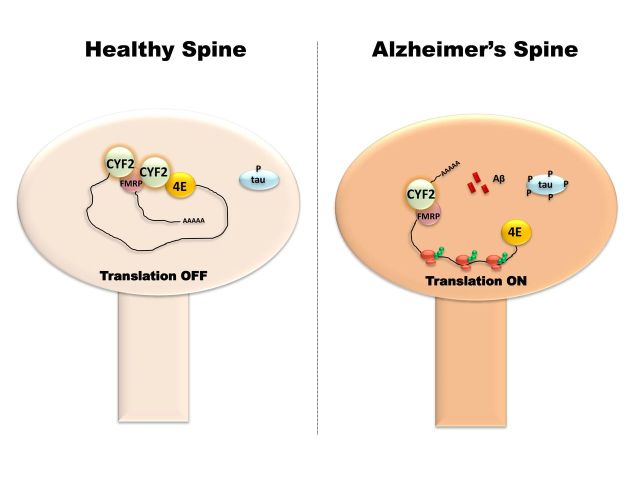
CYFIP2 is thought to regulate mRNA translation at synapses. Tiwari et al . reveal reduced CYFIP2 expression in post-mortem Alzheimer’s disease brains, and show that CYFIP2 reduction in mice causes abnormal amyloid production, tau hyperphosphorylation, and spatial memory loss. CYFIP2 could represent a molecular ‘hub’ with potential as a therapeutic target in Alzheimer’s disease.
Keywords: amyloid beta, BACE1, mRNA translation, spatial memory, tau phosphorylation
CYFIP2 is thought to regulate mRNA translation at synapses. Tiwari et al . reveal reduced CYFIP2 expression in post-mortem Alzheimer’s disease brains, and show that CYFIP2 reduction in mice causes abnormal amyloid production, tau hyperphosphorylation, and spatial memory loss. CYFIP2 could represent a molecular ‘hub’ with potential as a therapeutic target in Alzheimer’s disease.
Abstract
Characteristic features of Alzheimer’s disease are memory loss, plaques resulting from abnormal processing of amyloid precursor protein (APP), and presence of neurofibrillary tangles and dystrophic neurites containing hyperphosphorylated tau. Currently, it is not known what links these abnormalities together. Cytoplasmic FMR1 interacting protein 2 (CYFIP2) has been suggested to regulate mRNA translation at synapses and this may include local synthesis of APP and alpha-calcium/calmodulin-dependent kinase II, a kinase that can phosphorylate tau. Further, CYFIP2 is part of the Wiskott-Aldrich syndrome protein-family verprolin-homologous protein complex, which has been implicated in actin polymerization at synapses, a process thought to be required for memory formation. Our previous studies on p25 dysregulation put forward the hypothesis that CYFIP2 expression is reduced in Alzheimer’s disease and that this contributes to memory impairment, abnormal APP processing and tau hyperphosphorylation. Here, we tested this hypothesis. First, in post-mortem tissue CYFIP2 expression was reduced by ∼50% in severe Alzheimer’s hippocampus and superior temporal gyrus when normalized to expression of a neuronal or synaptic marker protein. Interestingly, there was also a trend for decreased expression in mild Alzheimer’s disease hippocampus. Second, CYFIP2 expression was reduced in old but not in young Tg2576 mice, a model of familial Alzheimer’s disease. Finally, we tested the direct impact of reduced CYFIP2 expression in heterozygous null mutant mice. We found that in hippocampus this reduced expression causes an increase in APP and β-site amyloid precursor protein cleaving enzyme 1 (BACE1) protein, but not mRNA expression, and elevates production of amyloid-β 42 . Reduced CYFIP2 expression also increases alpha-calcium/calmodulin-dependent kinase II protein expression, and this is associated with hyperphosphorylation of tau at serine-214. The reduced expression also impairs spine maturity without affecting spine density in apical dendrites of CA1 pyramidal neurons. Furthermore, the reduced expression prevents retention of spatial memory in the water maze. Taken together, our findings indicate that reduced CYFIP2 expression triggers a cascade of change towards Alzheimer’s disease, including amyloid production, tau hyperphosphorylation and memory loss. We therefore suggest that CYFIP2 could be a potential hub for targeting treatment of the disease.
Introduction
Alzheimer’s disease is a neurodegenerative disease that causes memory loss. The pathological hallmarks of Alzheimer’s disease are the occurrence of amyloid plaques and neurofibrillary tangles, next to substantial neuronal loss in forebrain ( Braak and Braak, 1991 ; Hardy and Selkoe, 2002 ). Amyloid plaques contain amyloid peptide amyloid-β 42 that has resulted from abnormal cleavage of amyloid precursor protein (APP). Neurofibrillary tangles contain hyperphosphorylated forms of the microtubule binding protein tau. Further, post-mortem brain analyses have established that synapse loss precedes neuronal loss in Alzheimer’s disease ( Arendt, 2009 ). This synaptic degeneration correlates best with the memory impairment ( deKosky and Scheff, 1990 ; Terry et al. , 1991 ; Coleman et al. , 2004 ). The initial cause of synaptic degeneration is thought to be formation of soluble amyloid-β 42 oligomers ( Marcello et al. , 2008 ). Amyloid oligomers increase translation of APP mRNA at synapses of cultured neurons ( Westmark et al. , 2011 ; Sadleir et al. , 2014 ), suggesting that increased APP expression contributes to amyloid toxicity ( Westmark, 2013 ). Fragile X mental retardation protein (FMRP, encoded by FMR1 ) binds to APP mRNA and prevents its translation ( Westmark and Malter, 2007 ; Lee et al. , 2010 ). Interestingly, FMRP also prevents local translation of mRNA encoding α-isoform of calcium/calmodulin-dependent kinase II (αCaMKII, encoded by CAMK2A ) ( Darnell et al. , 2011 ), a kinase implicated in tau hyperphosphorylation and Alzheimer’s disease ( Ghosh and Giese, 2015 ). FMRP binds to cytoplasmic FMRP-interacting proteins 1 and 2 (CYFIP1 and CYFIP2) ( Schenck et al. , 2001 ). Both of these proteins are expressed at synapses of hippocampal neurons ( Pathania et al. , 2014 ). CYFIP1 represses cap-dependent translation of mRNA by interacting with the initiation factor eIF4E ( Napoli et al. , 2008 ; Panja et al. , 2014 ; Genheden et al. , 2015 ). CYFIP2 has an identical eIF4E-binding motif as CYFIP1 ( Napoli et al. , 2008 ). In addition to regulating translation of mRNA, CYFIP1 and CYFIP2 are part of the Wiskott-Aldrich syndrome protein-family verprolin-homologous protein (WAVE) complex that regulates actin polymerization at synapses ( Eden et al. , 2002 ; De Rubeis et al. , 2013 ).
Our previous work showed that in the early stages of Alzheimer’s disease expression of cyclin-dependent kinase 5 (Cdk5) activator p25 is reduced ( Engmann et al. , 2011 ). This suggests that Cdk5-mediated phosphorylation of tau may be decreased in Alzheimer’s disease, whereas due to decreased inhibition by Cdk5 glycogen synthase kinase 3β-mediated tau phosphorylation may be increased ( Giese, 2014 ). Further, we showed that p25 overexpression upregulates CYFIP2 expression ( Engmann et al. , 2011 ). Therefore, we hypothesized that in early Alzheimer’s disease CYFIP2 expression is reduced and this would contribute to synaptic degeneration. Here, we tested this hypothesis by analysing CYFIP2 and CYFIP1 expression in post-mortem Alzheimer’s disease brain tissue. We found that CYFIP2 expression was reduced by ∼50%. This effect was modelled in mice and we found that reduced CYFIP2 expression leads to increased APP, β-site APP cleaving enzyme 1 (BACE1), and αCaMKII protein expression, abnormal amyloid-β 42 production, tau hyperphosphorylation at serine-214, alterations in dendritic spine morphology, and memory loss. Therefore, we suggest that CYFIP2 could be one of the key targets for prevention of Alzheimer’s disease.
Materials and methods
Post-mortem human brain samples
Human brain samples were obtained from the London Neurodegenerative Diseases Brain Bank, Institute of Psychiatry, Psychology and Neuroscience, King’s College London. They were received in two sets, which were treated separately. The first set contained hippocampal tissue from control subjects, subjects with mild Alzheimer’s disease (Braak stages I–II) and subjects with severe Alzheimer’s disease (Braak stages V–VI) ( n = 7 per group), as well as superior temporal gyrus (STG) samples from control and severe Alzheimer’s disease ( n = 7 and n = 9, respectively). The second set comprised hippocampus and STG samples from control, mild and severe Alzheimer’s disease patients ( n = 5 per group). The causes of death were not related to neurodegenerative disease in these control subjects ( Supplementary Table 1 ). All human tissue samples were handled according to the regulations of King’s College London Neurodegenerative Diseases Brain Bank and the Human Tissue Authority.
The frozen samples were lysed at 4°C in RIPA lysis buffer (Santa Cruz Biotechnology, Inc). The RIPA buffer contained 0.1% sodium dodecyl sulphate (SDS), 1% Nonidet P-40, 0.5% sodium deoxycholate and 0.004% sodium azide in Tris-buffered saline (TBS) (pH 7.5). Protease inhibitors cocktail, sodium orthovanadate, and α-toluenesulphonyl fluoride were added to the buffer, diluted to 1:100. The SDS concentration was increased by adding 0.25% SDS in the final volume of buffer. About 100 mg of brain tissue was lysed in 300 μl buffer. Samples were homogenized using a dounce homogenizer (12 strokes, 700 rpm) and centrifuged at 3000 rpm for 10 min. Supernatants were used for western blot analysis.
Mutant mice
APP Swe (Tg2576) mice, expressing mutant human APP (K670N/M671L) under the control of the hamster prion promoter ( Hsiao et al. , 1996 ) were obtained from Taconic farms. Mice were maintained by breeding Tg2576 males in C57BL/6 × SJL F1 genetic background with C57BL/6 × SJL F1 wild-type females, as recommended by the supplier. Mice were genotyped by polymerase chain reaction (PCR) using primer set 5′-CGACTCGACCAGGTTCTGGGT-3′, 5′-ATAACCCCTCCCCCAGCCTAGA-3′. The amplification conditions were as follows. PCR reaction mixture: 1× buffer, 2 mM MgCl 2 , 0.2 mM dNTP, 0.75 μM APP forward primer, 0.75 μM APP reverse primer, 0.025 μM Taq polymerase (Invitrogen), 2 μl/reaction. PCR program: (i) initialization: 94°C for 3 min; (ii) 35 cycles – denaturation: 94°C for 30 s, annealing: 60°C for 60 s, extension: 72°C for 60 s; (iii) final hold: 4°C. Cortico-hippocampal tissue from 4 month: ( n = 3) and 12-month-old ( n = 4) Tg2576 mutants as well as wild-type littermates (4 months, n = 4; 12 months, n = 4) were used for analysis by immunoblotting. Sexes of the animals were balanced.
Cyfip2 tm1a(EUCOMM)Wtsi/Wtsi mice (EM:05949) were generated in a C57BL/6N background by European Conditional Mouse Mutagenesis Program (EUCOMM). These mutants have an insertion of a promoter-driven neo gene in an intron of the Cyfip2 gene, which results in a null mutation ( Kumar et al. , 2013 ). Therefore, we designated these heterozygous mutants as Cyfip2+/− mice. Cyfip2+/− mice were maintained in the C57BL/6N background. Mice were genotyped by PCR. The mutants were detected by mutant allele specific 248 bp PCR product (forward CYFIP2 primer 5′-TTCCTTCCTTCCCTTGTCCC-3’, reverse CASR1 primer 5′-TCGTGGTATCGTTATGCGCC-3’) and wild-type mice had only the wild-type allele specific 461 bp PCR product (forward CYFIP2 primer 5′-TTCCTTCCTTCCCTTGTCCC-3’; reverse, CYFIP2 primer 5′-TGCCAGGAGAGACAGTGGTG-3’). The PCR reaction consisted of 1× buffer, 1 mM MgCl 2 , 0.2 mM dNTP, 10 µM primers and 0.125 μl Taq DNA polymerase (5 U/µl; Invitrogen). The PCR protocol consisted of a 2 min heating at 93°C, followed by 35 cycles of denaturation (30 s, 93°C), annealing (30 s, 56°C) and extension (30 s, 72°C). At the end of the PCR cycle, samples were heated for 10 min at 72°C and held at 4°C until recovered.
Mice were housed on 12 h light:12 h dark cycles with food and water available ad libitum . All animal procedures were conducted in accordance with the UK Animals Scientific Procedures Act 1986.
Protein preparation from mouse brain
Frozen tissue from Tg2576 mice was homogenized at 100 mg tissue/ml 2× sample buffer (0.5 M Tris-HCl, pH 6.8, 4.4% SDS, 20% glycerol, 2% 2-mercaptoethanol, 0.01% bromophenol blue, and complete mini-protease inhibitor cocktail) (Roche), using a mechanical homogenizer. Following brief sonication, homogenates were centrifuged at 25 000 g for 20 min at 4°C, and the supernatant was collected.
For preparation of crude synaptosomes from Cyfip2+/− mutants and wild-type littermates, frozen hippocampi were homogenized (10 strokes, 750 rpm) in 20 µl/mg homogenization buffer (0.32 M sucrose, 1 mM NaHCO 3 , 1 mM MgCl 2 , 10 mM HEPES pH 7.4). A small volume was collected for analysis of total hippocampal lysates. Following centrifugation at 381 g for 10 min, 4°C to remove nuclei and cell debris, the resulting supernatant was centrifuged at 16 089 g for 15 min to obtain a crude synaptosomal pellet. Synaptosomal pellets (P2 fraction) were resuspended in 300 µl homogenization buffer for each 50 mg of starting tissue.
A BCA-based protein quantification procedure (Thermo Fisher) was used to determine protein amounts for all mouse samples.
Primary neuronal cell culture
Cortical tissues were prepared from embryonic Day 16 mice as described ( Pooler et al. , 2012 ). The cells were plated on 6-well dishes coated with poly-D-lysine in neurobasal media (without L-glutamine; with phenol red) containing 2% B27 supplement (Invitrogen), 0.5 mM L-glutamine, and penicillin/streptomycin (50 U/ml / 50 μg/ml, Sigma). After 4 days the cells were cultured in selection media (to remove non-neuronal cells): neurobasal media (without L-glutamine; with phenol red) containing 2% B27 supplement (Invitrogen), 0.5 mM L-glutamine, penicillin/streptomycin, and 3 µM cytosine, β-D-arabinofuranoside. Cells were harvested as described below 21 days after plating.
Glial cell culture
Glial cells were isolated from postnatal Day 1–4 (P1–P4) mouse cerebral cortices, as previously described ( Williams and Price, 1995 ) and cultured on poly-D-lysine (25 μg/ml, Sigma) coated T75 flasks at a density of two to three cortices per flask in Dulbecco’s modified Eagle medium (Gibco), supplemented with 10% foetal bovine serum (Biosera) and penicillin/streptomycin (50 U/ml/50 μg/ml, Sigma). Once these cultures reached confluence (12–14 days) they were composed of a base layer of non-dividing astrocytes and an upper layer of dividing microglia and a few oligodendrocytes. For western blot analysis these cells were plated onto 6-well plates; they were harvested from each well using a sterilized scraper and in the presence of 20 µl 2× sample buffer: 0.5 M Tris-HCl, pH 6.8, 4.4% SDS, 20% glycerol, 2% 2-mercaptoethanol, 0.01% bromophenol blue, and 10% mini-protease inhibitor cocktail (Roche).
Western blot analysis
Protein samples were diluted and boiled at 95°C in Laemmli sample buffer. Comparable protein amounts were separated on 4–15% Criterion™ TGX™ precast gels (Bio-Rad) and then transferred onto a methanol-activated 0.2 μm polyvinylidene fluoride membrane (Bio-Rad). Membranes were blocked at room temperature for 1 h (5% milk in TBST pH 7.5) and then incubated in primary antibody diluted in blocking buffer overnight at 4°C. Primary antibodies were detected using horseradish peroxidase (HRP)-conjugated secondary antibodies (Dako, P0447/8) and chemiluminescent reagent (Thermo Scientific), and signals in the linear range obtained by exposing membranes to X-ray films (Amersham). Prior to probing with other primary antibodies, the membranes were washed in western blot stripping buffer (Santa Cruz Biotechnology). Primary antibodies used were against amyloid-β sequence in APP (Millipore, MABN10, 1:300), APP-CTF sequence in APP (1:10 000; a gift from Prof. C.C. Miller, King’s College London), BACE1 (Gene Tex, GTX62419, 1:5000), αCaMKII (Chemicon, MAB8699, 1:100 000), CYFIP1 (Millipore, 07-531, 1:1000), CYFIP2 (GeneTex, GTX124387, 1:1000), Lamin B1 (Abcam, ab133741, 1:5000), NSE (Millipore, AB951, 1:60 000), synaptophysin (Cell Signaling, 4329, 1:1000), α-synaptotagmin (Sigma, S2177, 1:30 000), phosphoSer214-tau (Abcam, AB10891, 1:250), total tau (Dako, A0024, 1:10 000) and β-tubulin III (Sigma, T8578, 1:1000). Signals were analysed with ImageJ software (NIH).
Amyloid-β 42 ELISA
Frozen hippocampal tissue was weighed and manually homogenized in 8× mass of cold 5 M guanidine hydrochloride/50 mM Tris HCl pH 8.0 using a small piston homogenizer. Homogenates were shaken at room temperature for 4 h and 5-fold diluted in cold BSAT-DPBS (5% bovine serum albumin, 0.03% Tween-20, 2.7 mM KCl, 1.5 mM KH 2 PO 4 , 136.9 mM NaCl, 8.1 mM Na 2 HPO 4 ) supplemented with protease inhibitor cocktail (Sigma). Diluted samples were centrifuged at 27 287 g for 30 min and the supernatants (containing GuHCl-soluble amyloid) were collected and stored on ice until use. Supernatants were further 10-fold diluted in standard diluent buffer (supplied) and the assay was carried out according to manufacturer’s instructions (Life Technologies, KMB3441). Samples were measured in duplicates and the entire assay performed twice. As a positive control, tissue from 12-month-old Tg2576 mice was prepared in the same way.
RNA isolation and quantitative PCR analysis
Total RNA was extracted from hippocampal tissue using TRIzol® (Life Technologies) and purified using RNeasy® spin columns (Qiagen). RNA (1.5 μg) from each sample was reverse transcribed using SuperScript™ II reverse transcriptase (Invitrogen). The cDNA obtained was diluted 10× and stored at −20°C until use. Quantitative RT-PCR experiments were performed using a Chromo4™ Continuous Fluorescence Detector (Bio-Rad DNA Engine®) with KAPA SYBR® FAST Master Mix as a reference dye (KAPA Biosystems). Specific exon primers were designed across introns to unambiguously distinguish spliced cDNA from genomic DNA contamination and purchased from Integrated DNA Technologies. Primers used were APP Forward 5′-ACTCTGTGCCAGCCAATACC-3’; APP Reverse 5′-GAACCTGGTCGAGTGGTCAG-3’; BACE1 Forward 5′-GACCACTCGCTATACACGGG-3’; BACE1 Reverse 5′-TCCTTGCAGTCCATCTTGAGA-3’; CaMKII Forward 5′-TCAGCATCCCAGCCCTAGTT-3’; CaMKII Reverse 5′-AAGGCTCCCTTTCCCAGTTC-3’; HPRT-L Forward 5′-GCTGGTGAAAAGGACCTCT-3’; HPRT-L Reverse 5′-CACAGGACTAGAACACCTGC-3’. Optimum primer and cDNA concentrations were empirically determined; each primer was used at a final concentration of 300 nM and all cDNA was further diluted 50×. The reaction was performed in Thermo-Fast® white 96-well plates capped with Ultra Clear caps (both Thermo Fisher). The reaction mixture was heated at 95°C for 10 min followed by 45 cycles (30 s at 95°C, 30 s at 60°C, 30 s at 72°C), and PCR product levels measured using Opticon Monitor version 3.1.32 (Bio-Rad). Samples were tested in duplicates or triplicates and the entire assay performed twice. mRNA levels were normalized to that of the housekeeping gene Hprt1 , and the Livak method (or ΔΔC t ) was used for analysis.
Golgi-Cox staining
Female litters aged 15 weeks of either Cyfip2+/− ( n = 4) or wild-type genotypes ( n = 5) were used to analyse spine density using modified Golgi-Cox staining method as described by the manufacturer (Rapid Golgi; FD NeuroTechnologies). Briefly, brains were removed from skull as quickly as possible, rinsed in Milli-Q® water and stored in Golgi impregnation solution at room temperature in the dark for 2 weeks. Tissue was then transferred into Golgi solution C for 1 week in the dark at room temperature. Coronal sections (150 µm) were obtained using a cryostat and mounted on 2% gelatine-coated glass slides. Sections were rinsed, stained with solution C, dehydrated, cleared of xylene, and were cover slipped and allowed to dry before quantitative analysis. Several pyramidal neurons impregnated with the Golgi solution were readily identified in the dorsal hippocampal region by their characteristic triangular soma shape and numerous dendritic spines. For spine quantification, a 100× oil-immersion objective was used to identify spines in distal dendrites (150 µm away from soma) longer than 10 µm. Straight branches were preferred for Z -stack reconstructions to have a clear presentation of spines. More than 50 dendritic segments were analysed for both groups in apical and basal dendrites. Spine densities were calculated as mean numbers of spines per micrometre per dendrite in individual mice per group using ImageJ (NIH). Spines were classified as long-thin, filopodia, or stubby/mushroom, as described ( Harris and Stevens, 1989 ; Harris et al. , 1992 ).
Morris water maze studies
The study with the Cyfip2+/− mice and control littermates was performed blind to genotype. Three to 4.5-month-old mutants (10 males) and wild-type mice (13 males) were tested in the hidden-platform version of the water maze. The diameter of the swimming pool was 1.5 m, the platform diameter 0.1 m. After handling the mice for 10 days, 2 min/day, the mice were tested with four trials per day for 10 days. The maximal trial length was 90 s and the intertrial interval was 60 s. Probe trials (60 s) were given at the end of training Day 10 and 5 days after the first probe trial. Four wild-type and three mutants were excluded from analysis due to floating during probe tests. Another group of mice ( Cyfip2+/− : eight males, wild-type: eight males) were tested with a visible platform using two trials with a 60 s intertrial interval after hidden platform training for 6 days. Video-tracking and EthoVision XT (Noldus) were used to analyse swimming speed and search strategies.
Social behaviour and marble burying behaviour
Social behaviour
The three-chamber social approach task was performed based on a published method ( Yang et al. , 2011 ). Briefly, the subject mouse was habituated to the centre chamber for 10 min and then all three chambers for 10 min. Sociability was tested for 10 min by assessing whether the subject mouse spent more time in the chamber containing a novel mouse (129S2/Sv strain) or in the chamber with an empty cup as novel object. Social novelty was tested for 10 min by assessing whether the subject mouse prefers the chamber containing the first novel mouse or a second novel mouse placed inside the previously empty object cup. Time spent in each chamber was measured using EthoVision XT tracking system (Noldus).
Marble burying
The experiments were performed based on a published method ( Deacon, 2006 ). Mice were individually placed for 30 min in animal cages (30 cm × 16 cm) containing 5 cm deep bedding and 18 marbles arranged in three evenly spaced rows of six marbles each. The number of marbles that remained unburied in each 10 min interval was counted. Marbles were counted as buried if they were at least covered two-thirds with bedding.
Statistical analysis
Unless otherwise specified, data were analysed with t -tests.
Post-mortem human brain studies
Statistical analysis for individual datasets was performed using unpaired t -tests. The pooling of data from two sets of post-mortem tissue samples that were separately obtained was performed using a linear regression model based on the following equation (see also, Tiwari et al. , 2015 ):
| (1) |
Where Χ1i is the categorical predictor coding for the group difference (e.g. Control versus Severe), and Χ2i is the categorical predictor coding for the different experiments (‘1st cohort’ versus ‘2nd cohort’)
This regression model allowed us to pool the CYFIP2 data from the two different sets of post-mortem tissues thereby eliminating potential variability resulting from differences in experimental conditions. Data analysis was performed using SPSS (version 20), which provides the output as a P score indicating the overall significance. The contribution and the significance of specific factors of interest, e.g. the disease pathology, are subsequently provided by t -test analysis. Hence, this analysis strategy helps to identify the significant change of altered CYFIP2 scores in diseased brain samples when compared to control tissues eliminating any effect induced by the processing of different sample sets. The level of significance for the analysis was 0.05 and the outliers were identified as being out with a threshold of mean ± 4 × standard deviation (SD).
Behavioural studies
One-way ANOVA, one-way ANOVA on ranks (if data were not distributed normally), two-way ANOVA with repeated measures, and Student-Newman-Keuls tests were used for statistical analysis as appropriate.
Results
Reduced CYFIP2 expression in post-mortem Alzheimer’s disease forebrain
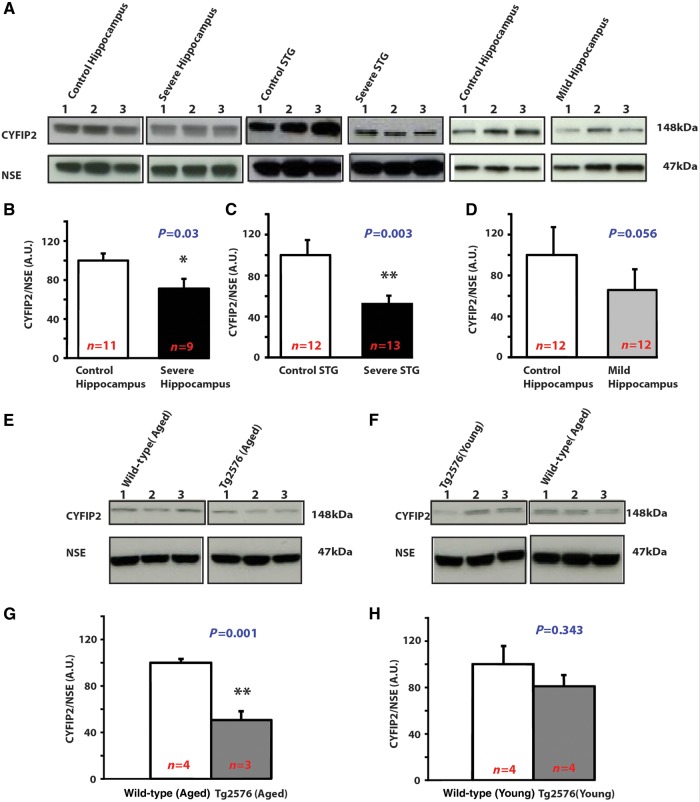
To assess whether regulation of local mRNA translation might be affected in Alzheimer’s disease, we investigated CYFIP2 protein levels in forebrain samples of severe disease cases and controls, all of which had short post-mortem delay ( Supplementary Table 1 and Supplementary material ). Antibodies that react specifically with CYFIP2 and neuron-specific enolase (NSE) protein, a neuronal marker ( McAleese et al. , 1988 ), were used in western blots ( Supplementary Fig. 1 ). Considering that CYFIP2 is not expressed in glial cells ( Supplementary Fig. 2 ), CYFIP2 expression normalized to NSE expression estimates CYFIP2 expression per neuron at the time of death. We found that CYFIP2 expression was significantly reduced by ∼40% in hippocampus of late Alzheimer’s disease (Braak stages V–VI) in comparison to expression in control subjects ( t = −2.33; P < 0.05; see Supplementary material ) (Fig. 1A and B).
CYFIP2 protein is expressed at synapses ( Pathania et al. , 2014 ). Therefore, reduced CYFIP2 expression in severe Alzheimer’s disease might be due to synapse loss that occurs before neuronal loss ( Arendt, 2009 ). Alternatively, reduced CYFIP2 expression may occur before synapses die in Alzheimer’s disease. We tested for this possibility by normalizing CYFIP2 expression to the presynaptic marker synaptophysin ( Sudhof et al. , 1987 ). We found that CYFIP2/synaptophysin expression was significantly decreased by ∼40% in severe Alzheimer’s disease hippocampus ( t = 2.43; P < 0.05; Supplementary Fig. 3 ), suggesting that CYFIP2 expression declines before synapse loss occurs.
We also studied CYFIP2 expression in the STG, which is affected to a lesser extent and at later stages than the hippocampus in Alzheimer’s disease ( Braak and Braak, 1991 ). CYFIP2 expression was found to be significantly downregulated in severe Alzheimer’s disease STG ( t = −3.28, P < 0.01; Fig. 1 C), supporting our finding of reduced CYFIP2 expression in the hippocampus.
Figure 1.
Reduced CYFIP2 expression in post-mortem Alzheimer’s disease forebrain and in a mouse model of familial Alzheimer’s disease. ( A–D ) CYFIP2 expression is decreased in forebrain of patients with severe Alzheimer’s disease. ( A ) Representative western blots. ( B ) CYFIP2 expression in hippocampal lysates from patients with severe Alzheimer’s disease ( n = 9) and control subjects ( n = 11). ( C ) CYFIP2 expression in lysates of STG from patients with severe Alzheimer’s disease ( n = 13) and control subjects ( n = 12). ( D ) CYFIP2 expression in hippocampal lysates of patients with mild Alzheimer’s disease ( n = 12) and control subjects ( n = 12). ( E–G ) Age-dependent decrease of CYFIP2 expression in forebrain of Tg2576 mice. ( E and F ) Representative western blots. ( G ) CYFIP2 expression in hippocampal–cortical lysates of 12-month-old wild-type mice ( n = 4) and Tg2576 ( n = 3). ( H ) CYFIP2 expression in hippocampal-cortical lysates of 4-month-old wild-type ( n = 4) and Tg2576 mice ( n = 4). In all panels CYFIP2 expression was normalized against NSE. Means ± standard error of the mean (SEM) are shown. * P < 0.05; ** P < 0.01.
To investigate if these dysregulations are already present in the early stages of the disease, we analysed hippocampal lysates from patients with mild Alzheimer’s disease (Braak stages I–II). There was a statistical trend towards a reduction of CYFIP2 expression by ∼50% in these cases ( t = −2.02, P = 0.056; Fig. 1 D). These data suggested that CYFIP2 downregulation may be an early event in Alzheimer’s disease.
CYFIP2 binds to FMRP, a regulator of local mRNA translation ( Schenck et al. , 2001 ). Therefore, we studied whether not only CYFIP2 expression but also FMRP expression would be altered in severe Alzheimer’s disease hippocampus ( Supplementary Fig. 4 ). However, we did not find any evidence that FMRP levels are changed in severe Alzheimer’s disease ( t = 0.33, P = 0.75), suggesting that the reduced CYFIP2 expression is specific.
Age-dependent reduction of CYFIP2 expression in an Alzheimer’s disease mouse model
We investigated if the reduced CYFIP2 expression observed in post-mortem Alzheimer’s disease brain ( Fig. 1 A–D) can also be found in a mouse model of a familial form of the disease. Tg2576 mice expressing APP with the Swedish mutations do not have amyloid plaques and spatial memory impairment at 4 months of age, whereas they have amyloid plaques and spatial memory deficits at 12 months of age ( Hsiao et al. , 1996 ; Stewart et al. , 2011 ). We performed a western blot analysis for CYFIP2 expression at these two ages. The comparison revealed significant differences between genotypes. CYFIP2 expression was significantly reduced by 50% in Tg2576 mice in comparison to wild-type mice at 12 months of age ( t = −6.56, P < 0.01; Fig. 1 E and G). The reduction in CYFIP2 expression is age-dependent, as it was not found for 4-month-old mice ( t = −1.03, P = 0.34; Fig. 1 F and H). Reduced CYFIP2 expression by ∼50% in 12-month-old Tg2576 mice resembles the reduced CYFIP2 expression in post-mortem Alzheimer’s disease forebrain. Further, it suggests that abnormal APP processing may be sufficient to cause CYFIP2 downregulation.
CYFIP1 expression is not changed in mild Alzheimer’s disease hippocampus and severe Alzheimer’s disease STG
As CYFIP1 has similar functions in vitro to CYFIP2 ( Abekhoukh and Bardoni, 2014 ), we wanted to know if CYFIP1 could be implicated in mild stages of Alzheimer’s disease. We performed western blot analysis with hippocampal protein from patients with mild Alzheimer’s disease and did not observe a downregulation of CYFIP1 expression ( t = −1.28, P = 0.22; Supplementary Fig. 5A and Supplementary Data ), contrary to the results for CYFIP2 expression ( Fig. 1 D). There was also no change of CYFIP1 expression in STG tissues from patients with severe Alzheimer’s disease ( t = −1.15, P = 0.26; Supplementary Fig. 5C ), as opposed to CYFIP2 reduction seen in severe Alzheimer’s disease STG ( Fig. 1 C). There appeared to be an upregulation in CYFIP1 levels in severe Alzheimer’s disease hippocampus ( t = 3.27, P < 0.01; Supplementary Fig. 5D ), contrary to the significant downregulation of CYFIP2 expression ( Fig. 1 B). These results indicate that CYFIP1 and CYFIP2 are regulated differently in Alzheimer’s disease hippocampus.
We further studied whether CYFIP1 expression is altered in old Tg2576 mice, which model early stages of Alzheimer’s disease. As for CYFIP2 ( Fig. 1 E–H), we performed western bot analysis for CYFIP1 expression using the same forebrain tissues of Tg2576 mice and controls. In 4-month-old Tg2576 mouse forebrain there was no change in CYFIP1 expression as compared to wild-type mice ( t = 0.97, P = 0.37; Supplementary Fig. 5H ). Unexpectedly, we found a downregulation of CYFIP1 expression by ∼60% in 12-month-old Tg2576 mice ( t = −5.49, P < 0.01; Supplementary Fig. 5G ), instead of an upregulation of CYFIP1 expression observed in post-mortem, severe Alzheimer’s disease hippocampus ( Supplementary Fig. 5D ).
CYFIP2 regulation is independent of CYFIP1 regulation
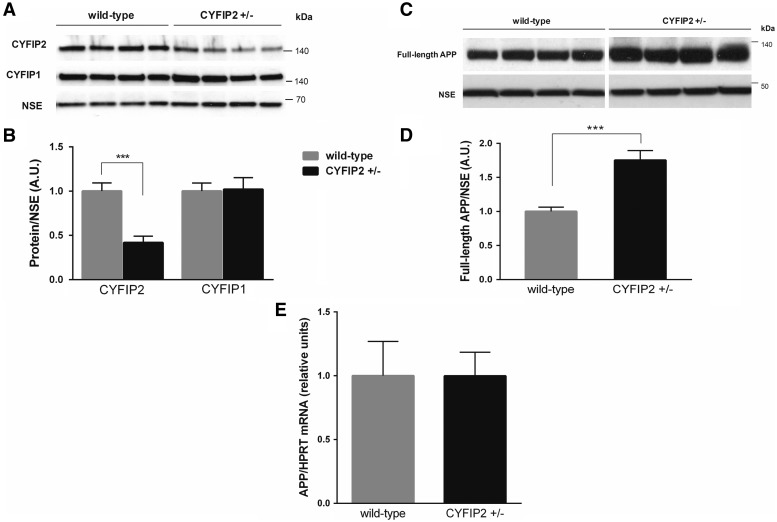
We studied the functional impact of reduced CYFIP2 expression in Cyfip2+/− mice. We confirmed earlier work ( Kumar et al. , 2013 ) that showed that these mutants have reduced CYFIP2 protein expression by ∼50% in hippocampal synaptosomes in comparison to wild-type littermates ( t = 4.90, P < 0.001; Fig. 2 A and B) (for specificity of hippocampal synaptosomes, see Supplementary Fig. 6 ), and ∼40% in total hippocampal lysates ( Supplementary Fig. 7A and Supplementary Data ). Further, we found that CYFIP1 expression was not altered in hippocampal synaptosomes from CYFIP2+/− mutants in comparison to wild-type littermates ( t = 0.13, P > 0.090; Fig. 2 A and B). Hence, we showed that CYFIP1 and CYFIP2 expression regulations are not dependent on each other.
Figure 2.
Reduced CYFIP2 expression does not affect CYFIP1 expression, but leads to increased APP expression at protein but not mRNA level in the hippocampus. ( A ) Representative western blots. ( B ) Quantification showed that CYFIP2 expression is reduced by ∼50% in hippocampal synaptosomes of Cyfip2+/− mice (black bar; n = 7) in comparison to wild-type littermates (grey bar, n = 8). CYFIP1 levels are not significantly changed in the same lysates. For quantification CYFIP1 and CYFIP2 expression were normalized to levels of NSE, a neuronal marker. ( C ) Representative western blots. ( D ) Quantification showed that APP protein expression is significantly increased ∼2-fold in hippocampal synaptosomes of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). ( E ) Quantitative PCR analysis showed that App mRNA levels are not elevated in hippocampi of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar) when normalized to the housekeeping gene Hprt ( n = 8 per genotype). Means ± SEM are shown. *** P < 0.001.
Reduced CYFIP2 leads to upregulation of APP protein, but not mRNA expression
At synapses APP mRNA is locally translated in an FMRP-dependent manner ( Westmark, 2013 ). As CYFIP2 interacts with FMRP ( Schenck et al. , 2001 ), we tested whether reduced CYFIP2 expression impacts on APP protein expression. Western blot analysis with an antibody directed against the C-terminus of APP showed that full-length APP protein expression was significantly elevated ∼2-fold in hippocampal synaptosomes of Cyfip2+/− mice in comparison to wild-type littermates ( t = 4.84, P < 0.001; Fig. 2 C and D). About a 2-fold upregulation of full-length APP protein expression in hippocampal synaptosomes from Cyfip2+/− mice was confirmed with an antibody reacting against an epitope in amyloid-β sequence (data not shown). We did not detect a significant increase of full-length APP expression in total hippocampal lysates ( Supplementary Fig. 7A and Supplementary Data ), suggesting that the increase occurs primarily at synapses. We studied whether or not the increased APP expression in Cyfip2+/− mice could be due to increased App mRNA expression. Quantitative RT-PCR (qPCR) analysis showed that the levels of App mRNA in hippocampus did not differ between Cyfip2+/− mutants and wild-type littermates ( t = 0.007, P > 0.99; Fig. 2 E). This result shows that reduced CYFIP2 expression impacts post-transcriptionally on APP expression.
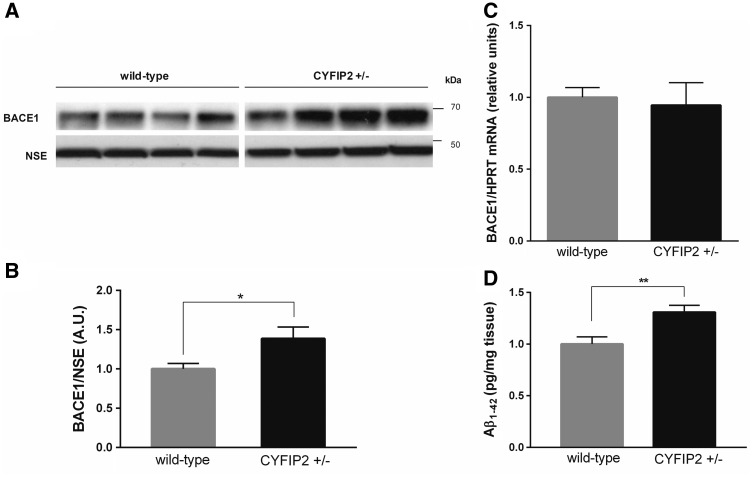
Reduced CYFIP2 leads to upregulation of BACE1 protein, but not mRNA, and abnormal production of amyloid-β 42
Translation of the mRNA encoding β-site APP cleaving enzyme 1 ( BACE1 ) is increased by amyloid-β 42 action on cultured neurons ( Sadleir et al. , 2014 ; Mamada et al. , 2015 ) and in an Alzheimer’s disease mouse model ( Caccamo et al. , 2015 ). As the elevated APP protein, but not mRNA, expression in Cyfip2+/− mutants ( Fig. 2 C–E) suggested that reduced CYFIP2 expression leads to increased translation of particular mRNAs, we tested whether BACE1 expression was altered. Western blot analysis showed that BACE1 protein expression was significantly elevated in hippocampal synaptosomes by ∼40% in Cyfip2+/− mice in comparison to wild-type littermates ( t = 2.36, P < 0.05; Fig. 3 A and B). This change was not observed in total hippocampal lysates ( Supplementary Fig. 7A and Supplementary Data ), possibly due to high levels of BACE1 expression in non-synaptic fractions (data not shown), suggesting BACE1 expression is mainly elevated at synapses in Cyfip2+/− mutants. Quantitative PCR analysis showed that the Bace1 mRNA expression level was not altered in the hippocampus of Cyfip2+/− mutants ( t = 0.48, P = 0.64; Fig. 3 C). Thus, reduced CYFIP2 expression impacts on the post-transcriptional expression of BACE1. In previous studies higher levels of BACE1 protein expression have been associated with abnormal APP cleavage and higher amyloid-β 42 production ( Caccamo et al. , 2015 ). Therefore, we studied whether amyloid-β 42 production is elevated in hippocampus of Cyfip2+/− mutants. As expected, an ELISA showed that amyloid-β 42 production was increased by ∼30% in hippocampus of Cyfip2+/− mutants in comparison to wild-type littermates ( t = 3.18, P < 0.01; Fig. 3 D). These findings indicate that reduced CYFIP2 expression affects APP processing.
Figure 3.
Reduced CYFIP2 expression leads to increased BACE1 expression at protein, but not mRNA level, and abnormal amyloid-β 42 production in the hippocampus. ( A ) Representative western blots. ( B ) Quantification showed that BACE1 protein expression is significantly upregulated by ∼40% in hippocampal synaptosomes of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). ( C ) Quantitative PCR analysis showed that Bace1 mRNA levels are not elevated in hippocampus of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). ( D ) An ELISA on whole hippocampi showed significantly elevated amyloid-β 42 levels by 30% in Cyfip2+/− mutants (black bar; n = 4) versus wild-type littermates (grey bar; n = 5). Means ± SEM are shown. * P < 0.05, ** P < 0.01.
Reduced CYFIP2 leads to upregulation of aCaMKII protein, but not mRNA, and increases tau phosphorylation
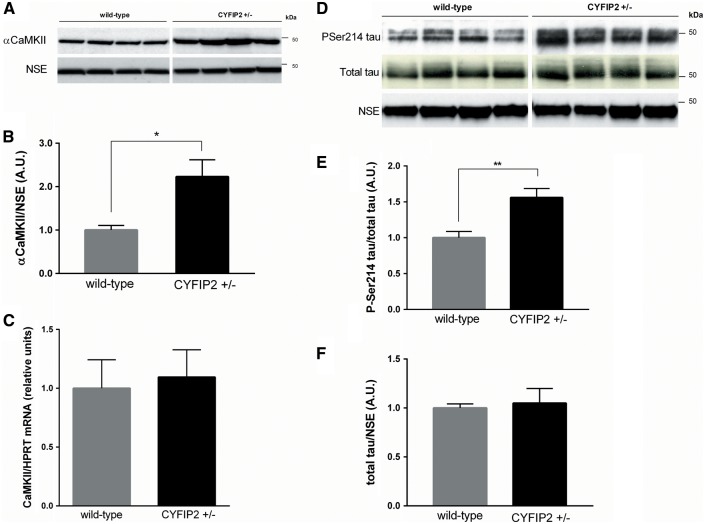
Like App mRNA, Camk2a (αCaMKII) mRNA is locally translated in an FMRP-dependent manner ( Napoli et al. , 2008 ). Moreover, recent studies have suggested that increased αCaMKII activity contributes to tau hyperphosphorylation in Alzheimer’s disease ( Ghosh and Giese, 2015 ). Therefore, we studied whether reduced CYFIP2 expression affects αCaMKII expression in the hippocampus. Western blot analysis showed that αCaMKII protein expression was significantly elevated ∼2.5-fold in hippocampal synaptosomes in Cyfip2+/− mice in comparison to wild-type littermates ( t = 3.05, P < 0.05; Fig. 4 A and B). This effect was not observed in total hippocampal lysates ( Supplementary Fig. 7A and Supplementary Data ). This is likely due to a dilution of synaptic signal in total lysates as αCaMKII is also highly expressed in somata and dendrites of hippocampal neurons ( Giese et al. , 1998 ). Quantitative PCR analysis showed that the Camk2a /αCaMKII mRNA expression level was not altered in the hippocampus of Cyfip2+/− mutants ( t = 0.39, P = 70; Fig. 4 C). This result suggests that CYFIP2 regulates translation of Camk2a /αCaMKII mRNA. We tested also whether the increased αCaMKII protein expression correlates with altered tau phosphorylation at serine-214 (S214), a site that is phosphorylated by CaMKII, and one of the key sites that are hyperphosphorylated in Alzheimer’s disease ( Lee et al. , 2001 ). The expression of total tau was not altered in Cyfip2+/− mutants in comparison to wild-type littermates ( t = 0.31, P = 076; Fig. 4 D and F). However, phosphorylation of tau at S214 was significantly increased by ∼60% in Cyfip2+/− mice in comparison to wild-type littermates ( t = 3.63, P < 0.01; Fig. 4 D and E).
Figure 4.
Reduced CYFIP2 expression leads to increased αCaMKII expression at protein but not mRNA level, and increased tau phosphorylation in the hippocampus. ( A ) Representative western blots. ( B ) Quantification showed that αCaMKII protein expression is significantly upregulated ∼2-fold in hippocampal synaptosomes of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). ( C ) Quantitative PCR analysis showed that Camk2a /αCaMKII mRNA levels are not elevated in hippocampus of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). ( D ) Representative western blots. ( E ) Quantification showed an ∼50% increase in phosphorylation of tau at serine-214 in hippocampal synaptosomes of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). ( F ) Quantification showed that total levels of tau did not differ in hippocampal synaptosomes of Cyfip2+/− mice (black bar) in comparison to wild-type littermates (grey bar; n = 8 per genotype). Means ± SEM are shown. * P < 0.05, ** P < 0.01.
Reduced CYFIP2 affects spine morphology on apical, but not basal, dendrites of hippocampal CA1 pyramidal neurons
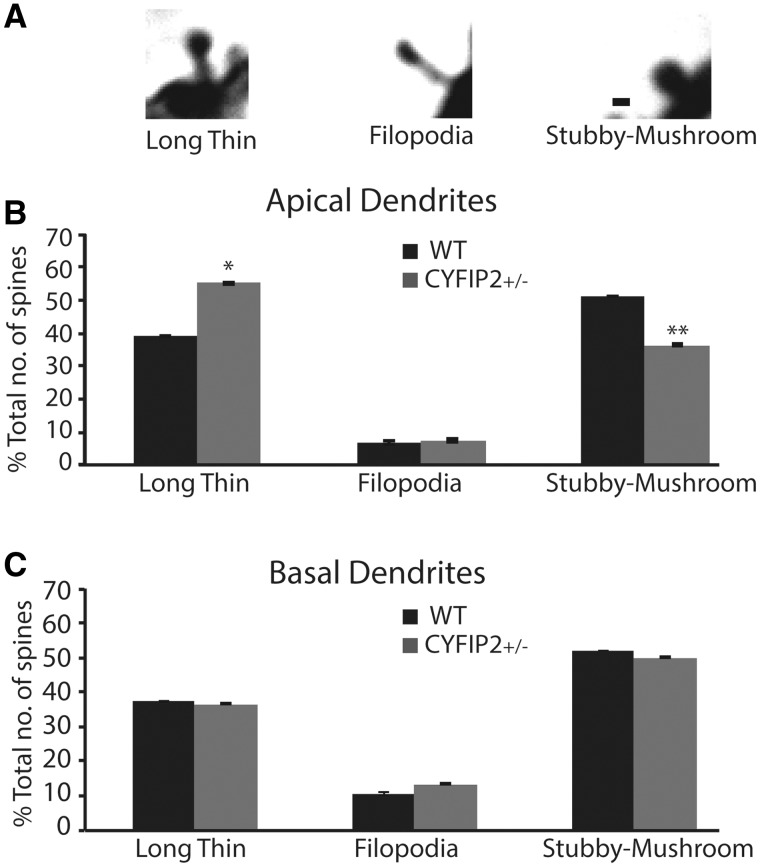
CYFIP2 not only binds to FMRP, it is also part of the WAVE complex ( Eden et al. , 2002 ), which regulates actin polymerization. As CYFIP1 is also part of the WAVE complex and because reduced CYFIP1 expression affects dendritic spine morphology of hippocampal CA1 pyramidal neurons ( De Rubeis et al. , 2013 ; Pathania et al. , 2014 ), we studied whether reduced CYFIP2 expression would also alter dendritic spines. Using Golgi-Cox staining, we analysed dendritic spines on apical and basal segments of CA1 pyramidal neurons of Cyfip2+/− mutants and wild-type littermates ( Supplementary Fig. 8 ). There was no significant difference in spine density between the genotypes for apical distal segments ( t = 0.095, P = 0.93) or for basal distal segments ( t = 0.10, P = 0.92) ( Supplementary Fig. 8C and Supplementary Data ). Next, we categorized the spines into three classes: long-thin, filopodia and stubby-mushroom spines according to widely used criteria ( Harris and Stevens, 1989 ; Harris et al. , 1992 ) ( Fig. 5 A). Comparison of these spine types on apical distal dendrites revealed a significant difference between genotypes ( Fig. 5 B). Compared to wild-type littermates, Cyfip2+/− mutants had a greater proportion of long-thin spines (55% for mutants versus 39% for wild-types; t = 3.45, P < 0.05) and a smaller proportion of stubby/mushroom spines (36% for mutants versus 51% for wild-types; t = 3.51, P < 0.01). The proportion of filopodia, however, remained constant between the two groups ( t = 0.61, P = 0.56). In contrast with the differences in apical spine morphology between genotypes, we found that the abundance of spine types on basal dendrites was not altered in Cyfip2+/− mutants (long-thin, t = 0.34, P = 0.75; filopodia, t = 0.99, P = 0.37; stubby/mushroom, t = 0.51, P = 0.63; Fig. 5 C). These findings show that reduced CYFIP2 expression affects spine morphology only in particular dendritic segments of CA1 pyramidal neurons.
Figure 5.
Reduced CYFIP2 expression leads to altered spine morphology in apical dendrites of CA1 pyramidal neurons. ( A ) Spines were classified as long-thin, filopodia, or stubby/mushroom, according to criteria set by Harris et al. (1992) . Spines were imaged under bright-field microscopy using a 100× oil-immersion objective. Scale bar = 0.5 µm. ( B ) Spines classified as a proportion of total number of spines for apical dendritic segments of dorsal CA1 pyramidal neurons. Wild-type (WT) controls (black bars, n = 5) were compared to Cyfip2+/− mutants (grey bars, n = 4) for long-thin spines (39% versus 55%), filopodia (6.7% versus 7.5%), and stubby/mushroom (51% versus 36%). Cyfip2+/− mutants have significantly more long thin spines, but less stubby-mushroom spines than wild-type mice. ( C ) Spines classified as a proportion of total number of spines for basal dendritic segments of dorsal CA1 pyramidal neurons. Wild-type controls (black bars, n = 5) were compared to Cyfip2+/− mutants (grey bars, n = 4) for long-thin, filopodia, and stubby/mushroom spines. No difference was found between the genotypes. Means ± SEM are shown. * P < 0.05; ** P < 0.01.
Reduced CYFIP2 impairs retention of spatial memory
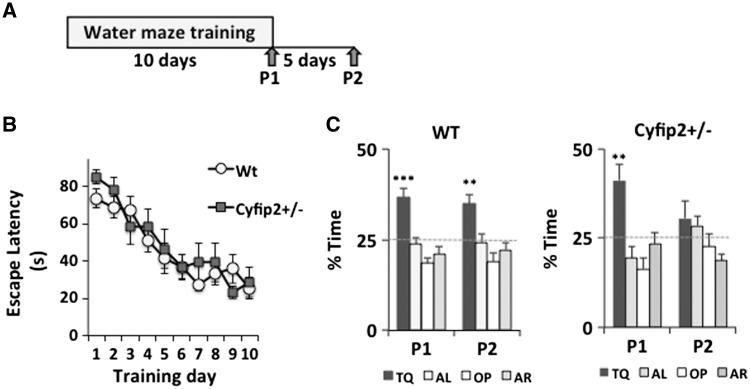
Hippocampus-dependent spatial memory is affected in the early stages of Alzheimer’s disease . To investigate whether reduced CYFIP2 expression affects spatial memory formation, we studied the Cyfip2+/− mutants and wild-type littermates in the hidden platform version of the water maze ( Fig. 6 ). The mice were trained with four trials per day for 10 days. The genotypes did not differ in latency to locate the platform [two-way ANOVA with repeated measures; effect of genotype F (1,14) = 0.29, P = 0.60; effect of training F (9,126) = 18.9, P < 0.001; genotype × training interaction F (9,126) = 0.91, P = 0.52] ( Fig. 6 B). Studies with a separate cohort of mice indicated that Cyfip2+/− mutants had normal visible platform learning (time to reach platform for wild-type mice, 9.6 ± 1.5 s; for mutants, 11.1 ± 2.7 s; one-way ANOVA on ranks H = 0.025, P = 0.88. This indicates that the Cyfip2+/− mutants were not impaired in swimming abilities, motivation, and vision. To assess hippocampus-dependent spatial memory, probe trials were performed. A probe trial given at the end of training Day 10 (P1) showed that Cyfip2+/− mutants formed normal spatial memory after training ( Fig. 6 C). However, a second probe trial given 5 days after first probe test (P2) revealed that Cyfip2+/− mutants were impaired in retention of spatial memory, in contrast with wild-type littermates ( Fig. 6 C). During these probe trials the average swimming speed did not significantly differ between genotypes [P1: wild-type mice, 21.7 ± 2.0 cm/s; mutants, 24.3 ± 2.7 cm/s; one-way ANOVA with genotype as variable, F (1,14) = 4.58, P = 0.051; P2: wild-type, 22.1 ± 1.60 cm/s; mutants, 24.1 ± 2.3 cm/s; one-way ANOVA with genotype as variable, F (1,14) = 3.90, P = 0.068]. Analysis of search time in the four quadrants during probe trial P1 showed that Cyfip2+/− mutants and wild-type littermates searched selectively [one-way ANOVA with analysis of the quadrant as variable; mutants, F (3,24) = 9.65, P < 0.001; wild-type mice, F (3,32) = 17.3, P < 0.001] ( Fig. 6 C). The mutants and wild-type mice spent more time searching in the target quadrant than in any other quadrant (Student-Newman-Keuls test; mutants, P < 0.001 target quadrant versus opposite quadrant and adjacent left quadrant, P = 0.002 target quadrant versus adjacent right quadrant; wild-type, P < 0.001 target quadrant versus opposite quadrant, adjacent left quadrant, adjacent right quadrant). On the other hand, during probe trial P2 the Cyfip2+/− mutants searched randomly, spending similar times in all quadrants [one-way ANOVA with quadrant as variable, F (3,24) = 2.55, P = 0.08], in contrast with wild-type littermates [one-way ANOVA with quadrant as variable, F (3,32) = 8.47, P < 0.001]. The wild-type mice spent more time searching in the target quadrant than in any other quadrant (Student-Newman-Keuls test; P < 0.001 target quadrant versus opposite quadrant, P = 0.002 target quadrant versus adjacent right quadrant, P = 0.004 target quadrant versus adjacent left quadrant). Taken together, these results indicate that Cyfip2+/− mutants are able to form spatial memories, but they cannot maintain these memories.
Figure 6.
Reduced CYFIP2 expression impairs the maintenance of spatial memory. ( A ) The experimental design for the hidden-platform training in the Morris water maze. Mice were trained with four trials per day for 10 days. Probe trials were given at the end of training Days 10 (P1) and 5 after the end of training (P2). ( B ) The time to reach the hidden platform did not differ between wild-type ( n = 9) and Cyfip2+/− mutant mice ( n = 7). Both groups improved with training. ( C ) Both wild-type and Cyfip2+/− mutants searched selectively target quadrant in a probe trial given immediately after training Day 10 (P1). However, Cyfip2+/− mutants did not search selectively in the second probe trial after 5 days of training (P2), in contrast to wild-type mice. Thus, the mutants cannot maintain spatial memory. TQ = target quadrant; OP = opposite quadrant; AL = adjacent left quadrant; AR = adjacent right quadrant. Means ± SEM; *** P < 0.001, ** P < 0.01.
CYFIP2 downregulation does not cause abnormal social and repetitive behaviour
In humans heterozygous microdeletion of chromosome 15, which includes the CYFIP1 gene, causes neurobehavioural disturbances including autism ( Abekhoukh and Bardoni, 2014 ; Cox and Butler, 2015 ). However, human genetic studies have not found a link between mutations of the CYFIP2 gene and autism. Further, CYFIP2 is expressed much later during cortical development in comparison with CYFIP1 ( Bonaccorso et al. , 2015 ). We studied whether reduced CYFIP2 expression in Cyfip2+/− mice affects social behaviour in the three-chamber task and repetitive behaviour in the marble burrowing task ( Supplementary Fig. 9 ), two behaviours that are impaired in mouse models of autism ( Silverman et al. , 2010 ; Santini et al. , 2013 ). We found that Cyfip2+/− mice did not significantly differ from wild-type littermates in these tasks. Thus, reduced CYFIP2 expression might not cause any neurodevelopmental abnormalities that lead to autism-like behaviours.
Discussion
Here, we show that expression of CYFIP2 protein is substantially reduced in Alzheimer’s disease forebrain and our functional studies in mice suggest that this CYFIP2 downregulation links together amyloid production, tau hyperphosphorylation and spatial memory loss.
Our previous studies on p25 dysregulation suggested that CYFIP2 expression could be reduced in Alzheimer’s disease ( Engmann et al. , 2011 ). Here, post-mortem expression analysis shows that this is the case. To assure that CYFIP2 expression is not simply reduced due to neuronal and synaptic loss in Alzheimer’s disease, we normalized CYFIP2 levels to expression of a neuronal housekeeping protein as well as to a synaptic vesicle protein. The decreased, normalized CYFIP2 expression in post-mortem Alzheimer’s disease forebrain indicates that CYFIP2 levels decline before neurons and synapses die. We suggest that CYFIP2 expression decreases already in the early stages of Alzheimer’s disease, as we found an ∼2-fold reduction in mild Alzheimer’s disease hippocampus (Braak stages I–II). Consistent with this idea, we found that CYFIP2 protein expression is reduced in an APP transgenic mouse line, the Tg2576 mouse, which models early Alzheimer’s disease ( Hsiao et al. , 1996 ; Stewart et al. , 2011 ). This finding also suggests that abnormal APP processing may be sufficient to decrease CYFIP2 expression. Follow-up investigations will be needed to characterize the mechanism by which abnormal APP processing downregulates CYFIP2 expression.
We also investigated CYFIP1 protein expression in the Alzheimer’s disease forebrain, as CYFIP1 and CYFIP2 are thought to have similar functions in vitro , although these two proteins do not compensate for each other in the haploinsufficient state in mice or humans ( De Rubeis et al. , 2013 ; Pathania et al. , 2014 ; Cox and Butler, 2015 ). In contrast to CYFIP2 expression, the level of CYFIP1 expression is not changed in post-mortem Alzheimer’s disease brain, with the exception that CYFIP1 expression is elevated in severe Alzheimer’s disease hippocampus. This particular increase in CYFIP1 expression in severe Alzheimer’s disease hippocampus, but not in severe Alzheimer’s disease superior temporal gyrus, may represent a late wave of further neurodegeneration in the disease, or alternatively a compensatory mechanism contributing to the survival of some hippocampal neurons in the disease ( West et al. , 1994 ). It is also important to note that a study of large rare copy number variants in Alzheimer’s disease among Caribbean Hispanics identified a nominal association between Alzheimer’s disease and a chromosomal duplication that includes the CYFIP1 gene ( Ghani et al. , 2012 ). This duplication was found in 10 Alzheimer’s disease cases (2.6%) versus three controls (0.8%).
Synapse loss and the resulting impact on synaptophysin expression is uneven within the Alzheimer’s disease hippocampus ( Heinonen et al. , 1995 ; Honer, 2003 ). Reduced synaptophysin expression is pronounced in area CA1, where most neuronal loss occurs ( West et al. , 1994 ). Additionally, reduced synaptophysin expression is thought to occur particularly in neurons with neurofibrillary tangles ( Coleman et al. , 2004 ; Arendt, 2009 ). However, in a western blot analysis where synaptophysin expression was normalized to expression of a neuronal marker protein we and others did not detect reduced synaptophysin expression in Alzheimer’s disease hippocampal homogenates ( Davidsson et al. , 1998 ; Tiwari et al. , 2015 ). This is in agreement with a recent study showing that in mammalian models of tauopathy there is little evidence that synaptophysin expression is lost when normalized to NSE expression in brain regions affected by significant neurodegeneration ( Bondulich et al. , 2016 ). This suggests that synaptophysin levels are not reduced in most neurons that have not yet died. A similar observation was made for synapsin I expression in severe Alzheimer’s disease cortex ( Kurbatskaya et al. , 2016 ), whereas expression of other synaptic markers is altered ( Honer, 2003 ; Kurbatskaya et al. , 2016 ).
We studied the functional impact of reduced CYFIP2 expression in heterozygous null mutant mice. We found that reduced CYFIP2 expression increases APP and αCaMKII expression in synapses at the post-transcriptional level, similar to previous findings with CYFIP1 ( Napoli et al. , 2008 ). FMRP binds to APP and CAMK2A /αCaMKII mRNAs and suppresses their translation ( Westmark and Malter, 2007 ; Lee et al. , 2010 ; Darnell et al. , 2011 ). Further, CYFIP2 binds to FMRP ( Schenck et al. , 2001 ) and has a binding motif for the translational initiator eIF4E ( Napoli et al. , 2008 ). Taken together, this suggests that CYFIP2 normally suppresses translation of APP and CAMK2A /αCaMKII mRNAs and that reduction of CYFIP2 expression is sufficient to increase translation of these mRNAs.
Studies with various models of Alzheimer’s disease have suggested that there is a feed-forward mechanism of amyloid-β 42 production that involves elevated translation of APP mRNA ( Westmark et al. , 2011 ; Westmark, 2013 ; Sadleir et al. , 2014 ; Caccamo et al. , 2015 ). In this scenario, increased APP protein expression would provide more material for abnormal APP cleavage resulting in further amyloid-β 42 production. The amyloid-β 42 -induced elevation of APP mRNA translation does not involve phosphorylation of the translation initiation factor eIF2α ( Sadleir et al. , 2014 ), and is not well understood at the mechanistic level. Our work suggests that reduction of CYFIP2 expression is an important step in upregulating APP mRNA translation in a feed-forward amyloid-β 42 production process. Additionally, recent studies have shown that amyloid-β 42 oligomers elevate BACE1 expression post-translationally, suggesting that not only increased APP protein expression, but also enhanced BACE1 protein expression may be part of a feed-forward mechanism for amyloid-β 42 production ( Sadleir and Vassar, 2012 ; Mamada et al. , 2015 ). Interestingly, we found that reduced CYFIP2 expression increases BACE1 expression at the post-transcriptional level. As BACE1 mRNA is not regulated by FMRP ( Buffington et al. , 2014 ), it is possible that this increased expression is secondary to elevated amyloid-β 42 production resulting from increased APP protein expression when CYFIP2 expression is reduced. Thus, reduction of CYFIP2 expression may be an essential step for increasing APP and BACE1 protein expression for the feed-forward production of amyloid-β 42 .
A fundamental question in Alzheimer’s disease research is how abnormal APP processing causes tau hyperphosphorylation, which is a prerequisite for neurofibrillary tangle formation. Our work suggests that reduced CYFIP2 expression links amyloid production with tau hyperphosphorylation. Reduced CYFIP2 expression is suggested to increase translation of mRNA encoding CAMK2A /αCaMKII, resulting in increased CaMKII activity, which leads to hyperphosphorylation of tau ( Sengupta et al. , 1998 ; Wang et al. , 2007 ). Tau is phosphorylated at many different sites and in this study we only investigated phosphorylation at S214, a site that is hyperphosphorylated in Alzheimer’s disease ( Lee et al. , 2001 ). Follow-up studies are needed to study the impact of reduced CYFIP2 expression on tau phosphorylation in much greater detail.
Our studies in mice have revealed that reduction of CYFIP2 expression leads to a characteristic spatial memory phenotype: spatial memory acquisition is not impaired but spatial memory cannot be retained and is lost within a few days in comparison to normal wild-type mice. To our knowledge there is only one other mutant mouse with a similar spatial memory phenotype, mice lacking the prion-like translational regulator CPEB3 ( Fioriti et al. , 2015 ). Thus, it is conceivable that CYFIP2 and CPEB3 regulate the same mRNA translation, which is needed for memory storage. Alternatively, reduced WAVE function may account for the spatial memory loss in CYFIP2 mutant mice, as the mutants have more immature spines and less mature (‘memory’) spines ( Bailey et al. , 2015 ). In any case, it is intriguing that reduced CYFIP2 expression links together spatial memory loss with amyloid production and tau hyperphosphorylation, three features and hallmarks of Alzheimer’s disease.
Recently, another laboratory has studied the phenotype of a newly generated Cyfip2 heterozygous null mutant mouse line ( Han et al. , 2015 ). These authors also showed that reducing CYFIP2 expression does not impact on CYFIP1 expression, but they suggested that CYFIP2 does not regulate APP and αCaMKII protein expression. One possible explanation for the discrepancy with our findings is that Han et al. (2015) studied APP and αCaMKII expression in cortical, and not hippocampal, neurons. In cortical neurons CYFIP2 expression is higher than in hippocampal neurons ( Han et al. , 2015 ) and a higher level of residual CYFIP2 expression may suppress a phenotype. However, Han et al. (2015) found a dendritic spine phenotype in cortical but not in hippocampal neurons in their CYFIP2 mutant mouse line. Therefore, it is conceivable that differences in genetic background account for the phenotypic differences. Our Cyfip2 heterozygotes were maintained on a C57BL/6N genetic background where the residual CYFIP2 has a S968F point mutation that reduces the half-life of CYFIP2 ( Kumar et al. , 2013 ), whereas Han et al. (2015) used Cyfip2 heterozygotes in a C57BL/6J genetic background. Applied to Alzheimer’s disease this could suggest that genetic modifiers may suppress the impact of reduced CYFIP2 expression on amyloid production, tau hyperphosphorylation and spatial memory loss in some patients.
In conclusion, to our knowledge this is the first report modelling a protein expression change detected in post-mortem Alzheimer’s disease brain that causes a combination of amyloid production, tau hyperphosphorylation and spatial memory loss, i.e. key features of the disease. This suggests that there are molecular ‘hubs’ that contribute to various aspects of Alzheimer’s disease, and that reduced CYFIP2 expression is one of these. Such ‘hubs’ could be ideal targets for pharmacological treatment where a multitude of neurodegenerative processes may be rectified.
Supplementary Material
Acknowledgements
We thank Dr Victoria Harris and Dr Ulrike Naumann (Department of Biostatistics, King’s College London) for their advice on statistical analysis. We are grateful to Dr Lotta Parviainen (Department of Neuroscience, King’s College London) for cultured glial cells.
Glossary
Abbreviations
- FMRP
fragile X mental retardation protein
- NSE
neuronal specific enolase
- STG
superior temporal gyrus
Funding
This work was supported by an international PhD scholarship from King’s College London to S.S.T. and the British Medical Research Council (MRC) to K.P.G. T.H. is supported by the Hungarian Brain Research Programme (KTIA_13_NAP-A-II/7). Human tissue samples were supplied by the London Neurodegenerative Disease Brain Bank, which receives funding from the MRC and as part of the Brains for Dementia Research programme, jointly funded by Alzheimer’s Research UK and Alzheimer Society.
Supplementary material
Supplementary material is available at Brain online.
References
- Abekhoukh S, Bardoni B . CYFIP family proteins between autism and intellectual disability: links with fragile X syndrome . Front Cell Neurosci 2014. ; 8 : 81 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt T . Synaptic degeneration in Alzheimer's disease . Acta Neuropathol 2009. ; 118 : 167 – 79 . [DOI] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Harris KM . Structural components of synaptic plasticity and memory consolidation . Cold Spring Harb Perspect Biol 2015. ; 7 : a021758 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondulich MK, Guo T, Meehan C, Manion J, Rodriguez-Martin T, Mitchell JC, et al. . Tauopathy induced by low level expression of a human brain-derived tau fragment in mice is rescued by phenylbutyrate . Brain 2016. ; 139 : 2290 – 306 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorso CM, Spatuzza M, DiMarco B, Gloria A, Barrancotto G, Cupo A, et al. . Fragile X mental retardation protein (FMRP) interacting proteins exhibit different expression patterns during development . Int J Dev Neurosci 2015. ; 42 : 15 – 23 . [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E . Alzheimer's disease affects limbic nuclei of the thalamus . Acta Neuropathol 1991. ; 81 : 261 – 8 . [DOI] [PubMed] [Google Scholar]
- Buffington SA, Huang W, Costa-Mattioli M . Translational control in synaptic plasticity and cognitive dysfunction . Annu Rev Neurosci 2014. ; 37 : 17 – 38 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Branca C, Talboom JS, Shaw DM, Turner D, Ma L, et al. . Reducing ribosomal protein S6 kinase 1 expression improves spatial memory and synaptic plasticity in a mouse model of Alzheimer’s disease . J Neurosci 2015. ; 35 : 14042 – 56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman P, Federoff H, Kurlan R . A focus on the synapse for neuroprotection in Alzheimer disease and other dementias . Neurology 2004. ; 63 : 1155 – 62 . [DOI] [PubMed] [Google Scholar]
- Cox DM, Butler MG . The 15q11.2 BP1-BP2 microdeletion syndrome: a review . Int J Mol Sci 2015. ; 42 : 15 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. . FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism . Cell 2011. ; 146 : 247 – 61 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsson P, Blennow K . Neurochemical dissection of synaptic pathology in Alzheimer’s disease . Int Psychogeriatr 1998. ; 10 : 11 – 23 . [DOI] [PubMed] [Google Scholar]
- Deacon RM . Digging and marble burying in mice: simple methods in vivo identification of biological impace . Nat Protocols 2006. ; 1 : 122 – 24 . [DOI] [PubMed] [Google Scholar]
- DeKosky ST, Scheff SW . Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity . Ann Neurol 1990. ; 27 : 457 – 64 . [DOI] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernandez E, Di Marino D, Buzzi A, et al. . CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation . Neuron 2013. ; 79 : 1169 – 82 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW . Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck . Nature 2002. ; 418 : 790 – 3 . [DOI] [PubMed] [Google Scholar]
- Engmann O, Hortobagyi T, Thompson AJ, Guadagno J, Troakes C, Soriano S, et al. . Cyclin-dependent kinase 5 activator p25 is generated during memory formation and is reduced at an early stage in Alzheimer's disease . Biol Psychiatry 2011. ; 70 : 159 – 68 . [DOI] [PubMed] [Google Scholar]
- Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, et al. . The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3 . Neuron 2015. ; 86 : 1433 – 48 . [DOI] [PubMed] [Google Scholar]
- Genheden M, Kenney JW, Johnston HE, Manousopoulou A, Garbis SD, Proud CG . BDNF stimulation of protein synthesis in cortical neurons requires MAP kinase-interacting kinase MNK1 . J Neurosci 2015. ; 35 : 972 – 84 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani M, Pinto D, Lee JH, Grinberg Y, Sato C, Moreno D, et al. . Genome-wide survey of large rare copy number variants in Alzheimer’s disease among Caribbean hispanics . G3 (Bethesda) 2012. ; 2 : 71 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Giese KP . Calcium/calmodulin-dependent kinase II and Alzheimer’s disease . Mol Brain 2015. ; 8 : 78 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese KP, Fedorov NB, Filipkowski RK, Silva AJ . Autophosphorylation at Thr 286 of the a calcium-calmodulin kinase II in LTP and learning . Science 1998. ; 279 : 870 – 3 . [DOI] [PubMed] [Google Scholar]
- Giese KP . Generation of the Cdk5 activator p25 is a memory mechanism that is affected in early Alzheimer’s disease . Front Mol Neurosci 2014. ; 7 : 36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Chen H, Gennarino VA, Richman R, Lu HC, Zoghbi HY . Fragile X-like behaviors and abnormal cortical dendritic spines in cytoplasmic FMR1-interacting protein 2-mutant mice . Hum Mol Genet 2015. ; 24 : 1813 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ . The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics . Science 2002. ; 297 : 353 – 6 . [DOI] [PubMed] [Google Scholar]
- Harris KM, Stevens JK . Dendritic spines of CA1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics . J Neurosci 1989. ; 9 : 2982 – 97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B . Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation . J Neurosci 1992. ; 12 : 2685 – 705 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen O, Soininen H, Sorvari H, Kosunen O, Paljärvi L, Koivistro E, Riekkinen PJ, Sr . Loss of synaptophysin-like immunoreactivity in the hippocmapal formation is an early phenomenon in Alzheimer’s disease . Neuroscience 1995. ; 64 : 375 – 84 . [DOI] [PubMed] [Google Scholar]
- Honer WG . Pathology of presynaptic proteins in Alzheimer’s disease: more than simple loss of terminals . Neurobiol Aging 2003. ; 24 : 1047 – 62 . [DOI] [PubMed] [Google Scholar]
- Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, et al. . Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice . Science 1996. ; 274 : 99 – 102 . [DOI] [PubMed] [Google Scholar]
- Kumar V, Kim K, Joseph C, Kourrich S, Yoo SH, Huang HC, et al. . C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response . Science 2013. ; 342 : 1508 – 12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurbatskaya K, Phillips EC, Croft CL, Dentoni G, Hughes MM, Wade MA, et al. . Upregulation of calpain activity precedes tau phosphorylation and loss of synaptic proteins in Alzehimer’s disease brain . Acta Neuropathol Commun 2016. ; 4 : 34 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Kim HH, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, et al. . hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies . Nat Struct Mol Biol 2010. ; 17 : 732 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ . Neurodegenerative tauopathies . Annu Rev Neurosci 2001. ; 24 : 1121 – 59 . [DOI] [PubMed] [Google Scholar]
- Mamada N, Tanokashira D, Hosaka A, Kametani F, Tamaoka A, Araki W . Amyloid ß-protein oligomers upregulate the ß-secretase, BACE1, through a post-translational mechanism involving its altered subcellular distrbution in neurons . Mol Brain 2015. ; 8 : 73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello E, Epis R, DiLuca M . Amyloid flirting with synaptic failure: towards a comprehensive view of Alzheimer’s disease pathogenesis . Eur J Pharmacol 2008. ; 585 : 109 – 18 . [DOI] [PubMed] [Google Scholar]
- McAleese SM, Dunbar B, Fothergill JE, Hinks LJ, Day IN . Complete amino acid sequence of the neurone-specific gamma isozyme of enolase (NSE) from human brain and comparison with the non-neuronal alpha form (NNE) . Eur J Biochem 1988. ; 178 : 413 – 17 . [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. . The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP . Cell 2008. ; 134 : 1042 – 54 . [DOI] [PubMed] [Google Scholar]
- Panja D, Kenney JW, D’Andrea L, Zalfa F, Vedeler A, Wilbrand K, et al. . Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK . Cell Rep 2014. ; 9 : 1430 – 45 . [DOI] [PubMed] [Google Scholar]
- Pathania M, Davenport EC, Muir J, Sheehan DF, Lopez-Domenech G, Kittler JT . The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines . Transl Psychiatry 2014. ; 4 : e374 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooler AM, Usardi A, Evans CJ, Philpott KL, Noble W, Hanger DP . Dynamic association of tau with neuronal membranes is regulated by phosphorylation . Neurobiol Aging 2012. ; 33 : 431.e27 – 38 . [DOI] [PubMed] [Google Scholar]
- Sadleir KR, Elmer WA, Kaufman RJ, Osten P, Vassar R . Genetic inhibition of phosphorylation of the translation initiation factor eIF2α does not block Aß-dependent elevation of BACE1 and APP levels or reduce amyloid pathology in a mouse model of Alzheimer’s disease . PLoS One 2014. ; 9 : e101643 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadleir KR, Vassar R . Cdk5 protein inhibition and Ab42 increase BACE1 protein level in primary neurons by a post-transcriptional mechanism: implications of Cdk5 as a therapeutic target for Alzheimer's disease . J Biol Chem 2012. ; 287 : 7224 – 35 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini R, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, et al. . Exaggerated translation causes synaptic and behavioural aberrations associated with autism . Nature 2013. ; 493 : 411 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN . Behavioural phenotyping assays for mouse models of autism . Nat Rev 2010. ; 11 : 490 – 502 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL . A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P . Proc Natl Acad Sci USA 2001. ; 98 : 8844 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Kabat J, Novak M, Wu Q, Grundke-Iqbal I, Iqbal K . Phosphorylation of tau at both Thr-231 and Ser-262 is required for maximal inhibition of its binding to microtubules . Arch Biochem Biophys 1998. ; 357 : 299 – 309 . [DOI] [PubMed] [Google Scholar]
- Stewart S, Cacucci F, Lever C . Which memory task for my mouse? A systematic review of spatial memory performance in the Tg2576 Alzheimer's mouse model . J Alzheimers Dis 2011. ; 26 : 105 – 26 . [DOI] [PubMed] [Google Scholar]
- Südhof TC, Lottspeich F, Greengard P, Mehl E, Jahn R . The cDNA and derived amino acid sequences for rat and human synaptophysin . Nucleic Acids Res 1987. ; 15 : 9607 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry RD, Masliah E, Salmon DP, Buters N, DeTeresa, Hill R, et al. . Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment . Ann Neurol 1991. ; 30 : 572 – 80 . [DOI] [PubMed] [Google Scholar]
- Tiwari SS, d’Orange M, Troakes C, Shurovi BN, Engmann O, Noble W, et al. . Evidence that the presynaptic vesicle protein CSPalpha is a key player in synaptic degeneration and protection in Alzheimer’s disease . Mol Brain 2015. ; 8 : 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, Grundke-Iqbal I, Iqbal K . Kinases and phosphatases and tau sites involved in Alzheimer neurofibrillary degeneration . Eur J Neurosci 2007. ; 25 : 59 – 68 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Coleman PD, Flood DG, Troncoso JC . Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease . Lancet 1994. ; 344 : 769 – 72 . [DOI] [PubMed] [Google Scholar]
- Westmark CJ . What's hAPPening at synapses? The role of amyloid beta-protein precursor and beta-amyloid in neurological disorders . Mol Psychiatry 2013. ; 18 : 425 – 34 . [DOI] [PubMed] [Google Scholar]
- Westmark CJ, Malter JS . FMRP mediates mGluR5-dependent translation of amyloid precursor protein . PLoS Biol 2007. ; 5 : e52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmark CJ, Westmark PR, O'Riordan KJ, Ray BC, Hervey CM, Salamat MS, et al. . Reversal of fragile X phenotypes by manipulation of AbetaPP/Abeta levels in Fmr1KO mice . PLoS One 2011. ; 6 : e26549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Price J . Evidence for multiple precursor cell types in the embryonic rat cerebral cortex . Neuron 1995. ; 14 : 1181 – 8 . [DOI] [PubMed] [Google Scholar]
- Yang M, Silverman JL, Crawley JN . Automated three-chambered social approach task for mice . Curr Protoc Neurosci 2011. ; Chapter 8: Unit 8.26.1–16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.