Abstract
Various micro-engineered tools or platforms have been developed recently for cell mechanics studies based on acoustic, magnetic, and optical actuations. Compared with other techniques for single cell manipulations, microfluidics has the advantages with simple working principles and device implementations. In this work, we develop a multi-layer microfluidic pipette aspiration device integrated with pneumatically actuated microfluidic control valves. This configuration enables decoupling of cell trapping and aspiration, and hence causes less mechanical perturbation on trapped single cells before aspiration. A high trapping efficiency is achieved by the microfluidic channel design based on fluid resistance model and deterministic microfluidics. Compared to conventional micropipette aspiration, the suction pressure applied on the aspirating cells is highly stable due to the viscous nature of low Reynolds number flow. As a proof-of-concept of this novel microfluidic technology, we built a microfluidic pipette aspiration device with 2 × 13 trapping arrays and used this device to measure the stiffness of a human breast cancer cell line, MDA-MB-231, through the observation of cell deformations during aspiration. As a comparison, we studied the effect of Taxol, a FDA-approved anticancer drug on single cancer cell stiffness. We found that cancer cells treated with Taxol were less deformable with a higher Young's modulus. The multi-layer microfluidic pipette aspiration device is a scalable technology for single cell mechanophenotyping studies and drug discovery applications.
INTRODUCTION
Cell stiffness and cell viscoelasticity have emerged as label-free biophysical markers for reporting physiological and pathological status of cells.1–3 For example, cancer cells have been shown to be more deformable than their healthy counterparts in both patient clinical samples and different cancer (e.g., breast, bladder, cervix, pancreas, and ovarian) cell lines.4–6 In addition, it is well recognized that cancer progression is accompanied by altered cytoskeletal architectures in cancer cells, causing them to be more deformable2 and, in part, facilitate their metastatic journey out of a tumor into the blood stream. Malaria-infected red blood cells (RBCs) or sickle cells tend to become stiffer than healthy RBCs.7 Depending on the physiological condition, many cells undergo morphological alterations with changes in cell deformability, which can bring insight into disease pathologies.
Study of single cell mechanics requires the development of specialized tools that match the size of biological cells, which is usually on the order of microns. Compared with other advanced micro-engineered platforms for cell mechanics studies8 based on optical,9 magnetic,10 and acoustic forces,11 microfluidic devices that rely on geometrical constrictions and fluid phenomena to exert hydrodynamic stress on single cells have the advantages of simplicity in implementations and high throughput.12 Through measuring cell deformation, the mechanical property of single cells can be extrapolated and quantified. Studies involving alteration of membrane tension and mechanotransduction through cytoskeletal network on gene expression can benefit from the development of novel microfluidic tools.
Microfluidic devices for cell deformability studies can be divided into non-contact and contact modes. In a non-contact approach, the geometry or the constriction of the microfluidic channel is manufactured to be slightly larger than the diameter of the biological cells. For example, Tse et al. utilize cross-flow to apply both compressive and shear forces in a deformability cytometry (DC) technique that enables rapid mechanophenotyping of single cells in suspension.13 Real-time DC also deforms cells purely by hydrodynamic interactions,14 and analytical derivation for the elastic deformation has been developed.15 Mechanical properties of a cell can be extracted from shape recovery rate after passing through narrow microfluidic channels.16 The non-contact mode has the advantages of very high throughput and less mechanical invasiveness to cells. However, in the regime of moderate Re number, as in inertial microfluidics, the inertial effect can cause bias in the measurement of mechanical properties which are intrinsically coupled with cell size.
In a contact approach, the geometry of the micro-constriction features is made to be comparable or smaller than the cell diameters. As such, a physical contact is imposed for the cells to deform. A microfluidic device with gap sizes about 3 μm was used to study mechanics of cell nucleus.17 Also, transit time of cells through microfluidic channels slightly smaller than the cell diameters has been shown to correlate with hematologic disease pathology.18 Cell stiffness and viscoelastic deformation characteristics can be leveraged to sort cells using periodic, diagonal ridges that compress and laterally translate cells.19
All the above microfluidic approaches for studying cell deformability are based on a flow through approach. This usually provides a one-time measurement of cell mechanical properties. Micropipette aspiration is the most classical approach to measure single cell mechanics, both for cells in suspension or attached cells.20,21 Analytical models based on continuum mechanics have been well developed to measure different mechanical properties of single cells for membrane tension, shear elastic modulus of the membrane, Young's modulus assuming the cell is a homogeneous solid, and viscosity of the cytoplasm of the cell.22,23
A novel approach combining micropipette aspiration with microfluidics and optical imaging was recently developed.24 Using a single syringe pump, the device performs parallel aspiration measurements on an array of autonomously trapped single cells. The microfluidic device works in the regime of low Re number, where the fluid flow is attached and viscous. A single meandering microfluidic channel with repetitive trapping structures based on a flow resistance deterministic model25 was implemented for trapping individual cells. In order to aspirate, volume flow rate was varied to generate pressure differences on trapped cells. A limitation in this approach is that cell trapping took several minutes and trapping and aspiration processes happened simultaneously. The cells would be aspirated immediately after they are trapped which is undesirable for attaining accurate mechanical measurements.
Our current microfluidic device has two significant improvements over the previous implementation. The main innovation of this generation is the incorporation of integrated microfluidic valves to control fluid passage and flow of cells along the main microfluidic channel using pressure-dependent deflection of a polydimethylsiloxane (PDMS) membrane as microvalves.26–28 This configuration allows decoupling of cell trapping and aspiration processes. In addition, the second improvement as a result of the microfluidic valves is that the time it takes for cell trapping is greatly reduced. As a proof-of-concept of this microfluidic device, we showed the effect of Taxol, an anti-cancer drug, on cell mechanical property.
DEVICE PRINCIPLES AND THEORETICAL MODELING
Theoretical modeling and device parameters
The microfluidic pipette aspiration system consists of 26 trapping cups. The initial inlet divides into two channels with 13 cups on each side of the channel (Figures 1(a), 1(c), and 1(d)). Having 26 trapping and aspiration cups allows for all trapping and aspiration events to be observed under a 20× magnification objective. The main microfluidic channel has a meandering structure that is connected by the square micropipettes (Figure 1(b)). The long main microfluidic channel creates flow resistance and pressure drop described by Fuerstman et al.29 We notice that this flow resistance model is arguably valid up to Re ∼1000, in which inertial effect starts to come into play. The pressure drop is modeled by the Darcy-Weisbach equation
| (1) |
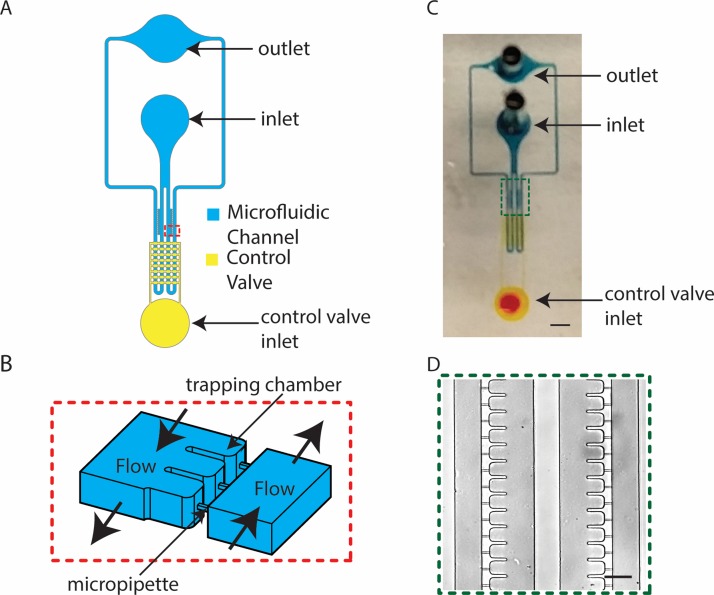
FIG. 1.
Overview of a microfluidic micropipette aspiration device. (a) Overall schematic of the multilayer device with main microfluidic channel (blue) and pneumatically actuated control valves (yellow). (b) Three-dimensional sketch of the zoomed-in red box in (a) showing trapping chambers and microchannels connecting the two main channels. (c) Photo of the device showing dye filled microchannels (scale bar is 2 mm). (d) Bright field image of the zoomed-in green box in (c) showing 2 × 13 trapping chambers (scale bar is 80 μm).
Here, C(α) is the laminar friction constant, which is a function of the cross section aspect ratio, α. α1 and α2 are defined by the smaller value of height (H)/width (W) or width/height, which are both equal to one for square cross sections for both the aspiration micropipette and the main microfluidic channels. Here, = 56.91 for = 1. ΔP is the change in pressure, and μ is the dynamic viscosity of the fluid, which in this case was assumed to be 0.001 Pa s at 20 °C for water. The subscripts under Q, L, W, and H denote the path in which 2 refers to the main channel (and 1 refers to the micropipette). After cells are trapped in the aspiration micropipette channels, takes the value of the volume flow rate set by the syringe pump by continuity of incompressible flow. Since the trapping cups are located along different lengths of the main channel, the change of pressure is summarized in Table I. A volume flow rate of 0–10 μl min−1 was used to generate pressure differences of several hundreds of Pascals across the aspiration micropipette channel. The cross section of an aspiration micropipette channel was designed to be 5 μm × 5 μm to robustly capture and aspirate cells into the chamber and yet allows small debris to flow through without clogging. The aspiration micropipette channel length was set to be 25 μm to provide sufficient length for the protrusion of cells during aspiration.
TABLE I.
Device parameters.
| Position | W1 (μm) | H1 (μm) | L1 (μm) | W2 (μm) | H2 (μm) | L2 (μm) | Q1/Q2 | ΔP (Pa) (Q = 1 μl/min) |
|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 25 | 80 | 40.9 | 4251 | 0.0121 | 114.4 |
| 2 | 5 | 5 | 25 | 80 | 40.9 | 4151 | 0.0118 | 111.7 |
| 3 | 5 | 5 | 25 | 80 | 40.9 | 4051 | 0.0115 | 109.0 |
| 4 | 5 | 5 | 25 | 80 | 40.9 | 3951 | 0.0112 | 106.3 |
| 5 | 5 | 5 | 25 | 80 | 40.9 | 3851 | 0.0109 | 103.7 |
| 6 | 5 | 5 | 25 | 80 | 40.9 | 3751 | 0.0106 | 101.0 |
| 7 | 5 | 5 | 25 | 80 | 40.9 | 3651 | 0.0103 | 98.2 |
| 8 | 5 | 5 | 25 | 80 | 40.9 | 3551 | 0.01 | 95.6 |
| 9 | 5 | 5 | 25 | 80 | 40.9 | 3451 | 0.0097 | 92.9 |
| 10 | 5 | 5 | 25 | 80 | 40.9 | 3351 | 0.0094 | 90.2 |
| 11 | 5 | 5 | 25 | 80 | 40.9 | 3251 | 0.0091 | 87.5 |
| 12 | 5 | 5 | 25 | 80 | 40.9 | 3151 | 0.0088 | 84.8 |
| 13 | 5 | 5 | 25 | 80 | 40.9 | 3051 | 0.0085 | 82.1 |
Conceptual design
The micropipette aspiration device works in the low Reynolds number regime. The operational principle stems from the least flow resistance theory. According to the least flow resistance path principle, the device was designed to guarantee that volume flow rate Q1 is higher than Q2. In other words, Q1/Q2 (denoted as Q ratio below, Equation (2)) should exceed 1. In this way, the flow resistance of path 1 should be less than that of path 2 such that the cell solution can be directed to the trapping cups. Although path 1 includes a much narrower constriction that increases the flow resistance significantly, the long length of path 2 compensates by increasing the flow resistance in path 2
| (2) |
Given the low velocity in typical microfluidic flows, the characteristic Stokes number of cells flowing across the trapping chamber is on the order of 0.001. Since the Stokes number is the ratio of the particle response time to the characteristic response time of the fluid, the cells will flow along the streamline of the velocity flow field and the cells' inertia is negligible. The operational control of the microfluidic device is straightforward and requires a pressure source for actuating pneumatically controlled valves and a syringe pump for aspiration (Figure 2(a)). The operation of the multi-layer microfluidic device can be divided into two parts: (1) trapping and (2) aspiration (Figure 2(b)). In the previous version of the microfluidic device, without the implementation of the microfluidic valves, trapping and aspiration are intrinsically coupled to each other.24 In the new design, the microfluidic device is composed of a control layer with an array of valves integrated with a deformable fluidic layer underneath. During cell trapping, pressure is applied to the control layer that blocks the fluidic layer, which increases the fluid flow resistance leading to an increase in Q ratio to increase trapping efficiency. Once the cells are trapped, the microfluidic valves are released and aspiration into the square micropipettes can begin using fluid flow.
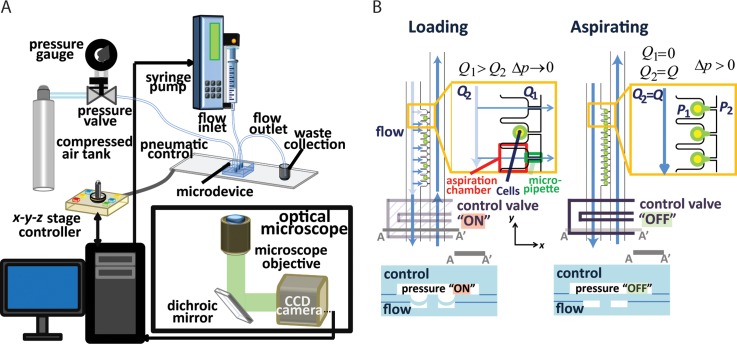
FIG. 2.
Operation principle of the microfluidic micropipette aspiration device. (a) Experimental setup for device operation. (b) Schematic of trapping (left) and aspiration (right) of cells using the control valves. When the control valves are on (depressed PDMS membrane), the pressure restricts the main microfluidic channel and increases flow into the micropipette channels (Q1 > Q2). When all the cells are trapped, the control valves are turned off to generate pressure differences across the micropipettes to begin cell aspiration.
MATERIALS AND METHODS
Device fabrication
The microfluidic micropipette aspiration device was fabricated using a polydimethylsiloxane (PDMS) multi-layer soft-lithography technique.30 The detailed process flow is described in Figure 3. The microfluidic device consists of a control substrate and a deformable microfluidic flow layer, which were aligned and bonded permanently on a PDMS-coated microscopic glass slide. Three transparency photomasks were produced by CAD/Art Services, Inc. (Bandon, OR, USA). These transparency photomasks were used to fabricate the silicon molds using photolithography. The first one defined the patterns of the microfluidic control layer of the integrated microvalve array. The second one defined the main microfluidic channel and the trapping structures for both the top-flow and bottom-flow layers. The third photomask defined patterns of the micropipette channel array. The mask layouts were designed in AutoCAD. Three silicon molds were patterned with SU-8 photoresist for PDMS casting for both the control and flow-bottom layers and PDMS spin-coating for the top-flow layers following the standard SU-8 patterning protocol available online (http://www.microchem.com/Prod-SU82000.htm). Four photolithography processes (two in step 1, step 2, and step 3) were administered for three silicon molds (Flow: MID + TOP, Flow: BOTTOM, and CONTROL) under a contact aligner (Karl Suss, MJB45). The thickness of SU-8 patterns of three silicon molds was measured with a profilometer (Veeco).
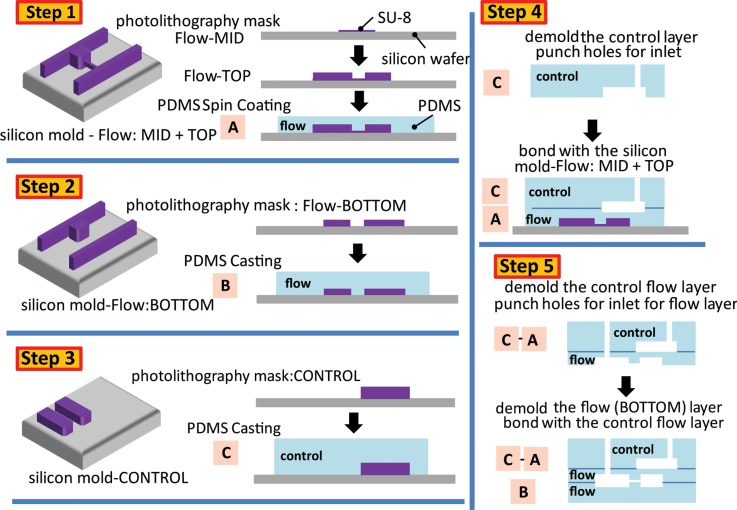
FIG. 3.
Device fabrication steps. Step 1—5 μm SU-8 was spin coated on a silicon wafer to develop the micropipette channels for the Flow-MID layer. Then, a second layer of SU-8 was spin coated to generate the Flow-TOP layer on top. After fabricating the mold, approximately 50 μm of PDMS was spin coated to generate block A. Step 2—The Flow-BOTTOM layer was generated after spin coating 17 μm of SU-8 on a silicon wafer. Once the mold was created, PDMS was poured on, cured, and demolded to form block B. Step 3—Another photomask was used to generate the control layer. After the mold was created, PDMS was poured on, cured, and demolded to form block C. Step 4—Block C was bonded to block A. Step 5—The combined blocks A and C was demolded from the silicon mold and hole punched for the inlet of the flow layer. Then the combined blocks A and C was aligned and bonded to block B. Refer to the main text for more detailed description.
The silicon mold of the top-flow layer involves two-layer of SU-8 patterning by wafer-level alignment (step 1). The first patterned layer defined the patterns of the micropipette channel array. While the second layer defined the trapping structures and the main microfluidic channel. For patterning the first layer of the top substrate, SU-8 2005 was spin coated on a silicon wafer at 1900 rpm for 90 s, having a thickness of 5 μm. One high-resolution chrome photomask (Photo Sciences Inc., Torrance, CA, USA), which defines the micropipette channel pattern for the Flow-MID layer, was purchased and the mold was generated using a contact photolithography machine (Karl Suss MA 45S). After patterning and development of the first SU-8 layer, the silicon wafer was hard-baked at 150 °C for 30 min to ensure that the SU-8 pattern is fully cured before the application of the second layer. For the second layer, SU-8 2025 was applied with a spinning speed of 2200 rpm for 90 s and gave a total thickness of 23.9 μm. The silicon mold was silanized with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (Sigma-Aldrich) in a desiccator before approximately 50 μm thick PDMS (Sylgard-184, Dow Corning) layer with a 20:1 monomer to curing agent ratio was spin coated on the flow layer Si mold at 1700 rpm for 1 min, degassed, and baked at 60 °C for 2 h to form block A.
We designed the transparency photomask for the main microfluidic channels such that Flow-TOP and Flow-BOTTOM form mirror image to each other. This fabrication method allowed us to manufacture symmetric features and reduced the number of transparency photomasks needed. For the bottom flow layer, a single layer of SU-8 2010 was patterned with a thickness of 17 μm under a spinning speed of 1400 rpm, and this layer defines trapping structures and the main microfluidic channel (step 2). The silicon mold was silanized, and PDMS with a mixing ratio of 10:1 (base: curing agent) was added to the wafers, degassed, cured at 60 °C overnight, and demolded to form block B.
The control layer PDMS substrate was molded by SU-8 patterned silicon wafer (step 3). SU-8 2025 was spin coated on the silicon wafer at 2000 rpm for 90 s, which gave the thickness of 25 μm. A 5 mm thick PDMS is made from the CONTROL mold following the standard procedures discussed above to form block C. Dices cut from the control PDMS layer were cleaned and treated with air plasma (Femto Science, Gyeonggi-Do, Korea). 1 mm diameter holes were punched on block C, which was then aligned and bonded under a customized alignment platform under an optical microscope (step 4). After baking at 60 °C for overnight to complete irreversible bonding, the PDMS stack A-C was peeled off from the Flow MID + TOP mold and hole-punched. The PDMS stack was treated with air plasma before bonded to block B (step 5). The final microfluidic chip was sealed using PDMS with 10:1 monomer to curing agent ratio and baked at 60 °C overnight to ensure complete PDMS polymerization and strengthened bonding between different PDMS layers.
Integrated microfluidic valves control and fluid control
The microfluidic valve array was connected to a pressure gauge (IMI Norgren, 0–60 psi) then to a compressed air source. For visualization, a solution of de-ionized (DI) water laden with 20 nm fluorescence microspheres (Invitrogen, 1:1000 dilution) was injected to the microfluidic flow channel. The volume flow rate was solely determined by the syringe pump. A 1 ml small syringe with a diameter of 4.7 mm was used to minimize the effect of peristalsis during liquid pumping. The syringe pump (Chemyx Fusion 400) exerts a linear force of 50 lb and has a step resolution of 0.016 μm.
Visualization of flow streamlines
1 μm Y-G microspheres (Invitrogen) were reconstituted in DI water. The PDMS microfluidic device was perfused with DI water to eliminate any trapped air bubbles before use. Fluorescent microspheres were flown into the device at 1 μl min−1 with 200 ms exposure time to generate streamlines to show the trajectory of the traveling fluid. Flow simulation was performed using COMSOL 4.4 (COMSOL Multiphysics), using an identical model and the conditions described previously.24
Human breast cancer cells culture, preparation, and treatment with Taxol
MDA-MB-231 cells were cultured and maintained in growth media RPMI1640 with 2 mM L-glutamine (Gibco) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 50 units/ml streptomycin (Invitrogen) 2.5 μg/ml Fungizone (Invitrogen), 5 μg/ml gentamicin (Invitrogen) at 5% CO2 and 37 °C until about 70% confluency. 0.05% trypsin-EDTA in phosphate buffered saline was used to detach cells for preparing cell suspension. Cells were suspended at 1.0 × 106 cells/ml to minimize cell clumping and possible pressure fluctuation during the experiment. For experiments involving Taxol, MDA-MB-231 cells were cultured with 10 μM of Taxol in the growth media for 22 h. Vybrant DiD cell-labeling solution (Thermo Fisher) that fluorescently labels cell membrane was added at 1:200 for 20 min to Taxol-treated and non-treated cells suspended in medium at a density of approximately 1 × 106 cells /ml to enhance cell visualization. The cells were washed three times with growth medium.
Imaging set up, analysis, and statistical analysis
The microfluidic device was set up on an optical microscope (Nikon, Ti Eclipse) for time lapse imaging under a 20× objective. The field of view was approximately 0.5 mm, which allowed us to visualize all 26 chambers without having to move the stage. The images were captured at 5-s intervals with a 50 ms exposure time using 647 nm laser to excite Vybrant DiD-labeled cell membrane. A custom written Matlab program was developed to align frames and correct for image drift during image acquisition. Protrusion lengths were measured manually using ImageJ (http://imagej.nih.gov/ij/), and Young's modulus was computed. Statistical analysis was performed in Matlab using independent two-sample t-test.
EXRERIMENTAL RESULTS AND DISCUSSIONS
Actuation of control valve facilitates cell trapping
We first characterized and validated the use of control valves for blocking flow in the main microfluidic channel. With fluorescent beads flowing through the main channel, the valves were depressed at 10 psi increment from 0 to 30 psi, and the corresponding bright field and fluorescence images are shown in Figure 4(a). It is clearly evident that as the pressure increased, the fluorescence level under the valve decreased (to about 30% of the original value), indicating that flow was obstructed (but not completely blocked off). This reduction in flow was reversible when the pressure was released. As a rough estimate, over 50% of the microfluidic device fabricated can sustain 30 psi of applied pressure without air bubble generation and PDMS device de-laminations. We noticed that given the variability of the microfabrication process, some of the microfluidic valves will only be completely closed at 50 psi of pressure.
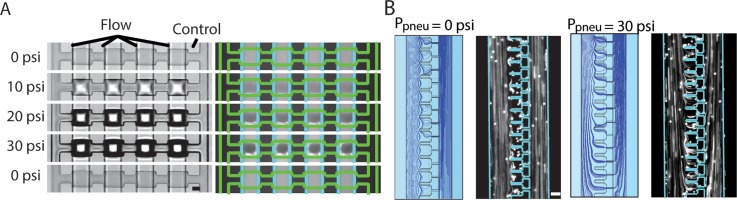
FIG. 4.
Experimental and modeling validation on the effectiveness of the control valves. (a) Images of fluorescent microspheres in the main channel with microfluidic control valve actuation from 0 to 30 psi and then back to 0 psi (scale bar is 40 μm). (b) Images and COMSOL simulation of fluid streamlines through the trapping chambers and the micropipette channel when the control valves are at 0 psi and 30 psi (scale bar is 40 μm).
When the integrated microfluidic valves were off at 0 psi, the streamlines only passed through the main microfluidic channels, as shown by both our flow simulation and experimental validation (Figure 4(b)). However, at 30 psi, the streamlines were redirected through the trapping structures. This redirection results from the significant increase in flow resistance in the main channel that effectively increases the Q ratio. Due to the low Stokes number, the suspended fluorescent particles closely followed the streamlines of the COMSOL simulations by visual comparison.
During cell loading, the syringe pump was set at a flow rate of 0.1 μl min−1 to minimize cell perturbation. With the integrated control valves, the efficiency of cell trapping improved dramatically in which 2 × 13 trapping cups were completely occupied within tens of seconds (Video S1, supplementary material) compared to minutes from the previously reported device.24 We ensured a 100% trapping efficiency, which also included conditions where large cell debris or cell clusters were trapped in the micropipette channel so that there was no pressure leakage through the micropipette channels during an experiment. In addition to more efficient cell trapping, the use of the integrated valves effectively decoupled cell trapping from aspiration since with flow in the main channel blocked, there is no pressure difference across the micropipette channel. We did not observe significant cell deformation during cell loading once a cell was trapped. During our experiments, we monitored cell trapping in real time and released the pressure as soon as all the chambers are filled up to prevent any pressure buildup. Together, these results demonstrate a significant improvement in the development of microfluidic micropipette aspiration.
Microfluidic micropipette aspiration revealed an increase in cell stiffness in Taxol-treated cells
An immediate application of our microfluidic pipette aspiration device is to quantify cell mechanical property. We examined the effect of anti-cancer drug Taxol on cell stiffness as a demonstration of our microfluidic device. Taxol is an anti-mitotic chemotherapeutic agent that stabilizes microtubules against depolymerization.31 Taxol is given intravenously as a combination therapy after surgery to reduce cancer relapse, and this treatment is accompanied by a host of side effects. This is presumably due to targeting cytoskeleton dynamics that is ubiquitous to both cancer and normal cells.
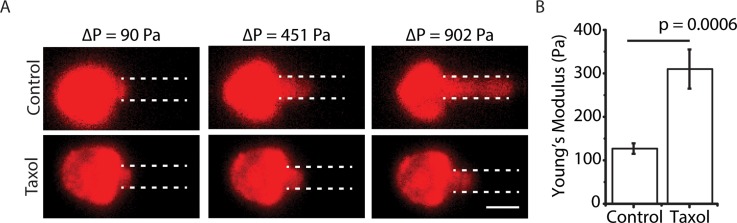
We aspirated breast cancer MDA-MB-231 cells with and without Taxol treatment in our microfluidic aspiration device. Once cells were trapped in the chambers, aspiration began with the volume flow rate of 1 μl min−1. Aspiration pressure can be easily controlled by changing the fluid flow rate.24 We increased the flow rate with increments of 1 μl min−1 for every 30 s, which allowed the protrusion lengths to come to equilibrium, until the volume flow rate reached 10 μl min−1. The maximum working threshold of 10 μl min−1 did not cause any leakage problem with the device, and the device was able to be reused for future experiments after flowing deionized water in reverse direction to clean out the cells and debris. The cells trapped in chambers were isolated from shear flow of the main microfluidic channel,24 and we did not observe shear-induced rotation or deformation of the cells. To facilitate visualization of protrusion length, cells were stained with Vybrant DiD, which labels cell membrane. With increasing aspiration pressure, cells were aspirated into the microchannel and we clearly observed a longer cell protrusion for control cells compared to Taxol-treated cells (Figure 5(a)), indicating a difference in mechanical properties. With Taxol treatment, we noticed some cells blebbed and presumably were undergoing apoptosis. These cells were excluded from further characterization.
FIG. 5.
Taxol-treated cells are stiffer. (a) Fluorescence images of DiD labeled (pseudo-colored) MDA-MB-231 cells treated with and without Taxol at various pressure differences (scale bar is 20 μm). (b) Young's modulus (mean ± standard error) of control cells (N = 20) and Taxol-treated cells (N = 30). p ∼ 0.0006 for Student's t-test.
As a versatile technique, micropipette aspiration measurements can inform both cortical tension and Young's modulus of a cell. The mechanical behavior of cells can be modeled as a drop of liquid encapsulated by a membrane or a piece of elastic solid. Here, we adopt an elastic solid model from Theret et al.,32 which relates Young's modulus to pressure and several geometric parameters
| (3) |
where E is the Young's modulus, and Φ is a geometric micropipette constant, which takes the value of 2.1. is the protrusion length and is the hydraulic radius of the micropipette. Since the cross section of the aspiration micropipette is a square, the hydraulic radius is equivalent to half of the side length. The square cross section minimizes pressure leakage by ensuring that the aspirated cells fully enclose the entrance of the micropipette. The pressure difference across the trapped cell was calculated from Eq. (2), which assumed a straight channel. (The bend in the channel is negligible.) Young's moduli of MDA-MB-231 cells treated with and without Taxol were calculated based on Eq. (3) to be 310 ± 45 Pa and 127 ± 12 Pa, respectively (Figures 5(b) and S3 in the supplementary material). The increase in cell stiffness with Taxol treatment is consistent with the previous work.17 The larger variability in cell stiffness with Taxol treatment indicates heterogeneity in cells' response to Taxol. Because our measurement is based on single cells, this offers the possibility to study chemoresistance using mechanical heterogeneity as a readout.
The Young's modulus of MDA-MB-231 was lower than the previously reported measurement,24 which could be due to the use of fluorescent membrane dye and/or due to changes to cell culture conditions. We believe the latter is the case. To test this, we performed conventional pipette aspiration experiments with MDA-MB-231 cells with and without the Vybrant DiD treatment. Young's moduli were calculated to be 124 ± 15 Pa and 138 ± 10 Pa with and without Vybrant DiD, respectively (Figure S1, supplementary material). These results closely match the Young's modulus obtained from the microfluidic device and also showed Vybrant DiD has a minimal effect on cell stiffness. We speculate that different cell culturing conditions (i.e., source of FBS in media) may have a direct impact on cell mechanical properties.
Improvement and future work
The microfluidic device used in this work for cell aspiration is significantly improved over conventional micropipette aspiration. The autonomous cell trapping eliminates the manual search of cells and is able to perform measurements in parallel. One of the major innovations of this work is the incorporation of PDMS microfluidic control valves that deflect under pressure actuation from a compressed air source to block the main channel. Implementation of the microfluidic control valves allowed efficient trapping in all 26 chambers on the order of seconds and effectively decoupled cell trapping from aspiration. Measurements of cell mechanical property in our current device take minutes. We envision that our microfluidic pipette aspiration device would increase the number of data points per experiment, thereby allowing more experimental conditions to be examined. Improvement of automated image processing and analysis would provide ease of use for others.
By using different flow rate ramp (including stopping flow), the device, in principle, can be expanded to measure viscoelasticity of cells. It will be interesting to study the relaxation of cell deformation from pressure removal. This is akin to serial constriction device that has been used to study cell transit times and cell relaxation in cell mechanics studies.17 The effects of Taxol and other anti-cancer drugs on cell viscoelasticity are not entirely understood. Our microfluidic micropipette aspiration device has the potential to provide an alternative to cell migration assays that can greatly reduce the time from hours to several minutes. It could also aid the development of novel strategies in cancer diagnosis, prognosis, drug discovery, and treatment.33
Finally, our main microchannel dimensions are sufficiently different compared to the previous design, which allowed us to access a much lower range of aspiration pressure that was previously not possible. This improvement brings the possibility of mechanical measurement of other types of biological samples. Compared to single cells, lipid bilayer vesicles are more fragile and have a lytic tension of ∼3–10 mN/m.34 Using giant unilamellar vesicles (GUVs) formed by electroformation as a test case, our device could stably trap and aspirate on GUVs (Figure S2, supplementary material). However, GUV solution has a lot of lipid debris that easily clogged the microchannels. Additional development is needed to further explore this possibility. Nonetheless, this preliminary demonstration shows that the current device reported could be used for versatile applications.
CONCLUSION
Compared with other advanced measurement techniques in cell studies such as atomic force microscopy or traditional micropipette aspiration system, the improved microfluidics system for micropipette aspiration provides a simple and direct approach to measure mechanical properties of the cell. We reported a multilayer microfluidic device fabricated by PDMS soft-lithography. As a proof-of-concept, we measured the Young's moduli of metastatic breast cancer (MDA-MB-231) wild type cells and MDA-MB-231 cells treated with Taxol, and observed increased cell stiffness in Taxol-treated cells. Compared to the previous device,24 this device decouples cell trapping from cell aspiration. In addition, the use of control layer valves significantly decreases the time it requires to trap cells. We believe that this microfluidic tool can facilitate more rapid drug screening for cancer therapy based on cell mechanics measurements. Overall, our microfluidic micropipette aspiration device presented here has numerous potential applications for mechanobiology and cell mechanics research.
SUPPLEMENTARY MATERIAL
See supplementary material for the following: Supplementary Figure 1. Young's moduli of MDA-MB-231 cells with and without Vybrant DiD using conventional micropipette aspiration. Supplementary Figure 2. Aspiration of electroformed GUVs. Supplementary Figure 3. Histogram of Young's moduli of MDA-MB-231 cells. Video S1. Movie of MDA-MB-231 cells being captured with the control valves turned on.
ACKNOWLEDGMENTS
This work was supported by the NIH Director's New Innovator Award (DP2 HL117748-01). J.W.L. was supported by the NIH's Microfluidics in the Biomedical Sciences Training Program: NIH NIBIB T32 EB005582. Danielle Chase thanks the NINN REU program. We acknowledge the technical support from Lurie Nanofabrication Center (LNF) at the University of Michigan. We thank Zhou Yuan and Professor Cheng Zhu from Georgia Institute of Technology for providing us micropipettes and Megan White for her assistance with the pipette aspiration experiments.
References
- 1. Ekpenyong A. E., Whyte G., Chalut K., Pagliara S., Lautenschlager F., Fiddler C., Paschke S., Keyser U. F., Chilvers E. R., and Guck J., PloS One (9), e45237 (2012). 10.1371/journal.pone.0045237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suresh S., Acta Biomater. (4), 413–438 (2007). 10.1016/j.actbio.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lautenschlager F., Paschke S., Schinkinger S., Bruel A., Beil M., and Guck J., Proc. Natl. Acad. Sci. U. S. A. (37), 15696–15701 (2009). 10.1073/pnas.0811261106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swaminathan V., Mythreye K., O'Brien E. T., Berchuck A., Blobe G. C., and Superfine R., Cancer Res. (15), 5075–5080 (2011). 10.1158/0008-5472.CAN-11-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu W., Mezencev R., Kim B., Wang L., McDonald J., and Sulchek T., PloS One (10), e46609 (2012). 10.1371/journal.pone.0046609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katira P., Zaman M. H., and Bonnecaze R. T., Phys. Rev. Lett. (2), 028103 (2012). 10.1103/PhysRevLett.108.028103 [DOI] [PubMed] [Google Scholar]
- 7. Suresh S., Spatz J., Mills J. P., Micoulet A., Dao M., Lim C. T., Beil M., and Seufferlein T., Acta Biomater. (1), 15–30 (2005). 10.1016/j.actbio.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 8. Bao G. and Suresh S., Nat. Mater. (11), 715–725 (2003). 10.1038/nmat1001 [DOI] [PubMed] [Google Scholar]
- 9. Zhang H. and Liu K. K., J. R. Soc. Interface (24), 671–690 (2008). 10.1098/rsif.2008.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang N. and Ingber D. E., Biochem. Cell Biol. (7–8), 327–335 (1995). 10.1139/o95-041 [DOI] [PubMed] [Google Scholar]
- 11. Heureaux J., Chen D., Murray V. L., Deng C. X., and Liu A. P., Cell. Mol. Bioeng. (3), 307–319 (2014). 10.1007/s12195-014-0337-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho K. K. Y., Lee L. M., and Liu A. P., Sci. Rep. , 32912 (2016). 10.1038/srep32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin H. and Marshall D., Curr. Opin. Biotechnol. (1), 110–119 (2012). 10.1016/j.copbio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 14. Tse H. T., Gossett D. R., Moon Y. S., Masaeli M., Sohsman M., Ying Y., Mislick K., Adams R. P., Rao J., and Di Carlo D., Sci. Transl. Med. (212), 212ra163 (2013). 10.1126/scitranslmed.3006559 [DOI] [PubMed] [Google Scholar]
- 15. Otto O., Rosendahl P., Mietke A., Golfier S., Herold C., Klaue D., Girardo S., Pagliara S., Ekpenyong A., Jacobi A., Wobus M., Topfner N., Keyser U. F., Mansfeld J., Fischer-Friedrich E., and Guck J., Nat. Methods (3), 199–202, 194, 202 (2015). 10.1038/nmeth.3281 [DOI] [PubMed] [Google Scholar]
- 16. Mietke A., Otto O., Girardo S., Rosendahl P., Taubenberger A., Golfier S., Ulbricht E., Aland S., Guck J., and Fischer-Friedrich E., Biophys. J. (10), 2023–2036 (2015). 10.1016/j.bpj.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng Y., Chen J., Cui T., Shehata N., Wang C., and Sun Y., Lab Chip (3), 577–583 (2014). 10.1039/C3LC51151K [DOI] [PubMed] [Google Scholar]
- 18. Mak M., Reinhart-King C. A., and Erickson D., Lab Chip (3), 340–348 (2013). 10.1039/C2LC41117B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rosenbluth M. J., Lam W. A., and Fletcher D. A., Lab Chip (7), 1062–1070 (2008). 10.1039/b802931h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang G., Crawford K., Turbyfield C., Lam W., Alexeev A., and Sulchek T., Lab Chip (2), 532–540 (2015). 10.1039/C4LC01150C [DOI] [PubMed] [Google Scholar]
- 21. Zhu C., Bao G., and Wang N., Annu. Rev. Biomed. Eng. , 189–226 (2000). 10.1146/annurev.bioeng.2.1.189 [DOI] [PubMed] [Google Scholar]
- 22. Tan X., Heureaux J., and Liu A. P., Integr. Biol. (9), 1033–1043 (2015). 10.1039/C5IB00111K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hochmuth R. M., J. Biomech. (1), 15–22 (2000). 10.1016/S0021-9290(99)00175-X [DOI] [PubMed] [Google Scholar]
- 24. Lee L. M. and Liu A. P., J. Nanotechnol. Eng. Med. (4), 040902 (2014). 10.1115/1.4029936 [DOI] [Google Scholar]
- 25. Lee L. M. and Liu A. P., Lab Chip (1), 264–273 (2015). 10.1039/C4LC01218F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan W. H. and Takeuchi S., Proc. Natl. Acad. Sci. U. S. A. (4), 1146–1151 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park W., Han S., and Kwon S., Lab Chip (20), 2814–2817 (2010). 10.1039/c005173j [DOI] [PubMed] [Google Scholar]
- 28. Doh I. and Cho Y. H., Lab Chip (14), 2070–2075 (2009). 10.1039/b821524c [DOI] [PubMed] [Google Scholar]
- 29. Fuerstman M. J., Lai A., Thurlow M. E., Shevkoplyas S. S., Stone H. A., and Whitesides G. M., Lab Chip (11), 1479–1489 (2007). 10.1039/b706549c [DOI] [PubMed] [Google Scholar]
- 30. Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science (5463), 113–116 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 31. Arnal I. and Wade R. H., Curr. Biol. (8), 900–908 (1995). 10.1016/S0960-9822(95)00180-1 [DOI] [PubMed] [Google Scholar]
- 32. Theret D. P., Levesque M. J., Sato M., Nerem R. M., and Wheeler L. T., J. Biomech. Eng. (3), 190–199 (1988). 10.1115/1.3108430 [DOI] [PubMed] [Google Scholar]
- 33. Kramer N., Walzl A., Unger C., Rosner M., Krupitza G., Hengstschlager M., and Dolznig H., Mutat. Res. (1), 10–24 (2013). 10.1016/j.mrrev.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 34. Rawicz W., Olbrich K. C., McIntosh T., Needham D., and Evans E., Biophys. J. (1), 328–339 (2000). 10.1016/S0006-3495(00)76295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the following: Supplementary Figure 1. Young's moduli of MDA-MB-231 cells with and without Vybrant DiD using conventional micropipette aspiration. Supplementary Figure 2. Aspiration of electroformed GUVs. Supplementary Figure 3. Histogram of Young's moduli of MDA-MB-231 cells. Video S1. Movie of MDA-MB-231 cells being captured with the control valves turned on.