 13Cα carbons at ~175 and ~60 ppm, respectively (Figure 2b). It is notable that NMR peaks in the amyloid are broader than those of the native state. However, our recent solid-state NMR experiments showed that TTR amyloid used in this study is a uniform protein assembly consisting of a single monomeric core conformer. 20
Native-like CBEF β-sheet
Two-dimensional proton-driven spin-diffusion (PDSD)38 experiments were conducted to probe the dipolar-coupled spin pairs in the native, and also in the amyloid state(s) (Figure 2c). The three spin pairs (F33-Y69, F87-Y114, and F95-Y69) were observed in the 2D spectrum of the native tetrameric TTR (Figure 2c), which is consistent with the crystal structure of TTR. The two cross-peaks for F33-Y69 and F95-Y69 were also detected in TTR amyloid, while the F87-Y114 cross-peak was not observed (Figure 2c, the assignment of the cross-peaks for the amyloid state were confirmed by using F33V and Y69H mutant amyloids, Supplemental Figure S2 and S3). The residues F87 and Y114 are located in EF and GH loops, respectively (Supplemental Figure S4), and the cross-peak between the two residues may be broadened by conformational disorder of the loop regions and/or the two loops may not be in close proximity in the amyloid state of TTR.
The relative intensities of the cross-peaks with respect to the diagonal peaks in the native TTR spectrum (Figure 2c) are about 2 %, 6 %, and 9 ± 0.5 % for the F87-Y114 (6.0 Å), F95-Y69 (4.9 Å), and F33-Y69 (4.4 Å) spin pairs, respectively. The relative intensities of the cross-peaks in the amyloid spectra recorded for all of the WT and mutant TTRs were all within 3 – 6 %, as demonstrated in Supplemental Figure S2, suggesting that the distances of the spin pairs investigated in this study are within 6 Å.
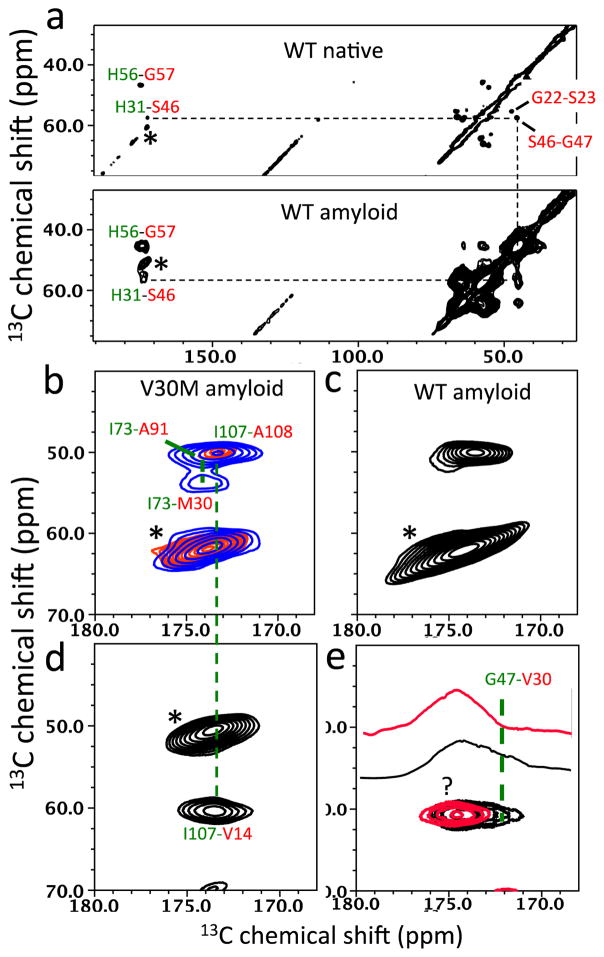
The two cross-peaks from the dipolar-coupled spin pairs (F33-Y69 and F95-Y69) indicate that TTR amyloid may contain the native-like BEF β-sheet substructure, which is line with our recent solution NMR results that showed soluble amyloidogenic precursor states contain native-like CBEF β-sheet structure. 20 Additional residues (13CO-His, 13Cα-Gly and 13Cα-Ser) were labeled to probe CB substructure using the solid-state NMR experiments (Figure 2a and Figure 3a). The cross-peak for the H31-S46 in the CB native structure was detected for both native tetrameric and amyloid states of TTR, corroborating the native CB-strand structure in the amyloid state.
Figure 3.
2D PDSD spectra of native and amyloid states of TTR with a contour level of 1.5 % obtained using various labeling schemes. A mixing time of 500 ms was used, unless indicated. (a) 13CO-His, 13Cα-Gly, and 13Cα-Ser. (b) 13CO-Ile, 13Cα-Met, and 13Cα-Ala labeled V30M amyloid PDSD spectra using mixing times of 100 ms (red) and 500 ms (blue) (c) 13CO-Ile, 13Cα-Met, and 13Cα-Ala labeled WT amyloid. (d) 13CO-Ile and 13Cα-Val labeled V30M amyloid. (e) 13CO-Gly and 13Cα-Val labeled WT (black) and V30M amyloid (red) with 1D slices at 60.5 ppm. * denotes spinning sidebands. The cross-peak marked by ? could not be assigned in this study.
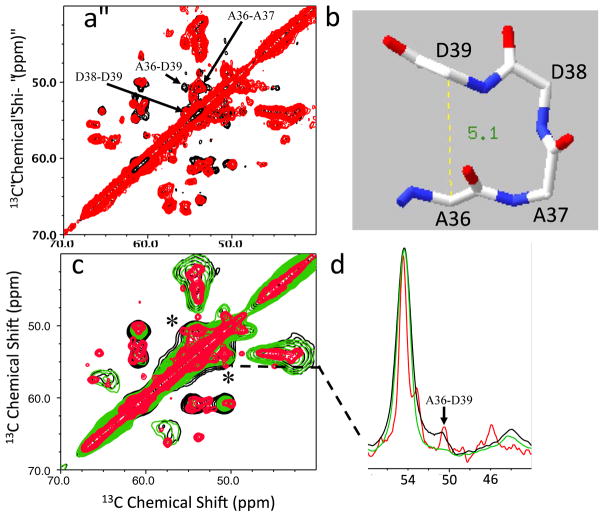
The native-like CB substructure was also confirmed with 13C-13C correlation experiments on selectively labeled TTR where only four amino acids (A, D, L, and V) were uniformly 13C-labeled (Figure 4). The cross-peaks in the native PDSD spectra in Figure 4 were unambiguously assigned based on the previous assignment (BMRB 5507) and sequential labeling of the alanines and aspartic acids. The cross-peak from the distant residues A36-D39 was observed only in the 500 ms PDSD spectrum in the native state (Figure 4a). The two distant residues also give rise to the cross-peak only in the longer mixing time amyloid spectrum (black in Figure 4c and 4d), indicating the presence of the BC turn in the amyloid state as well.
Figure 4.
(a) Overlay of the 2D PDSD spectra for the native TTR acquired using two mixing times of 100 ms (red) and 500 ms (black) at a 1H frequency of 800 MHz. The contour level was set to 3.0 % of the diagonal peaks. Individual spectra with an expanded spectra width are also shown in Supplemental Figure S5. (b) Crystal structure for the BC turn. (c) Overlay of the 2D PDSD spectra for the native TTR acquired at 500 ms mixing time (red), and amyloid state spectra obtained using two mixing times of 100 ms (green) and 500 ms (black). (d) One-dimensional slices along the cross-peak for A36-D39 (* in Figure S5c). In order to simplify the solid-state NMR spectra, four amino acids (A, D, L, and V) were uniformly 13C-labeled, while other amino acids were not 13C-labeled.
Additional evidence for the BEF native structure in amyloid state was obtained with 2D PDSD spectra for 13CO-Ile, 13Cα-Met, and 13Cα-Ala labeled WT and V30M TTR amyloid samples (Figure 2a, and Figure 3b and 3c). In the V30M mutant, the cross-peak from the I73-M30 spin pair that is not detected in WT amyloid was clearly observed (Figure 3b and 3c). The NMR resonance for the I73-A91 spin pair appears to be overlapped with that of the strongly-coupled I107-A108 (2.4 Å) in the 500 ms PDSD spectrum (blue in Figure 3b). In the shorter mixing time PDSD spectrum (100 ms, red in Figure 3b), the cross-peak from the I107-A108 spin pair is more clearly observed, which allows us to distinguish the two-cross peaks.
In order to obtain additional evidence for the CB β-structure, 13CO-Gly and 13Cα-Val residues were labeled for the 2D PDSD experiments (Figure 2a and Figure 3e). Two cross-peaks were observed in WT amyloid (black), while one cross-peak was detected in V30M mutant amyloid (red), indicating that the residue V30 is spatially connected to glycine. One of the two cross-peaks (G47-V30 in CB) was assigned on the basis of preserved native CBEF structure in amyloid state, as described above, and additional 2D experiments (resonance assignments in Supporting Information).
Native-like DAGH β-sheet
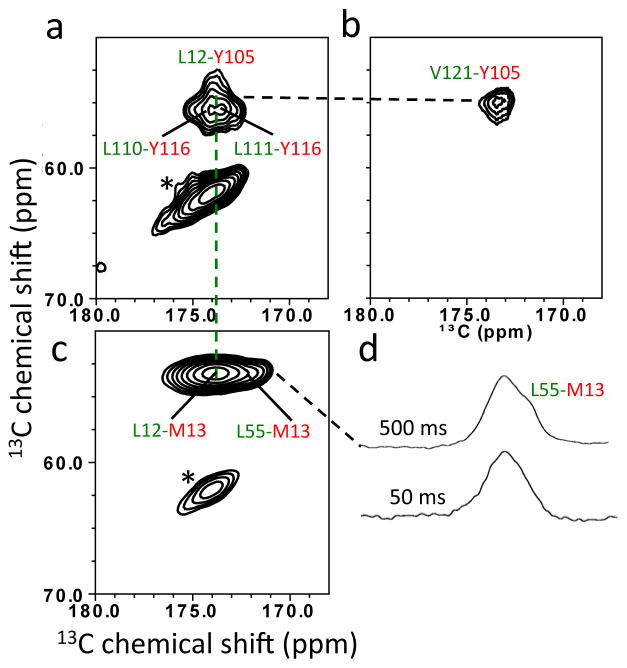
The solid-state NMR experiments were extended to investigate the DAGH β-structure (Figure 2a) in amyloid state by employing 13CO-Leu and 13Cα-Tyr (Figure 5a), 13CO-Val and 13Cα-Tyr (Figure 5b), and 13CO-Leu and 13Cα-Met (Figure 5c and 5d) labeling schemes. The residue Y105 is in close proximity to residues L12 and V121 in AGH structure (Figure 2a, 5a and 5b), and the residue Y116 gives rise to the cross-peaks with two leucines (Figure 2a and 5a). In the TTR sequence, there is only one neighboring, strongly coupled L-M spin pair (13CO-L12 and 13Cα-M13: 2.4 Å), which is observed in the PDSD spectrum with a shorter mixing time of 50 ms (Figure 5c and 5d), facilitating assignments of the tyrosine and thus leucine residues, and the L55-M13 spin pair in Figure 5 (the assignment was confirmed with additional solid-state NMR experiments using mutant forms of TTR, L55P and Y116S, Supplemental Figure S6 and S7). The long-range 13C-13C correlations strongly indicate the presence of the native-like DAGH β-sheet structure in the TTR amyloid state.
Figure 5.
2D PDSD spectra of amyloid states of WT TTR with a contour level of 1.5 % obtained using different labeling schemes. A mixing time of 500 ms was used, unless indicated. (a) 13CO-Leu and 13Cα-Tyr. (b) 13CO-Val and 13Cα-Tyr. (c) 13CO-Leu and 13Cα-Met. (d) 1D slices for the 13CO-Leu and 13Cα-Met PDSD spectra with two mixing times of 50 ms and 500 ms. * denotes spinning sidebands.
Discussion
TTR quickly forms moderately ordered amyloid at pH 4.4, leading to the broad NMR resonances with a full-width at half-maximum of ~ 2.5 ppm. The linewidth is somewhat broader than those from previously reported homogeneous amyloid fibrils (< 1 ppm). 33,39,40 However, the strong NMR signals of TTR amyloid in the dipolar based solid-state NMR are comparable to or narrower than those from Aβ oligomers 36 and β-microglubulin aggregates 39 formed at pH 3.6 (2–4 ppm) that are shown to be still homogeneous aggregates. In addition, a single set of the cross-peaks in the extensive 2D correlation spectra described above strongly indicates that TTR amyloid used in this study is a uniform protein assembly consisting of a single monomeric conformer. The broad NMR resonances most likely result from some inhomogeneity of interfacial interactions in subunit-subunit interfaces and/or local dynamic processes, which was observed even for highly ordered amyloid fibrils derived from a short peptide fragment TTR(105-115) 28.
The resonance assignment of the 2D PDSD spectra using 3D correlation MAS NMR experiments would be extremely challenging due to the relatively low NMR resolution for the TTR amyloid samples. Thus we used various single-point mutagenesis to confirm the assignment of the NMR resonances in the amyloid PDSD spectra (Table 1). Structural studies of various mutant forms of TTR by X-ray crystallography revealed that the mutations do not induce major structural changes of the β-sheet structures irrespective of the site of mutations. 41,42 For example, V30M TTR amyloid was used for the structural studies of the BEF substructure (Figure 2a and Table 1). The pathogenic mutation in strand B is known to facilitate the tetramer dissociations without inducing structural changes of the β-sheet structures, 43 suggesting that V30M amyloid has a similar β-sheet structure to that of WT amyloid. Indeed, our additional 2D PDSD experiments on F/Y labeled V30M amyloid revealed an identical correlation spectrum to that of WT amyloid (Supplemental Figure S10), indicating that the mutant TTR amyloid also retains at least the native-like BEF substructure like WT amyloid. The other mutant amyloids such as L55P, Y69H, and Y116S (Table 1) were used to confirm the resonance assignment for L55, Y69, and Y116, respectively. The single-point mutations most likely affect the vicinity of the mutation, rather than disrupt the whole β-sheet structure, as shown in Y116S mutant amyloid (Supplemental Figure S7). The cross-peak for Y116-L110 disappears in the Y116S PDSD spectrum, whereas the cross-peak for L12-Y105 is still observed, suggesting the AG substructure remains unchanged in the mutant amyloid. Structural studies using EPR with extensive site-directed mutagenesis15 and H/D exchange NMR13 have, therefore, employed various mutant TTRs including Y114C for the structural studies of TTR amyloid.
Previous biophysical studies of TTR amyloid proposed various structural models with different native-like structural features. The common feature in the proposed structural models is that part of the native β-structures undergoes conformational changes, while rest of the β-structures remains unchanged in TTR amyloid. For example, H/D exchange experiments of TTR amyloid suggested that strands C and D are exposed to solvent, proposing the two strands unfold from the β-structures during amyloid formation. 15 Recent protease digestion studies of TTR amyloid, however, suggested that strand D remains in amyloid core, while strands B and C undergo substantial unfolding from the β-structure.18 The discrepancies between the previous experiments and our NMR results might originate from different experimental conditions. It was recently shown that TTR amyloid in solution is in a dynamic equilibrium with soluble intermediate states,44 suggesting that the previous H/D exchange and protease digestion experimental results might be obtained from a mixture of TTR amyloid and soluble intermediate states.
In summary, the 13C-13C correlation solid-state NMR experiments utilizing selectively 13CO-and 13Cα-labeled TTR described here show that TTR amyloid adopts a native-like CBEF β-structure. The solid-state NMR results are consistent with our recent solution NMR experiments that showed the native-like CBEF β-structure is retained in soluble amyloidogenic precursor states of TTR. 20 Our recent NMR studies also demonstrated that the DAGH β-sheet becomes less stable and more dynamic on millisecond time scales due to the disruption of the interactions between the short β-strand in the AB loop and strand A, exposing strand A that can be available for intermolecular associations. 20 The solid-state NMR experiments described here, however, indicate that the main DAGH β-sheet structure is still maintained in TTR amyloid, while strand A might be exposed for intermolecular interactions. Our combined solid-state NMR results suggest that the native-like CBEF and DAGH β-structures are preserved in the amyloid state, while the AB loop regions that interact with strand A become unfolded and disordered, rendering the strand A unprotected. The structural information about the β-sheet structures provides the basis for developing quaternary structural models for TTR amyloid, which is of great challenge for the 127-residue amyloidogenic polypeptide.
For example, only outer strands in the native CBEF and DAGH β-sheet can be involved in intermolecular associations. Strand A that is exposed because of the structural perturbation of the AB loop can also play an important role in TTR aggregation. Recent studies of TTR aggregation mechanisms suggested that strands H and F may play a causative role in TTR amyloid formation.45 Strand A was also shown to have a high aggregation propensity. 45 In addition, our recent solution NMR studies showed the CBEF β-sheet does not undergo conformational exchanges between the native monomer and the amyloidogenic precursor states of TTR.19,20 These findings indicate that the DAGH β-sheet, particularly strands H and A, might be involved in the intermolecular associations to form cross-β structure in TTR amyloid. The structural model is in line with recent solid-state NMR results that ruled out parallel in-register β-sheet conformations in TTR amyloid.18 Although the native-like CBEF β-structure in the structural model appears contradicted from that proposed by the EPR signals between strands B and B’,17 the EPR signals may also originate from interactions between protofibrils that contain native-like CBEF β-sheet conformations.
Finally, we demonstrated that the solid-state NMR experiments using selective labeling schemes can be of great use for investigating structural models for the tertiary β-sheet structure within amyloid.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by NIH Grants NS084138 (KHL), DK34909 (PEW), AG10770 (DEW), DK46335 (JWK) and the Skaggs Institute of Chemical Biology (PEW and JWK). The solid-state NMR spectra were acquired at the National High Magnetic Field Laboratory, which is supported by NSF Cooperative Agreement No. DMR-1157490 and the State of Florida.
We thank Prof. Jon Kenney (East Carolina University) for his assistance during the CD experiments.
ABBREVIATIONS
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single-quantum coherence
- MAS
magic-angle spinning
- CP
cross-polarization
- WT
wild-type
- TTR
transthyretin
- PDSD
proton-driven spin-diffusion
- TEM
transmission electron microscopy
- ThT
thioflavin T
- Leu
leucine
- Val
Valine
- Pro
proline
- H/D
hydrogen/deuterium
- CD
circular dichroism
Footnotes
Notes
The authors declare no competing financial interests.
Supporting Information. Resonance assignments. 2D PDSD NMR spectra for the mutant TTR amyloids. Schematic diagrams of monomeric TTR displaying labeled amino acids for the solid-state NMR experiments. The following files are available free of charge.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 3.Sunde M, Blake CC. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q Rev Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 4.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 5.Jahn TR, Radford SE. Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai ZH, Colon W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 7.Saraiva MJ. Transthyretin mutations in health and disease. Hum Mutat. 1995;5:191–196. doi: 10.1002/humu.1380050302. [DOI] [PubMed] [Google Scholar]
- 8.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Pardo J, Grana-Montes R, Fernandez-Mendez M, Ruyra A, Roher N, Aviles FX, Lorenzo J, Ventura S. Amyloid formation by human carboxypeptidase D transthyretin-like domain under physiological conditions. J Biol Chem. 2014;289:33783–33796. doi: 10.1074/jbc.M114.594804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bemporad F, Vannocci T, Varela L, Azuaga AI, Chiti F. A model for the aggregation of the acylphosphatase from Sulfolobus solfataricus in its native-like state. Biochim Biophys Acta. 2008;1784:1986–1996. doi: 10.1016/j.bbapap.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc Natl Acad Sci U S A. 2006;103:10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soldi G, Bemporad F, Torrassa S, Relini A, Ramazzotti M, Taddei N, Chiti F. Amyloid formation of a protein in the absence of initial unfolding and destabilization of the native state. Biophys J. 2005;89:4234–4244. doi: 10.1529/biophysj.105.067538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 14.Blake CC, Geisow MJ, Oatley SJ, Rerat B, Rerat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. J Mol Biol. 1978;121:339–356. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 15.Olofsson A, Ippel JH, Wijmenga SS, Lundgren E, Ohman A. Probing solvent accessibility of transthyretin amyloid by solution NMR spectroscopy. J Biol Chem. 2004;279:5699–5707. doi: 10.1074/jbc.M310605200. [DOI] [PubMed] [Google Scholar]
- 16.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. A glimpse of a possible amyloidogenic intermediate of transthyretin. Nat Struct Biol. 2000;7:754–757. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 17.Serag AA, Altenbach C, Gingery M, Hubbell WL, Yeates TO. Arrangement of subunits and ordering of beta-strands in an amyloid sheet. Nat Struct Biol. 2002;9:734–739. doi: 10.1038/nsb838. [DOI] [PubMed] [Google Scholar]
- 18.Bateman DA, Tycko R, Wickner RB. Experimentally derived structural constraints for amyloid fibrils of wild-type transthyretin. Biophys J. 2011;101:2485–2492. doi: 10.1016/j.bpj.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim KH, Dyson HJ, Kelly JW, Wright PE. Localized structural fluctuations promote amyloidogenic conformations in transthyretin. J Mol Biol. 2013;425:977–988. doi: 10.1016/j.jmb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim KH, Dasari AK, Hung I, Gan Z, Kelly JW, Wemmer DE. Structural Changes Associated with Transthyretin Misfolding and Amyloid Formation Revealed by Solution and Solid-State NMR. Biochemistry. 2016;55:1941–1944. doi: 10.1021/acs.biochem.6b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Aca Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNeill SA, Gor’kov PL, Shetty K, Brey WW, Long JR. A low-E magic angle spinning probe for biological solid state NMR at 750 MHz. J Magn Reson. 2009;197:135–144. doi: 10.1016/j.jmr.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raleigh DP, Levitt MH, Griffin RG. Chem Phys Lett. 1988;147:71–76. [Google Scholar]
- 24.Seidel K, Lange A, Becker S, Hughes CE, Heise H, Baldus M. Protein solid-state NMR resonance assignments from (13C,13C) correlation spectroscopy. Phys Chem Chem Phys. 2004;6:5090–5093. [Google Scholar]
- 25.Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 26.Tang M, Comellas G, Rienstra CM. Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils. Acc Chem Res. 2013;46:2080–2088. doi: 10.1021/ar4000168. [DOI] [PubMed] [Google Scholar]
- 27.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debelouchina GT, Bayro MJ, Fitzpatrick AW, Ladizhansky V, Colvin MT, Caporini MA, Jaroniec CP, Bajaj VS, Rosay M, Macphee CE, Vendruscolo M, Maas WE, Dobson CM, Griffin RG. Higher order amyloid fibril structure by MAS NMR and DNP spectroscopy. J Am Chem Soc. 2013;135:19237–19247. doi: 10.1021/ja409050a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 30.Helmus JJ, Surewicz K, Apostol MI, Surewicz WK, Jaroniec CP. Intermolecular alignment in Y145Stop human prion protein amyloid fibrils probed by solid-state NMR spectroscopy. J Am Chem Soc. 2011;133:13934–13937. doi: 10.1021/ja206469q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linser R, Dasari M, Hiller M, Higman V, Fink U, Lopez del Amo JM, Markovic S, Handel L, Kessler B, Schmieder P, Oesterhelt D, Oschkinat H, Reif B. Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angew Chem Int Ed Engl. 2011;50:4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- 32.Barbet-Massin E, Ricagno S, Lewandowski JR, Giorgetti S, Bellotti V, Bolognesi M, Emsley L, Pintacuda G. Fibrillar vs crystalline full-length beta-2-microglobulin studied by high-resolution solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:5556–5557. doi: 10.1021/ja1002839. [DOI] [PubMed] [Google Scholar]
- 33.Daebel V, Chinnathambi S, Biernat J, Schwalbe M, Habenstein B, Loquet A, Akoury E, Tepper K, Muller H, Baldus M, Griesinger C, Zweckstetter M, Mandelkow E, Vijayan V, Lange A. beta-Sheet core of tau paired helical filaments revealed by solid-state NMR. J Am Chem Soc. 2012;134:13982–13989. doi: 10.1021/ja305470p. [DOI] [PubMed] [Google Scholar]
- 34.Hong M, Schmidt-Rohr K. Magic-angle-spinning NMR techniques for measuring long-range distances in biological macromolecules. Acc Chem Res. 2013;46:2154–2163. doi: 10.1021/ar300294x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan S, Suiter CL, Hou G, Zhang H, Polenova T. Probing structure and dynamics of protein assemblies by magic angle spinning NMR spectroscopy. Acc Chem Res. 2013;46:2047–2058. doi: 10.1021/ar300309s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parthasarathy S, Inoue M, Xiao Y, Matsumura Y, Nabeshima Y, Hoshi M, Ishii Y. Structural Insight into an Alzheimer’s Brain-Derived Spherical Assembly of Amyloid beta by Solid-State NMR. J Am Chem Soc. 2015;137:6480–6483. doi: 10.1021/jacs.5b03373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weingarth M, Baldus M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc Chem Res. 2013;46:2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- 38.Szeverenyi NM, Sullivan MJ, Maciel GE. J Magn Reson. 1982;47:462–475. [Google Scholar]
- 39.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Magic angle spinning NMR analysis of beta2-microglobulin amyloid fibrils in two distinct morphologies. J Am Chem Soc. 2010;132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hornberg A, Eneqvist T, Olofsson A, Lundgren E, Sauer-Eriksson AE. A comparative analysis of 23 structures of the amyloidogenic protein transthyretin. J Mol Biol. 2000;302:649–669. doi: 10.1006/jmbi.2000.4078. [DOI] [PubMed] [Google Scholar]
- 42.Palaninathan SK. Nearly 200 X-ray crystal structures of transthyretin: what do they tell us about this protein and the design of drugs for TTR amyloidoses? Curr Med Chem. 2012;19:2324–2342. doi: 10.2174/092986712800269335. [DOI] [PubMed] [Google Scholar]
- 43.Trivella DB, Bleicher L, de Palmieri LC, Wiggers HJ, Montanari CA, Kelly JW, Lima LM, Foguel D, Polikarpov I. Conformational differences between the wild type and V30M mutant transthyretin modulate its binding to genistein: implications to tetramer stability and ligand-binding. J Struct Biol. 2010;170:522–531. doi: 10.1016/j.jsb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Groenning M, Campos RI, Hirschberg D, Hammarstrom P, Vestergaard B. Considerably Unfolded Transthyretin Monomers Preceed and Exchange with Dynamically Structured Amyloid Protofibrils. Sci Rep. 2015;5:11443. doi: 10.1038/srep11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saelices L, Johnson LM, Liang WY, Sawaya MR, Cascio D, Ruchala P, Whitelegge J, Jiang L, Riek R, Eisenberg DS. Uncovering the Mechanism of Aggregation of Human Transthyretin. J Biol Chem. 2015;290:28932–28943. doi: 10.1074/jbc.M115.659912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.