Abstract
The demand to study the cellular localization of specific lipids has led to recent advances in lipid probes and microscopy. Alkyne lipids bear a small, noninterfering tag and can be detected upon click reaction with an azide-coupled reporter. Fluorescent alkyne lipid imaging crucially depends on appropriate azide reporters and labeling protocols that allow for an efficient click reaction and therefore a sensitive detection. We synthesized several azide reporters with different spacer components and tested their suitability for alkyne lipid imaging in fixed cells. The implementation of a copper-chelating picolyl moiety into fluorescent or biotin-based azide reagents strongly increased the sensitivity of the imaging routine. We demonstrate the applicability and evaluate the performance of this approach using different lipid classes and experimental setups. As azide picolyl reporters allow for reduced copper catalyst concentrations, they also enable coimaging of alkyne lipids with multiple fluorescent proteins including enhanced green fluorescent protein. Alternatively, and as we also show, microscopy of alkyne lipids can be combined with protein detection by immunocytochemistry. In summary, we present a robust, sensitive, and highly versatile protocol for the labeling of alkyne lipids with azide-coupled reporters for fluorescence microscopy that can be combined with different protein detection and imaging techniques.
Keywords: click chemistry, lipid imaging, copper-catalyzed azide-alkyne cycloaddition, immunohistochemistry, fluorescent proteins, fluorescence microscopy, molecular imaging, chemical synthesis, lipid biochemistry
A plethora of lipid species and a delicate network of lipid-lipid and lipid-protein interactions contribute to the form and function of biological membranes (1). It is still challenging to analyze the transport and localization of lipids in intact membranes by fluorescence microscopy. In contrast to macromolecules such as proteins, a direct fluorescent tagging of lipids [reviewed in (2)] remains difficult. Any tag may alter the structural and physical characteristics of these small, hydrophobic molecules and may therefore influence their localization (3), spatial dynamics (4), and metabolism (5). Furthermore, most lipids lack reactive functionalities toward aldehydes and hence fixation procedures cannot completely abolish lipid mobility (6). Therefore great care should be taken when interpreting data using any lipid probe except isotope-labeled lipids.
Generally the impact of the tag on the lipid characteristics should be kept as small as possible. Because isotope-labeled lipids (7–10) are not detectable by fluorescence microscopy, the need for small tags has brought alkyne lipids into the focus of intense research. The small nonfluorescent alkyne tag is introduced into the lipid structure and allows for its detection after conjugation to a reporter molecule bearing an azide. This chemical reaction can be performed in biological environments and under physiological conditions using the copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC) (11, 12), a type of click reaction (13) from a set of useful bioorthogonal labeling strategies (14).
Apart from imaging (15–19), alkyne lipids have been successfully used as lipid probes in a variety of applications, such as protein lipidation [reviewed in (20)], lipid-protein interaction studies (21, 22), metabolic tracing (23), and in vitro enzymatic assays (24). This broad applicability further allows the experimenter to gain high-content data regarding metabolism and localization of the lipid with different experimental methods from one individual sample.
As for the positioning of the alkyne moiety within the lipid molecule, several approaches have been taken. The tag has been synthetically introduced at the omega-terminal position of fatty acyl chains (23, 25), in sphingoid bases (24), as part of phospholipid headgroup precursors (16), within sterol side chains (18, 26, 27), or at a cholesterol methyl group (19). Given its size and properties, the alkyne group does not alter the polarity and only minimally changes the structure and hence biochemical characteristics of the lipid (24, 28).
While copper-free reactions of strain-promoted alkynes and azides have been described (29), a main strength of CuAAC is its speed and performance at physiological reaction conditions. However, the requirement for the copper catalyst accounts for the limited compatibility of this type of click reaction with studies in living biological specimens, as the catalyst is cytotoxic [discussed in (30)].
For imaging of alkyne lipids in biological membranes, the feasibility of CuAAC highly depends on azide reporters that are able to penetrate the hydrophobic interior of the membrane in order to meet the alkyne reaction partner and simultaneously engage with the copper catalyst. Thus, the characteristics of the azide detection reagent are likely to have a direct influence on labeling sensitivity. For labeling of cell surface proteins or biomolecules inside the cell (31, 32), the introduction of copper-chelating moieties into the azide reporter have greatly enhanced the reaction rate of CuAAC and thus increased its sensitivity. This approach also represents a promising strategy for alkyne lipid detection in membranes.
Here we describe advanced azide reporters for lipid localization studies using alkyne lipids as tracers. Having optimized the reporter and reaction procedures, we present a set of robust and convenient protocols for the subcellular detection of alkyne lipids in fixed cells by CuAAC and fluorescence microscopy. Specific emphasis has been given to i) high sensitivity and ii) compatibility of the procedures with colocalizations studies involving immunocytochemistry and fluorescent protein markers.
MATERIALS AND METHODS
Chemical synthesis
The syntheses of the azide detection reagents azido-PEG3-biotin (AP3Btn), 8-(5-azidopentyl)-4,4-difluor-1,3,5,7-tetramethyl-4-bora-3a,4a-s-indacene-2,6-disulfonic acid disodium salt (ASBDP), and of all alkyne lipids used here were described earlier (18, 23, 24). The syntheses of the reagents azido-PEG6-biotin (AP6Btn), azido-PEG10-biotin (AP10Btn), azido-picolyl-PEG2-biotin (APpic2Btn), and azido-picolyl-sulfo-tetramethyl-BODIPY (ApicSBDP) are provided in the supplemental data. Azido-picolyl-sulfo-Cy5 (ApicSCy5; #CLK-AZ118) and azido-sulfo-Cy5 (ASCy5; #CLK-1177) were from Jena Bioscience, Germany. The molecular structures of all reagents are available in supplemental Fig. S1.
Plasmids
pcDNA3.1 (Invitrogen) was used to construct the plasmids for colocalization studies in alkyne lipids and mitochondria. All plasmids contain the native presequence (MLRAALSTARRGPRLSRLL) of rat alcohol dehydrogenase (ALDH) followed by the first 17 amino acids of ALDH (SAAATSAVPAPNQQPEV), a linker (8 amino acids; FCNQIFSR), and one of the fluorescent proteins enhanced green fluorescent protein (EGFP), enhanced yellow fluorescent protein (EYFP), tdTomato, mCherry, mPlum (all from Clonetech), monomeric teal fluorescent protein 1 (mTFP1; Allele Biotechnology), monomeric red fluorescent protein 1 (mRFP1; Roger Y. Tsien laboratory), Emerald GFP (EmGFP; Invitrogen), or monomeric Kusabira Orange 1 (mKO1; MBL). All constructs were sequence verified. Phylogenetic tree analysis was performed using the ClustalW2 website.
Antibodies
The antibodies, their dilutions, and incubation conditions used in this study were as follows: Anti-Tom20 (Santa Cruz #sc-11415, 1:1,000, 1 h room temperature), anti-PDI (Stressgen #SPA891, 1:200, 1 h room temperature), and anti-Lamp1 (Abcam #ab25245, 1:1,000, 1 h room temperature). Secondary antibodies were labeled with Alexa555 (Invitrogen).
Cell culture
HuH7 hepatoma cells were grown in RPMI 1640 (Gibco #72400) supplemented with 0.1 mM nonessential amino acids and 10% FCS (Gibco #10270). A172 glioma cells and MEF (mouse embryonic fibroblast) cells were cultured in DMEM medium (Gibco #31966) containing 10% fetal calf serum and 1% penicillin/streptomycin (for MEF cells, Gibco). All cells were maintained at 37°C and 5% (v/v) CO2.
Click labeling and sample preparation for microscopy
A robust, basic protocol for alkyne lipid imaging in fixed cells has been described before (18). Based on the published protocol we have now developed three optimized protocol variants to enhance the sensitivity of alkyne lipid microscopy (variant A), and/or to combine imaging of alkyne lipids and fluorescent fusion proteins (variant B), or to allow for parallel protein detection by immunocytochemistry (variant C).
Click-labeling protocol (variant A).
Cells were grown on glass coverslips and incubated with medium containing FCS and 10 or 2.5 μM alkyne lipid for 16 h and subsequently washed with 1% delipidated BSA in PBS, and with PBS alone. Cells were fixed in 3.7% formalin in PBS for at least 16 h and washed twice with PBS. Generally, all wash steps of fixed cells were performed for 10 min while gently agitating the samples. After washing once with 500 mM Tris/HCl, pH 7.4 and twice with click buffer (100 mM HEPES/KOH, pH 7.4), the samples were placed in a water bath at 43°C. The buffer was removed and 1 ml of prewarmed click buffer containing the azide detection reagent (generally at 10 μM concentration, but see figure legends for occasional exceptions) was added, followed instantly by the injection of 20 μl of 10 mM tetrakis(acetonitrile)copper(I) fluoroborate (CuTFB) in acetonitrile into the reagent solution (final concentrations: 200 μM CuTFB and 2% acetonitrile in the reaction mix) and brief agitation. After 15 min incubation, another 10 μl of copper catalyst solution was added. After another 15 min, the reagent solution was removed and samples were washed with click buffer two times. Samples probed with biotinylated azide reagents were washed sequentially with PBS and blocking buffer (PBS containing 2% BSA), and then incubated upside down on parafilm in a 30 μl drop of 1:120 streptavidin-Alexa488 (Dianova) in blocking buffer for 60 min, followed by two wash steps with blocking buffer. These steps were left out for samples probed with fluorescent azide reporters. Finally, all samples were washed with PBS six times and once with water before mounting in Mowiol 4-88 (Calbiochem).
Click-labeling protocol (variant B) for coimaging with fluorescently labeled proteins.
Cells were transfected with vectors containing cDNA coding for fluorescent proteins using Lipofectamine2000 (Invitrogen) according to the manufacturers protocol. After 16 h, cells were incubated for 2 h with 1 μM (2S,3R)-2-aminooctadec-17-yn-1,3-diol (alkyne-sphinganine) added to the growth medium from ethanolic stock solutions. Cells were fixed by adding 1 ml of 3.7% formalin in click buffer and incubation for at least 16 h. Samples were washed sequentially with click buffer, 155 mM ammonium acetate, and click buffer before being placed in a water bath at 43°C. The buffer was removed and 1 ml of prewarmed click buffer containing 10 µM ApicSCy5 or ASBDP was added per well. Instantly, the reaction was started by addition of 20 µl of 10 mM or 100 mM CuTFB in acetonitrile (final concentrations: 200 μM or 2 mM, respectively), followed by brief agitation. After 15 min incubation, another 10 μl of copper catalyst solution was added. After a total of 30 min, the reagent solution was removed and the samples were washed with click buffer six times. Samples were mounted in Mowiol 4-88.
Click-labeling protocol (variant C) for the combination with immunofluorescence and cell organelle stains.
Cells were grown on glass coverslips and incubated with medium containing FCS and 1 µM nonadec-9-cis-en-18-ynoic acid (alkyne-oleate) for 24 h and washed with PBS. Samples were fixed in 3.7% formalin in PBS for 10 min and washed sequentially with PBS, 155 mM ammonium acetate, and PBS.
For immunofluorescence, antibody staining was performed before the click reaction. In detail, cells were permeabilized for 15 min in permeabilization buffer A [PBS/1% cold fish gelantine/0.01% saponin (Applichem #A2542)] for permeabilization of plasma membrane only or permeabilization buffer B (PBS/1% cold fish gelantine/0.1% saponin) for permeabilization of all cellular membranes. Cells were incubated with the primary and secondary antibodies in the corresponding permeabilization buffer for 1 h. Before the click reaction, cells were washed with click buffer before placing in a water bath at 43°C. The buffer was removed and 1 ml of prewarmed click buffer containing 10 µM ASBDP was added per well. Instantly, the reaction was started by addition of 20 µl of 100 mM CuTFB in acetonitrile (final concentration 2 mM) and brief agitation. After 1 h incubation, the reagent solution was removed and the samples were washed sequentially with click buffer, 20 mM EDTA, 155 mM ammonium acetate, and three times with PBS. Coverslips were mounted in Mowiol 4-88.
Mitotracker CMXRos (Molecular Probes #M7512) staining was performed according to the manufacturer’s instructions before the fixation and click reaction. In brief, living cells were incubated with 50 nM Mitotracker probe in growth medium for 15 min before washing and fixation as above. Before the click reaction, cells were washed with click buffer before placing in a water bath at 43°C. The buffer was removed and 1 ml of prewarmed click buffer containing 10 µM ASBDP was added per well. Instantly, the reaction was started by addition of 20 µl of 100 mM CuTFB in acetonitrile (final concentration 2 mM), followed by brief agitation. After 1 h incubation, the reagent solution was removed and the samples were washed sequentially with click buffer, 20 mM EDTA, 155 mM ammonium acetate, and three times with PBS. Coverslips were mounted in Mowiol 4-88.
Staining of cellular nuclei by 4’,6-diamidino-2-phenylindole (DAPI) and lipid droplets by 4,4-difluoro-2.3,5.6-bis-tetramethylene-4-bora-3a,4a-diaza-s-indacene (LD540) (33) was performed directly before mounting.
Microscopy
Epifluorescence microscopy was performed using a Zeiss Observer.Z1 microscope (Carl Zeiss) equipped with a CoolSNAP K4 (Photometrics) or Orca-Flash4.0 (Hamamatsu) camera (see figure legends for details), a Polychrome V 150 W xenon lamp (Till Photonics), and a Plan-Apochromate 63× (1.40 NA) DIC Oil or a Fluar 40× Oil (1.30 NA) objective (Zeiss). Optical sectioning was performed using Zeiss ApoTome.
Image processing and data analysis
Images were processed using Fiji (34) or Adobe Photoshop 6.0. Maximum intensity projections were calculated from Z-stacks using Zeiss Zen Blue software. For comparing the sensitivity of the different azide detection reagents, all samples were processed in parallel and images were taken at the same exposure times and postprocessed in the same way using Fiji. If indicated, images were corrected for illumination by dividing pixel values by those of an image displaying illumination inhomogeneities of the microscope setup. Cell outlines were manually marked as regions of interest (ROIs) and the mean alkyne lipid signal of each ROI was measured. See figure legends for details on the quantification.
Calculations and statistical analyses were carried out using Microsoft Excel 2011 and Graphpad Prism 6 software. One-way ANOVA was performed to determine whether values were different from each other with statistical significance. Asterisks refer to P values computed for the data: **** P < 0.0001; *** 0.0001< P < 0.001; ** 0.001< P < 0.01; * 0.01 < P < 0.05; n.s. (not significant), P > 0.05.
Molecular structures were drawn using ChemDraw software (CambridgeSoft Corporation, Version 8.0 Ultra), or Chem Doodle software (iChemLabs, Version 7.0). To obtain 3D models of molecules, structures were energetically optimized in Avogadro software, Version 1.1.1 (35), using the MMFF94 force-field protocol to minimize energy. Optimized structures were visualized and intramolecular distances measured using PyMOL software (Schrödinger, LLC). The dipalmitoylphosphatidylcholine (DPPC) bilayer model was kindly provided by Jeffrey Klauda, University of Maryland, as a free download file [323.15 K, 72 lipids, CHARMM36 force field simulation, tensionless ensemble (NPT) (36)].
RESULTS
We have previously developed a CuAAC-based protocol for fluorescence microscopy imaging of alkyne lipids in fixed cells (18). The established protocol uses 2 mM CuTFB and either a directly fluorescent azide reporter (ASBDP) or a biotin-conjugated azide reporter (AP3Btn) with subsequent binding of a fluorescent streptavidin probe [see Fig. 1 for structural representations of all azide reporters used here and previously (18)].
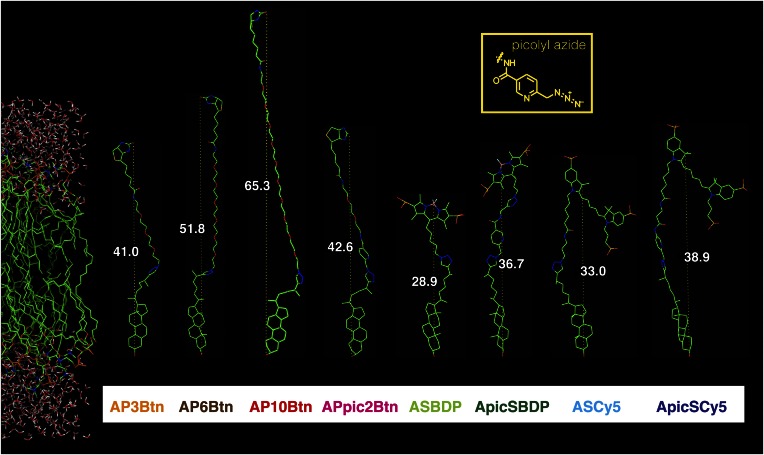
Fig. 1.
Structures of azide reporters used in this study, after click reaction with (25R)-25-ethinyl-26-nor-3β-hydroxycholest-5-en (alkyne-cholesterol) and compared with a DPPC bilayer. The azide reporter reagents tested in this study, each click-reacted to alkyne-cholesterol, are depicted next to a DPPC bilayer [left side (36)], including tightly associated water molecules, as a model membrane. The conjugates are displayed to scale to each other and to the membrane. The hydroxyl group of cholesterol has been lined up with the bilayer surface to give a realistic approximation of the possible orientation of the conjugates inside the bilayer, under the assumption that they are in a stretched conformation. Azido-biotin reagents with different polyethylene glycol (PEG) spacer lengths were included in the study (AP3Btn, APpic2Btn, AP6Btn, and AP10Btn), as well as sulfonated reagents based on the fluorescent dyes tetramethyl-BODIPY (ASBDP and ApicSBDP) and Cy5 (ASCy5 and ApicSCy5). Three of the reagents feature a copper-chelating picolyl moiety (pic) in their spacer component. Lengths of the click-conjugates are given in ångström (white numbers and dotted lines).
Structural optimization of azide reporters for alkyne lipid imaging
In order to optimize the established imaging protocol, we aimed to enhance the sensitivity of the detection by optimizing the azide reporter for efficient penetration into the cellular membranes. Accessibility issues of the reporter to the tracer may be minor for alkyne lipids that carry the tag at the headgroup placing it outside of the membrane (Fig. 2). However, they are critical for lipids where the alkyne moiety is positioned at the terminus of the hydrocarbon chain and thus buried within the membrane. Moreover, access to more rigid lipid molecules such as the cholesterol analog alkyne-cholesterol is particularly challenging. Rather bulky or hydrophilic azide reporters without linker groups will thus yield only a low signal intensity when used in the conventional protocol.
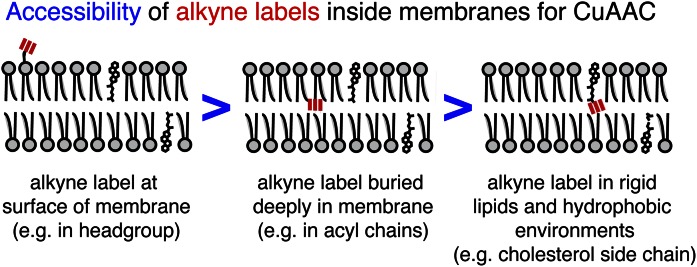
Fig. 2.
Schematic representation of alkyne label accessibility as a function of its position in a bilayer. The accessibility of the alkyne label (red) incorporated in a lipid molecule depends on its localization in biological membranes. Alkyne moieties on the surface of a bilayer show highest accessibility for click reaction with an azide reporter and the copper catalyst. The embedding of the alkyne label within the hydrophobic part of membranes yields lower accessibility, especially if rigid molecules are tagged.
Therefore, we synthesized a set of different biotin-conjugated azide detection reagents that possess the same fundamental structure as AP3Btn. In detail, the azide moiety in AP3Btn is connected to the biotin via a PEG spacer with three ethylene glycol units (Fig. 1). Compared with AP3Btn, the newly synthesized reagents differ in spacer length (AP6Btn and AP10Btn) or the additional capability to chelate copper (APpic2Btn). With this set of reagents, we addressed two questions: First, what is the optimal length of the spacer component? Second, does our click reaction protocol benefit from copper chelation by the azide reporter?
To create a more challenging setup compared with the established protocol (18), we used a reduced copper concentration (200 μM instead of 2 mM CuTFB) and a lower concentration of azide reporter (10 μM instead of 50 µM). In detail, A172 cells were incubated for 16 h with 10 μM of different alkyne lipids. Subsequently, cells were fixed and subjected to the click reaction under the more challenging conditions. We then evaluated the alkyne lipid staining visually and quantitatively for each combination of alkyne lipid and azide reporter (Fig. 3). In general, the signal for alkyne-oleate was stronger than that of the other lipids, so different exposure times were used, 50 ms for alkyne-oleate (Fig. 3A, B) and 100 ms for alkyne-cholesterol and propargylcholine, respectively (Fig. 3C, D). Alkyne-oleate showed the highest signal-to-noise ratios, whereas alkyne-cholesterol gave the least signal, proving itself as the most challenging lipid of those tested. The choice of the azide reagent greatly influenced the signal-to-noise ratio. In detail, the azide reporter containing the copper chelating picolyl moiety, APpic2Btn, gave the brightest staining. Therefore, a sample with a reduced concentration of the APpic2Btn reagent (2 μM) was additionally included and also gave a prominent labeling. Generally, the background signal in samples without alkyne lipid was very low. In the challenging setup, however, the background signal became relevant for those samples in which a lipid that is particularly challenging to access, namely alkyne-cholesterol, was probed with an azide reagent that does not contain a picolyl moiety.
Fig. 3.
At low azide reporter and copper concentration, only a picolyl-containing azido-biotin-based reporter yields a signal significantly above background. A172 cells were incubated with medium supplemented as indicated with 10 μM alkyne-oleate, alkyne-cholesterol, or propargylcholine, or in medium without lipid supplementation for 16 h. After fixation, cells were click-labeled using 10 μM or 2 μM azide detection reagent and 200 μM CuTFB. Epifluorescence images were taken with 50 ms (alkyne-oleate; A, B) or 100 ms (alkyne-cholesterol or propargylcholine; C, D) exposure. Cells were identified by staining the nuclei with DAPI. To demonstrate signal-to-noise ratios, the micrographs shown here are adjusted to the same display level in every column (control lacking alkyne lipid compared with lipid-incubated sample), but not in every row. Color-coding of the micrographs (A, C) corresponds to the diagrams in B and D and to Figs. 1, 4, and 5. Scale bars, 20 μm. E: Schematic representation of the general workflow used in quantification of the signal intensity of alkyne lipids click-reacted with various azide detection reagents in microscopy. In multiple images or large stitched tile images, cells were identified by their morphological boundaries and marked as ROIs. Within every ROI, the mean gray value was calculated as the sum of the gray values of all pixels of the ROI divided by the number of pixels. Data points in the diagrams (B and D; Figs. 4 and 5) thus represent the mean signal of individual cells. In addition, the mean value and the standard deviation for all cells of a sample are given in the graphs. In detail, in this figure the mean signal per cell for the detection of alkyne-oleate (50 ms exposure; B), alkyne-cholesterol or propargylcholine (100 ms exposure; D), and control samples lacking alkyne lipid is shown, determined after click labeling with various azide reporters (compare example pictures in A and C). For quantification, images were taken at six defined positions of every sample [Fluar 40× Oil (1.30 NA) objective (Zeiss), CoolSNAP K4 camera (Photometrics)] and corrected for illumination. Cell outlines were marked as ROIs and the mean signal for every individual cell (50–100 cells in six images per sample) measured. Asterisks designate levels of statistical significance (one-way ANOVA test). If no level of statistical significance is given for a sample and its corresponding control (no lipid), the difference is statistically not significant (n.s.).
To quantify our observations on imaging sensitivity, the mean signal derived from the alkyne lipids in every individual cell was measured in six images taken at defined positions of the sample (Fig. 3E). In the challenging setup used here, all alkyne lipids tested were detected very sensitively with 2 or 10 μM APpic2Btn, resulting in a signal statistically highly significant above background, compared with the signal in control samples lacking alkyne lipid (Fig. 3B, D). The use of the other biotin-based reagents (AP3Btn, AP6Btn, and AP10Btn), as well as of the fluorescent ASBDP, resulted in a much lower signal and hence lower detection sensitivity. While their signal is clearly visible in micrographs and is associated with cellular features (Fig. 3A, C), the mean signal intensity is not above background levels in the challenging setup (Fig. 3B, D). Interestingly, when probing with APpic2Btn, we noticed a differential effect of the used concentration on sensitivity for different lipids. For headgroup-tagged phosphatidylcholine (PC), in which the propargylcholine label is present at the surface of the bilayer, no significant increase in detection sensitivity was observed when using 10 μM compared with 2 μM of APpic2Btn (Fig. 3D). In contrast, the higher reporter concentration greatly improved the detection levels of the metabolites of alkyne-oleate (Fig. 3B), and especially alkyne-cholesterol (Fig. 3D). This indicates a saturated detection of propargylcholine-bearing PC, but not of alkyne-cholesterol metabolites, presumably due to differences in accessibility to the alkyne moiety. Additionally, by complexation of copper, the picolyl moiety directly facilitates the access of the catalyst to the membrane. Thus, the increased imaging sensitivity at higher concentrations of APpic2Btn probably reflects a more efficient delivery of copper to the alkyne label in the interior part of the membrane bilayer.
Introduction of a picolyl moiety into any azide reporter greatly increases the sensitivity of alkyne lipid imaging
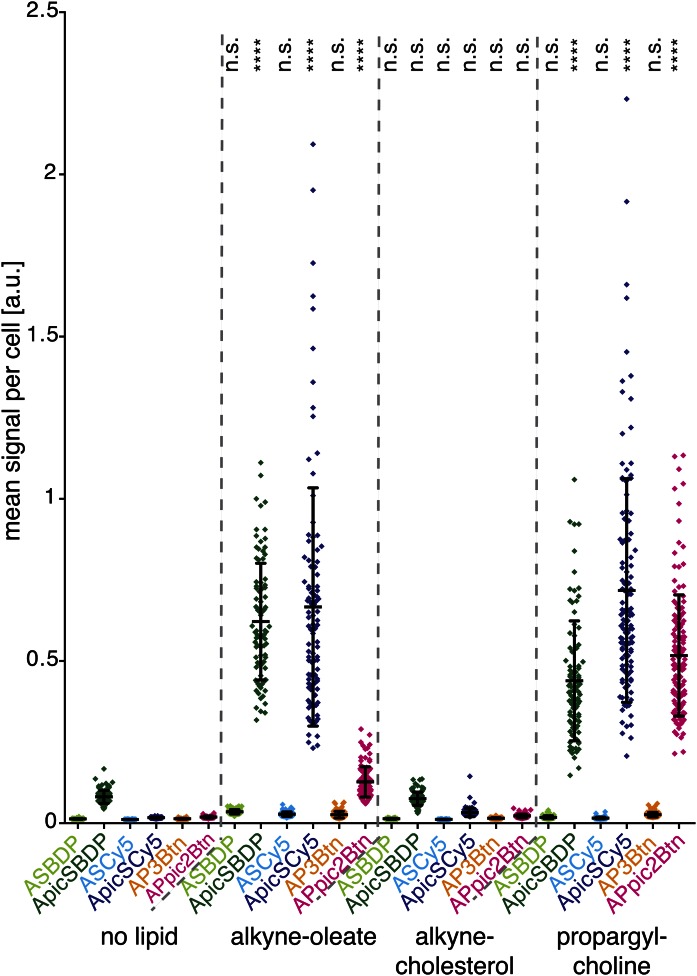
The introduction of a copper-chelating picolyl moiety into the biotin-conjugated reporters led to a strong increase in sensitivity for all lipids tested (Fig. 3). To assess whether this observation is transferable to azide reporters with intrinsic fluorescence, we performed similar experiments comparing detection reagents with or without picolyl group. For this, we synthesized ApicSBDP (as an analog to ASBDP) and acquired ASCy5 and ApicSCy5 from a commercial source. We tested the sensitivity of alkyne lipid imaging with the three pairs of azide detection reagents AP3Btn, ASBDP, ASCy5, or their picolyl-modified counterparts APpic2Btn, ApicSBDP, or ApicSCy5, respectively (Fig. 4). To apply very challenging conditions here too, alkyne lipid imaging was performed using again only 200 μM CuTFB catalyst and 10 μM of the fluorescent azide reporters. In contrast to the previous experiment, however, we also reduced the amount of available alkyne lipid by supplementing the cells with only 2.5 μM instead of 10 µM alkyne lipid. Our results show that alkyne-oleate- and propargylcholine-based lipids were detectable with a high sensitivity by using the picolyl-containing azide reagents, whereas their counterparts lacking the copper-chelating moiety failed to deliver signal intensities significantly above background noise. Remarkably, while propargylcholine-derived PC is equally well detected by all picolyl-containing reagents, the detection of alkyne-oleate metabolites gave weaker signal with APpic2Btn than with the other picolyl-bearing reagents. Steric requirements may account for this observation. Like in the previous experiment (Fig. 3), alkyne-cholesterol generally gave the lowest mean signal per cell, probably because of its rigid structure and hence limited accessibility inside the membrane.
Fig. 4.
Picolyl-containing azide reporters generally increase the sensitivity of alkyne lipid imaging. Quantification of signal intensities of alkyne lipids in HuH7 cells labeled with different azide detection reagents: HuH7 cells were incubated with medium supplemented with 2.5 μM alkyne-oleate, alkyne-cholesterol, or propargylcholine, or in medium without lipid supplementation for 16 h. After fixation, cells were click-labeled using 10 μM azide detection reagent and 200 μM CuTFB. Epifluorescence images were taken with 50 ms exposure, a high dynamic range camera (Orca-Flash4.0) and an Apochromate 63× (1.40 NA) DIC Oil objective (excluding the edges of the field of view to assure even illumination) and stitched to one large image of 1,000 × 1,000 μm for every sample. In these images, the mean signal of 80 to 160 cells was determined as described in Fig. 3E. Data points correspond to the mean signal of individual cells. In addition, the mean value and the standard deviation for all cells of a sample are given in the graph. Asterisks designate levels of statistical significance (one-way ANOVA test) for a difference in the mean of a sample and its corresponding control (no lipid). Color-coding of the data corresponds to that in Figs. 1, 3, and 5.
In summary, the introduction of a picolyl moiety in the linker of any azide detection reagent lead to a substantial increase in the sensitivity of alkyne lipid imaging, regardless of the alkyne lipid or the azide reporter type. This will be of special value for the detection of lipids that are less abundant or less accessible for CuAAC. Importantly, the use of picolyl-containing reporters allows for a tenfold reduction in the concentration of the copper catalyst.
Sensitive detection of alkyne lipids with various conventional azide reporter using a less stringent detection regime
Our results highlight the influence of the azide reporter on imaging sensitivity and suggest that click reactions do not proceed as unhindered inside a membrane bilayer as outside of it. Hence, depending on the azide reporter available as well as the alkyne lipid of interest and its respective cellular concentration, the optimal experimental conditions like the reagent concentrations may need to be adjusted. Therefore, the sensitivity of the click reaction was reexamined using increased concentrations for the different biotin-based detection reagents and longer exposure times at the microscope (Fig. 5), but otherwise the same conditions as in Fig. 3. Here, all reporters were used at 50 μM concentration, except for the very potent APpic2Btn, which was used at 5 μM or 0.5 μM. Micrographs (Fig. 5A) and their quantitative analysis (Fig. 5B) revealed that a sensitive detection of alkyne-oleate (i.e., a mean signal significantly higher than that of the control) was achieved with all reagents tested (AP3Btn, APpic2Btn, AP6Btn, and AP10Btn). For propargylcholine detection, AP3Btn, APpic2Btn, and AP10Btn met this criterion, while for alkyne-cholesterol, only APpic2Btn led to a signal that was about as strong as for the other lipids and statistically significant from controls. Hence, while alkyne-cholesterol can be reliably detected using conventional azide reporters without a copper-chelating moiety at 2 mM CuTFB (18), the detection of smaller amounts of alkyne-cholesterol or applications that require a reduced catalyst concentration (see below and Fig. 6) demand the usage of a picolyl-containing azide reporter for detection of the lipid with significant sensitivity. Our data further demonstrate that increasing the concentration of detection reagent per se leads to a gain in imaging sensitivity and could hence be a useful strategy when for example no picolyl-containing azide reagent is available.
Fig. 5.
Increased azide reporter concentrations significantly improve the sensitivity of alkyne lipid imaging. A172 cells were incubated in medium supplemented with 10 μM alkyne-oleate, alkyne-cholesterol, or propargylcholine, or in medium without lipid supplementation for 16 h. After fixation, cells were click-labeled using an optimized concentration (0.5–50 μM) of each azide detection reagent and 200 μM CuTFB. A: Epifluorescence images. Images are shown at the optimal display level each; display levels are not matching in either rows or columns. Cells were identified by staining the nuclei with DAPI. Color-coding of the micrographs corresponds to the quantification shown in B and Figs. 1, 3, and 4. B: Mean signal per cell for the detection of alkyne-oleate, alkyne-cholesterol, or propargylcholine, and of control samples lacking alkyne lipid, determined after click labeling with given azide reporters. For quantification, images were taken at six defined positions of every coverslip sample [Fluar 40× Oil (1.30 NA) objective (Zeiss), CoolSNAP K4 camera (Photometrics)] and corrected for illumination. For all reporters, an exposure time of 400 ms was chosen to achieve optimal signal-to-noise ratios (squares). However, for labeling with APpic2Btn, this sometimes led to overexposed images, in which case imaging was performed at 200 ms exposure (triangles). Cell outlines were marked as ROIs and the mean signal for every individual cell (50–120 cells in six images per sample) measured. Data points correspond to the mean signal of single cells. In addition, the mean value and the standard deviation for all cells of a sample are given in the graph. Asterisks designate levels of statistical significance (one-way ANOVA test). If no level of statistical significance is given for a sample and its corresponding control (no lipid), the difference is statistically not significant (n.s.). Scale bars, 20 μm.
Fig. 6.
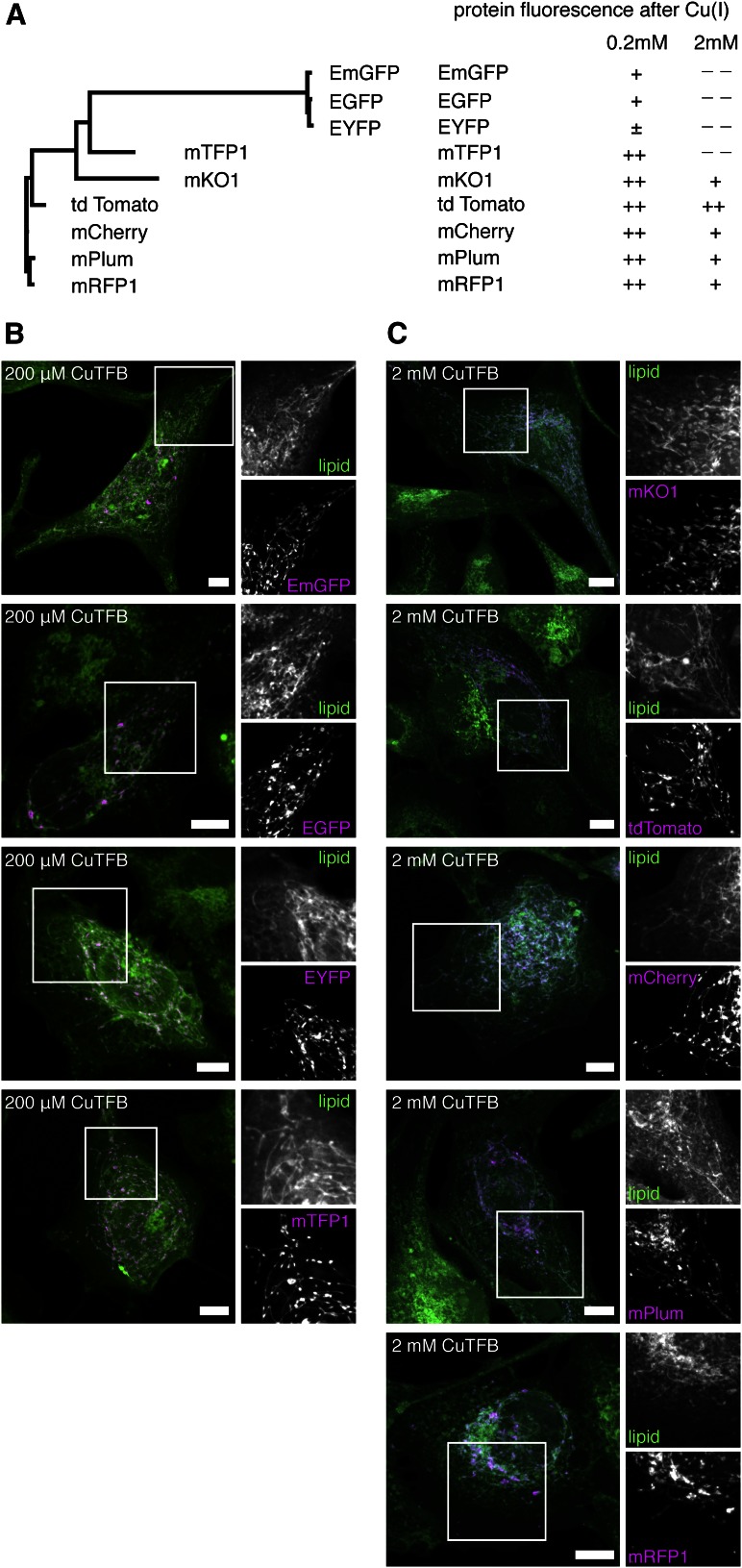
Alkyne lipid imaging is compatible with fluorescent protein detection. A: An overview displaying the relationship of selected fluorescent proteins (phylogenetic tree) and their fluorescence after exposure to various concentrations of copper during the click reaction. Protein fluorescence was subjectively evaluated by experienced scientists using a five-level scale (– – poor; – fair; ± satisfactory; + good; ++ very good). B, C: A172 cells heterologously expressing fluorescent proteins targeted to mitochondria were incubated in medium supplemented with 1 μM alkyne-sphinganine for 2 h. After fixation, cells were click-labeled using either 10 μM of ApicSCy5 detection reagent and 200 μM CuTFB (B) or 10 μM of ASBDP detection reagent and 2 mM CuTFB (C). Micrographs were recorded using structured illumination. Maximum image projections of z-stacks are shown, depicting alkyne lipids (green) and mitochondria (magenta, fluorescent protein marker) as color-merged overviews. Magnifications show alkyne lipids or mitochondria as gray scale images. All images were acquired using the Orca-Flash4.0 camera and are shown at the optimal display level each. Scale bars, 10 μm.
Furthermore, for all alkyne lipids tested, our results show that the length of the spacer has an influence on detection sensitivity as AP3Btn gave a significantly higher signal compared with AP6Btn or AP10Btn.
Compatibility of alkyne lipid imaging with detection of different fluorescent proteins
A variety of fluorescent proteins is successfully used to mark specific biological features ranging from distinct cells in tissues, organelles within cells, or even individual protein molecules. These fluorescent proteins can be grouped into families derived from different natural precursors, which have been engineered to generate improved versions covering a wide range of spectral properties (37–39). A phylogenetic tree analysis (Fig. 6A, left) of selected fluorescent proteins shows the close relationship of EGFP, EYFP, and EmGFP, all originating from a protein first isolated from Aequorea victoria. A second group consisting of mRFP1, mPlum, mCherry, and tdTomato is based on the natural protein DsRed of Discosoma striata (40), whereas mTFP1 and mKO1 were engineered from proteins first found in Clavularia and Fungia concinna, respectively. Here, we generated fusion constructs of nine fluorescent proteins that are targeted to mitochondria when expressed in mammalian cells. To assess the quality of the fluorescent protein signal upon CuAAC, we compared two click reaction protocols using either 200 μM or 2 mM copper catalyst in combination with azide reporters containing or not containing picolyl moieties. As a lipid probe, we chose alkyne-sphinganine, as we previously established its efficient targeting to mitochondria (15). Because alkyne-sphinganine carries the alkyne moiety at the terminal end of the hydrocarbon chain, it will be deeply embedded in the targeted membrane. Therefore, our lipid choice represents a more challenging setup than, for example, the headgroup-labeled propargyl-PC.
Using routine sample preparations and microscopy procedures the intensity of protein fluorescence was subjectively evaluated by experienced scientists (Fig. 6A, right). This analysis primarily determines the survival of the protein fluorescence signal after CuAAC but naturally is somewhat influenced by color perception and background characteristics. Albeit not strictly being quantitative, this reading has proved valuable and quite useful for planning experiments in the laboratory.
We found that EGFP, its close relatives EYFP and EmGFP, and mTFP1 cannot be reliably detected by our microscopy setup after exposure to 2 mM copper catalyst. However, their signal is well retained after click reaction using 200 μM copper. All tested DsRed-derived proteins and mKO1 are compatible with both 2 mM or 200 µM copper, although usually the protein signal intensity is higher after exposure to the reduced concentration of catalyst. To illustrate these findings, images of alkyne lipids colocalizing to mitochondria marked by the different fluorescent proteins are provided (Fig. 6B, C), acquired using optimal copper concentrations as indicated in the figure.
With carefully adjusted settings, these experiments demonstrate that alkyne lipid imaging after click reaction is compatible with the detection of various fluorescent proteins. Such colocalization experiments usually benefit from the use of the picolyl-containing reporters that allow for reduced concentrations of the copper catalyst.
Compatibility of alkyne lipid imaging with other fluorescence microscopy methods
The use of antibodies recognizing specific protein epitopes is a widely used technique in many laboratories. In fixed cells, fluorescently labeled antibodies allow for localization of a protein by fluorescence microscopy. To facilitate access of the bulky antibody to the epitope, the use of mild detergents during sample processing is common. However, for concomitant localization of lipids that often are only partially immobilized during fixation, the use of detergent is generally disadvantageous. Hence, our protocols introduced so far avoided detergents altogether.
However, sometimes the experimental benefits from using an antibody outbalance the trade-offs caused by the necessary detergent application. Therefore, we sought to establish a protocol for the combination of alkyne lipid detection by fluorescence microscopy and antibody based immunofluorocytochemistry. We optimized the fixation time for epitope preservation and varied the detergent concentration in the permeabilization steps. We further compared the performance of different protocols, altering the sequence of the labeling steps (antibody incubation and click reaction) and washing procedures. These screening efforts yielded the following refinements: Cells were only briefly fixed (10 min) and cells were subsequently permeabilized using low concentrations of saponin. To permeabilize the plasma membrane only or all cellular membranes, 0.01% or 0.1% saponin was used, respectively, depending on the localization of the protein epitope of interest. After the detection of the proteins by antibodies, the click reaction was performed as this sequence allowed for best epitope preservation.
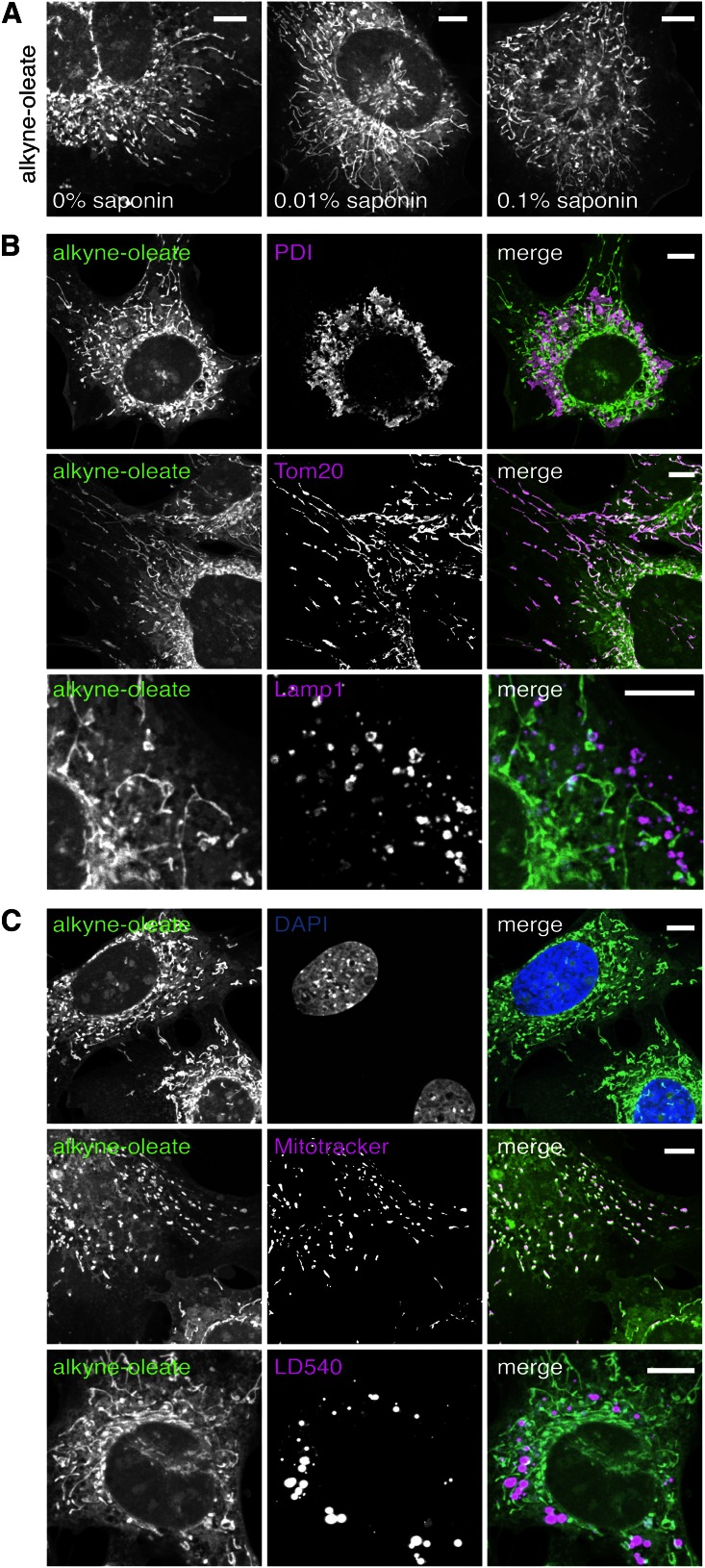
Importantly, the permeabilization by saponin prior to the click reaction did not cause apparent mislocalization of the alkyne lipids nor did it diminish the lipid signal (Fig. 7A). Using this protocol, we were able to simultaneously image alkyne lipids and organelles, identified by antibodies recognizing established marker proteins as shown exemplarily in Fig. 7B. A strong colocalization of endoplasmic reticulum (PDI) and mitochondria (Tom20) with lipids derived from alkyne-oleate was observed, while in contrast, lysosomes (Lamp1) showed only a weak lipid signal under the conditions applied. Finally, we also tested the compatibility of alkyne lipid detection and CuAAC with the use of common cellular stains. Costainings with the established dyes DAPI, mitotracker CMXRos, and the lipid droplet dye LD540 (33) are well possible, as all these dyes also proved insensitive to the sample preparation procedure using 2 mM copper catalyst (Fig. 7C).
Fig. 7.
Alkyne lipid imaging is compatible with immunofluorescence and commonly used cellular stains. MEF cells were treated for 24 h with 1 µM alkyne-oleate and fixed for 10 min. A: Prior treatment with saponin for membrane permeabilization did not diminish or change subsequent click-labeling of cellular structures. Cells were treated for 2 h with 0%, 0.1%, or 0.01% saponin before subjecting them to the click reaction using 2 mM CuTFB and 10 µM ASBDP. B: Immunofluorescence labeling is preserved during the CuAAC click procedure. Cells were first subjected to immunofluorescence staining for commonly used organelle marker proteins of three different cellular compartments: endoplasmic reticulum (PDI), mitochondria (Tom20), and lysosomes (LAMP1). Subsequently, cells were subjected to the click reaction using 2 mM CuTFB and 10 µM ASBDP. C: Click labeling of alkyne lipids by CuAAC (using 2 mM CuTFB and 10 µM ASBDP) is compatible with commonly used fluorescence stains. Cells were stained in situ using mitotracker CMXRos (50 nM, 15 min) before fixation and subsequent click labeling or stained with LD540 or DAPI after the click labeling. All images were acquired using the Orca-Flash4.0 camera and are shown at the optimal display level each. Scale bars, 10 μm.
DISCUSSION
Linker length of the azide reporter influences the sensitivity of alkyne lipid imaging
Fluorescent alkyne lipid imaging is a powerful tool to study the subcellular localization of lipids. This study clearly demonstrates an influence of structural attributes of azide reporters used in CuAAC for the sensitivity of alkyne lipid detection in fluorescence microscopy.
We hypothesized that an increased spacer length linking the azide and biotin moieties of a biotin-based azide reporter could increase imaging sensitivity because a longer portion of the spacer would reside outside the bilayer potentially resulting in less steric hindrance and more flexibility in streptavidin binding. Upon binding to streptavidin, biotin is buried deeply within an open barrel of the protein, which is then covered by a surface loop (41, 42). Hence, a certain steric requirement was expected. However, our results obtained were contradictory to this expectation. In detail, AP3Btn, the shortest biotin-based azide reporter tested, showed higher sensitivity compared with AP6Btn and AP10Btn. Assuming a stretched conformation, in a click-conjugate of AP3Btn and alkyne-cholesterol the biotin moiety would be situated just outside the membrane (Fig. 1). It would therefore still be positioned within the hydration sphere of a DPPC bilayer consisting of water molecules that directly interact with the phospholipid headgroups and are of functional importance for biological membranes (43, 44). In the AP6Btn conjugate, the biotin is able to move more freely because itself and part of the PEG linker would be situated outside of the bilayer. This also holds true for AP10Btn conjugated to alkyne-cholesterol, but here the spacer outside the membrane is longer. Hence, our results suggest that streptavidin binding likely is equally efficient just above the bilayer surface, and that longer linkers may lead to lower CuAAC reaction/ streptavidin binding efficiency due to the formation of conformations of the reactants that are unfavorable for CuAAC and/ or the biotin-streptavidin interaction.
A main asset of using biotin as a detection reagent is that it allows for full flexibility regarding the spectral properties of the fluorophore. The wide selection of commercially available dyes coupled to streptavidin includes fluorophores of excellent photostability and virtually every emission color. Difficulties using the biotin-based reporters arise though, when a direct numerical correlation of alkyne lipid molecules to streptavidin-dye signal is needed in an application. Here the tetrameric nature of this protein has to be considered. Due to its D2 (tetrahedron-like) symmetry (42), two binding pockets can simultaneously face the bilayer. If two biotin moieties are appropriately positioned, one molecule of streptavidin could bind both of them, possibly complicating quantification. This could potentially be overcome by the use of monovalent forms of streptavidin (45). Furthermore, it would be necessary to use an excessive amount of streptavidin conjugate if an application would require unequivocal labeling of every click-reacted alkyne lipid molecule with the conjugate. Hence, depending on the experimental approach, labeling alkyne lipids directly with an azide-coupled dye can be advantageous.
Azide reporters with picolyl moieties enable highly sensitive imaging of alkyne lipids
The introduction of a picolyl moiety into the azide reporter improved the detection of various click-reacted lipids in biological membranes with an increase of signal intensity of up to 42-fold (compare Fig. 4, mean signal per cell of propargylcholine-tagged PC labeled with ApicSCy5 or ASCy5). Recently, other groups have used picolyl-containing biotin-azide reporters (31, 46) or other multidentate chelators (32) for the highly sensitive labeling of various alkyne-bearing biomolecules and a further development of copper chelators can be expected. It has been shown that a copper ion that is bound very tightly to the azide reporter, and thereby shielded to prevent the generation of reactive oxygen species [see (30)], can be introduced as a complex into living cells (32). This development could potentially pave the way for CuAAC application in live cell imaging. However, live cell imaging with CuAAC is further complicated by the cytotoxicity of the copper catalyst. While labeling of lipids in the plasma membrane and in internal membranes (47) of living cells with classical CuAAC has been reported, copper shows significant toxicity at micromolar concentrations and has a profound impact on lipid metabolism, especially if present in loosely bound form (48, 49). More biocompatible copper catalysts (50) or additional ligands (48) for CuAAC, and copper-free coupling of azides to strain-promoted alkynes [strain-promoted azide-alkyne cycloaddition (SPAAC)] (29), have been developed to overcome these difficulties. SPAAC has successfully been used for labeling of lipids in the plasma membrane and intracellular membranes (17). In SPAAC, the use of a spring-loaded cyclooctyne avoids copper-derived cytotoxicity, on the cost of a bulkier label with a stronger tendency for unspecific side reactions (51). Furthermore, during the prolonged SPAAC reaction time in living cells, the unreacted lipid probe will undergo further trafficking and metabolism, whereas the already reacted pool carrying a large reporter impacting the lipids structure and hydrophobicity will experience more or less unconventional cellular processing. The functionalization of cyclooctynes leads to a significant increase in reaction rate of SPAAC [discussed in (30)], but they are laborious to synthesize (52) and hence not widely available. In general, as the interest in lipid imaging is growing and enormous improvements to click labeling have been made in the last years, the limitations to both SPAAC and CuAAC for life cell imaging will hopefully be overcome in the near future.
The introduction of a copper chelating moiety greatly increases sensitivity in alkyne lipid imaging. While conventional azide reporters are fully feasible for most approaches with easily accessible alkyne tags, a picolyl-containing reporter is highly advantageous for alkyne-lipids that are hard to access when embedded in a bilayer or are present at only very low concentrations. An alternative approach for optimizing the access to the alkyne label for the reporter and catalyst is the use of alkyne lipids or their precursors carrying the tag at a more accessible position, for example, at the headgroup or a point near the surface when embedded in a biomembrane. However, such positioning of the label is likely to significantly influence relevant properties of the lipid, as exemplified by a lyso-PC labeled with the alkyne at the headgroup that displays slightly different kinetics in enzymatic reactions compared with its tail-labeled counterpart (24). Thus, it appears likely that a higher accessibility of the alkyne label in many cases comes at the cost of a less optimal representation of the natural lipid by the alkyne lipid probe.
Fluorescence microscopy of proteins and alkyne lipid imaging are compatible
In addition to the gain in sensitivity, the usage of picolyl-containing reporters and consequently the possibility of reducing the copper concentration allows for high-content imaging in approaches where CuAAC-based alkyne imaging is combined with a copper-sensitive method. While we found many antibody-antigen interactions, a basis for immunofluorescence microscopy, and some widely used fluorescent organelle stains to be compatible with the use of 2 mM CuTFB during CuAAC, the fluorescence of some commonly used fluorescent proteins is strongly impacted. While this particularly effected the most popular of such proteins, EGFP and its derivatives, other fluorescent proteins proved more stable when exposed to copper and therefore represent a better choice for colocalization studies. However, all nine fluorescent proteins tested here, including EGFP, yielded useful fluorescent signals when CuAAC with 200 µM catalyst and picolyl-containing reporters were used for lipid detection. This will be of importance for many laboratories that have engineered fusion constructs and reporter animals using EGFP and the other fluorescent proteins tested here.
In summary, lipid tracing by microscopy has been challenging due to methodological limitations. Here, we describe an improved method for lipid fluorescence microscopy using alkyne lipids and CuAAC and provide an optimized protocol that can be adapted to the specific needs of the questions under study. These could include imaging of low abundance alkyne lipids in special experimental setups (e.g., pulse chase experiments with short pulse-times or using cytotoxic lipids) or localization studies in alkyne lipids whose rigid molecular structure reduces the access of the click reactants to the alkyne group deeply embedded in a bilayer (Fig. 2). We further present variants of the protocol allowing for the combination with fluorescent protein or immunofluorescence imaging. Importantly, our method is easy to perform and requires only standard laboratory equipment or microscopy setup. For the convenience of the experimenter, we provide a general guide how to adapt the protocol for specific needs (Table 1) and a straightforward trouble shooting guide to use when accounting problems with the method (supplemental data). Herewith, we offer a guideline for successful CuAAC-based alkyne-lipid imaging and believe it will be helpful for many researchers wishing to analyze subcellular lipid localization by fluorescence microscopy in a robust and convenient fashion.
TABLE 1.
Guideline for choosing labeling conditions in alkyne lipid imaging
| Alkyne Label Accessibility | Alkyne Lipid Amount | Coimaging with Proteins | Azide Reporter and Concentration | Copper Catalyst (CuTFB) |
| Easy | Medium to high | No | Any (50 μM conventional or 10 μM picolyl reporter) | 200–2,000 μM or 20–200 μM |
| Easy | Low | No | 10 μM Picolyl reporter | 200–2,000 μM |
| Less | Medium to high | No | 10 μM Picolyl reporter | 200 μM |
| Less | Low | No | 10 μM Picolyl reporter | 200–2,000 μM |
| Any | Any | Yes, fluorescent proteins | 10 μM Picolyl reporter | 200 μMa |
| Any | Any | Yes, antibody stainingb | Any (alkyne lipid dependent) | 200–2,000 μM |
Recommended reagents and concentrations are presented and depend on the alkyne lipid under study and a possible combination with protein imaging. Suggestions represent a starting point for individual optimization.
The tolerable copper concentration depends on the fluorescent protein.
Use short fixation, permeabilize, and perform antibody labeling before click reaction.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Henry Weiner (Purdue University, Lafayette) for providing the plasmid encoding for rat ALDH.
Footnotes
Abbreviations:
- ALDH
- alcohol dehydrogenase
- alkyne-cholesterol
- (25R)-25-ethinyl-26-nor-3β-hydroxycholest-5-en
- alkyne-oleate
- nonadec-9-cis-en-18-ynoic acid
- alkyne-sphinganine
- (2S,3R)-2-aminooctadec-17-yn-1,3-diol
- AP3Btn
- azido-PEG3-biotin
- AP6Btn
- azido-PEG6-biotin
- AP10Btn
- azido-PEG10-biotin
- ApicSBDP
- azido-picolyl-sulfo-tetramethyl-BODIPY
- ApicSCy5
- azido-picolyl-sulfo-Cy5
- APpic2Btn
- azido-picolyl-PEG2-biotin
- ASBDP
- 8-(5-azidopentyl)-4,4-difluor-1,3,5,7-tetramethyl-4-bora-3a,4a-s-indacene-2,6-disulfonic acid disodium salt
- ASCy5
- azido-sulfo-Cy5
- CuAAC
- copper (I)-catalyzed azide-alkyne cycloaddition
- CuTFB
- tetrakis(acetonitrile)copper(I) tetra fluoroborate
- DAPI
- 4’,6-diamidino-2-phenylindole
- DPPC
- dipalmitoylphosphatidylcholine
- EGFP
- enhanced green fluorescent protein
- EmGFP
- Emerald green fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- LD540
- 4,4-difluoro-2.3,5.6-bis-tetramethylene-4-bora-3a,4a-diaza-s-indacene
- MEF
- mouse embryonic fibroblast
- mKO1
- monomeric Kusabira Orange 1
- mTFP1
- monomeric teal fluorescent protein 1
- PC
- phosphatidylcholine
- PEG
- polyethylene glycol
- ROI
- region of interest
- SPAAC
- strain-promoted azide-alkyne cycloaddition
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB-TRR83 and SFB 645, and by the German Federal Ministry of Science and Education (Virtual Liver Network).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.van Meer G., Voelker D. R., and Feigenson G. W.. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maier O., Oberle V., and Hoekstra D.. 2002. Fluorescent lipid probes: some properties and applications (a review). Chem. Phys. Lipids. 116: 3–18. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser R. D., and London E.. 1998. Determination of the depth of BODIPY probes in model membranes by parallax analysis of fluorescence quenching. Biochim. Biophys. Acta. 1375: 13–22. [DOI] [PubMed] [Google Scholar]
- 4.Solanko L. M., Honigmann A., Midtiby H. S., Lund F. W., Brewer J. R., Dekaris V., Bittman R., Eggeling C., and Wustner D.. 2013. Membrane orientation and lateral diffusion of BODIPY-cholesterol as a function of probe structure. Biophys. J. 105: 2082–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naylor B. L., Picardo M., Homan R., and Pownall H. J.. 1991. Effects of fluorophore structure and hydrophobicity on the uptake and metabolism of fluorescent lipid analogs. Chem. Phys. Lipids. 58: 111–119. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka K. A. K., Suzuki K. G. N., Shirai Y. M., Shibutani S. T., Miyahara M. S. H., Tsuboi H., Yahara M., Yoshimura A., Mayor S., and Fujiwara T. K.. 2010. Membrane molecules mobile even after chemical fixation. Nat. Methods. 7: 865–866. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg A., and Pricer W. E.. 1953. Enzymatic synthesis of the coenzyme A derivatives of long chain fatty acids. J. Biol. Chem. 204: 329–343. [PubMed] [Google Scholar]
- 8.Ho J. K., Duclos R. I., and Hamilton J. A.. 2002. Interactions of acyl carnitines with model membranes a 13C-NMR study. J. Lipid Res. 43: 1429–1439. [DOI] [PubMed] [Google Scholar]
- 9.Li J., Hoene M., Zhao X., Chen S., Wei H., Häring H-U., Lin X., Zeng Z., Weigert C., and Lehmann R.. 2013. Stable isotope-assisted lipidomics combined with nontargeted isotopomer filtering, a tool to unravel the complex dynamics of lipid metabolism. Anal. Chem. 85: 4651–4657. [DOI] [PubMed] [Google Scholar]
- 10.Matthäus C., Krafft C., Dietzek B., Brehm B. R., Lorkowski S., and Popp J.. 2012. Noninvasive imaging of intracellular lipid metabolism in macrophages by Raman microscopy in combination with stable isotopic labeling. Anal. Chem. 84: 8549–8556. [DOI] [PubMed] [Google Scholar]
- 11.Rostovtsev V. V., Green L. G., Fokin V. V., and Sharpless K. B.. 2002. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 41: 2596–2599. [DOI] [PubMed] [Google Scholar]
- 12.Tornøe C. W., Christensen C., and Meldal M.. 2002. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 67: 3057–3064. [DOI] [PubMed] [Google Scholar]
- 13.Kolb H. C., Finn M. G., and Sharpless K. B.. 2001. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40: 2004–2021. [DOI] [PubMed] [Google Scholar]
- 14.Prescher J. A., and Bertozzi C. R.. 2005. Chemistry in living systems. Nat. Chem. Biol. 1: 13–21. [DOI] [PubMed] [Google Scholar]
- 15.Kuerschner L., and Thiele C.. 2014. Multiple bonds for the lipid interest. Biochim. Biophys. Acta. 1841: 1031–1037. [DOI] [PubMed] [Google Scholar]
- 16.Jao C. Y., Roth M., Welti R., and Salic A.. 2009. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. USA. 106: 15332–15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neef A. B., and Schultz C.. 2009. Selective fluorescence labeling of lipids in living cells. Angew. Chem. Int. Ed. Engl. 48: 1498–1500. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann K., Thiele C., Schott H. F., Gaebler A., Schoene M., Kiver Y., Friedrichs S., Lutjohann D., and Kuerschner L.. 2014. A novel alkyne cholesterol to trace cellular cholesterol metabolism and localization. J. Lipid Res. 55: 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jao C. Y., Nedelcu D., Lopez L. V., Samarakoon T. N., Welti R., and Salic A.. 2015. Bioorthogonal probes for imaging sterols in cells. ChemBioChem. 16: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charron G., Wilson J., and Hang H. C.. 2009. Chemical tools for understanding protein lipidation in eukaryotes. Curr. Opin. Chem. Biol. 13: 382–391. [DOI] [PubMed] [Google Scholar]
- 21.Smith M. D., Sudhahar C. G., Gong D., Stahelin R. V., and Best M. D.. 2009. Modular synthesis of biologically active phosphatidic acid probes using click chemistry. Mol. Biosyst. 5: 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haberkant P., and Holthuis J. C.. 2014. Fat & fabulous: bifunctional lipids in the spotlight. Biochim. Biophys. Acta. 1841: 1022–1030. [DOI] [PubMed] [Google Scholar]
- 23.Thiele C., Papan C., Hoelper D., Kusserow K., Gaebler A., Schoene M., Piotrowitz K., Lohmann D., Spandl J., Stevanovic A., et al. 2012. Tracing fatty acid metabolism by click chemistry. ACS Chem. Biol. 7: 2004–2011. [DOI] [PubMed] [Google Scholar]
- 24.Gaebler A., Milan R., Straub L., Hoelper D., Kuerschner L., and Thiele C.. 2013. Alkyne lipids as substrates for click chemistry-based in vitro enzymatic assays. J. Lipid Res. 54: 2282–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne S. B., Tallman K. A., Serwa R., Rouzer C. A., Armstrong M. D., Marnett L. J., Lukehart C. M., Porter N. A., and Brown H. A.. 2010. Capture and release of alkyne-derivatized glycerophospholipids using cobalt chemistry. Nat. Chem. Biol. 6: 205–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulce J. J., Cognetta A. B., Niphakis M. J., Tully S. E., and Cravatt B. F.. 2013. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods. 10: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Windsor K., Genaro-Mattos T. C., Kim H. Y., Liu W., Tallman K. A., Miyamoto S., Korade Z., and Porter N. A.. 2013. Probing lipid-protein adduction with alkynyl surrogates: application to Smith-Lemli-Opitz syndrome. J. Lipid Res. 54: 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beavers W. N., Serwa R., Shimozu Y., Tallman K. A., Vaught M., Dalvie E. D., Marnett L. J., and Porter N. A.. 2014. omega-Alkynyl lipid surrogates for polyunsaturated fatty acids: free radical and enzymatic oxidations. J. Am. Chem. Soc. 136: 11529–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyce M., and Bertozzi C. R.. 2011. Bringing chemistry to life. Nat. Methods. 8: 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKay C. S., and Finn M. G.. 2014. Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem. Biol. 21: 1075–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uttamapinant C., Tangpeerachaikul A., Grecian S., Clarke S., Singh U., Slade P., Gee K. R., and Ting A. Y.. 2012. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed. Engl. 51: 5852–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevilacqua V., King M., Chaumontet M., Nothisen M., Gabillet S., Buisson D., Puente C., Wagner A., and Taran F.. 2014. Copper-chelating azides for efficient click conjugation reactions in complex media. Angew. Chem. Int. Ed. Engl. 53: 5872–5876. [DOI] [PubMed] [Google Scholar]
- 33.Spandl J., White D. J., Peychl J., and Thiele C.. 2009. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 10: 1579–1584. [DOI] [PubMed] [Google Scholar]
- 34.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E., and Hutchison G. R.. 2012. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klauda J. B., Venable R. M., Freites J. A., O’Connor J. W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D. J., and Pastor R. W.. 2010. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 114: 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsien R. Y. 2009. Constructing and exploiting the fluorescent protein paintbox (Nobel lecture). Angew. Chem. Int. Ed. Engl. 48: 5612–5626. [DOI] [PubMed] [Google Scholar]
- 38.Shimomura O. 2009. Discovery of green fluorescent protein (GFP) (Nobel lecture). Angew. Chem. Int. Ed. Engl. 48: 5590–5602. [DOI] [PubMed] [Google Scholar]
- 39.Chalfie M. 2009. GFP: lighting up life (Nobel lecture). Angew. Chem. Int. Ed. Engl. 48: 5603–5611. [DOI] [PubMed] [Google Scholar]
- 40.Shaner N. C., Steinbach P. A., and Tsien R. Y.. 2005. A guide to choosing fluorescent proteins. Nat. Methods. 2: 905–909. [DOI] [PubMed] [Google Scholar]
- 41.Hendrickson W. A., Pahler A., Smith J. L., Satow Y., Merritt E. A., and Phizackerley R. P.. 1989. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl. Acad. Sci. USA. 86: 2190–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freitag S., Le Trong I., Klumb L., Stayton P. S., and Stenkamp R. E.. 1997. Structural studies of the streptavidin binding loop. Protein Sci. 6: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitter J., Lechner R. E., and Dencher N. A.. 1999. Interactions of hydration water and biological membranes studied by neutron scattering. J. Phys. Chem. B. 103: 8036–8050. [Google Scholar]
- 44.Nagle J. F., and Tristram-Nagle S.. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469: 159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim K. H., Huang H., Pralle A., and Park S.. 2013. Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnol. Bioeng. 110: 57–67. [DOI] [PubMed] [Google Scholar]
- 46.Jiang H., Zheng T., Lopez-Aguilar A., Feng L., Kopp F., Marlow F. L., and Wu P.. 2014. Monitoring dynamic glycosylation in vivo using supersensitive click chemistry. Bioconjug. Chem. 25: 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dauner M., Batroff E., Bachmann V., Hauck C. R., and Wittmann V.. 2016. Synthetic glycosphingolipids for live-cell labeling. Bioconjug. Chem. 27: 1624–1637. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy D. C., McKay C. S., Legault M. C., Danielson D. C., Blake J. A., Pegoraro A. F., Stolow A., Mester Z., and Pezacki J. P.. 2011. Cellular consequences of copper complexes used to catalyze bioorthogonal click reactions. J. Am. Chem. Soc. 133: 17993–18001. [DOI] [PubMed] [Google Scholar]
- 49.Krishnamoorthy L., Cotruvo J. A. J., Chan J., Kaluarachchi H., Muchenditsi A., Pendyala V. S., Jia S., Aron A. T., Ackerman C. M., Wal M. N., et al. 2016. Copper regulates cyclic-AMP-dependent lipolysis. Nat. Chem. Biol. 12: 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soriano Del Amo D., Wang W., Jiang H., Besanceney C., Yan A. C., Levy M., Liu Y., Marlow F. L., and Wu P.. 2010. Biocompatible copper(I) catalysts for in vivo imaging of glycans. J. Am. Chem. Soc. 132: 16893–16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Geel R., Pruijn G. J., van Delft F. L., and Boelens W. C.. 2012. Preventing thiol-yne addition improves the specificity of strain-promoted azide-alkyne cycloaddition. Bioconjug. Chem. 23: 392–398. [DOI] [PubMed] [Google Scholar]
- 52.Sletten E. M., de Almeida G., and Bertozzi C. R.. 2014. A homologation approach to the synthesis of difluorinated cycloalkynes. Org. Lett. 16: 1634–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.