Abstract
Adenosine deaminases that act on RNA (ADARs) convert adenosine residues to inosine in double-stranded RNA. In vivo, ADAR1 is essential for the maintenance of hematopoietic stem/progenitors. Whether other hematopoietic cell types also require ADAR1 has not been assessed. Using erythroid- and myeloid-restricted deletion of Adar1, we demonstrate that ADAR1 is dispensable for myelopoiesis but is essential for normal erythropoiesis. Adar1-deficient erythroid cells display a profound activation of innate immune signaling and high levels of cell death. No changes in microRNA levels were found in ADAR1-deficient erythroid cells. Using an editing-deficient allele, we demonstrate that RNA editing is the essential function of ADAR1 during erythropoiesis. Mapping of adenosine-to-inosine editing in purified erythroid cells identified clusters of hyperedited adenosines located in long 3’-untranslated regions of erythroid-specific transcripts and these are ADAR1-specific editing events. ADAR1-mediated RNA editing is essential for normal erythropoiesis.
Hematopoiesis describes the process of blood cell formation from hematopoietic stem cells (HSCs). It has proven a highly tractable system for understanding the role of proteins/genes in the maintenance of an organ system and in lineage commitment in vivo. There is now a detailed understanding of the roles of a large number of transcription factors, epigenetic regulators, and signaling cascades in hematopoiesis; however, one mechanism of gene regulation that has not been explored fully are the requirements and consequences of direct modification of RNA. It is now becoming apparent that RNA sequences can be modified widely by RNA editing.
The deamination of genomically encoded adenosine to inosine (A-to-I editing) in double-stranded RNA (dsRNA) by adenosine deaminases that act on RNA (ADAR) enzymes is a highly abundant form of RNA base modification in higher eukaryotes. Hundreds of thousands to millions of A-to-I editing sites have been reported in the human transcriptome [1–3]. In the mouse, only tens of thousands of editing events are reported [2,4]. A-to-I editing most frequently occurs within repetitive elements such as short interspersed nuclear element (SINE; nonprimate mammals), including Alu repeats (primate specific) [1,5,6]. RNA editing within retroelements occurs when proximal paired inverted complementary repeats hybridize to form dsRNA hairpins after transcription [7,8]. Long dsRNAs with few or no mismatches can be subjected to hyperediting, in which up to 50% of the adenosines are deaminated in a single transcript [7,9]. Despite the prevalence of A-to-I editing and the rapid advances in the appreciation of the extent of editing, our knowledge of the in vivo cell types and physiological requirements for ADARs is relatively limited [10–12].
Three ADAR proteins are present in mammals, ADAR (ADAR1), ADARB1 (ADAR2), and ADARB2 (ADAR3).
ADAR1 and ADAR2 have editing activity in vitro, but not ADAR3 [13]. ADAR2 and ADAR3 are highly expressed in the CNS, whereas ADAR1 is expressed broadly throughout mammalian cell types. Although they demonstrate a similar preference for editing sites [14], murine Adar1 and Adar2 knockouts have revealed striking differences in in vivo requirements. Adar2−/− mice die shortly after birth from seizures and the lack of editing of a single adenosine within the Gria2 transcript causes the phenotype of the Adar2−/− animals [15,16]. In contrast, no substrate(s) has been identified that accounts for the Adar1−/− phenotype.
Adar1 encodes two isoforms, a short, constitutive, nuclear-restricted ADAR1p110 and a longer interferon (IFN)-inducible ADAR1p150, which can be both nuclear and cytoplasmic [17]. The first mutant mouse model reported suggested a role for ADAR1 in embryonic erythropoiesis; however, this model reported a heterozygous phenotype that was likely an artifact of the targeting strategy because subsequent models did not confirm this phenotype [18]. Homozygous germline deletion of both Adar1 isoforms [10,19] or only the Adar1p150 isoform [20], resulted in lethality at approximately embryonic day 12 (E12), indicating an essential requirement for ADAR1, especially ADAR1p150, in development. In all of these models, heterozygous animals have been normal. A prominent feature of the Adar1 deficiency was failed fetal liver (FL) hematopoi-esis [10,19]. ADAR1 is dispensable for B-lymphopoiesis [21]. HSC-restricted deletion of Adar1 resulted in failed hematopoiesis and a profound up-regulation of IFN-stimulated gene (ISG) signatures [22,23]. These studies showed that ADAR1 is essential for the maintenance of both the fetal and adult HSC compartment in a cell-autonomous fashion and that it is a critical negative regulator of the IFN response. It is unclear why not all hematopoietic cell types respond to a loss of ADAR1 in the same manner, and there has not been a detailed assessment of the role of ADAR1 within more committed hematopoietic lineages [21–23].
Although A-to-I editing is presumed to be the primary function of ADAR1, recent reports have proposed editing-independent functions, including microRNA (miRNA) biogenesis and RISC loading, through direct protein–protein interactions with DICER1 [24]. Because it is plausible that different cell types may use different functions of ADAR1, we have assessed the role of ADAR1 systematically in the myeloid and erythroid lineages using in vivo lineage-restricted murine deletion models. These studies demonstrate a specific requirement for ADAR1 in erythropoiesis and show that the primary in vivo function of ADAR1 is A-to-I editing.
Materials and methods
Animals
All animal experiments were approved by the AEC (AEC#030/14; St. Vincent’s Hospital, Melbourne, Australia). Epor-Cre knock-in mice [25], Rosa26-enhanced yellow fluorescent protein (eYFP) reporter mice (strain #006148; The Jackson Laboratory [26]), LysM-Cre knock-in mice (strain #004781; The Jackson Laboratory [27]), Adar1+E861A [28] (point mutant knock-in of a Glu861 →Ala change E861A homologous to human E912A and referred to herein as Adar1E861A/+), Adar1+/− mice [10] (PGK-neocassette replacement of exons 2–13 referred to herein as Adar1+/−), and Adar1fl/fl mice [10,23] (loxP sites introduced flanking exons 7–9 referred to herein as Adar1fl/fl ) have been described previously. Ifnar−/− [29] and Ifngr−/− (strain #003288; The Jackson Laboratory) lines were generously provided by Prof. M. Smyth (Peter MacCallum Cancer Centre) on a C57BL/6 background. All lines were on a backcrossed C57BL/6 background. Congenic B6.SJL-PtprcaPep3b/BoyJArc mice were purchased form the Animal Resources Centre (Canning Vale, Western Australia).
Where indicated “–” refers to the germline-null allele (PGK-neocassette replacement of exons 2–13); “fl” refers to a loxP flanked, nondeleted allele (loxP sites introduced flanking exons 7–9); “ Δ “ refers to a recombination of the floxed allele, resulting in a deletion of exons 7–9; and “E861A” refers to the knock-in point mutation which is expressed in both ADAR1 p110 and p150 isoforms.
Embryo dissection and genotyping
Embryos were derived from timed matings, with noon of the vaginal plug day defined as E0.5, and carefully staged by somite counts in combination with morphological criteria. Embryos were genotyped by polymerase chain reaction (PCR) of Adar1 allele-specific oligonucleotide combinations (Supplementary Table E1, online only, available at www.exphem.org) of genomic DNA (gDNA) isolated from tail snips (ISOLATE II Genomic DNA Kit, Bioline or DNeasy Blood & Tissue Kit, Qiagen).
Flow cytometry analysis and fluorescent-activated cell sorting (FACS)
Peripheral blood (PB) was analyzed on a hematological analyzer (Sysmex KX-21N, Roche Diagnostics). Single-cell FL suspensions were prepared by passing through a 23-gauge needle. Bones were flushed, spleens crushed, and single-cell suspensions were prepared [30]. Antibodies against murine Ter119, CD71, B220, IgM, Mac-1, Gr1, F4/80, CD43, CD19, CD4, CD8, CD44, Sca-1, c-Kit, CD34, FLT3, FcγR (CD16/32), CD41, CD48, and CD51, either biotinylated or conjugated with FITC, phycoerythrin, phycoerythrin-Cy5, peridinin chlorophyll protein-Cy5.5, phycoerythrin-Cy7, allophycocyanin, or allophycocyanin eFluor780, were all obtained from eBioscience. CD105 and CD150 were from Bio-Legend. Biotinylated antibodies were detected with streptavidin conjugated with Brilliant Violet-605 (BioLegend) [26]. AnnexinV (BD) and 7-aminoactinomycin D (7-AAD, Molecular Probes) was used to assess viability as described previously [23]. Cells were analyzed on a BD LSRII Fortessa (BD Biosciences). Results were analyzed with FlowJo software version 10.0 (TreeStar).
For microarrays, miRNA and identification of novel editing sites analyses, CD71hiTer119− (R2), CD71hiTer119+ (R3), CD71medTer119+ (R4), CD71loTer119+ (R5), and YFP+ (all 7-AAD−) erythrocytes [31] were isolated from Epor-Adar1Δ/– and Epor-Adar1Δ/+ E14.5 FL. For microfluidics-based multiplex PCR and deep sequencing (mmPCR-seq) and qRT-PCR analyses, hematopoietic fractions were purified without lineage depletion according to the following immunophenotypes: CD3−CD4−CD5−CD8a−CD11b−Gr1−B220−Ter119− (lineage−) c-Kit+Sca1+ (LKS+) CD34−CD135−, long-term HSC (LT-HSC); LKS+CD34+CD135−, short-term HSC (ST-HSC); LKS+CD34+ CD135+, multipotent progenitor (MPP); lineage−c-Kit+Sca1−(LKS−) CD34−CD16/32−, megakaryocyte–erythroid progenitor (MEP); LKS−CD34+CD16/32−, common myeloid progenitor (CMP); LKS−CD34+CD16/32+, granulocyte–macrophage progenitor (GMP); LKS−CD150+CD41+, megakaryocyte progenitor (MkP); LKS−CD41−CD16/32−CD150−CD105−, Pre-GM; LKS−CD41−CD16/32−CD150+CD105−, pre-megakaryocyte-erythroid (Pre-Meg-E); LKS−CD41− CD16/32−CD150+CD105+, pre-colony-forming unit erythroid (Pre-CFU-E); LKS−CD41−CD16/ 32−CD150−CD105+, CFUE; CD11b+Gr1lo, Gr1lo; CD11b+Gr1hi, Gr1hi; CD71hiTer119−, R2; CD71hiTer119+, R3; CD71medTer119+, R4. CD71 loTer119+, R5 [31,32] were isolated from the bone marrow (BM) of independent wild-type C57B6/J mice. Cells were sorted with either a FACSAria or Influx cell sorter (BD Biosciences).
FL transplantation
A total of 1 × 10 unfractionated E14.5 FL cells (CD45.2) were competitively transplanted competitively with 2 × 105 wild-type BM cells (CD45.1/CD45.2) into lethally irradiated (5 Gy split dose 3 hours apart; total of 10 Gy) recipients (CD45.1) through intravenous injection. A total of four recipients per genotype were transplanted and the experiment was performed in duplicate. For quantification of serum erythropoietin (Epo) levels, ELISA was performed according to the manufacturer’s protocol on PB serum obtained from recipients at 16 weeks after transplantation (Quantikine Mouse Epo Kit, R&D Systems).
Differential gene expression analysis
Complementary DNA (cDNA) library preparations and Affymetrix Mouse Gene 1.0 ST Array were performed at the Ramaciotti Centre for Genomics, University of North South Wales. Data were RMA normalized in GenePattern using NormalizeAffimetrixST. Genes were considered differentially expressed if the false discovery rate was <0.1 and p < 0.05. Quantitative Set Analysis for Gene Expression (QuSAGE) was used to determine gene set enrichment [33] against the MSigDB collection [34]. The gene ontology (GO) term biological phase (GO:0044848) was used to determine enrichment of biological process from our array dataset. Datasets are deposited in the Gene Expression Omnibus (GEO Accession #GSE710417).
qRT-PCR
LT-HSC, ST-HSC, MPP, CMP, GMP, MEP, Pre-Meg-E, Pre-CFU-E, CFU-E, R2, R3, R4, and R5 hematopoietic fractions isolated by FACS from three independent 8- to 12-week-old C57BL/6 biological replicates. R2, R3, and R4 erythrocytes were isolated by FACS from three independent Epor-Adar1Δ/− and Epor-Adar1Δ/+ E14.5 FL. RNA was extracted as per the manufacturer’s instructions (RNeasy Mini Kit, Qiagen). For E14.5 Ifnαr−/− Ifnγr−/− Adar1+/+ and Ifnαr−/− ifnγr−/− Adar1−/− embryos, total RNA was extracted with TRIsure (Bioline) as per the manufacturer’s instructions from unfractionated FL. cDNA was synthesized using AffinityScript QPCR cDNA Synthesis Kit (Agilent Technologies). Real-time PCR was done in duplicate with Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies) and primers (5 µM) from IDT (Supplementary Table E1).
mmPCR-seq identification of A-to-I editing sites
MPP, CMP, MEP, Pre-Meg-E, Pre-CFU-E, CFU-E, R3, R4, R5, and B-lymphocyte hematopoietic fractions were isolated by FACS from three independent C57BL/6 biological replicates. RNA was isolated using TRIsure (Bioline) as per the manufacturer’s instructions. cDNA was synthesized using AffinityScript QPCR cDNA Synthesis Kit (Agilent Technologies). A total of 200 ng of cDNA was used for mmPCR-seq [35]. Briefly, 557 loci containing 11,103 known A-to-I editing sites were PCR amplified using multiplexed primer pools. PCR amplicons were used for high-throughput sequencing and A-to-I editing frequencies were determined as described by Zhang et al. [35].
RNA-seq, mapping, variant calling, and identification of A-to-I (G) edited sites
Viable committed erythrocytes (YFP+7-AAD−) were isolated by FACS from two independent Epor-Cre Rosa26eYFP E14.5 FL. Total RNA was extracted by TRIsure (Bioline) according to the manufacturer’s instructions. cDNA library preparations and RNA-seq were performed at the Ramaciotti Centre for Genomics (University of New South Wales). Datasets were deposited in GEO (Accession #GSE710417).
RNA-seq reads were aligned to the MM9/NCBIM37 reference genome with BWA [36]. A-to-I (G) editing sites were determined as follows: paired-end reads were mapped separately to the mm9 reference genome and exonic sequences encompassing known splicing junctions. Samtools rmdup [37] was used to remove identical reads (PCR duplicates). Unique reads were subjected to local realignment and base score recalibration using the Genome Analysis ToolKit (GATK) [38]. Subsequently, variants were called with the UnifiedGenotyper in GATK and reported at lenient call criteria (stand_call_conf 0, stand_emit_conf 0). Variants were removed from the resulting candidate list if they: (1) overlapped with known genomic variation [39]; (2) had a variant call quality < 20; or (3) were located in the first six bases of a read, within 4 bp of a splice junction, in homopolymer runs, or in simple repeats. The resulting list of variants was combined with editing sites discovered in previous studies and annotated using ANNO-VAR [40] based on Gencode, RefSeq, and UCSC gene models. The editing level at each of these sites was quantified using the samtools mpileup. Sites that were covered by >20 reads in the RNA-seq data of all Adar1+/+ and Adar1E861A/E861A triplicates were tested for differential editing using ANOVA and reported as significant for p < 0.1.
To find hyperedited A-to-I (G) sites, all reads were aligned to the MM9/NCBIM37 reference genome using BWA aln 0.6.2 (default parameters) and mem 0.7.4 (minimum seed length 50) [7]. Reads with potential sequencing errors, long stretches of simple repeats, or an average Phred quality <25 were removed. To be able to realign reads with a large number of mismatches caused by A-to-G editing sites, all “A’s” were transformed to “G’s” in both the remaining RNA reads and in the genome. Transformed reads were aligned to the transformed genome, again using BWA aln 0.6.2. The original read sequences that aligned (after the transformation) to multiple locations were treated separately by selecting the location with the largest fraction of A-to-G to all mismatches. Low-quality/mismapped reads having: (1) a too dense A-to-G mismatch cluster (length < 10% of the read length); (2) a cluster too close to the ends of the read (completely contained within either the first or last 20% of the read; or (3) a cluster with a particularly large percentage (> 60%) of a single nucleotide were discarded. Edited PE reads (which were treated throughout the pipeline as two separate single-end reads) also required that the mate read was mapped to a nearby region (within 500 kbp) and in an opposite orientation. Reads that passed all filters were finally designated as hyperedited.
RNA secondary structure prediction
Genomic sequences from Ensembl Genes 75, Mus musculus genes (GRCm38.p2) were used to predict RNA secondary structure of lnc-Tbccd1-IT1 using the mfold web server as described previously [41], RNAfold version 3.0 with default settings. lnc-Tbccd1-IT1 sequence was determined by region of Tbccd1 intron 1 with read coverage ≥5.
miRNA bead-based detection assay
Total RNA was extracted (TRIzol, Invitrogen) from Epor-Adar1Δ/− and Epor-Adar1Δ/+ E14.5 FL R3 and R5 erythrocytes. miRNA expression profiling was performed using the plate capture method [42,43]. Ninety-six-well PCR plates with an N-oxysuccinimide surface (DNA-BIND plates, Corning Costar) were coated at room temperature for 1 hour with a 5 µM mixture of 5′-amino-antisense oligonucleotides at 20 µL per well according to the manufacturer’s protocol. Coated plates were washed successively with 100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA and 10 mM Tris-HCl, pH 8.0, 1 mM EDTA. Total RNA (10 ng or higher) was diluted to 20 µL in 50 mM Tris-HCl, pH 8.0, 1 M NaCl, 1 mM EDTA, and 1 × RNAsecure (Am-bion) containing precontrol synthetic miRNA mixture at the ratio described previously [42]. miRNAs were captured in the coated wells by denaturing at 80° C for 5 minutes and gradually cooling to room temperature for 1.5 hours in a PCR machine, followed by three 2× SSC washes. 3′- and 5′-adaptor ligations were carried out as described previously [42] in 20 µL reaction volumes with four 2× SSC washes after each ligation. Ligated miRNA were denatured for 5 minutes at 80°C in 20 µL of water with 2 µM adaptor-specific RT primer, chilled on ice, and reverse transcribed as described previously [42]. RT products were denatured at 95°C for 5 minutes before two rounds of PCR amplification with previously described conditions [42] for 26 and 27 cycles, respectively, to incorporate biotin labels for low-input RNA profiling. Labeled miRNAs were hybridized to a bead-based detection platform [42]. Median fluorescence intensities were quantitated on a Luminex 100S machine.
Data were normalized as described previously [42] with modifications. Average readings from five water-only labeled samples were used for probe-specific background subtraction. Linear normalization among different bead sets for the same sample was performed using readings from two postcontrol probes with equal contribution. Sample normalization was subsequently performed assuming equal total fluorescence readings. To identify markers, all Epor-Adar1Δ/− samples were compared with all Epor-Adar1Δ/+ samples with a median-based t test.
Statistical analysis
For biological experiments, the significance of the results was analyzed using the unpaired two-tailed Student’s t test on the basis that highly inbred genetically identical mice have been used in all experiments unless otherwise stated. p < 0.05 was considered significant. All data are presented as mean ± SEM.
Results
Adar1 is highly expressed in mature granulocytes and throughout erythropoiesis
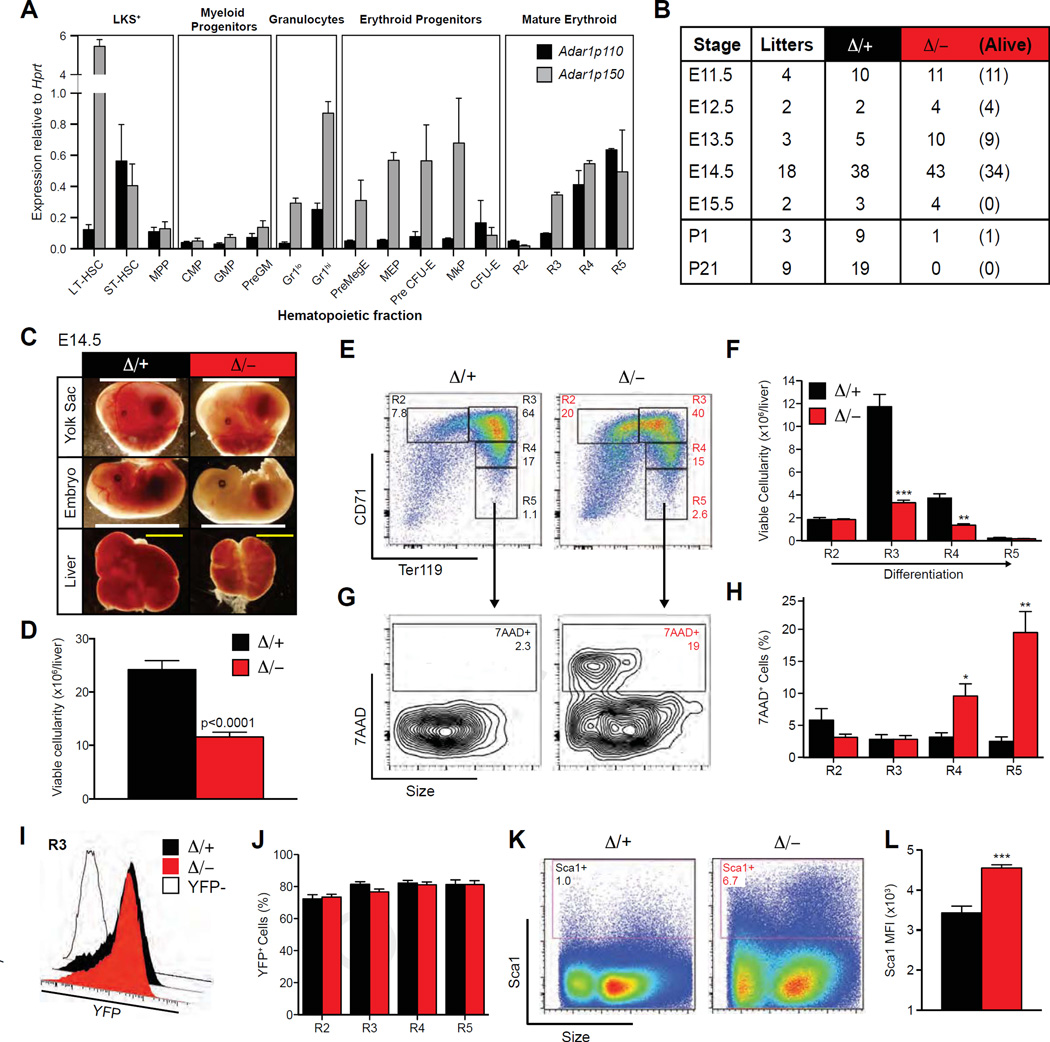
To assess which cell types express ADAR1, Adar1p110 and Adar1p150 expression was mapped in purified hematopoietic populations by qRT-PCR. Adar1 was highly expressed in HSCs (Fig. 1A), consistent with our previous findings [23]. Within the myeloid lineage, Adar1 was expressed at low levels in the progenitor fractions, but at much higher levels in mature granulocytes, whereas both erythroid progenitors and mature erythrocytes expressed high levels of Adar1 (Fig. 1A). The Adar1p150 isoform was more highly expressed than the Adar1p110 isoform, particularly in longterm HSCs, granulocytes, and the erythroid lineage (Fig. 1A).
Figure 1.
Erythroid-specific deletion of Adar1 causes embryonic death. (A) qRT-PCR of Adar1p110 and Adar1p150 normalized to Hprt in purified hematopoietic fractions. (B) Survival of Epor-Adar1Δ/− (Δ/−) embryos. All Epor-Adar1Δ/+ (Δ/+) were viable at indicated time points. (C) Representative images at E14.5. Scale bar: Yolk sac/embryo proper = 1 cm; FL = 2 mm. (D–L) E14.5 FL of Δ/− and D/+. (D) Total viable cellularity. Representative FACS plots (E) and enumeration of erythroblast populations (F). Representative FACS plots of 7-AAD staining within R5 (G) and frequency in R2–R5 (H) erythroblasts. Representative histogram plots of eYFP expression within R3 (open histogram is eYFP control) (I) and frequencies of viable (7-AAD−) YFP+ erythrocytes (J). Representative FACS plots (K) and mean fluorescence intensity (MFI) (L) of Sca1 expression in unfractionated FL. Results are mean ± SEM (Δ/+ n = 14 and Δ/− n = 18). *p< 0.05, **p< 0.005, and ***p< 0.0005 compared with Δ/+.
ADAR1 is essential for erythropoiesis, but is dispensable for myelopoiesis
Because Adar1 transcript expression increased in parallel with differentiation during granulopoiesis (Fig. 1A), we crossed the conditional Adar1 allele (Adar1fl) to LysM-Cre, resulting in deletion in the myeloid lineage [27]. We assessed cohorts of both LysM-Cre Adar1fl/fl and LysM-Cre Adar1fl/−, with neither displaying any changes in PB parameters compared with either heterozygous or wild-type controls (Table 1). These data demonstrate that ADAR1 is not required for homeostatic maintenance of myeloid cells.
Table 1.
PB analysis of mice with a myeloid-restricted deletion of ADAR1
| Population | +/+ | Δ/+ | Δ/Δ | Δ/− | p value |
|---|---|---|---|---|---|
| WBC (× 103/µL) | 11.1 ± 2.1 | 11.1 ± 3.6 | 11.2 ± 1.3 | 14.2 ± 3.8 | 0.155 |
| RBC (× 10 6/ µL) | 8.0 ± 0.5 | 8.4 ± 1.0 | 9.3 ± 0.9 | 8.7 ± 1.0 | 0.646 |
| Hgb (g/dL) | 15.1 ± 0.8 | 15.2 ± 1.1 | 15.7 ± 1.6 | 15.3 ± 1.0 | 0.880 |
| Hct (%) | 45.0 ± 1.9 | 42.3 ± 4.3 | 44.7 ± 4.6 | 42.9 ± 2.6 | 0.767 |
| MCH (pg) | 18.9 ± 1.0 | 18.0 ± 0.9 | 16.9 ± 0.1 | 17.6 ± 1.2 | 0.501 |
| MCHC (g/dL) | 33.5 ± 1.3 | 36.0 ± 2.7 | 35.2 ± 0.6 | 35.6 ± 1.4 | 0.707 |
| Plt (× 103/µL) | 749.3 ± 289.3 | 1169.3 ± 634.6 | 1321.5 ± 193.8 | 1367.4 ± 587.8 | 0.571 |
| Gran (× 10 2/ µL) | 11.0 ± 5.4 | 12.4 ± 7.4 | 9.3 ± 8.8 | 12.1 ± 5.7 | 0.958 |
| N | 3 | 4 | 6 | 7 |
Genotypes were LysM-Cre Adar1+/+(+/+), LysM-Cre Adar1Δ/+(Δ/+), LysM-Cre Adar1Δ/Δ (Δ/Δ), and LysM-Cre Adar1Δ/− (Δ/−). Data are expressed as mean ± SD; the number (n) of animals is indicated. Statistical significance (p value) between Δ/− and Δ/+ was calculated by unpaired t test. Gran = Granulocytes; Hct = hematocrit; Hgb = hemoglobin; MCH = mean corpuscular hemoglobin; MCHC = MCH concentration; Plt = platelet; WBC = white blood cells.
Because Adar1 was also highly expressed in erythroid cells (Fig. 1A), we investigated whether ADAR1 was required for erythropoiesis. Germline Adar1 deficiency resulted in a nearly complete absence of mature erythrocytes in the FL [10]; however, it is not possible to differentiate the failed erythropoiesis in the Adar1−/− FL from the cell-intrinsic requirement for ADAR1 in HSC/progenitors. Therefore, we used an erythroid-specific deletion approach. Mice expressing Cre knocked in to the Epor locus (Epor-Cre) [25,26] were crossed with Adar1fl/fl mice [10,23]. Epor-CreKI/+ Rosa26-eYFPKI/+ Adar1fl/− (Epor-Adar1Δ/−) mice were compared with Epor-CreKI/+ Rosa26-eYFPKI/+ Adar1fl/+ (Epor-Adar1 Δ/+) control littermates (Supplementary Figure E1 A, online only, available at www.exphem.org) [26]. Epor-Adar1Δ/− mice died in utero at E14.5, ~2 days later than the germline-deficient animals, and no viable embryos were recovered at E15.5 (Fig. 1B). Surprisingly, one Epor-Adar1Δ/− pup survived to birth, but was smaller and pale relative to littermate controls (Supplementary Figure E1B, online only, available at www.exphem.org) and died shortly thereafter, with analysis revealing inefficient excision of the Adar1fl allele. No Epor-Adar1Δ/− mice survived until weaning (Fig. 1B). Both the embryo proper and the yolk sac of Epor-Adar1Δ/− were pale compared with controls (Fig. 1C). The FL of viable Epor-Adar1 Δ/− embryos atE14.5 was small with decreased viable cellularity (Fig. 1C and 1D).
Analysis of FL erythropoiesis revealed a loss of cells after the phenotypic polychromatophilic erythroblast stage of erythroid differentiation in E14.5 Epor-Adar1 Δ/− FL compared with littermate controls (Fig. 1E and 1F). Adar1 -deficient erythroid cells had a significant increase in the proportion of dead cells (7-AAD+) within the R4 and R5 fractions, encompassing reticulocytes and red blood cells (RBCs; Fig. 1G and 1H). Within the FL erythroid lineage, Cre-recombinase activity was comparable between genotypes, as indicated by eYFP expression (Fig. 1I and 1J).
The proportion of FL cells expressing Sca-1, a known cell-surface-expressed ISG, was increased in Epor-Adar1Δ/–FL across all erythroid populations (Fig. 1K and 1L; Supplementary Figure E1C, online only, available at www.exphem.org), suggestive of an active IFN response within the ADAR1-null erythrocytes.
Adar1 is required for adult erythropoiesis
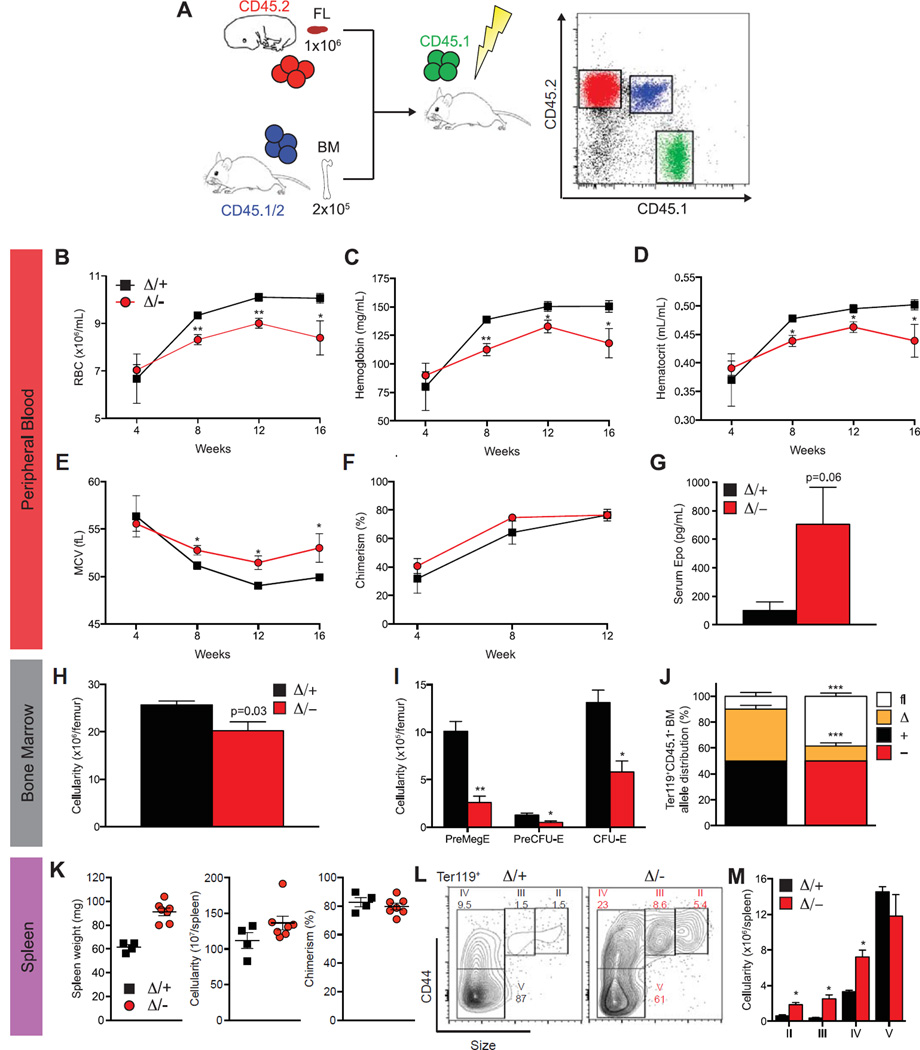
The ontogeny and transcriptional regulation of fetal and postnatal erythropoiesis are disparate [25]. To understand ADAR1 function in adult erythropoiesis, FL transplantations were performed. Unfractionated FL from E14.5 Epor-Adar1Δ/– and control littermates (both CD45.2+) was transplanted competitively into lethally irradiated con-genic (CD45.1+) recipients (Fig. 2A). Recipients of Epor-Adar1Δ/− FL cells developed a macrocytic anemia (Fig. 2B and 2E). PB leukocyte chimerism (CD45.2+) was comparable between genotypes from 4 to 12 weeks after transplantation (Fig. 2F). However, mature erythroid cells do not express CD45.2 because this marker is lost upon enucleation. Given the high level of leukocyte chimerism, the majority of RBCs found in the periphery of reconstituted recipients were likely to have stemmed from CD45.2+ donor cells (Fig. 2F). No leukopenia or thrombocytopenia was observed in Epor-Adar1Δ/– recipients (Supplementary Figure E2, online only, available at www.exphem.org). Consistent with anemia and stress erythropoiesis, there were increased serum erythropoietin levels in Epor-Adar1Δ/− recipients (Fig. 2G).
Figure 2.
ADAR1 is essential for definitive erythropoiesis in transplanted adult mice. (A) Schematic representation of transplant experiments. PB enumeration of RBCs (B) and hemoglobin (C), hematocrit (D), mean corpuscular volume (MCV) (E), chimerism (CD45.2+) (F), and serum Epo levels (G) at 16 weeks after transplantation. BM cellularity (H), enumeration of erythroid progenitors (I), and allele distribution (determined by gDNA semi-qPCR) (J). (K) Spleen weight, cellularity, and chimerism. Representative FACs plots of Ter119+ erythrocytes (L) and quantitation of mature erythroid fractions (M) determined by size and CD44 expression. Results are mean ± SEM (Δ/+ n = 4 and Δ/− n = 7, pooled from two experiments). *p < 0.05 and **p< 0.005 compared with Δ/+.
Analysis of nucleated CD45.2 cells in the BM revealed significantly decreased cellularity in Epor-Adar1Δ/− reconstituted recipients (Fig. 2H). The Pre-Meg-E population is the first to express Epor-Cre [26] and therefore would be the first populations to be Adar1 deficient. Pre-Meg-E, Pre-CFU-E, and CFU-E phenotypic populations were all decreased significantly in Epor-Adar1Δ/− recipients (Fig. 2I). Analysis of Adar1 allelic distribution in CD45.1−Ter119+ BM cells revealed a strong selection against excision of the Adar1fl allele in recipients of Epor-Adar1Δ/− FL (Fig. 2J), indicating that erythropoiesis could not be maintained in the absence of ADAR1. We have reported this previously in HSCs [23]. Anemic recipients, reconstituted with Epor-Adar1Δ/− FL donor cells, also had splenomegaly and a nonsignificant increase in cellularity despite having chimerism comparable to controls at 16 weeks after transplantation (Fig. 2K). Analysis of the erythroid compartment using Ter119 and CD44 surface markers [44] revealed a twofold to threefold increase in basophilic erythroblasts through to reticulocytes (Figs. 2L and 2M) as evidence of extramedullary erythropoiesis in the spleen. Taken together, these data demonstrate that ADAR1 is required for both fetal and adult erythropoiesis in a cell-autonomous manner.
Adar1 suppresses the dsRNA-IFN response in erythroid cells
We showed previously that ADAR1 loss induced ISGs in hematopoietic stem and progenitor cells (HSPCs) [23]. Transcriptional profiling was performed on purified CD71+Ter119− (R2) and CD71+Ter119+ (R3) erythroid populations isolated from independent Epor-Adar1Δ/− and Epor-Adar1Δ/+ E14.5 embryos to assess gene expression changes specifically in committed erythroid cells. QuSAGE [33] revealed that gene sets involved in various immune responses were enriched in Epor-Adar1Δ/− R2 and R3 erythrocytes (Fig. 3A). In addition to immune-related signatures, hypoxia-induced genes were enriched within Epor-Adar1Δ/− R3 erythrocytes (Fig. 3A); however, this may be a secondary consequence of failed erythropoiesis.
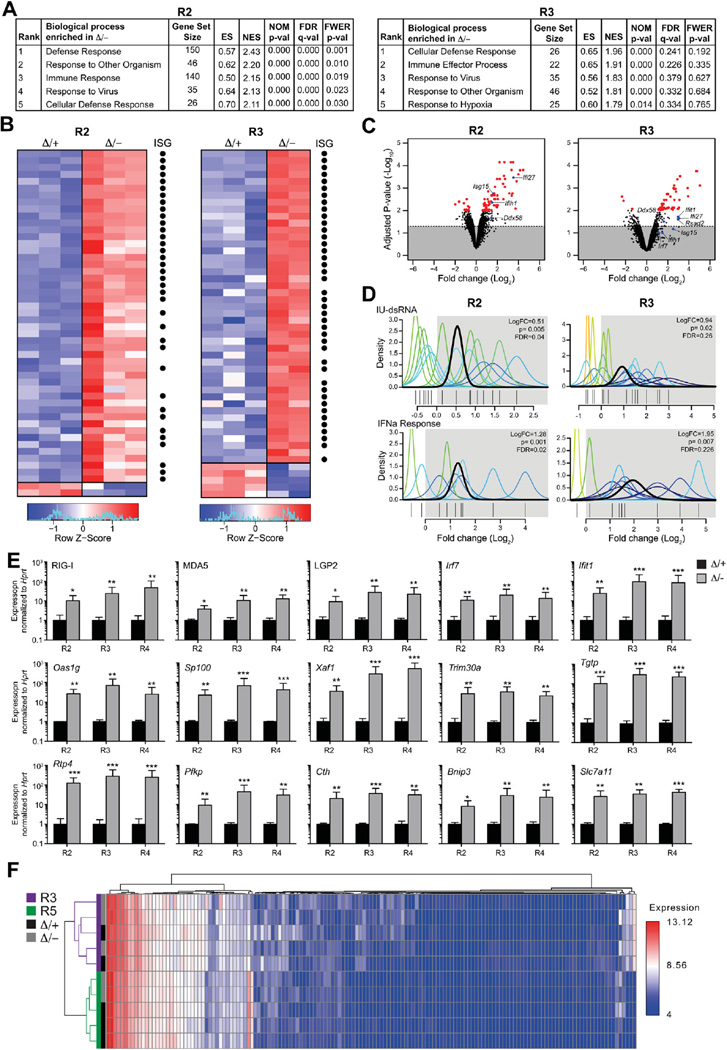
Figure 3.
Upregulation of viral responsive genes in Adar1-null erythrocytes. Gene expression analysis of EpoR-Adar1Δ/− (Δ/−) and EpoR-Adar1Δ/+ (Δ/+) E14.5 FL erythrocytes. (A) Pathways analysis of R2 (left) and R3 (right) erythroid populations. (B) Heat map of the top 50 most differentially expressed genes in R2 (left) and R3 (right) erythroblast fractions. Black dots indicate ISGs. (C) Volcano plot analysis of differentially expressed genes. Genes above dotted line are significant and red dots are p< 0.01. (D) QuSAGE analysis of IU-dsRNA stimulated (top) and IFNα responsive (top) gene sets. (R2 n = 3, R3 Δ/+ n = 3 and R3 Δ/− n = 2). (E) qRT-PCR validation of genes that are upregulated in Δ/− R2, R3, and R4 erythroblasts. Results are mean ± SD (n = 3). *p< 0.05, **p< 0.005, and ***p< 0.0005 compared with D/+. (F) miRNA profiles of R3 (purple) and R5 (green) erythroblasts from Δ/− (grey) and Δ/+ (black). Expression was clustered with samples in rows and miRNAs in columns.
We next analyzed the individual gene-level deregulation. The majority of upregulated (75% and 91% for R2 and R3 fractions, respectively), but not downregulated, genes were ISGs (Fig. 3B) [45]. The significant enrichment of ISGs in Epor-Adar1Δ/− erythroblasts compared with controls is comparable in direction and magnitude to the IFN signature observed in Adar1 -deficient HSCs [23] and mouse embryonic fibroblasts [46] (Fig. 3B; Supplementary Table E2, online only, available at www.exphem.org). There was a significant upregulation of genes typically induced upon the presence of dsRNA in the cytosol (Fig. 3C). Interestingly, the cytosolic sensors of viral dsRNA, retinoic acid inducible gene I (RIG-I, Ddx58), and melanoma-differentiation-associated protein 5 (MDA5, Ifih1) were significantly upregulated in Adar1-null erythroid cells (Fig. 3C). Furthermore, transcripts downstream of MDA5 and RIG-I, including Irf7 and Ifit1, were upregulated in the absence of ADAR1 (Fig. 3C). Therefore, ADAR1 regulation of ISGs is conserved across multiple hematopoietic cell types.
ADAR1 can suppress the induction of ISGs via a RIG-I like receptor (RLR)-mediated axis [46]. We assessed changes in expression of IFN- and dsRNA-responsive gene sets. IFNα-responsive signatures were enriched in both Adar1-null erythroid fractions (Fig. 3D). Genes induced by dsRNA that are also suppressed by inosine-uracil mismatched RNA (IU-dsRNA), representative of endogenous hyperediting [47], were significantly enriched within Epor-Adar1Δ/− R2 and R3 erythroid cells (Fig. 3D). The significant overlap between the IU-dsRNA gene set and ADAR1 deficiency in erythropoiesis supports the concept that endogenous dsRNA could stimulate the RLR pathway and activate downstream ISGs in the absence of inosine generated by ADAR1 (Supplementary Table E3, online only, available at www.exphem.org). qRT-PCR validation of genes represented within the RLR pathway and downstream ISGs confirmed that these genes were significantly increased in Epor-Adar1Δ/− R2, R3, and R4 erythroid cells (Fig. 3E). The most striking feature of ADAR1-deficient erythropoiesis was not the number of ISGs, but rather the magnitude by which these genes were upregulated (up to 508-fold; Fig. 3E). However, the mechanism by which ADAR1 regulated this process in erythroid cells was unclear.
In addition to the known roles for ADAR1 (A-to-I editing), it was proposed that ADAR1 is a key mediator of miRNA biogenesis through a direct protein–protein interaction with DICER1 [24]. Therefore, to investigate the requirement for ADAR1 in erythropoiesis, we assessed systematically the contributions of miRNA biogenesis, IFN signaling, and A-to-I editing specifically to the phenotype.
ADAR1 is not required for mature miRNA biogenesis during erythropoiesis
The levels and expression of mature miRNAs were quantitated in Adar1-deficient erythroid cells. Polychromatophilic erythroblasts (R3) and reticulocytes/RBCs (R5) were isolated from independent Epor-Adar1Δ/− and Epor-Adar1 Δ/+ E14.5 FL and profiled. Of the 435 miRNAs analyzed, 201 were expressed in at least one sample of each genotype. Hierarchical clustering resulted in samples clustering by cell type, not genotype (Fig. 3F). Only 10 miRNAs were expressed differentially between Epor-Adar1Δ/− and Epor-Adar1Δ/+, all of which had fold changes of <20% (Supplementary Table E4, online only, available at www.exphem.org). The altered miRNAs are not implicated in the regulation of erythropoiesis or ISGs [48]. Therefore, ADAR1 does not contribute significantly to the generation of mature miRNA in erythropoiesis and this is not likely to contribute to the phenotype.
Type I and type II IFN receptor signaling are not required for the transcriptional response in Adar1−/−erythroid cells
Loss of ADAR1 led to a deregulation of ISGs and the production of type I and II IFN, but not type III IFN, in vivo [23,46] (Fig. 3). Although it has been demonstrated recently that the loss of the type I IFN receptor (Ifnar) genetically does not rescue the phenotype of ADAR1 deficiency [46], it is possible that type II IFN signaling pathways may contribute in Ifnαr−/−Adar1−/− embryos. To determine the role of signaling from both type I and type II IFN receptors directly, we generated Ifnαr−/−Ifnγr−/−Adar1−/− triple knockout (TKO) mice. Ifnar−/−Ifnγr−/−Adar1+/− intercrosses yielded no viable TKO neonates (Fig. 4A). Analysis of embryogenesis revealed that TKO animals were dying in utero at E14.5–E15.5 (Figure 4A), 2–3 days later than Adar1−/− embryos [10,19]. The extended survival was similar to that recently reported for Ifnαrr−/−Adar1−/− embryos, but no hematopoietic analysis was reported [46].
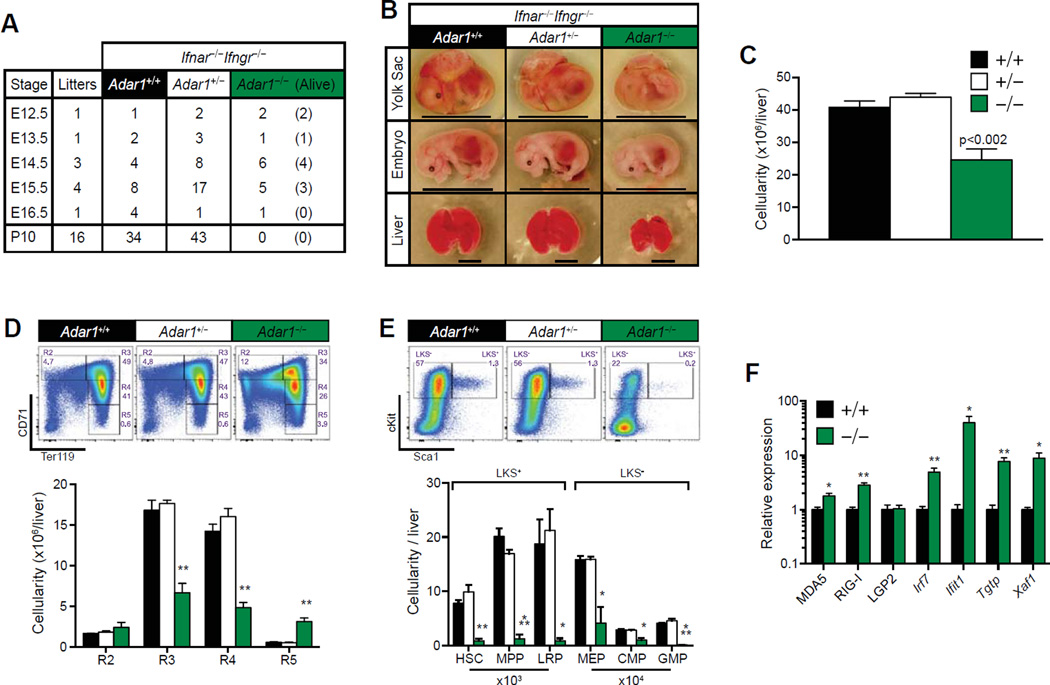
Figure 4.
Absence of IFN receptor signaling partially rescues Adar1−/− mice. (A) Survival table of Ifnar−/− Ifngr−/− Adar1−/− embryos. (B) Representative images at E15.5. Genotypes indicated. Scale bar: Yolk sac/embryo = 1 cm; FL = 4 mm. (C–E) FL analysis at E15.5. All embryos are Ifnar−/− Ifngr−/− and Adar1 genotype indicated. (C) Total viable (7-AAD) FL cellularity. Representative FACs plots (top) and enumeration (bottom) of (D) erythrocytes and (E) HSC, MPP, and lineage-restricted progenitors (LRP). Results are mean ± SEM (+/+ n = 8, +/− n = 17 and −/− n = 3). (F) qRT-PCR of ISGs from +/+ and −/− E14.5 FL. Expression was normalized to Hprt. Relative expression refers to fold change of −/− compared with +/+. Results are mean ± SEM (+/+ n = 4, +/− n = 8, and −/− n = 4). *p< 0.05, **p< 0.005, and ***p< 0.0005 compared with +/+.
At E14.5, TKO embryos, FL size, cellularity, and hema-topoiesis were comparable to Ifnar−/−Ifnγr−/−Adar1+/− and Ifnar−/−Ifnγr−/−Adar1+/+ littermates (Supplementary Figure E3, online only, available at www.exphem.org). By E15.5, viable TKO yolk sac and embryo proper were small and pale compared with controls (Fig. 4B). Consistent with Adar1−/− embryos, TKO animals appeared to be developmentally delayed prior to death, with significantly reduced FL size and cellularity (Fig. 4B and 4C) [10]. There was a loss of polychromatophilic erythroblasts through to reticulocyte populations in the TKO FL (Fig. 4D). A marked increase in the total number of TKO R5 erythrocytes (Fig. 4D) was observed despite reduced total FL cellularity within these mice (Fig. 4C). Fractionation of the LKS+ and LKS− fractions revealed a nearly complete absence of HSCs and both early and late HSPCs in TKO FL compared with controls (Fig. 4E). The manifestations of TKO embryos are highly similar to Adar1−/− mice, albeit with a delayed presentation of the phenotype.
Interestingly, RLR pathway genes were still significantly activated in E14.5 TKO FL cells compared with ADAR1 wild-type controls (Ifnαr−/− Ifnγr−/− Adar1+/+; Fig. 4F), albeit at reduced levels compared with Adar1 -deficient erythroid cells with intact IFN receptors (Fig. 3E). Transcript levels of the RLRs Ifih1 (MDA5) and Ddx58 (RIG-I), but not Dhx58 (LGP2), were significantly upregulated in TKO embryos (Fig. 4F). Irf7 and its downstream targets Ifit1, Tgtp1m and Xaf1 were also upregulated compared with controls (Fig. 4F). Interestingly, Sca1 expression was normalized in the TKO FL (Fig. 4E), providing evidence for a role in IFN signaling in the manifestations of the ADAR1-null phenotype. These data establish that: (1) ADAR1 is required to suppress the RLR pathway independently of IFN receptor signaling in a cell-intrinsic fashion and (2) IFN signaling amplifies transcriptional deregulation in the absence of ADAR1.
ADAR1-mediated RNA editing is required for erythropoiesis
Because we could not attribute a role for miRNA biogenesis nor a necessity for type I and II IFN receptor signaling in the ADAR1-deficient phenotype, we focused on the requirement for A-to-I editing. We used mice with the erythroid-restricted expression of an editing inactive Adar1 allele, ADAR1E861A (Epor-Adar1Δ/E861A) [28]. This approach resulted in the deletion of the wild-type Adar1fl allele, leaving only the catalytically inactive allele expressed in erythroid lineage cells. Epor-Adar1Δ/E861A animals were compared with Epor-Adar1Δ/+ littermate controls.
Epor-Adar1Δ/E861A embryos were underrepresented from ~E15.5 (Fig. 5A), 1 day later than Epor-Adar1Δ/−mice (Fig. 1B). This is consistent with the extended survival of the editing deficient compared with the germline-deficient allele [28]. Similar to Epor-Adar1Δ/−animals, at E15.5, both the yolk sac and Epor-Adar1Δ/E861A viable embryos were pale and FL cellularity was significantly reduced compared with controls (Fig. 5B and 5C). Analysis of E15.5 erythropoiesis revealed a significant loss of cells from the polychromatophilic erythroblast stage in Epor-Adar1Δ/E861A embryos, likely due to increased cell death (Fig. 5D, 5E, and 5F). Sca1 expression was elevated (Fig. 5G), indicative of an active IFN response and exceedingly similar to the Epor-Adar1Δ/− mice. Interestingly, there was a marked reduction in the frequency of eYFP+ R2-R4 erythroid cells in Epor-Adar1Δ/E861A FL at E15.5 compared with controls, suggesting selection against cells expressing only ADAR1E861A (Fig. 5H). This was confirmed by gDNA semiquantitative PCR of whole FL from E15.5 to E17.5 (Fig. 5I). These analyses demonstrate that the A-to-I editing function of ADAR1 is essential for fetal erythropoiesis in a cell-autonomous manner.
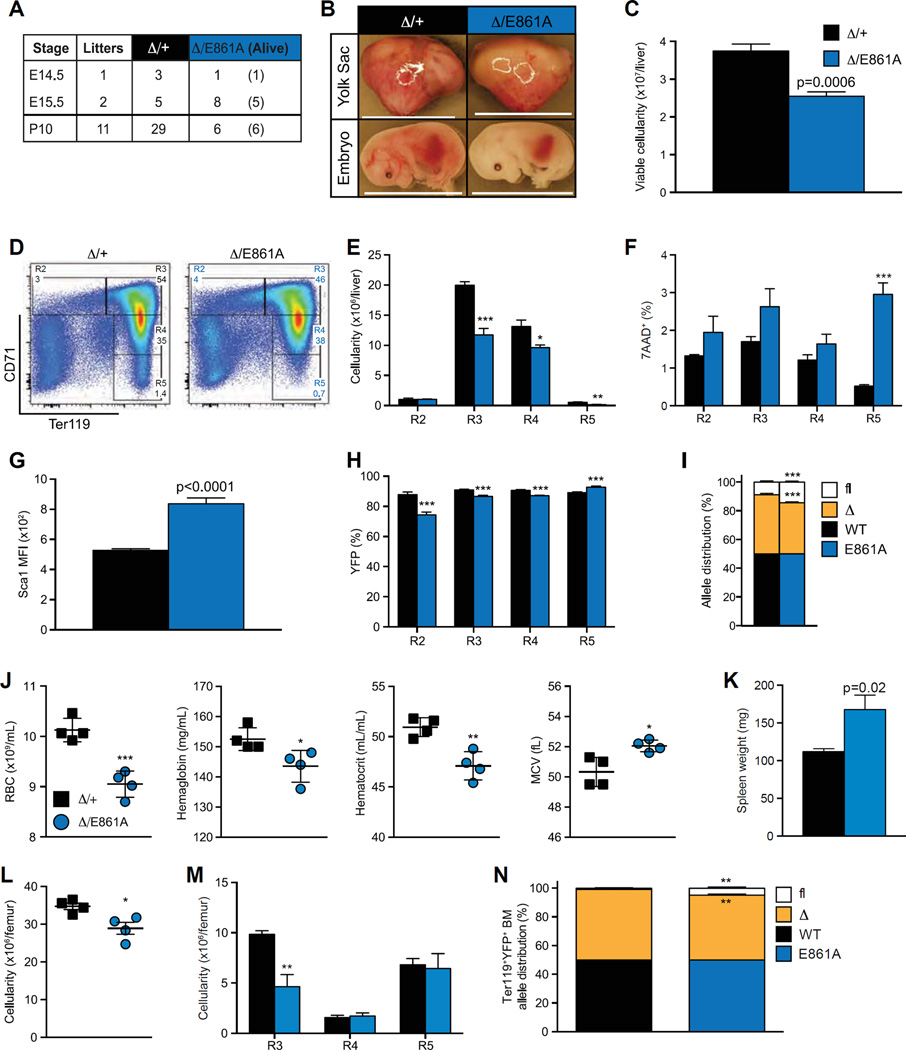
Figure 5.
A-to-I RNA editing by ADAR1 is required for fetal erythropoiesis. (A) Survival of EpoR-Adar1Δ/E861A (Δ/E861A) embryos compared with EpoR-Adar1Δ/+ littermate controls (Δ/+). (B) Representative images at E15.5. Scale bar: 1.2 cm. (C-H) FL analysis of Δ/E861A and Δ/+ at E15.5. (C) Total viable (7-AAD−) FL cellularity. (D) Representative FACS plots. (E) Enumeration of erythroid cells. Frequency of (F) dead (7-AAD+) and (G) YFP+ R2-R5 erythroid cells. (H) Sca1 MFI. (I) Allele distribution of Adar1flAdar1Δ, and Adar1+ or Adar1E861A of FL cells at E15.5–17.5 as determined by gDNA semi-qPCR. (J–N) Analysis of ≥ 16-week-old Δ/E861A and Δ/+ mice. (J) PB RBC counts, hematocrit, and hemoglobin levels and mean RBC volume. (K) Spleen weights. Cellularity of whole BM (L) and erythroid fractions (M). (N) Adar1 allele distribution of FACs isolated Ter1 19+YFP+ BM. Results are mean ± SD (E15.5, n = 5; E16.5, D/+ n = 2 and Δ/ E861A n = 1; E17.5, Δ/+ n = 1 and Δ/E861A n = 2; ≥ 16 week, n = 4). *p< 0.05, **p< 0.005, and ***p< 0.0005 compared with Δ/+.
A small number of Epor-Adar1Δ/E861A mice survived after birth (20% of the expected number; six Epor-Adar1Δ/E861A compared with 29 controls, Fig. 5A). The surviving adult Epor-Adar1Δ/E861A animals had macrocytic anemia with an ~ 10% reduction in RBCs, hemoglobin, and hematocrit levels and increased mean corpuscular volume (Fig. 5J). Epor-Adar1Δ/E861A animals had significantly larger spleens than littermate controls (Fig. 5K). There was decreased total cellularity and a significant reduction in the number polychromatophilic erythroblasts in the BM of Epor-Adar1Δ/E861A compared with Epor-Adar1Δ/+ (Fig. 5L and 5M). Collectively, the peripheral anemia, splenomegaly, and reduced BM cellularity observed in Epor-Adar1Δ/E861A adults are equivalent to the transplanted Epor-Adar1Δ/− FL recipient phenotype (Fig. 2). To determine whether postnatal survival of Epor-Adar1Δ/E861A animals is explained by incomplete excision of the Adar1fl allele, BM erythroid cells that had expressed Cre were isolated (Ter119+YFP+). A proportion of Ter119+YFP+ cells from EpoR-Adar1Δ/E861A animals retained an unexcised Adar1fl allele (Fig. 5N). Collectively, these data indicate that normal erythropoiesis cannot occur in the absence of ADAR1-mediated A-to-I editing and that RNA editing is the primary physiological function of ADAR1 in erythroid cells.
Abundant editing of erythroid-specific transcripts containing retrotransposons
To date, no described editing sites have been able to explain the lethality of ADAR1-null animals, in part due to the lack of analysis of specific cell types that require ADAR1 editing. The suppression of ISGs by ADAR1 is common to HSPCs and differentiating erythroid cells, but not B-lymphocytes and granulocytes. Therefore, we hypothesized that, in the absence of ADAR1 editing, cell-type-specific endogenous transcripts are immunogenic; that is, self-RNA results in the upregulation of ISGs [28]. We considered two scenarios. First, we considered that erythroid cells may have a requirement for an increased level of A-to-I editing compared with other cell types that do not require ADAR1, such as B-lymphocytes. If this were the case, then we would expect differential editing levels in erythroid cells than in B-lymphocytes. The alternative possibility was that cell-type-specific transcripts expressed in HSCs and/or erythroid cells, but not B-lymphocytes and granulocytes, require ADAR1 editing to reduce or suppress their immunogenicity.
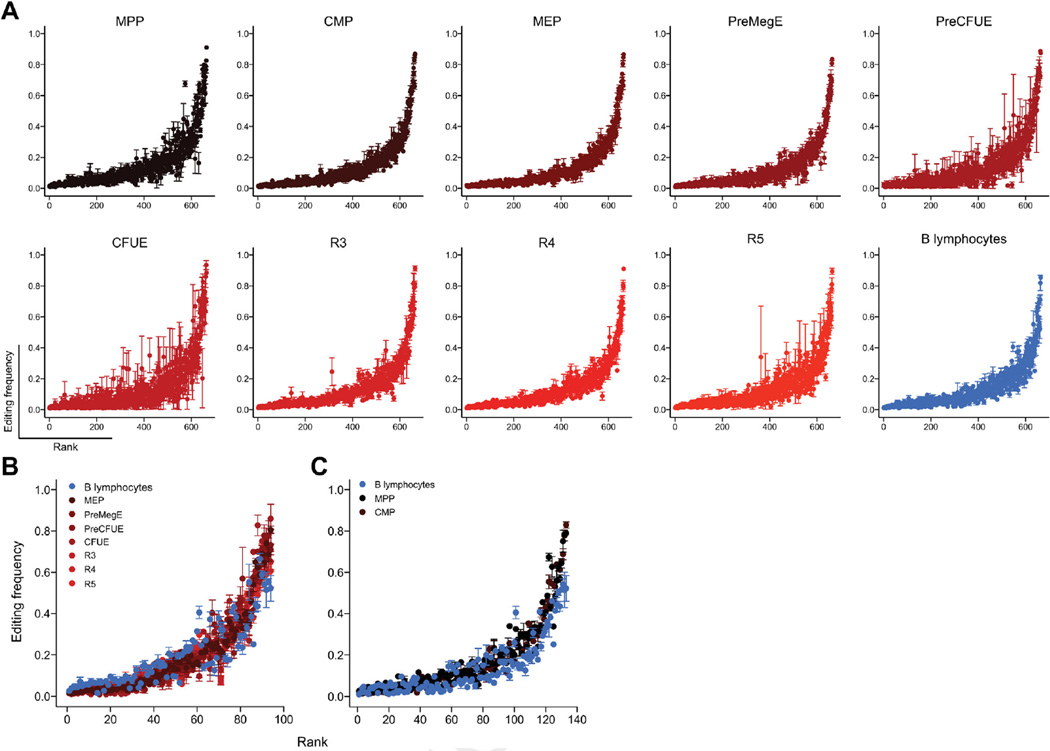
To address the first model, editing levels were compared between purified progenitors, erythroid, and B cells using mmPCR-seq [35]. We identified 664 adenosines edited at > 1% frequency across 150 transcripts that were expressed in all cell types (Fig. 6A; Supplementary Table E5, online only, available at www.exphem.org). When assessing the frequency of editing at these loci, a consistent pattern of A-to-I editing was observed between all cell types independently of a cell type’s requirement for ADAR1 (Fig. 6A; Supplementary Figure E4, online only, available at www.exphem.org). A total of 94 and 133 A-to-I sites were edited at significantly different levels between erythroid cells and B cells (Fig. 6B) and hematopoietic progenitors and B cells (Fig. 6C), respectively. The observed editing frequencies differed by <7%, on average, and targeted substrates are not known to be critical for hematopoiesis or the regulation of RLR/ISG pathways. Therefore, A-to-I editing of known substrates does not differ substantially between cells that require ADAR1 function and those that do not.
Figure 6.
Few RNA editing differences of known editing sites between diverse cell types (A) Editing frequencies plotted on y-axis with each editing site ranked from the lowest (rank 0 on the x-axis) to the highest editing frequency (rank 664 on the x-axis) based on averages at each site across each population. Within each cell type, editing sites are ranked in the same order. Editing frequencies of (B) 94 of 664 sites differentially edited between erythrocytes (MEP to R5) and B-lymphocytes and (C) 133 of 664 sites differentially edited between hematopoietic progenitors (MPP and CMP) and B-lymphocytes.
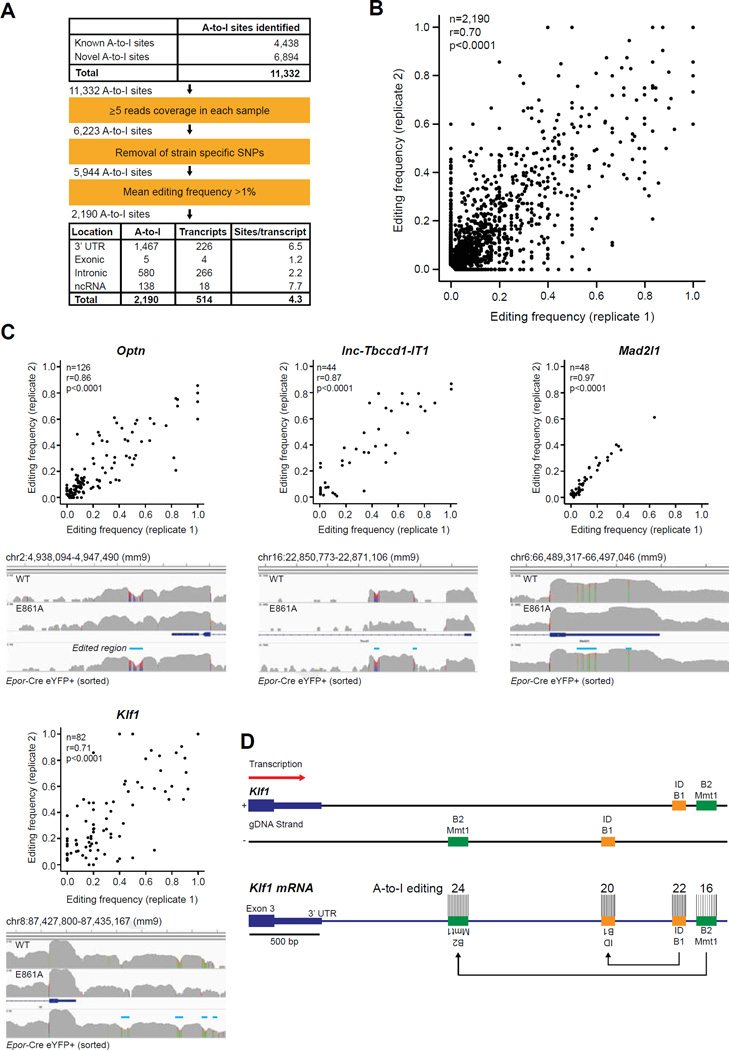
To address the second model of a cell-type-specific requirement for ADAR1, we performed RNA-seq of viable committed FL erythroid cells (YFP+) of two independent Epor-Cre R26-eYFP E14.5 littermates. We mapped A-to-I mismatches comprehensively, including hyperedited regions and editing within repetitive elements, from these data [1,7]. A total of 11,332 A-to-I editing sites were identified, 6,894 of which were novel (Fig. 7A). In parallel studies, we had determined that >95% of A-to-I editing in whole FL is ADAR1 specific [28]. After filtering out low coverage regions, strain-specific single-nucleotide polymorphisms, and editing sites with a mean editing frequency of < 1%, 2,190 high-confidence A-to-I editing sites located across 514 transcripts remained (Fig. 7A). The majority of editing was located within 3’-untranslated regions (3′-UTRs; 1,467, 67%, Fig. 7A). Editing was most dense in 3′-UTRs and noncoding RNAs (ncRNAs), with an average of 6.5 and 7.7 editing sites per targeted substrate respectively (Fig. 7A). The editing frequencies of the 2,190 sites in each biological replicate significantly correlated with one another (Pearson correlation coefficient of 0.70, p < 0.0001; Fig. 7B). Notably, the majority of editing occurred at low frequencies with only 4% edited at ≥50% (94 of 2,190 sites; Fig. 7B; Supplementary Table E6, online only, available at www.exphem.org).
Figure 7.
Novel editing sites within long UTRs of erythroid-specific transcripts. A-to-I (G mismatches in YFP+ cells isolated from two independent Epor-Adar1Δ/+ E14.5 FL. (A) Summary of A-to-I editing site analysis. (B) Scatter plot of 2190 high-confidence A-to-I editing frequencies from each replicate. (C) Scatter plots (top) and representative screenshots (bottom) of four transcripts with hyperedited 3′-UTRs (edited regions are indicated by blue line). See Supplementary Table E6 (online only, available at www.exphem.org) for other genes with hyperedited 3′-UTRs. n refers to the number of A-to-I sites within each gene. Correlations and statistics were determined by two-tailed Pearson correlation coefficients. (D) Scaled schematic representation of hyperedited clusters within the 3′-UTR of Klf1. Two tandem duplicated SINEs, ID_B1 (orange) and B2_Mmt1 (green), are located on opposite DNA strands downstream of the annotated Klf1 3′-UTR. The number of A-to-I editing sites within each SINE is depicted.
Only 5 of 2,190 defined sites were in exonic regions, resulting in predicted nonsynonymous coding changes of 4 genes (Cog3, Cdk13, Flna and Blcap), none of which could explain the observed phenotype of ADAR1-deficient erythroid cells (Supplementary Table E6). A potential consequence of A-to-I editing within introns is perturbed splicing [49]. However, none of the 580 sites located within introns were edited at known acceptor splice sites. 138 A-to-I editing sites were found in 18 ncRNAs (Fig. 7A). Fourty four of these sites were identified in a novel lncRNA within the first intron of Tbccd1, which we termed lnc-Tbccd1-IT1 (Fig. 7C). Furthermore, all 44 editing sites were restricted to the long terminal repeat (LTR) ORR1A (RepeatMasker; Supplementary Table E6), which is tan-demly duplicated on opposite DNA strands and therefore is capable of forming a 396bp dsRNA structure when transcribed (Supplementary Figure E5, online only, available at www.exphem.org).
A similar pattern of editing was observed within 3’-UTRs: 511 of 1,467 A-to-I sites identified were largely located in SINEs of only 10 hyperedited transcripts (Supplementary Table E6). A total of 82 and 126 A-to-I sites were located in long 3′-UTRs of Klf1 and Optn, respectively (Fig. 7C), both of which are restricted in expression to the erythroid lineage (Supplementary Table E6; Supplementary Figure E6, online only, available at www.exphem.org) [50]. Editing within these erythroid-specific transcripts, including lnc-Tbccd1-IT1, is more extensive than other hyperedited transcripts such as Mad2l1 (Fig. 7C), which is expressed in multiple cell types. All 82 editing sites in the 3′-UTR of Klf1 are restricted to the duplicated SINEs B2_Mmt1 and ID_B1. These SINEs are only transcribed in the long 3′-UTR of Klf1 (Fig. 7D), which is expressed at ~ 15-fold less than the annotated 3′-UTR (data not shown). Taken together, these findings link the cell-type-specific requirement for ADAR1 with the extensive hyperediting of retrotransposons in cell type-specific transcripts. Therefore, we propose that ADAR1-mediated editing of these repetitive elements within cell-type-specific transcripts is required to prevent their inappropriate activation of the RLR pathway.
Discussion
The requirement for ADAR1 within different mature hematopoietic lineages has not been explored in detail outside of the HSC/progenitor populations [22,23]. ADAR1 function is required in both mouse and human HSPCs [51]. B-lymphopoiesis was reported to be normal is the absence of ADAR1, although no hematological analysis was provided [21]. Through assessing the requirement for ADAR1 systematically using lineage-restricted deletion in vivo, we define an essential requirement for ADAR1 in erythropoiesis, but not for basal myelopoiesis, in a cell-autonomous manner. The complete absence of ADAR1 or the editing-deficient ADAR1 allele produced comparable erythroid phenotypes, demonstrating that the most physiologically relevant function of ADAR1 in erythropoiesis is RNA editing, consistent with our recent report in the whole animal [28]. The deletion of Adar1 from committed erythroid cells was accompanied by the activation of transcriptional signatures associated with IFN and viral infection, a highly conserved response to loss of ADAR1 function in diverse cell types [22,23,28,46].
Our data demonstrate that erythroid cells are capable of eliciting an innate immune response in a cell-autonomous manner. Because cells of the erythroid lineage do not express cell surface MHC, they may be poised to initiate cell death and IFN pathway activation in response to viral infection as a means to prevent the spread of infection. B cells and granulocytes may not harbor these preprogrammed states because they are key immune effector cells. Therefore, the inappropriate activation of the innate immune response and subsequent transcriptional changes are the most plausible cause of cell death in both ADAR1-null and editing-deficient erythroid cells, but not B cells and granulocytes.
The concurrent deletion of both Ifnar (type I) and Ifngr (type II) IFN receptors extended the survival of Adar1−/− embryos to E15.5, consistent with the recent report showing that Ifnar−/−Adar1−/− embryos die at E14.5 [46] and restored Sca1 expression but did not prevent intracellular activation of RLR signaling. The data demonstrate that ADAR1 is required to suppress RLR activation and that the activation of these proteins leads to the production of ISGs, which in turn activate IFN signaling. We interpret this as meaning that IFN-receptor signaling amplifies and accelerates the manifestations of the Adar1 -deficient phenotype, but that IFN does not initiate the signaling. Consistent with this model, ADAR1-editing-deficient mice were rescued, both embryonically and as adults, by deletion of Ifih1 (MDA5) [28]. Mannion et al. reported that concurrent deletion of Mavs rescued the Adar1−/− to the first day of birth [46]. MAVS is the common downstream adaptor of MDA5/RIG-I signaling. Pestal et al. recently independently confirmed our findings and elegantly demonstrated that the MDA5–MAVS axis is the only pathway involved in the response to unedited endogenous dsRNA [52]. We propose that ADAR1 be defined as a suppressor of activation of the innate immune system, focused on MDA5-MAVS, rather than as a suppressor of ISGs.
Editing-independent functions for ADAR1 have been proposed, most prominently in miRNA biogenesis. The data regarding the participation of ADAR1 in miRNA biogenesis are confounding. One report proposed that an editing-independent function of ADAR1, through a direct protein–protein interaction with DICER1, was critical in generating normal levels of mature miRNAs [24]. Studies from other groups have suggested a more prominent role for ADAR2 in miRNA biology, with only relatively subtle effects of ADAR1 deficiency on miRNA biogenesis [53]. We did not detect changes in the levels of mature miRNAs in the Epor-Cre Adar1Δ/−. Consistent with our findings, it was reported that Epor-Cre Dicerfl/fl animals were viable and had largely normal erythropoiesis, which suggests that, at least within erythroid cells, these two proteins function independently [54]. In contrast, we demonstrate that A-to-I editing is the primary physiological role of ADAR1 in erythropoiesis based on the Epor-Adar1Δ/E861A phenotype. ADAR1-mediated RNA editing, not functions in miRNA biogenesis, is required to prevent the activation of the innate immune response and subsequent cell death during erythropoiesis.
Clusters of hyperediting sites were identified that were restricted to tandemly duplicated SINEs within 3′-UTRs of a number of transcripts, including Klf1, Optn, and lnc-Tbccd1-IT1. KLF1 is a key erythroid transcription factor [55]. A similar pattern of hyperediting was observed in the long 3′-UTR of Optn, which, like Klf1, appears restricted in expression to the erythroid lineage, thus providing a potential link to the cell-type-specific requirement of ADAR1 editing. Nearby duplicated retrotransposons and LTRs are able to form dsRNA structures when transcribed [56]. Repetitive, double-stranded regions of endogenous RNAs are also enriched in editing sites in Cae-norhabditis elegans, indicating that the requirement for hyperediting such sequences is evolutionarily conserved [57]. Distinct dsRNA structures are detected in the cytosol of infected cells by RLRs such as RIG-I, MDA5, and LGP2. IU-dsRNA was shown to inhibit ISG transcription by binding to RIG-I and MDA5 and preventing IRF3 phosphorylation [47]. It has now been demonstrated that the key cytosolic mediator of the response in ADAR1-editing-deficient cells is MDA5 signaling, not RIG-I signaling, via MAVS [28,52]. These results focus attention on a particular class of dsRNAs that are critical in the phenotype because MDA5 has a preference for longer dsRNAs of ~ 0.5–7 kb and its signaling is enhanced by perfectly paired substrates [58,59].
We propose that, in the absence of ADAR1 editing, duplicated SINEs and LTRs in endogenous transcripts form self-dsRNAs capable of activating MDA5 and subsequent signaling [28,60,61]. Retrotransposons located within a gene must reside in either the CDS or 3′-UTR in order to persist in the cytosol. The majority (>90%) of transcribed duplicate SINEs are located within introns and, although they are subjected to hyperediting by ADARs [7], editing at these sites are probably not essential because they will be removed by splicing in the nucleus. The cell-type-specific requirement for ADAR1 would depend on the coordinated expression of endogenous dsRNA transcripts containing regions of perfectly paired sequence that would normally be hyperedited together with MDA5. Tissues that require ADAR1 function, such as intestine [62] and bone [63], coordinately express MDA5 and dsRNA transcripts that are normally subjected to ADAR1 editing that we have not identified here. MDA5 is most highly expressed in HSPCs, T-lymphocytes, and erythroid progenitors [50], consistent with the in vivo hematopoietic requirement for ADAR1. Such a model is consistent with results from ADAR1-deficient 293T cells, which are human and express low/negligible basal levels of MDA5 [52]. Overexpression of MDA5, but not RIG-I, activated an IFN response in ADAR 1-deficient cells, demonstrating that this is a species conserved response to a lack of ADAR 1-mediated editing that is not dependent on the presence of the primate-specific Alu repetitive elements. Our data indicate that the physiological role of ADAR1 is to “detoxify” endogenous dsRNA transcripts in an RNA-editing-dependent manner.
Supplementary Material
Acknowledgments
The authors thank M. Smyth for Ifnar−/− and Ifngr−/−; the SVH BioResources Centre; and the SVI Flow Cytometry Core Facility (M. Thomson). This work was supported by grants from the Leukaemia Foundation (C.R.W.); the Leukaemia Foundation (doctoral scholarship to B.J.L.); the National Health and Medical Research Council (NHMRC Project Grant APP1021216, NHMRC Career Development Award APP559016 to C.R.W.; NHMRC Research Fellowship Grant #1003339 to L.E.P); the German Academic Exchange Service (postdoctoral fellowship); a Stanford Dean’s Fellowship to R.P; Stanford Genome National Institutes of Health Grant R01GM102484, the Ellison Medical Foundation; Stanford University Department of Genetics (J.B.L.); National Institutes of Health (Grant R01HL116364 to J.P); and the Victorian State Government OIS Program (to St. Vincent’s Institute). S.H.O. is an investigator at the Howard Hughes Medical Institute. C.R.W. was the Leukaemia Foundation Phillip Desbrow Senior Research Fellow.
P.H.S. and J.C.H. generated the knock-in mouse line; B.J.L., J.C.H., R.P, G.R., A.M.C, P.D.K., M.W., J.P, J.L., J.B.L., and C.R.W. performed experiments and analyzed and interpreted data; B.J.L., J.C.H., V.G.S., M.W., L.E.P, P.H.S., J.P, S.H.O., J.L., J.B.L., and C.R.W. provided intellectual input and conceptual advice; and B.J.L. and C.R.W. wrote the manuscript.
Footnotes
Conflict of interest disclosure
J.C.H. was a paid employee of Taconic Biosciences. Taconic Bioscience had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The remaining authors declare no competing financial interests.
Supplementary data
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.exphem.2016.06.250.
References
- 1.Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazak L, Haviv A, Barak M, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danecek P, Nellaker C, McIntyre RE, et al. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levanon EY, Eisenberg E, Yelin R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Bio-technol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 6.Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porath HT, Carmi S, Levanon EY. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun. 2014;5:4726. doi: 10.1038/ncomms5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci U S A. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 10.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, See-burg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 11.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 12.Horsch M, Seeburg PH, Adler T, et al. Requirement of the RNA-editing enzyme ADAR2 for normal physiology in mice. J Biol Chem. 2011;286:18614–18622. doi: 10.1074/jbc.M110.200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi M, Maas S, Single FN, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 16.Brusa R, Zimmermann F, Koh DS, et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 17.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Miyakoda M, Yang W, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 20.Ward SV, George CX, Welch MJ, et al. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci USA. 2011;108:331–336. doi: 10.1073/pnas.1017241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Chendrimada TP, Wang Q, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.XuFeng R, Boyer MJ, Shen H, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci USA. 2009;106:17763–17768. doi: 10.1073/pnas.0903324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ota H, Sakurai M, Gupta R, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinrich AC, Pelanda R, Klingmuller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 2004;104:659–666. doi: 10.1182/blood-2003-05-1442. [DOI] [PubMed] [Google Scholar]
- 26.Singbrant S, Russell MR, Jovic T, et al. Erythropoietin couples eryth-ropoiesis, B-lymphopoiesis, and bone homeostasis within the bone marrow microenvironment. Blood. 2011;117:5631–5642. doi: 10.1182/blood-2010-11-320564. [DOI] [PubMed] [Google Scholar]
- 27.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysM-cre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 28.Liddicoat BJ, Piskol R, Chalk AM, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang SY, Hertzog PJ, Holland KA, et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smeets MF, DeLuca E, Wall M, et al. The Rothmund-Thomson syndrome helicase RECQL4 is essential for hematopoiesis. J Clin Invest. 2014;124:3551–3565. doi: 10.1172/JCI75334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 32.Pronk CJ, Rossi DJ, Månsson R, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 2013;41:e170. doi: 10.1093/nar/gkt660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R, Li X, Ramaswami G, et al. Quantifying RNA allelic ratios by mi-crofluidic multiplex PCRand sequencing. Nat Methods. 2014;11:51–54. doi: 10.1038/nmeth.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 43.Lu J, Guo S, Ebert BL, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci USA. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusinova I, Forster S, Yu S, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mannion NM, Greenwood SM, Young R, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitali P, Scadden AD. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Flygare J, Wong P, Lim B, Lodish HF. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 2011;25:119–124. doi: 10.1101/gad.1998711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 50.Seita J, Sahoo D, Rossi DJ, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci USA. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43:933–944. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vesely C, Tauber S, Sedlazeck FJ, von Haeseler A, Jantsch MF. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 2012;22:1468–1476. doi: 10.1101/gr.133025.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Byon JC, Padilla SM, Papayannopoulou T. Deletion of Dicer in late erythroid cells results in impaired stress erythropoiesis in mice. Exp Hematol. 2014;42:852–856.e1. doi: 10.1016/j.exphem.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 56.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whipple JM, Youssef OA, Aruscavage PJ, et al. Genome-wide profiling of the C. elegans dsRNAome. RNA. 2015;21:786–800. doi: 10.1261/rna.048801.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peisley A, Jo MH, Lin C, et al. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci USA. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu B, Peisley A, Richards C, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 60.Hundley HA, Krauchuk AA, Bass BL, Celegans H. sapiens mRNAs with edited 3’ UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.George CX, Ramaswami G, Li JB, Samuel CE. Editing of cellular self RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem. 2016;291:6158–6168. doi: 10.1074/jbc.M115.709014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu W, Wang X, Buchanan M, et al. ADAR1 is essential for intestinal homeostasis and stem cell maintenance. Cell Death Dis. 2013;4:e599. doi: 10.1038/cddis.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu S, Sharma R, Nie D, et al. ADAR1 ablation decreases bone mass by impairing osteoblast function in mice. Gene. 2013;513:101–110. doi: 10.1016/j.gene.2012.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.