Abstract
Proteoglycans are major constituents of the extracellular matrices as well as the cell surfaces and basement membranes. They play key roles in supporting the dynamic extracellular matrix by generating complex structural networks with other macromolecules and by regulating cellular phenotypes and signaling. It is becoming evident, however, that proteolytic enzymes are required partners for matrix remodeling and for modulating cell signaling via matrix constituents. Proteinases contribute to all stages of diseases, particularly in cancer development and progression, and contextually participate in either the removal of damaged products or in the processing of matrix molecules and signaling receptors. Indeed, the dynamic interplay between proteoglycans and proteolytic enzymes is a crucial biological step that contributes to the pathophysiology of cancer and inflammation. Moreover, proteoglycans are implicated in the expression and secretion of proteolytic enzymes and often modulate their activities. In this review we present emerging biological roles of proteoglycans and proteinases with special emphasis on their complex interplay. We critically evaluate this important proteoglycan-proteinase interactome and discuss future challenges of potentially targeting this axis in the treatment of cancer.
Keywords: matrix metalloproteinases, proteoglycans, glycosaminoglycans, cathepsins, syndecans, serglycin, versican
Graphical Abstract
Proteolysis is a highly regulated and specific process. Here, we highlight recent findings, which suggest that PGs/GAG interplay regulates proteinases’ activity, stability, substrate specificity and localization. The interaction between proteinases and proteoglycans may occur either through the GAG chains or the core protein of PGs. Such findings introduce a new avenue for specific cancer therapeutics.
INTRODUCTION
Extracellular matrix (ECM) is a highly dynamic and functional network, which is assembled from a variety of molecules. This network creates the scaffold where cells are hosted forming certain tissues and organs. ECM varies in composition among tissues and is continuously remodeled under physiological and pathological conditions. The changes in the expression of matrix molecules and the compositional alterations among them markedly affect the assembly of ECM and its ability to regulate a multitude of cellular functions. Among major ECM components are proteoglycans (PGs) as well as fibrillar proteins, like collagens and elastin, and other (glyco)proteins [1]. In cancer, PG expression is often altered in the stroma and this might contribute to disease progression or do quite the opposite. ECM remodeling is also a hallmark of cancer progression and in this process matrix proteinases play a central role [2]. Indeed, proteinases contribute to all stages of cancer development and progression. The intracellular proteases participate in the removal of damaged or undesirable products, whereas the extracellular proteases are actively involved in tumor progression and metastasis by degrading the majority of ECM macromolecules. Moreover, there are transmembrane proteases, which mostly target transmembrane receptors and signaling molecules [2–4]. Thus the interaction of PG and proteases is complex and often multifactorial, and generally operates in a cell- and tissue-contextual manner.
PROTEINASES: STRUCTURE, FUNCTIONS AND TARGETING IN CANCER
The term “proteinase” encompasses a large family of enzymes that catalyze the hydrolytic breakdown of proteins into peptides or amino acids at their terminal ends (exopeptidases) or inside the peptide chain (endopeptidases) [5]. According to the MEROPS database, the human degradome contains at least 569 proteinases distributed intra- and extracellularly, and classified based on the chemical moiety that participates in the hydrolysis (aspartic, cysteine, threonine, serine, and metallo-proteinases) [6]. This is in contrast to the PG family, which contains less than fifty genes. Thus a single PG would be expected to interact and modulate, on the average, more than ten proteases.
It is well recognized that proteinases contribute to all stages of tumor progression including tumor growth and survival, angiogenesis, cell invasion, cell adhesion, migration, EMT, immune surveillance and are produced not only by the tumor cells themselves, but also by the tumor microenvironment [2, 7, 8]. Metalloproteinases and cathepsins are among the major families of proteinases implicated in cancer cell biology. These two categories in terms of structure, function and possibilities for their targeting are described in detail below.
Metalloproteinases
The metzincin family of metalloproteinases is characterized by the presence of the conservative methionine residue at the active site and the use of a zinc ion in the enzymatic reaction [9]. This family comprises matrix mettalloproteinases (MMPs), a disintegrin and metalloproteinases (ADAMs) [4] and ADAMs with thrombospondin motifs (ADAMTSs) [10], the bacterial serralysins, and proteases such as the astacins (including the meprins) [10]. The most well studied representatives of this super-family are MMPs, as well as ADAMs and ADAMTSs. All members of the metalloproteinases family share domain structure similarities. The current MMP classification, based on substrate specificity, sequence similarity and domain organization categorize MMPs into six groups; collagenases, gelatinases, stromelysins, matrilysins, membrane type MMPs (MT-MMPs) and other MMPs [11]. A typical MMP structure involves several conserved distinct domains involving a N-terminal signal peptide responsible for secretion or plasma membrane insertion and a propeptide (containing highly conserved PRCGVPDV sequence) that following its proteolytic cleavage the enzyme is activated. The catalytic domain is defined by a 3-His motif (HEXXHXXGXXH) to which the catalytic zinc ion is held followed by a hemopexin domain responsible for the substrate specificity (except in MMP-7), and the interactions with other MMPs and tissue inhibitors of metalloproteinases (TIMPs) [12]. Furthermore, most ADAMs are type I transmembrane proteins that possess disintegrin, cysteine-rich, and EGF domains instead of MMP hemopexin domain, whereas ADAMTSs are secreted proteins that contain thrombospondin I motifs instead of the EGF and transmembrane domain [13].
MMPs represent one of the most-well studied classes and the main group of regulating proteinases in ECM. MMPs are responsible for the turnover and degradation of almost all ECM components [14], including collagens, fibronectin and laminins etc, as well as non-ECM cell regulators such as numerous cell-surface receptors, integrins, kinases, chemokines and cytokines [15, 16]. Thus, they are central regulators not only in physiological processes, such as embryogenesis, remodeling, wound healing and angiogenesis, but also in pathological conditions.
Along with ADAMS, MMPs exert their actions in cancer progression not only by degrading the physical barriers that cancer cells encounter but also by increasing the availability of signaling effectors like growth factors and cytokines to the tumor milieu. They also act on ECM macromolecules such as collagen, PGs, integrins, cell surface receptors, generating regulatory molecules and modulating cell-cell and cell-matrix interactions [2, 17, 18]. However, some MMPs are anchored to the cell surface [i.e. MT-MMPs, ADAMS], following their intracellular activation, whereas most are produced as secreted proteins and activated extracellularly. As mentioned above, secreted metalloproteinases are produced as inactive zymogens that generally need to be proteolytically activated, involving removal of the propeptide by other proteinases or by autoactivation. It is well documented that pro-forms of metalloproteinases may be activated through allosteric interactions with other molecules “revealing” the active site form the propeptide. In the case of membrane bound metalloproteinases, a furin recognition sequence between their propeptide and catalytic domains allows the cleavage and activation by furin convertase enzymes in the Golgi apparatus [19].
ADAMTSs exert various functions such as angiogenesis inhibition, degradation of proteoglycans; aggrecan, versican and brevican, collagen processing, and blood coagulation, among others [20]. In the case of cancer, ADAMTSs may have dual effect either promoting or suppressing tumor growth. ADAMTS -1, -2 and -8 exert antiangiogenic effects and suppression of ADAMTS -1 stimulates breast cancer cell migration and invasion [20, 21]. Also ADAMTS -9 is down-regulated in liver metastasis of colorectal tumors [22]. On the other hand, ADAMTS -4 and -5 are up-regulated in gliomas [23].
Based on the critical importance of metalloproteinases in cancer progression, numerous metalloproteinases inhibitors have been developed with goal to target their synthesis, secretion, activation and enzymatic activity. Many strategies have been employed involving synthetic MMPIs like peptidomimetics, mimicking metalloproteinases substrates and functioning as competitive inhibitors, non-peptidomimetic compounds interacting with the catalytic zinc ion and inhibiting enzymatic activity, as well as chemical derivatives of tetracycline group of drugs which lack antimicrobial action to antibody-based inhibitors and tetracycline like derivatives. The use of natural MMPs’ inhibitors like genistein contradicted the high toxicity profile observed with the synthetic MMPIs. In that line, antibody-based MMP inhibitors, as well as novel mechanism-based inhibitors were introduced to the field. Last, several off-target inhibitors such as bisphosphonates are documented in the literature to inhibit indirectly MMPs [3, 4, 24–28]. Bisphosphonates are used in clinical practice for treatment of osteoporosis and bone metastasis. It has been found that zolendronic acid, a third generation bisphosphonate used in clinical practice, can modulate expression of ECM genes [29, 30].
The various targeting strategies developed though time involve at first the generation of wide-spectrum inhibitors reaching nowadays to more selective targeting. Batimastat (BB94) and Ilomastat (GM-6001) peptidomimetics, followed by improved second-generation inhibitors Marimastat (BB2516), Prinomastat (AG3340), BAY12-9566) were the first anticancer MMP inhibitors that entered cancer clinical trials. Several clinical trials of such inhibitors have been carried tested alone and in combination with standard chemotherapeutic drugs. The outcomes were controversial resulting in the discontinuation of Phase III clinical trials due to lack of efficacy or severe musculoskeletal side effects in some patients [2, 25, 31–33]. The clinical failures are attributed to the multiple and in some cases dual roles of metalloproteinases at different stages of cancer progression, such as tumor stage, site, enzyme localization and substrate profile [34]. Despite significant clinical interest, it has proven difficult to achieve selective inhibition of metalloproteinases due their common active site [35]. However, the emerging evolution of proteinases degradomics leads to novel inhibition strategies of metalloproteinases based on blocking exosite-mediated cell surface interactions and activation as well as multifunctional inhibitors able to interact with both the active site and exosites, unique to individual metalloproteinases [36–39]. Although ADAMTSs have a structure similar to MMPs, unlike the other metalloproteinases, they have a narrow substrate specificity [40]. This characteristic might give an advantage to ADAMTSs inhibitors vis-à-vis other metalloproteinase inhibitors. Although targeting of ADAMTSs to fight cancer is promising, more mechanistic studies need to be done to fully understand how each ADAMTS is implicated in cancer development and progression [41]. Current efforts focus on designing inhibitors of ADAMTS-4 and -5 that could fight arthritis [40]. Pentosan polysulfate, a chemically-sulfated xylanopyranose, is a multifaceted exosite inhibitor of ADAMTS-4 and -5. It interacts with the noncatalytic spacer domain of ADAMTS-4 and the cysteine-rich domain of ADAMTS-5, blocking their activity. It also mediates the formation of a high-affinity trimolecular complex with ADAMTS-5 and TIMP-3 improving the efficacy of TIMP-3 as an aggrecanase inhibitor [42, 43]. Notably, some inhibitors of ADAMs used in clinical cancer trials were interrupted because of liver toxicity. Despite this handicap, ADAMs inhibitors remain a promising target for cancer treatment. Selective inhibitors, such as INCB3619 [44] G1254023X [45] and KB-R7785 [46], have generated promising results in pre-clinical studies.
Cathepsins
Cathepsins belong to a large family of proteases found in mammals as well as other organisms. Depending on their structure, their catalytic type and the proteins they cleave, cathepsins are classified in three different groups: (I) Aspartic proteases, including cathepsins D and E; (II) Serine proteases, including cathepsins A and G; and (III) Cysteine proteases, consisting of 11 papain-like lysosomal cathepsins (B, C, F, H, L, K, O, S, V, X and W) [6]. The majority of cathepsins and more specifically cysteine cathepsins are optimally active efficient in acidic compartments, and, thus, are often located within lysosomes and acidic endosomes [47, 48]. However, cathepsins are also present within secretory vesicles, cytosol and nuclei [49–53]. Several cathepsins. i.e., cathepsins B, H, L, X, are ubiquitously expressed, whereas cathepsins S, V, F, C and W are specifically expressed by particular tissues or cell types [54]. For instance, cathepsin K is strongly expressed in osteoclasts [55] and cathepsin W is mainly expressed in CD8 and natural killer cells [56]. As mentioned above, the structure and the mechanism of action of the cathepsins differ among the three groups.
All papain-like cysteine cathepsins consist of three functional regions. The signal peptide, the propeptide, which contributes in several functions such as the proper folding of the active enzyme and finally the catalytic site of the enzyme [54]. Their catalytic mechanism is in common with other cysteine proteases. Through their proteolytic action, cathepsins regulate vital processes in both physiological and pathological conditions. Therefore, cathepsins are involved in processes such as the collagen turnover in bone and cartilage [57, 58], hormone production [59] and the regulation of antigen presentation in the immune system [60]. However, a variety of cathepsins play a regulatory role in several diseases. For example, cathepsins K, L, S and B are expressed in patients with bone and cartilage diseases such as rheumatoid arthritis and osteoarthritis, contributing to bone destruction [61–64]. In addition, cathepsins are also implicated in cardiovascular diseases, immune defects, lung diseases and cancer. Particularly in cancer, the role of cathepsins in the progression of the diseases has been reviewed [65, 66]. Although it has been suggested that a variety of cathepsins are implicated in cancer, most studies focus on the action of cathepsins B, S and L. Cathepsins demonstrate elevated expression and activity levels in many tumor cells contributing to cancer cell invasion, migration and metastasis, commonly through ECM degradation or by promoting the availability of several growth factors. For example, cathepsin B is over-expressed in many tumor cells including breast, colon, brain, lung and prostate [67–77]. Among its oncogenic effects, cathepsin B leads in enhanced MMPs activity through degradation of TIMPs [78] and is suggested to assist in the activation of oncogenic signaling of TGF-β [79]. In breast cancer cells, activation of ErbB2 receptor induces cathepsin B expression promoting invasiveness [75]. Cathepsin B over-expression increases ECM degradation fostering collective cell invasion into adjacent tissue [68]. Cathepsin B and pro-uPA are located on tumor cells surface together with their receptors in a caveolin-1 dependent manner thus mediating cell-surface proteolytic events associated with invasion of colon cancer cells [69]. The importance of uPAR and cathepsin B in the regulation of malignant stem cell self-renewal through regulation of hedgehog signaling components, Bmi1 and Sox2, has been also highlighted [72]. Selective inhibition of cathepsin B leads to limited breast to bone metastasis [77] and reduced invasion of breast cancer cells [76]. Moreover, the inhibition of cathepsin S significantly reduced the migration of aggressive lung adenocarcinoma and melanoma cells [80].
Several inhibitors specifically targeting particular cathepsins have been developed. The majority of them are peptidic or peptidomimetic molecules which covalently interact with the active site of the cysteine cathepsins. Apart from general inhibitor of cysteine cathepsins, inhibitors of higher specificity need to developed and tested for their efficiency in targeting particular enzymes of this family. The most recent inhibitors against cathepsin K include balicatib and odanacatib, which are nitrile-based inhibitors, relacatib, a non-basic azepanone analogue, as well as ONO-5334 and MIV-711. These inhibitors have been used in preclinical and clinical trials inhibiting bone resorption and increasing bone mineral density [65, 81, 82]. Targeting cathepsin B may be valuable for treatment of cancer progression. CA-074, a small molecule inhibitor of cathepsin B, reduces metastasis in tumor-bearing animals indicating a pro-metastatic role for cathepsin B in breast cancer and therefore illustrating the therapeutic benefits of its inhibition in vivo [77]. Similarly, CA-074 also prevents human melanoma growth as wells as lung metastasis [83]. The majority of cathepsins inhibitors target the active site of the enzyme blocking their catalytic activity. As cathepsins are ubiquitously expressed, direct inhibition of a given cathepsin could affect its physiological action in other tissues, causing significant off-target effects. In addition, the selectivity of the inhibitor is crucial for the appearance of side effects. For example two cathepsin K inhibitors, relicatib and balicatib were excluded from clinical trial due to lack of specificity. Balicatib is selective for cathepsin K in enzyme assays, while it is not in whole-cells assays. Furthermore, balicatib may cause skin rashes and morphea-like skins incidences [84].
PROTEOGLYCANS: BIOLOGICAL ROLES, THEIR RELATIONS WITH PROTEINASES AND TARGETING IN CANCER
PGs are complex macromolecules, which are composed of a core protein that carries one or more covalently linked glycosaminoglycan (GAG) chains. GAGs are linear polysaccharides, which are composed of repeating disaccharides of hexosamines (N-acetyl-galactosamine or N-acetyl-glucosamine) and uronic acids (D-glucuronic acid or L-iduronic acid) being sulfated at various positions. Keratan sulfate (KS) is the only GAG to be comprised of repeating disaccharides containing N-acetyl-glucosamine and galactose. Notably, hyaluronan (HA) is the only GAG which is not covalently bound to PG protein core, and it has been shown that its synthesis is epigenetically regulated [85]. The number and the type of GAG chains as well as the specific structure of each GAG chain covalently linked to protein cores may greatly differ [86, 87]. These variations in the overall PG structure may be cell- and tissue-specific but may also depend on the differentiation stage and the action of various stimuli on the cells. The structural diversity of PGs determines their functional heterogeneity making them biological chameleons [86, 88]. According to their localization, PGs are categorized as ECM-secreted, cell surface associated and intracellular ones. They can interact with almost all proteins in ECM with different affinities. Their GAGs chains are mainly implicated in these interactions although their protein cores are sometimes involved. Apart from their participation in the organization of ECM and regulation of its mechanical properties, PGs interact with growth factors, cytokines and chemokines protecting them from degradation and form effective gradients of these components in ECM [86]. Furthermore, PGs act as co-receptors for these molecules promoting their signaling. The proven ability of PGs to form complexes with growth factor receptors results in the regulation of their signaling properties. PGs regulate cell behavior and phenotype. They are involved in cell proliferation, adhesion, migration and invasion. In this context, certain PGs affect the expression of bioactive molecules, their secretion, localization and activity [86, 88]. PGs are well established as key players in the regulation of physiological and pathological conditions, such as cardiovascular diseases including myocardial dysfunction and failure as well as cancer development and progression [86, 89]. The major PG families involved in cancer progression and have been also related to certain proteinases are presented below.
Versican
Versican is a chondroitin sulfate (CS)/dermatan sulfate (DS) proteoglycan (CS/DSPG), which is present in ECM of many tissues. It belongs to subfamily of hyalectans, for hyaluronan and lectin binding proteoglycans [90]. Versican is essentially composed of three modular domains: an N-terminal domain with the ability to bind hyaluronan (HA) through its globular domain (G1), a central domain that harbors CS or/and DS, and a C-terminal globular domain (G3), which exhibits lectin-like activity, located in the C-terminal. Versican exist as a full-length proteoglycan (named V0) and three splice variants (V1, V2 and V3). They differ in the central domain and carry different number of GAG chains (V0, V1 and V2), whereas the third splice variant V3 lacks completely the GAG attachment region and it should be considered as a protein and not as true PG [86, 91, 92]. Versican is highly expressed in most malignancies and has been associated with cancer relapse and poor patient outcome in breast, prostate and other cancer types [86, 88, 93–99]. Versican is predominantly secreted by the activated peritumoral stromal cells in malignancies in response to various stimuli. Its expression is up-regulated in cancer associated fibroblasts upon TGF-β treatment and promotes the motility and invasion of ovarian cancer cells by activating the NF-κB signaling pathway and by up-regulating expression of CD44, MMP-9, and the HA-mediated motility receptor [100]. However, some cancer cells can also secrete versican [86, 101–105]. Versican modulates the adhesion of cancer cells to ECM and increases their motility, proliferation and metastasis [86]. Versican is highly expressed in sporadic clear cell renal cell carcinoma [101]. Its expression is associated with the expression of CD26 on T-anaplastic large cell lymphoma. Abrogation of versican expression results in decreased levels of MT1-MMP and CD44 and marked suppression of T-cell adhesion and invasion [102]. Versican is substrate for ADAMTS and the use of antibodies against to ADAMTS specific versican cleavage site inhibits glioma cell migration [106]. ADAMTS are often over-expressed in various tumors and the formation of neo-epitopes of versican fragments within tumor stroma may be used for targeted therapy. In this context, a novel versican isoform V4 is highly expressed in breast cancer [107], whereas versican is also differentially glycosylated in breast cancer since it contains more sialic acid [108]. This alternative splice variant of versican or the presence of unusual glycosylation may be possible targets for therapeutic intervention with antibody-related agents. Furthermore, disruption of the HA–CD44 interaction with HA oligomers may be also used for targeting tumor progression. HA oligomers can block the acquisition of a HA-versican pericellular matrix by ovarian cancer cells that increases their metastatic potential and are promising inhibitors of cancer dissemination [109].
Decorin
Another important class of matrix PGs is the small leucine-rich PG (SLRP) family, with decorin being the most studied member of this family with well-documented implication in cancer [110]. Decorin is a CS/DSPG that plays an important role in tumor progression by affecting both the composition of the tumor stroma as well as tumor cell growth [86, 88, 94–96, 98, 99, 111]. Specifically, decorin protein core has a potent antitumor effect by interfering with several signaling pathways and repressing and attenuating tumor cell survival, growth, migration and angiogenesis. Soluble decorin is a robust pan-RTK inhibitor targeting not only EGFR, but also Met, IGF-IR, VEGFR2 and PDGFR [112–117]. Indeed, soluble decorin can be considered a tumor repressor [118] that possesses anti-oncogenic and anti-metastatic [119] properties and, thus, it has been proposed to be a “guardian from the matrix” [120]. Decorin protein core binds to EGFR leading to rapid caveolar endocytosis and lysosomal degradation [121]. This interaction also promotes ERK1/2 signaling and transcription of p21 leading to tumor cell growth arrest [122] as well as caspase-3 activation and induction of apoptosis [123]. Notably, decorin inhibits angiogenesis [120] by directly antagonizing the pro-angiogenic Met and simultaneous suppression of HIF-1α and VEGFA transcription [124, 125]. Decorin favors angiostasis by rapidly inducing the secretion of thrombospondin-1 [126], a key antiangiogenic factor [127, 128]. Thus, soluble decorin is an endogenous anti-oncogenic agent [86, 110], a notion supported by robust genetic evidence indicating that targeted deletion of the decorin gene is permissive for tumorigenesis [129, 130]. Adenoviral-mediated gene delivery of decorin or systemic administration human recombinant decorin or decorin protein core to various tumor xenograft models (breast and prostate carcinomas), suppress tumor growth [131–135]. The recent discoveries that decorin is pro-inflammatory and interacts with Toll-like receptors [136] together with induction of autophagy in endothelial cells [137] and mitophagy in breast cancer cells [138] indicate that decorin can affect both the tumor stroma and the tumor proper in a variety of ways. As decorin interacts with various metalloproteases, directly or indirectly by modulating their production, it is likely that this important endogenous factor might be involved in the turnover not only of matrix molecules, but, most importantly, in the cleavage and processing of cell surface receptors.
Syndecans
Syndecans are type 1 transmembrane heparan sulfate (HS) PGs (HSPGs) that have important roles during development, wound healing, neural and glioma stem cell differentiation as well as in tumor progression [139–141]. There are four syndecans in mammals with different distribution among tissues. Syndecan-1 is expressed at early stages during development, in epithelial and cancer cells in adults as well as in some leukocytes. Syndecan-2 is synthesized in mesenchymal tissues, fibroblasts, liver and neuronal cells. Syndecan-3 is present in neural and musculoskeletal tissues, whereas syndecan-4 is ubiquitously distributed. Syndecan-1 and syndecan-3 may also carry CS apart from HS chains. Syndecans interact with numerous ECM molecules, growth factors, chemokines and cytokines primarily via their HS chains. Thus, they influence cell proliferation, differentiation, adhesion and migration often in co-operation with other cell surface receptors [139, 140].
Syndecans are expressed in various tumor types possessing diverse roles according to tumor type and stage of the disease, acting either as promoters or inhibitors of tumor progression [139]. Synstatin, a syndecan-derived peptide inhibitor, interferes with syndecan-1/αvβ3 integrin interaction and indirectly attenuates angiogenesis and impairs mammary tumor growth in a mouse tumor model suggesting that syndecan-1 might be involved in regulating angiogenesis and tumorigenesis [142, 143]. Syndecan-1 plays important role in supporting α2β1 integrin-mediated adhesion to collagen. The formation of syndecan-1/α2β1 integrin contributes to proper organization of cortical actin and enhances the transcription of MMP-1 in response to collagen binding [144]. Activation of K-Ras in tumor cells correlates with increased expression of α2β1 integrin, MT1-MMP, syndecan-1, and syndecan-4. α2β1 integrin and MT1-MMP are positive regulators of invasion, whereas syndecans inhibit cell invasion into 3D collagen matrix [145]. Syndecan-1 serves as collagen receptor in squamous cancer cells in addition to α2β1 integrin and promotes strong cell adhesion. Lowering syndecan-1 levels results in enhanced cell spreading and motility on collagen I by modulating RhoA and Rac activity, but does not affect cell motility on other ECM substrates [146]. Syndecan levels at the cell surface are regulated by a fine-tuned proteolytic cleavage, which is called shedding. Syndecans are cleaved by various proteases such as MMPs, ADAMTS and ADAMS, in their extracellular domain near the plasma membrane [139, 147]. This process is regulated by a variety of stimuli such as growth factors, chemokines, oxidative stress, heparanase and others [139, 147]. Shed syndecan ectodomains are implicated in several steps of cancer progression. For example soluble syndecan-1 may bind growth factors and chemokines protecting them from inactivation and promote their signaling in tumor microenvironment or may sequester inhibitory signals for tumor cells [86, 139].
Syndecans may be potential targets for tumor therapy due to their well-documented implication in tumorigenesis. A possibility is to directly target syndecans by using therapeutic antibodies in malignancies where these molecules are highly expressed [86, 139, 148–150]. Another option is to target the formation of functional syndecan complexes with other receptors such as integrins and growth factor receptors by using synthetic molecules such as synstatin [151]. Blocking of syndecans shedding either by inhibiting heparanase activity using specific inhibitors or proteinase inhibitors may be also of interest [152–154]. It is worth noticing that many drugs used in cancer therapy can regulate the expression levels of syndecans and may benefit patients [29, 30, 155, 156]. Furthermore, syndecans’ function in cancer is regulated by miRNAs and this open a new area of research [157].
CSPG4/NG2
Neuron-glial antigen 2 (NG2) also known as chondroitin sulfate proteoglycan 4 (CSPG4) or melanoma associated CSPG, is a transmembrane PG, which is up-regulated in several tumor types and play important role in tumor cell proliferation, survival, migration and tumor progression [158]. Expression of CSPG4 enhances integrin-mediated cell spreading, FAK phosphorylation, and activation of ERK1/2 through independent mechanisms [159]. Furthermore, CSPG4 is implicated in the activation of pro-MMP-2 by MT3-MMP [160, 161]. CSPG4 mediates tumor cell adhesion and spreading on collagen VI by cooperating with α2β1 integrin and activating PI-3K [162]. CSPG4 is an attractive target for antibody-based cancer immunotherapy because of its proven role in tumor cell biology, its high expression on tumor and cancer stem cells and its restricted distribution in normal tissues [158, 163]. The development and use of several therapeutic antibodies give promising results in cancer treatment [158, 163–166]. Furthermore, T lymphocytes genetically modified with a CSPG4-specific chimeric antigen receptor controlled tumor growth in vitro and in vivo in mice engrafted with various tumor cells. This suggests that CSPG4-redirected T cells should provide an effective treatment modality for a variety of solid tumors [167].
Serglycin
Serglycin is an intracellular PG that is highly expressed in hematopoietic cells. It is involved in the proper formation of secretory granules and vesicles as well as the storage and secretion of several components in ECM [168]. Serglycin can carry HS, heparin, CS and DS chains and its glycanation depends on the cellular source of serglycin [168]. Several studies have demonstrated the involvement of serglycin in tumorigenesis [169–174]. Serglycin with different GAG chains and sulfation patterns is expressed in several hematopoietic and non-hematopoietic tumors [168]. The role of serglycin in malignancies is rather intriguing since it seems to participate in tumor development in a manner that at least partially requires interactions between tumor cells and their microenvironment [175]. In mast cells, serglycin is present in intracellular granules where is implicated in the storage of granule-localized proteases such as chymases, tryptases and carboxypaptidase A [176, 177]. Mast cell proteases exert dual roles in the regulation of inflammation and tumorigenesis. Several pro-inflammatory chemokines and cytokines are substrates of mast cell proteases and their cleavage can result in either activation or inactivation of inflammatory mediators. Serglycin is over-expressed and is constitutively secreted in multiple myeloma carrying CS side chains [174, 178]. It is also localized on the cell surface where is attached through its CS-4 chains promoting the adhesion of myeloma cells to collagen I as well as the biosynthesis and secretion of MMP-9 and MMP-2 [173]. Serglycin is involved in the biosynthesis and secretion of proteases. For example, Madin-Darby canine kidney cells stably transfected with serglycin express elevated levels of MMP-9 and urokinase plasminogen activator (uPA) [179]. Recently it has been shown that serglycin bearing CS chains is the major PG that is synthesized and secreted from aggressive breast cancer tumor cells [170]. Ectopic over-expression of serglycin in low aggressive MCF-7 breast cancer cells promotes their anchorage-independent growth, migration and invasion [170] via up-regulation of biosynthesis of proteolytic enzymes (unpublished data). The tumor promoting properties of serglycin are dependent on the overexpression and secretion of glycanated serglycin [170]. Altered biosynthesis of CS chains has been demonstrated in various cancer types [180]. CHST11 gene that specifically mediates 4-O sulfation of CS is highly expressed in MDA-MB-231 breast cancer cells and breast cancer tissues together with CSPG4 another candidate carrier molecule for CS-4. CS-4 chains mediate the binding of breast cancer cells to P-selectin and facilitate the formation of metastasis [181].
INTERPLAY OF MATRIX PROTEINASES WITH PROTEOGLYCANS AND GAGs
Proteolysis is a highly regulated and specific process. Apart from the interactions between the substrate and catalytic domain, non-catalytic sites, called exosites, contribute to substrate cleavage by presenting the substrate to the catalytic domain [37, 39, 182]. In addition, proteinases can form complexes with other macromolecules, like proteins and PGs/GAG in allosteric sites, regulating the activity, stability, substrate specificity and localization of the enzymes. The interacting PGs/GAGs with certain proteinases and the biological roles affected by such interactions are presented in Table 1. An emerging concept is the compartmentalization of proteinases to secretory vesicles and cell membranes. Specifically, the concentrations of both the enzyme and substrate are maintained at catalytically-favorable levels within the pericellular space and are utilized upon cell stimuli rather than diffuse into the extracellular space [183].
Table 1.
PGs / GAGs as binding partners for matrix proteinases
| Proteinase | Domain | Interacting PG/GAGs |
Biological roles [Ref] |
|---|---|---|---|
| MMP-2 | Pro-form and active form |
Syndecan-2 (HS) | Inhibits activation of pro- MMP-2 [191] |
| MMP-2 | Hemopexin | CSPG4 (CS-4) | Activation of MMP-2, increased cell migration and invasion [160] |
| MMP-7 | ? | HSPGs/heparin | Increase activity [184] |
| MMP-7 | Pro-form (higher affinity) and active form (less affinity) |
Heparin/CS-E/DS | Autocatalytic activation and increase activity [185, 186, 218] |
| MMP-7 | Pro-form | Syndecan-2 (core protein) |
Activation and cell migration [37, 187] |
| MMP-9 | ? | HSPGs | Cell migration and invasion [188] |
| MMP-9 | Pro-form (hemopexin / fibronectin-like module) |
Serglycin / Versican (core proteins |
Modulates the activation of proMMP-9 and the binding to substrates [194, 195] |
| MMP-13 | hemopexin | Syndecan-4, serglycin, decorin (core proteins) |
Unknown [198] |
| MT3-MMP | ? | CSPG4 (CS-4) | Cell invasion [160, 161] |
| ADAMTS-4 | ? | Syndecan-1 (HS/CS) | Cell surface localization and activation [189] |
| ADAMTS-5 | ? | Syndecan-4 | Activation and MMP-3 expression [190] |
| ADAM-12 | Pro-form and catalytic form |
HSPGs (HS) | Inhibition or activation against specific substrates [192] |
| Cathepsin K | GAG binding sites | HS / DS | Inhibit the activity [206] |
| Cathepsin K | GAG binding sites | CS-4 / KS | Enhance activity [206] |
|
Cathepsins V and K |
GAG binding sites | CS-4 / CS-6 / DS / heparin |
Inhibit elastolytic activity [213] |
| Cathepsin S | GAG binding sites | CS-4 | Inhibits activity [209] |
| Cathepsin B | GAG binding sites | Heparin / HS | Enhance activity and stability [210] |
| Cathepsin X | GAG binding sites | Heparin / HS / HSPGs |
Enhance activity, cell surface localization [212] |
|
Pro cathepsin D |
GAG binding sites (catalytic / pro-form) |
Heparin / HS / CS-E | Enhance activity [214] |
It has been shown that MMP-7 co-localizes with HSPGs and heparin increases MMP-7 activity [184]. It is well documented that there is a correlation between GAG sulfation and the regulation of MMP-7 activity. Highly-sulfated GAGs, such as heparin, chondroitin-4,6-sulfate (CS-E) and DS, enhance MMP-7 activity against specific substrates to a greater extent than low-sulfated forms of CS [185, 186]. Thus, specific GAGs may function as allosteric physiological activators of MMPs. Syndecan-2 can act as a docking site for pro-MMP-7 at the cell surface, and in turn such surface localization is pivotal for its ability to regulate carcinogenesis [37, 187]. MMP/HSPG complexes (MMP-9/HSPG) are also concentrated at the leading edge of highly-metastatic cells, mediating their migration and invasion [188]. However, GAG chains could simultaneously interact with an active MMP and substrate, bringing them together to act as highly-coordinated trimeric complex. For example, trimeric complexes of pro-MMP-2 and MMP-16 (MT3-MMP) and cell-surface CSPG4 can activate MMP-2 and increase cell migration, invasion, and metastasis [160, 161]. In addition, binding of ADAMTS-4 and MMP-17 (MT4-MMP) to syndecan-1 induces the activation of ADAMTS-4. Then, the activated form is located to the cell membrane interacting with the GAG chains of syndecan-1 [189]. Syndecan-4 binding to ADAMTS-5 results in its activation, regulating mitogen-activated protein kinase (MAPK)-dependent synthesis of MMP-3 [190]. On the other hand, syndecan-2 via the HS chains may compete for the binding of pro-MMP-2 interfering with the proper formation of MT1-MMP/TIMP-2/pro-MMP-2 complex that leads to its activation. In this case, syndecan-2 inhibits the activation of pro-MMP-2 suppressing metastasis in mice [191]. Another example of negative regulation is that of HSPGs and pro-ADAM-12. These interactions results in the suppression of ADAM-12 catalytic activity in some substrates but increase the activity of the enzyme against others, such as IGFBP3 [192]. This finding suggests that such inhibition is mediated by the binding of the GAG to basic amino acids in the pro- and catalytic domains of ADAM-12 creating a unique molecular switch [192].
The regulation of the activation/activity of MMPs can be achieved in various ways. The activation mechanism (known as the ‘cysteine switch’) involves proteolytic cleavages of the propeptide causing a destabilization of the cysteine zinc interaction [193]. In the latent state of MMPs, the propeptide “protects” the catalytic site, thereby blocking any interaction with the substrates. Bypassing the common mechanism of activation, the cysteine zinc interaction may be disrupted by an allosteric interaction between the prodomain and the GAG chains of a PG, leading to an active MMP and then the propeptide is either inter- or intra-molecularly cleaved [185]. However, the binding of MMPs to PGs occurs either through specific interactions with the GAG chains or the protein core of the PG. Serglycin and versican isolated from various sources form complexes with proMMP-9 in vivo and in vitro [194, 195]. Both the hemopexin-like (PEX) domain and the fibronectin-like (FnII) module of proMMP-9 are involved in the interaction with core proteins of PGs. The formation of the complexes alters the mode of activation of proMMP-9 and the interaction of the enzyme with its substrates [194, 196]. ProMMP-9 associated with PGs is activated in the presence of Ca2+ and may be important for the activation of pro-enzyme in pathological situation, which are associated with increasing concentrations of this cation such as tumor-induced bone disease [197]. For MMP-13, the PEX domain is responsible for the binding to the protein core of syndecan-4, serglycin, and decorin in chondrocytes [198]. Serglycin interacts with mast cell proteases regulating their activity. Heparin present on serglycin in mast cells forms complexes with chymase and promotes the binding of heparin-binding substrates to the enzyme thus presenting them to chymase and enhancing their proteolysis [199]. Heparin significantly blocks the inhibition of chymase by natural inhibitors such as α1-protease inhibitor, α1-antichymotrypsin, α2-macroglobulin and soybean trypsin inhibitor [200, 201]. Heparin is also implicated in the formation of active tryptase tetramers [202, 203]. Chymase can activate various MMPs, whereas both tryptase and chymase directly degrade ECM components. Chymase cleaves vitronectin and procollagen, while tryptase degrades collagen IV and both degrade fibronectin [168].
GAGs also interact with cathepsins controlling their activation, stability and activity. It has been shown that the ability of cathepsin K to cleave collagen is highly dependent on the formation of an oligomeric complex with GAGs, in particular, with CS-4. This GAG has no effect on the activity of cathepsin L and MMP-1 [204, 205]. Some GAGs such as KS also enhances cathepsin K activity, while HS and DS inhibit the activity of the protease. These data suggested that the activity of some cathepsins is significantly enhanced by specific GAG chains [206]. Complex formation with GAGs is unique for cathepsin K among other papain-like proteases [205, 206]. It is remarkable that monomeric cathepsin K has no significant collagenase activity [205]. The specific binding of CS-4 to cathepsin K is required to allow the formation of an oligomeric complex capable to digest triple-helical collagen. It is suggested that the main function of this complex is the partial unfolding of triple-helical collagen that increases the accessibility of the scissile bond to the active site of cathepsin K [207].
Recently it has been reported the presence of three positively charged clusters at the bottom part of the protease opposing the active site, which are involved in alternative GAG binding sites. These may play other roles in the formation of collagenolytically active protease complexes, or contribute in a yet unknown manner to the specific binding to collagen [208]. Therefore, any disruption of the complex formation between cathepsin K and GAGs might be of therapeutic value in collagen degradation-related diseases such as cancer. Contrary, CS-4 inhibits the action of cathepsin S and may modulate its collagenase activity in vivo [209]. Notably, the interaction of cathepsin B specifically with heparin or HS induces its activity protecting it from alkaline pH [210]. Heparin and HS accelerate the autocatalytic removal of the propeptide, thus activating cathepsin B [211]. In this line of evidence, heparin and HS specifically bind to cathepsin X enhancing its activity [212]. Cathepsin X also binds to cell surface HSPGs in wild-type CHO cells but not in CHO-745 cells, which are deficient in GAG synthesis. In addition, cathepsin X is only endocytosed in CHO cells suggesting that HSPGs may also regulate its cellular trafficking [212]. Although CS-4 significantly enhances the collagenase activity of cathepsin K, the same GAG as well as others, specifically inhibits the potent elastolytic activity of cathepsins V and K [213]. In another study, the activity of pro-cathepsin D is significantly increased in the presence of heparin, HS and CS-E. The regulation of the activity by heparin depends on the sulfation groups and the length of the GAG chain, since more sulfated groups or larger fragments exhibit enhanced activity [214].
Squamous cell carcinoma antigens (SCCA-1 and SCCA-2), members of the high-molecular weight serine proteinase inhibitor (serpin) family, bind to heparin and this significantly enhances the inhibition of cathepsin L-like proteolytic activity secreted from breast and melanoma cancer cell. This finding raises the possibility that the anticancer properties of heparin may be due, at least partly, to enhanced inhibition of pro-metastatic proteases [215]. The above data highlight the fact that PGs/GAGs are able to control and regulate the activity of many cathepsins. However, this is a complex regulation, since specific GAGs can activate or inhibit specific cathepsins and in addition, a particular GAG may have opposite effects in different cathepsins. Moreover, the length of the GAG and its specific sulfation seems to be of significance.
CONCLUDING REMARKS
Given the central role played by matrix molecules and their processing enzymes, we propose that the interplay between proteoglycans and proteinases could represent a new avenue for specific cancer therapeutics. This hypothesis envisages a novel molecular targeting for treating cancer achieving allosteric inhibition of proteolytic enzyme activity and concurrent compartmentalization of proteinases via proteoglycan modulation. The main axis of regulatory interactions between proteoglycans and proteinases is summarized in Figure 1. Τhe use of modified GAGs or GAG mimetics utilized to modulate GAG-protein interactions [216] alone, or in conjunction with specific proteinases exosites may introduce a new era in cancer therapeutics. One such approach is targeting the exosites of particular cathepsins since GAG chains interact with the exosites of specific cathepsins modulating their catalytic activity. Thus, targeting the exosites could block the proteolytic activity against specific substrates of enzyme without disturbing other processes. Selent et al. [217] demonstrated that negatively charged molecules are able to inhibit only the collagenase activity of cathepsin K without affecting the overall proteolytic functions of the enzyme. The inhibitors are negatively charged molecules such as poly-Asp and poly-Glu with ionic properties similar to those of CS-4. These molecules exert their inhibitory effect mainly by interfering with the formation of cathepsin K/CS-4 complex. Therefore, we suggest that the targeting of proteinase exosites, which interact with GAGs, could be a novel mechanism of inhibition. Currently, our knowledge of this field is relatively sparse. Therefore, future targeted studies should be done especially focusing on the development of novel inhibitors, which should selectively inhibit specific proteinase activities and possibly bypass the detrimental side effects caused by present inhibitors.
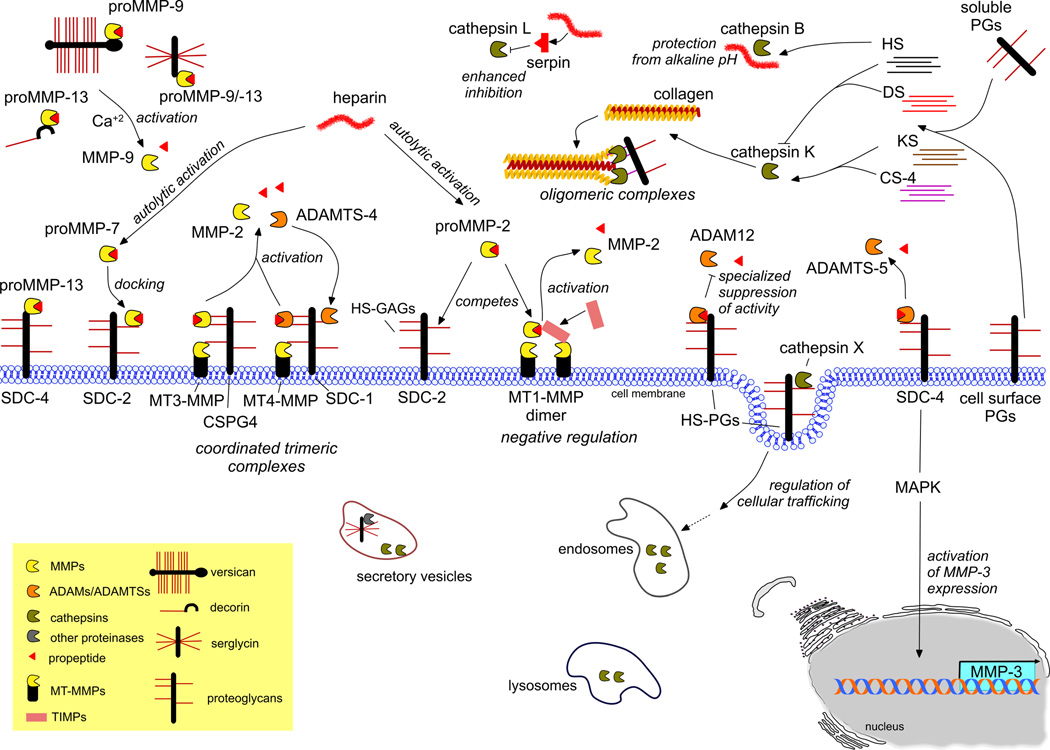
Figure 1. Main axis of proteinases regulation by proteoglycans action.
The interaction between proteinases and proteoglycans occurs either through the GAG chains or the core protein of PGs. One main outcome of such interactions is the compartmentalization of proteinases confined to secretion vesicles and especially to cell membrane as well as stored in confined spaces within the extracellular space. Thus, proMMPs (proMMP-9, proMMP-13) bind to the core protein of several PGs secreted or membrane bound, like serglycin, decorin, versican and SDC-4 or interact with the GAG chains of SDC-2 acting as a docking site (top-left panel). HSPGs also represent a binding site for cathepsins at the cell surface, implicating them also to their cellular trafficking in the case of cathepsin X. The activation and activity of proteinases is strongly regulated by GAGs in either positive or negative mode. CS-4 chains of cell-surface CSPG4 are able to simultaneously interact with active MT3-MMP and its substrate pro-MMP-2, leading to the formation of a coordinated trimeric complex and subsequent activation of MMP-2 (left-middle panel). Similarly, the complex of ADAMTS-4/MT4-MMP/SDC-1 induces the activation of ADAMTS-4, further located to the cell membrane via interacting with the GAG chains of SDC-1 (left-middle panel). On the other hand, SDC-2 HS chains compete with MT1-MMP for the binding of pro-MMP2, which prevents the activation of the proMMP2 via cell membrane ternary complex, which also involves MT1-MMP dimer and TIMP-2 (middle panel). Another example of negative regulation is that of HSPGs and pro-ADAM-12, resulting in the specialized suppression of ADAM-12 catalytic activity against some substrates (right-middle panel). SDC-4 is also able to control the ADAMTS-5 activation through direct interaction, leading also to MAPK-mediated activation of MMP-3 expression (right panel). In the case of cathepsins, the formation of oligomeric complexes with GAGs for example CS-4 with cathepsin K renders cathepsin capable to unfold and further digest the triple-helical collagen. In coordination, HS and KS chains also enhance cathepsin K activity, while HS and DS inhibit it (top-right panel). Heparin, a highly sulfated glycosaminoglycan, enhanced the autolytic activation of proMMP-2 and proMMP-7. Heparin, along with HS chains, protects cathepsin B action from alkaline pH, whereas members of serpin family bind to heparin and this significantly enhances the inhibition of cathepsin L-like proteolytic activity (top-right / middle panel).
Acknowledgments
This research has been co-financed by the European Union (European Social Fund — ESF) and Greek National Funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) Research Funding Program: Thales. Investing in knowledge society through the European Social Fund.
Abbreviations
- ADAM
a disintegrin and metalloproteinase
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- CS
chondroitin sulfate
- DS
dermatan sulfate
- ECM
extracellular matrix
- GAG
glycosaminoglycan
- HA
hyaluronan
- HS
heparan sulfate
- MMP
matrix metalloproteinase
- MT-MMP
membrane type-MMP
- PG
proteoglycan
- TIMP
tissue inhibitors of MMPs
References
- 1.Theocharis A, Gialeli C, Hascall V, Karamanos Nikos K. 1.1 Extracellular matrix: a functional scaffold. Extracellular Matrix: Pathobiology and Signaling. 2012 [Google Scholar]
- 2.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. The FEBS journal. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nature reviews Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 4.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagase H. Metalloproteases. Current protocols in protein science / editorial board, John E Coligan [et al] 2001;Chapter 21(Unit 21):4. doi: 10.1002/0471140864.ps2104s24. [DOI] [PubMed] [Google Scholar]
- 6.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuman Moss LA, Jensen-Taubman S, Stetler-Stevenson WG. Matrix metalloproteinases: changing roles in tumor progression and metastasis. Am J Pathol. 2012;181:1895–1899. doi: 10.1016/j.ajpath.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curran S, Murray GI. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur J Cancer. 2000;36:1621–1630. doi: 10.1016/s0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 9.Sterchi EE. Special issue: metzincin metalloproteinases. Mol Aspects Med. 2008;29:255–257. doi: 10.1016/j.mam.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Blobel CP, Wolfsberg TG, Turck CW, Myles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- 11.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 12.Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.van Goor H, Melenhorst WB, Turner AJ, Holgate ST. Adamalysins in biology and disease. J Pathol. 2009;219:277–286. doi: 10.1002/path.2594. [DOI] [PubMed] [Google Scholar]
- 14.Vihinen P, Kahari VM. Matrix metalloproteinases in cancer: prognostic markers and therapeutic targets. Int J Cancer. 2002;99:157–166. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 15.Butler GS, Overall CM. Updated biological roles for matrix metalloproteinases and new "intracellular" substrates revealed by degradomics. Biochemistry. 2009;48:10830–10845. doi: 10.1021/bi901656f. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez D, Morrison CJ, Overall CM. Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim Biophys Acta. 2010;1803:39–54. doi: 10.1016/j.bbamcr.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Pytliak M, Vargova V, Mechirova V. Matrix metalloproteinases and their role in oncogenesis: a review. Onkologie. 2012;35:49–53. doi: 10.1159/000336304. [DOI] [PubMed] [Google Scholar]
- 18.McCawley LJ, Matrisian LM. Matrix metalloproteinases: they're not just for matrix anymore! Curr Opin Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 19.Nagase H. Activation mechanisms of matrix metalloproteinases. Biol Chem. 1997;378:151–160. [PubMed] [Google Scholar]
- 20.Dunn JR, Reed JE, du Plessis DG, Shaw EJ, Reeves P, Gee AL, Warnke P, Walker C. Expression of ADAMTS-8, a secreted protease with antiangiogenic properties, is downregulated in brain tumours. British journal of cancer. 2006;94:1186–1193. doi: 10.1038/sj.bjc.6603006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freitas VM, do Amaral JB, Silva TA, Santos ES, Mangone FR, Pinheiro Jde J, Jaeger RG, Nagai MA, Machado-Santelli GM. Decreased expression of ADAMTS-1 in human breast tumors stimulates migration and invasion. Molecular cancer. 2013;12:2. doi: 10.1186/1476-4598-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleivi K, Lind GE, Diep CB, Meling GI, Brandal LT, Nesland JM, Myklebost O, Rognum TO, Giercksky KE, Skotheim RI, Lothe RA. Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Molecular cancer. 2007;6:2. doi: 10.1186/1476-4598-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Held-Feindt J, Paredes EB, Blomer U, Seidenbecher C, Stark AM, Mehdorn HM, Mentlein R. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. International journal of cancer Journal international du cancer. 2006;118:55–61. doi: 10.1002/ijc.21258. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhary AK, Pandya S, Ghosh K, Nadkarni A. Matrix metalloproteinase and its drug targets therapy in solid and hematological malignancies: an overview. Mutation research. 2013;753:7–23. doi: 10.1016/j.mrrev.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 26.Skandalis Spyros S, Gialeli C, Afratis N, Aletras Alexios J, Tsegenidis T, Theocharis Achilleas D, Karamanos Nikos K. 8.5 Pharmacological targeting of proteoglycans and metalloproteinases: an emerging aspect in cancer treatment. In: Karamanos N, editor. Extracellular Matrix: Pathobiology and Signaling. DE GRUYTER; 2012. pp. 785–801. [Google Scholar]
- 27.Devy L, Dransfield DT. New Strategies for the Next Generation of Matrix-Metalloproteinase Inhibitors: Selectively Targeting Membrane-Anchored MMPs with Therapeutic Antibodies. Biochemistry research international. 2011;2011:191670. doi: 10.1155/2011/191670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, Chang E, Tao Q, Vanhove M, Lejeune A, van Gool R, Sexton DJ, Kuang G, Rank D, Hogan S, Pazmany C, Ma YL, Schoonbroodt S, Nixon AE, Ladner RC, Hoet R, Henderikx P, Tenhoor C, Rabbani SA, Valentino ML, Wood CR, Dransfield DT. Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer research. 2009;69:1517–1526. doi: 10.1158/0008-5472.CAN-08-3255. [DOI] [PubMed] [Google Scholar]
- 29.Dedes PG, Gialeli C, Tsonis AI, Kanakis I, Theocharis AD, Kletsas D, Tzanakakis GN, Karamanos NK. Expression of matrix macromolecules and functional properties of breast cancer cells are modulated by the bisphosphonate zoledronic acid. Biochimica et biophysica acta. 2012;1820:1926–1939. doi: 10.1016/j.bbagen.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Dedes PG, Kanakis I, Gialeli C, Theocharis AD, Tsegenidis T, Kletsas D, Tzanakakis GN, Karamanos NK. Preclinical evaluation of zoledronate using an in vitro mimetic cellular model for breast cancer metastatic bone disease. Biochimica et biophysica acta. 2013;1830:3625–3634. doi: 10.1016/j.bbagen.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Shi ZG, Li JP, Shi LL, Li X. An updated patent therapeutic agents targeting MMPs. Recent patents on anti-cancer drug discovery. 2012;7:74–101. doi: 10.2174/157489212798357976. [DOI] [PubMed] [Google Scholar]
- 32.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 33.Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000;19:6642–6650. doi: 10.1038/sj.onc.1204097. [DOI] [PubMed] [Google Scholar]
- 34.Dufour A, Overall CM. Missing the target: matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol Sci. 2013;34:233–242. doi: 10.1016/j.tips.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Morrison CJ, Butler GS, Rodriguez D, Overall CM. Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol. 2009;21:645–653. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- 37.Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 38.Overall CM, Kleifeld O. Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br J Cancer. 2006;94:941–946. doi: 10.1038/sj.bjc.6603043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sela-Passwell N, Rosenblum G, Shoham T, Sagi I. Structural and functional bases for allosteric control of MMP activities: can it pave the path for selective inhibition? Biochim Biophys Acta. 2010;1803:29–38. doi: 10.1016/j.bbamcr.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Tortorella MD, Malfait F, Barve RA, Shieh HS, Malfait AM. A review of the ADAMTS family, pharmaceutical targets of the future. Current pharmaceutical design. 2009;15:2359–2374. doi: 10.2174/138161209788682433. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Rao N, Ge R. Emerging Roles of ADAMTSs in Angiogenesis and Cancer. Cancers. 2012;4:1252–1299. doi: 10.3390/cancers4041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Troeberg L, Fushimi K, Khokha R, Emonard H, Ghosh P, Nagase H. Calcium pentosan polysulfate is a multifaceted exosite inhibitor of aggrecanases. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:3515–3524. doi: 10.1096/fj.08-112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Troeberg L, Mulloy B, Ghosh P, Lee MH, Murphy G, Nagase H. Pentosan polysulfate increases affinity between ADAMTS-5 and TIMP-3 through formation of an electrostatically driven trimolecular complex. The Biochemical journal. 2012;443:307–315. doi: 10.1042/BJ20112159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q, Lo Y, Zhou BB, Pan M, Thomas SM, Grandis JR, Zhuo J, Yao W, Newton RC, Friedman SM, Scherle PA, Vaddi K. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:1892–1902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
- 45.Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Combinatorial chemistry & high throughput screening. 2005;8:161–171. doi: 10.2174/1386207053258488. [DOI] [PubMed] [Google Scholar]
- 46.Shimura T, Kataoka H, Ogasawara N, Kubota E, Sasaki M, Tanida S, Joh T. Suppression of proHB-EGF carboxy-terminal fragment nuclear translocation: a new molecular target therapy for gastric cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:3956–3965. doi: 10.1158/1078-0432.CCR-07-4794. [DOI] [PubMed] [Google Scholar]
- 47.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. The EMBO journal. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Current pharmaceutical design. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 49.Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Molecular cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 50.Puri N, Roche PA. Mast cells possess distinct secretory granule subsets whose exocytosis is regulated by different SNARE isoforms. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2580–2585. doi: 10.1073/pnas.0707854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reiser J, Oh J, Shirato I, Asanuma K, Hug A, Mundel TM, Honey K, Ishidoh K, Kominami E, Kreidberg JA, Tomino Y, Mundel P. Podocyte migration during nephrotic syndrome requires a coordinated interplay between cathepsin L and alpha3 integrin. The Journal of biological chemistry. 2004;279:34827–34832. doi: 10.1074/jbc.M401973200. [DOI] [PubMed] [Google Scholar]
- 52.Sever S, Altintas MM, Nankoe SR, Moller CC, Ko D, Wei C, Henderson J, del Re EC, Hsing L, Erickson A, Cohen CD, Kretzler M, Kerjaschki D, Rudensky A, Nikolic B, Reiser J. Proteolytic processing of dynamin by cytoplasmic cathepsin L is a mechanism for proteinuric kidney disease. The Journal of clinical investigation. 2007;117:2095–2104. doi: 10.1172/JCI32022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wartmann T, Mayerle J, Kahne T, Sahin-Toth M, Ruthenburger M, Matthias R, Kruse A, Reinheckel T, Peters C, Weiss FU, Sendler M, Lippert H, Schulz HU, Aghdassi A, Dummer A, Teller S, Halangk W, Lerch MM. Cathepsin L inactivates human trypsinogen, whereas cathepsin L-deletion reduces the severity of pancreatitis in mice. Gastroenterology. 2010;138:726–737. doi: 10.1053/j.gastro.2009.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lecaille F, Kaleta J, Bromme D. Human and parasitic papain-like cysteine proteases: their role in physiology and pathology and recent developments in inhibitor design. Chemical reviews. 2002;102:4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 55.Salminen-Mankonen HJ, Morko J, Vuorio E. Role of cathepsin K in normal joints and in the development of arthritis. Current drug targets. 2007;8:315–323. doi: 10.2174/138945007779940188. [DOI] [PubMed] [Google Scholar]
- 56.Linnevers C, Smeekens SP, Bromme D. Human cathepsin W, a putative cysteine protease predominantly expressed in CD8+ T-lymphocytes. FEBS letters. 1997;405:253–259. doi: 10.1016/s0014-5793(97)00118-x. [DOI] [PubMed] [Google Scholar]
- 57.Deal C. Potential new drug targets for osteoporosis. Nature clinical practice Rheumatology. 2009;5:20–27. doi: 10.1038/ncprheum0977. [DOI] [PubMed] [Google Scholar]
- 58.Stoch SA, Wagner JA. Cathepsin K inhibitors: a novel target for osteoporosis therapy. Clinical pharmacology and therapeutics. 2008;83:172–176. doi: 10.1038/sj.clpt.6100450. [DOI] [PubMed] [Google Scholar]
- 59.Funkelstein L, Toneff T, Mosier C, Hwang SR, Beuschlein F, Lichtenauer UD, Reinheckel T, Peters C, Hook V. Major role of cathepsin L for producing the peptide hormones ACTH, beta-endorphin, and alpha-MSH, illustrated by protease gene knockout and expression. The Journal of biological chemistry. 2008;283:35652–35659. doi: 10.1074/jbc.M709010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nature reviews Immunology. 2003;3:472–482. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- 61.Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, Okamoto K, Nishikawa K, Latz E, Golenbock DT, Aoki K, Ohya K, Imai Y, Morishita Y, Miyazono K, Kato S, Saftig P, Takayanagi H. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- 62.Dejica VM, Mort JS, Laverty S, Percival MD, Antoniou J, Zukor DJ, Poole AR. Cleavage of type II collagen by cathepsin K in human osteoarthritic cartilage. The American journal of pathology. 2008;173:161–169. doi: 10.2353/ajpath.2008.070494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schurigt U, Hummel KM, Petrow PK, Gajda M, Stockigt R, Middel P, Zwerina J, Janik T, Bernhardt R, Schuler S, Scharnweber D, Beckmann F, Saftig P, Kollias G, Schett G, Wiederanders B, Brauer R. Cathepsin K deficiency partially inhibits, but does not prevent, bone destruction in human tumor necrosis factor-transgenic mice. Arthritis and rheumatism. 2008;58:422–434. doi: 10.1002/art.23224. [DOI] [PubMed] [Google Scholar]
- 64.Svelander L, Erlandsson-Harris H, Astner L, Grabowska U, Klareskog L, Lindstrom E, Hewitt E. Inhibition of cathepsin K reduces bone erosion, cartilage degradation and inflammation evoked by collagen-induced arthritis in mice. European journal of pharmacology. 2009;613:155–162. doi: 10.1016/j.ejphar.2009.03.074. [DOI] [PubMed] [Google Scholar]
- 65.Fonovic M, Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbagen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 66.Joyce JA, Hanahan D. Multiple roles for cysteine cathepsins in cancer. Cell cycle. 2004;3:1516–1619. doi: 10.4161/cc.3.12.1289. [DOI] [PubMed] [Google Scholar]
- 67.Aggarwal N, Sloane BF. Cathepsin B: Multiple Roles in Cancer. Proteomics Clinical applications. 2014 doi: 10.1002/prca.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bengsch F, Buck A, Gunther SC, Seiz JR, Tacke M, Pfeifer D, von Elverfeldt D, Sevenich L, Hillebrand LE, Kern U, Sameni M, Peters C, Sloane BF, Reinheckel T. Cell type-dependent pathogenic functions of overexpressed human cathepsin B in murine breast cancer progression. Oncogene. 2013 doi: 10.1038/onc.2013.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cavallo-Medved D, Mai J, Dosescu J, Sameni M, Sloane BF. Caveolin-1 mediates the expression and localization of cathepsin B, pro-urokinase plasminogen activator and their cell-surface receptors in human colorectal carcinoma cells. Journal of cell science. 2005;118:1493–1503. doi: 10.1242/jcs.02278. [DOI] [PubMed] [Google Scholar]
- 70.Chan AT, Baba Y, Shima K, Nosho K, Chung DC, Hung KE, Mahmood U, Madden K, Poss K, Ranieri A, Shue D, Kucherlapati R, Fuchs CS, Ogino S. Cathepsin B expression and survival in colon cancer: implications for molecular detection of neoplasia. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:2777–2785. doi: 10.1158/1055-9965.EPI-10-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gong F, Peng X, Luo C, Shen G, Zhao C, Zou L, Li L, Sang Y, Zhao Y, Zhao X. Cathepsin B as a potential prognostic and therapeutic marker for human lung squamous cell carcinoma. Molecular cancer. 2013;12:125. doi: 10.1186/1476-4598-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gopinath S, Malla R, Alapati K, Gorantla B, Gujrati M, Dinh DH, Rao JS. Cathepsin B and uPAR regulate self-renewal of glioma-initiating cells through GLI-regulated Sox2 and Bmi1 expression. Carcinogenesis. 2013;34:550–559. doi: 10.1093/carcin/bgs375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nalla AK, Gorantla B, Gondi CS, Lakka SS, Rao JS. Targeting MMP-9, uPAR, and cathepsin B inhibits invasion, migration and activates apoptosis in prostate cancer cells. Cancer gene therapy. 2010;17:599–613. doi: 10.1038/cgt.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nouh MA, Mohamed MM, El-Shinawi M, Shaalan MA, Cavallo-Medved D, Khaled HM, Sloane BF. Cathepsin B: a potential prognostic marker for inflammatory breast cancer. Journal of translational medicine. 2011;9:1. doi: 10.1186/1479-5876-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rafn B, Nielsen CF, Andersen SH, Szyniarowski P, Corcelle-Termeau E, Valo E, Fehrenbacher N, Olsen CJ, Daugaard M, Egebjerg C, Bottzauw T, Kohonen P, Nylandsted J, Hautaniemi S, Moreira J, Jaattela M, Kallunki T. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Molecular cell. 2012;45:764–776. doi: 10.1016/j.molcel.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 76.Victor BC, Anbalagan A, Mohamed MM, Sloane BF, Cavallo-Medved D. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast cancer research : BCR. 2011;13:R115. doi: 10.1186/bcr3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K, Sloane BF, Anderson RL, Bogyo MS, Parker BS. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer research. 2012;72:1199–1209. doi: 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kostoulas G, Lang A, Nagase H, Baici A. Stimulation of angiogenesis through cathepsin B inactivation of the tissue inhibitors of matrix metalloproteinases. FEBS Lett. 1999;455:286–290. doi: 10.1016/s0014-5793(99)00897-2. [DOI] [PubMed] [Google Scholar]
- 79.Somanna A, Mundodi V, Gedamu L. Functional analysis of cathepsin B-like cysteine proteases from Leishmania donovani complex. Evidence for the activation of latent transforming growth factor beta. The Journal of biological chemistry. 2002;277:25305–25312. doi: 10.1074/jbc.M203034200. [DOI] [PubMed] [Google Scholar]
- 80.Tsai JY, Lee MJ, Chang MD, Wang HC, Lin CC, Huang H. Effects of novel human cathepsin S inhibitors on cell migration in human cancer cells. Journal of enzyme inhibition and medicinal chemistry. 2013 doi: 10.3109/14756366.2013.823957. [DOI] [PubMed] [Google Scholar]
- 81.Boonen S, Rosenberg E, Claessens F, Vanderschueren D, Papapoulos S. Inhibition of cathepsin K for treatment of osteoporosis. Current osteoporosis reports. 2012;10:73–79. doi: 10.1007/s11914-011-0085-9. [DOI] [PubMed] [Google Scholar]
- 82.Helali AM, Iti FM, Mohamed IN. Cathepsin K inhibitors: a novel target but promising approach in the treatment of osteoporosis. Current drug targets. 2013;14:1591–1600. doi: 10.2174/13894501113149990202. [DOI] [PubMed] [Google Scholar]
- 83.Matarrese P, Ascione B, Ciarlo L, Vona R, Leonetti C, Scarsella M, Mileo AM, Catricala C, Paggi MG, Malorni W. Cathepsin B inhibition interferes with metastatic potential of human melanoma: an in vitro and in vivo study. Molecular cancer. 2010;9:207. doi: 10.1186/1476-4598-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Runger TM, Adami S, Benhamou CL, Czerwinski E, Farrerons J, Kendler DL, Mindeholm L, Realdi G, Roux C, Smith V. Morphea-like skin reactions in patients treated with the cathepsin K inhibitor balicatib. Journal of the American Academy of Dermatology. 2012;66:e89–e96. doi: 10.1016/j.jaad.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 85.Passi A. The FEBS journal. 2014 doi: 10.1111/febs.13106. SAME MRS. [DOI] [PubMed] [Google Scholar]
- 86.Theocharis AD, Skandalis SS, Tzanakakis GN, Karamanos NK. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010;277:3904–3923. doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 87.Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavao MS, Tzanakakis GN, Karamanos NK. Glycosaminoglycans: key players in cancer cell biology and treatment. The FEBS journal. 2012;279:1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 88.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ida L. The FEBS journal. 2014 SAME MRS. [Google Scholar]
- 90.Iozzo RV, Murdoch AD. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996;10:598–614. [PubMed] [Google Scholar]
- 91.Theocharis AD. Versican in health and disease. Connective tissue research. 2008;49:230–234. doi: 10.1080/03008200802147571. [DOI] [PubMed] [Google Scholar]
- 92.Wight TN, Kinsella MG, Evanko SP, Potter-Perigo S, Merrilees MJ. Versican and the regulation of cell phenotype in disease. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbagen.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Labropoulou VT, Theocharis AD, Ravazoula P, Perimenis P, Hjerpe A, Karamanos NK, Kalofonos HP. Versican but not decorin accumulation is related to metastatic potential and neovascularization in testicular germ cell tumours. Histopathology. 2006;49:582–593. doi: 10.1111/j.1365-2559.2006.02558.x. [DOI] [PubMed] [Google Scholar]
- 94.Skandalis SS, Labropoulou VT, Ravazoula P, Likaki-Karatza E, Dobra K, Kalofonos HP, Karamanos NK, Theocharis AD. Versican but not decorin accumulation is related to malignancy in mammographically detected high density and malignant-appearing microcalcifications in non-palpable breast carcinomas. BMC cancer. 2011;11:314. doi: 10.1186/1471-2407-11-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skandalis SS, Theocharis AD, Papageorgakopoulou N, Vynios DH, Theocharis DA. The increased accumulation of structurally modified versican and decorin is related with the progression of laryngeal cancer. Biochimie. 2006;88:1135–1143. doi: 10.1016/j.biochi.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 96.Theocharis AD. Human colon adenocarcinoma is associated with specific post-translational modifications of versican and decorin. Biochimica et biophysica acta. 2002;1588:165–172. doi: 10.1016/s0925-4439(02)00161-8. [DOI] [PubMed] [Google Scholar]
- 97.Theocharis AD, Tsara ME, Papageorgacopoulou N, Karavias DD, Theocharis DA. Pancreatic carcinoma is characterized by elevated content of hyaluronan and chondroitin sulfate with altered disaccharide composition. Biochimica et biophysica acta. 2000;1502:201–206. doi: 10.1016/s0925-4439(00)00051-x. [DOI] [PubMed] [Google Scholar]
- 98.Theocharis AD, Vynios DH, Papageorgakopoulou N, Skandalis SS, Theocharis DA. Altered content composition and structure of glycosaminoglycans and proteoglycans in gastric carcinoma. The international journal of biochemistry & cell biology. 2003;35:376–390. doi: 10.1016/s1357-2725(02)00264-9. [DOI] [PubMed] [Google Scholar]
- 99.Tsara ME, Theocharis AD, Theocharis DA. Compositional and structural alterations of proteoglycans in human rectum carcinoma with special reference to versican and decorin. Anticancer research. 2002;22:2893–2898. [PubMed] [Google Scholar]
- 100.Yeung TL, Leung CS, Wong KK, Samimi G, Thompson MS, Liu J, Zaid TM, Ghosh S, Birrer MJ, Mok SC. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer research. 2013;73:5016–5028. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dondeti VR, Wubbenhorst B, Lal P, Gordan JD, D'Andrea K, Attiyeh EF, Simon MC, Nathanson KL. Integrative genomic analyses of sporadic clear cell renal cell carcinoma define disease subtypes and potential new therapeutic targets. Cancer research. 2012;72:112–121. doi: 10.1158/0008-5472.CAN-11-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Havre PA, Dang LH, Ohnuma K, Iwata S, Morimoto C, Dang NH. CD26 expression on T-anaplastic large cell lymphoma (ALCL) line Karpas 299 is associated with increased expression of versican and MT1-MMP and enhanced adhesion. BMC cancer. 2013;13:517. doi: 10.1186/1471-2407-13-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li D, Wang X, Wu JL, Quan WQ, Ma L, Yang F, Wu KY, Wan HY. Tumor-produced versican V1 enhances hCAP18/LL-37 expression in macrophages through activation of TLR2 and vitamin D3 signaling to promote ovarian cancer progression in vitro. PloS one. 2013;8:e56616. doi: 10.1371/journal.pone.0056616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Said N, Sanchez-Carbayo M, Smith SC, Theodorescu D. RhoGDI2 suppresses lung metastasis in mice by reducing tumor versican expression and macrophage infiltration. The Journal of clinical investigation. 2012;122:1503–1518. doi: 10.1172/JCI61392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xia L, Huang W, Tian D, Zhang L, Qi X, Chen Z, Shang X, Nie Y, Wu K. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59:958–973. doi: 10.1002/hep.26735. [DOI] [PubMed] [Google Scholar]
- 106.Arslan F, Bosserhoff AK, Nickl-Jockschat T, Doerfelt A, Bogdahn U, Hau P. The role of versican isoforms V0/V1 in glioma migration mediated by transforming growth factor-beta2. British journal of cancer. 2007;96:1560–1568. doi: 10.1038/sj.bjc.6603766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kischel P, Waltregny D, Dumont B, Turtoi A, Greffe Y, Kirsch S, De Pauw E, Castronovo V. Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. International journal of cancer Journal international du cancer. 2010;126:640–650. doi: 10.1002/ijc.24812. [DOI] [PubMed] [Google Scholar]
- 108.Tian Y, Esteva FJ, Song J, Zhang H. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Molecular & cellular proteomics : MCP. 2012;11 doi: 10.1074/mcp.M111.011403. M111 011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ween MP, Hummitzsch K, Rodgers RJ, Oehler MK, Ricciardelli C. Versican induces a pro-metastatic ovarian cancer cell behavior which can be inhibited by small hyaluronan oligosaccharides. Clinical & experimental metastasis. 2011;28:113–125. doi: 10.1007/s10585-010-9363-7. [DOI] [PubMed] [Google Scholar]
- 110.Nastase MV, Iozzo RV, Schaefer L. Key roles for the small leucine-rich proteoglycans in renal and pulmonary pathophysiology. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbagen.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Iozzo RV, Schaefer L. Proteoglycans in health and disease: novel regulatory signaling mechanisms evoked by the small leucine-rich proteoglycans. The FEBS journal. 2010;277:3864–3875. doi: 10.1111/j.1742-4658.2010.07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goldoni S, Humphries A, Nystrom A, Sattar S, Owens RT, McQuillan DJ, Ireton K, Iozzo RV. Decorin is a novel antagonistic ligand of the Met receptor. J Cell Biol. 2009;185:743–754. doi: 10.1083/jcb.200901129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schonherr E, Sunderkotter C, Iozzo RV, Schaefer L. Decorin, a novel player in the insulin-like growth factor system. J Biol Chem. 2005;280:15767–15772. doi: 10.1074/jbc.M500451200. [DOI] [PubMed] [Google Scholar]
- 114.Iozzo RV, Buraschi S, Genua M, Xu SQ, Solomides CC, Peiper SC, Gomella LG, Owens RC, Morrione A. Decorin antagonizes IGF receptor I (IGF-IR) function by interfering with IGF-IR activity and attenuating downstream signaling. J Biol Chem. 2011;286:34712–34721. doi: 10.1074/jbc.M111.262766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khan GA, Girish GV, Lala N, Di Guglielmo GM, Lala PK. Decorin is a novel VEGFR-2-binding antagonist for the human extravillous trophoblast. Mol Endocrinol. 2011;25:1431–1443. doi: 10.1210/me.2010-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schaefer L, Iozzo RV. Small leucine-rich proteoglycans, at the crossroad of cancer growth and inflammation. Curr Opin Genet Dev. 2012;22:56–57. doi: 10.1016/j.gde.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 117.Baghy K, Horvath Z, Regos E, Kiss K, Schaff Z, Iozzo RV, Kovalszky I. Decorin interferes with platelet-derived growth factor receptor signaling in experimental hepatocarcinogenesis. FEBS J. 2013;280:2150–2164. doi: 10.1111/febs.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hausser H, Hoppe W, Rauch U, Kresse H. Endocytosis of a small dermatan sulphate proteoglycan. Identification of binding proteins. Biochem J. 1989;263:137–142. doi: 10.1042/bj2630137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goldoni S, Seidler DG, Heath J, Fassan M, Baffa R, Thakur ML, Owens RT, McQuillan DJ, Iozzo RV. An antimetastatic role for decorin in breast cancer. Am J Pathol. 2008;173:844–855. doi: 10.2353/ajpath.2008.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neill T, Schaefer L, Iozzo RV. Decorin: a guardian from the matrix. Am J Pathol. 2012;181:380–387. doi: 10.1016/j.ajpath.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu JX, Goldoni S, Bix G, Owens RT, McQuillan DJ, Reed CC, Iozzo RV. Decorin evokes protracted internalization and degradation of the epidermal growth factor receptor via caveolar endocytosis. The Journal of biological chemistry. 2005;280:32468–32479. doi: 10.1074/jbc.M503833200. [DOI] [PubMed] [Google Scholar]
- 122.Csordas G, Santra M, Reed CC, Eichstetter I, McQuillan DJ, Gross D, Nugent MA, Hajnoczky G, Iozzo RV. Sustained down-regulation of the epidermal growth factor receptor by decorin. A mechanism for controlling tumor growth in vivo. The Journal of biological chemistry. 2000;275:32879–32887. doi: 10.1074/jbc.M005609200. [DOI] [PubMed] [Google Scholar]
- 123.Seidler DG, Goldoni S, Agnew C, Cardi C, Thakur ML, Owens RT, McQuillan DJ, Iozzo RV. Decorin protein core inhibits in vivo cancer growth and metabolism by hindering epidermal growth factor receptor function and triggering apoptosis via caspase-3 activation. The Journal of biological chemistry. 2006;281:26408–26418. doi: 10.1074/jbc.M602853200. [DOI] [PubMed] [Google Scholar]
- 124.Neill T, Painter H, Buraschi S, Owens RT, Lisanti MP, Schaefer L, Iozzo RV. Decorin antagonizes the angiogenic network: concurrent inhibition of Met, hypoxia inducible factor 1alpha, vascular endothelial growth factor A, and induction of thrombospondin-1 and TIMP3. J Biol Chem. 2012;287:5492–5506. doi: 10.1074/jbc.M111.283499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grant DS, Yenisey C, Rose RW, Tootell M, Santra M, Iozzo RV. Decorin suppresses tumor cell-mediated angiogenesis. Oncogene. 2002;21:4765–4777. doi: 10.1038/sj.onc.1205595. [DOI] [PubMed] [Google Scholar]
- 126.Neill T, Jones HR, Crane-Smith Z, Owens RT, Schaefer L, Iozzo RV. Decorin induces rapid secretion of thrombospondin-1 in basal breast carcinoma cells via inhibition of Ras homolog gene family, member A/Rho-associated coiled-coil containing protein kinase 1. FEBS J. 2013;280:2353–2368. doi: 10.1111/febs.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murphy-Ullrich JE, Iozzo RV. Thrombospondins in physiology and disease: new tricks for old dogs. Matrix Biol. 2012;31:152–154. doi: 10.1016/j.matbio.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sweetwyne MT, Pallero MA, Lu A, Van Duyn Graham L, Murphy-Ullrich JE. The calreticulin-binding sequence of thrombospondin 1 regulates collagen expression and organization during tissue remodeling. Am J Pathol. 2010;177:1710–1724. doi: 10.2353/ajpath.2010.090903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bi X, Tong C, Dockendorff A, Bancroft L, Gallagher L, Guzman G, Iozzo RV, Augenlicht LH, Yang W. Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis. 2008;29:1435–1440. doi: 10.1093/carcin/bgn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bi X, Pohl NM, Qian Z, Yang GR, Gou Y, Guzman G, Kajdacsy-Balla A, Iozzo RV, Yang W. Decorin-mediated inhibition of colorectal cancer growth and migration is associated with E-cadherin in vitro and in mice. Carcinogenesis. 2012;33:326–330. doi: 10.1093/carcin/bgr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reed CC, Gauldie J, Iozzo RV. Suppression of tumorigenicity by adenovirus-mediated gene transfer of decorin. Oncogene. 2002;21:3688–3695. doi: 10.1038/sj.onc.1205470. [DOI] [PubMed] [Google Scholar]
- 132.Tralhao JG, Schaefer L, Micegova M, Evaristo C, Schonherr E, Kayal S, Veiga-Fernandes H, Danel C, Iozzo RV, Kresse H, Lemarchand P. In vivo selective and distant killing of cancer cells using adenovirus-mediated decorin gene transfer. FASEB J. 2003;17:464–466. doi: 10.1096/fj.02-0534fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reed CC, Waterhouse A, Kirby S, Kay P, Owens RT, McQuillan DJ, Iozzo RV. Decorin prevents metastatic spreading of breast cancer. Oncogene. 2005;24:1104–1110. doi: 10.1038/sj.onc.1208329. [DOI] [PubMed] [Google Scholar]
- 134.Hu Y, Sun H, Owens RT, Wu J, Chen YQ, Berquin IM, Perry D, O'Flaherty JT, Edwards IJ. Decorin suppresses prostate tumor growth through inhibition of epidermal growth factor and androgen receptor pathways. Neoplasia. 2009;11:1042–1053. doi: 10.1593/neo.09760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Buraschi S, Neill T, Owens RT, Iniguez LA, Purkins G, Vadigepalli R, Evans B, Schaefer L, Peiper SC, Wang ZX, Iozzo RV. Decorin protein core affects the global gene expression profile of the tumor microenvironment in a triple-negative orthotopic breast carcinoma xenograft model. PLoS One. 2012;7:e45559. doi: 10.1371/journal.pone.0045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, Lemarchand P, Pfeilschifter J, Schaefer RM, Iozzo RV, Schaefer L. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci Signal. 2011;4:ra75. doi: 10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, Schaefer L, Torres A, Iozzo RV. Decorin causes autophagy in endothelial cells via Peg3. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2582–E2591. doi: 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Neill T, Torres A, Buraschi S, Owens RT, Hoek JB, Baffa R, Iozzo RV. Decorin induces mitophagy in breast carcinoma cells via peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha) and mitostatin. J Biol Chem. 2014;289:4952–4968. doi: 10.1074/jbc.M113.512566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barbouri D, Afratis N, Gialeli C, Vynios DH, Theocharis AD, Karamanos NK. Syndecans as Modulators and Potential Pharmacological Targets in Cancer Progression. Frontiers in oncology. 2014;4:4. doi: 10.3389/fonc.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Choi Y, Chung H, Jung H, Couchman JR, Oh ES. Syndecans as cell surface receptors: Unique structure equates with functional diversity. Matrix Biol. 2011;30:93–99. doi: 10.1016/j.matbio.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 141.Forsberg K. The FEBS journal. 2014 SAME MRS. [Google Scholar]
- 142.Beauvais DM, Ell BJ, McWhorter AR, Rapraeger AC. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. The Journal of experimental medicine. 2009;206:691–705. doi: 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rapraeger AC, Ell BJ, Roy M, Li X, Morrison OR, Thomas GM, Beauvais DM. Vascular endothelial-cadherin stimulates syndecan-1-coupled insulin-like growth factor-1 receptor and cross-talk between alphaVbeta3 integrin and vascular endothelial growth factor receptor 2 at the onset of endothelial cell dissemination during angiogenesis. The FEBS journal. 2013;280:2194–2206. doi: 10.1111/febs.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vuoriluoto K, Jokinen J, Kallio K, Salmivirta M, Heino J, Ivaska J. Syndecan-1 supports integrin alpha2beta1-mediated adhesion to collagen. Experimental cell research. 2008;314:3369–3381. doi: 10.1016/j.yexcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 145.Vuoriluoto K, Hognas G, Meller P, Lehti K, Ivaska J. Syndecan-1 and-4 differentially regulate oncogenic K-ras dependent cell invasion into collagen through alpha2beta1 integrin and MT1-MMP. Matrix biology : journal of the International Society for Matrix Biology. 2011;30:207–217. doi: 10.1016/j.matbio.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 146.Ishikawa T, Kramer RH. Sdc1 negatively modulates carcinoma cell motility and invasion. Experimental cell research. 2010;316:951–965. doi: 10.1016/j.yexcr.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Manon-Jensen T, Multhaupt HA, Couchman JR. Mapping of matrix metalloproteinase cleavage sites on syndecan-1 and syndecan-4 ectodomains. The FEBS journal. 2013;280:2320–2331. doi: 10.1111/febs.12174. [DOI] [PubMed] [Google Scholar]
- 148.Ikeda H, Hideshima T, Fulciniti M, Lutz RJ, Yasui H, Okawa Y, Kiziltepe T, Vallet S, Pozzi S, Santo L, Perrone G, Tai YT, Cirstea D, Raje NS, Uherek C, Dalken B, Aigner S, Osterroth F, Munshi N, Richardson P, Anderson KC. The monoclonal antibody nBT062 conjugated to cytotoxic Maytansinoids has selective cytotoxicity against CD138-positive multiple myeloma cells in vitro and in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:4028–4037. doi: 10.1158/1078-0432.CCR-08-2867. [DOI] [PubMed] [Google Scholar]
- 149.Orecchia P, Conte R, Balza E, Petretto A, Mauri P, Mingari MC, Carnemolla B. A novel human anti-syndecan-1 antibody inhibits vascular maturation and tumour growth in melanoma. European journal of cancer. 2013;49:2022–2033. doi: 10.1016/j.ejca.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 150.Rousseau C, Ferrer L, Supiot S, Bardies M, Davodeau F, Faivre-Chauvet A, Baumgartner P, Wijdenes J, Lacombe M, Barbet J, Guillaume T, Moreau P, Harousseau JL, Kraeber-Bodere F, Cherel M. Dosimetry results suggest feasibility of radioimmunotherapy using anti-CD138 (B-B4) antibody in multiple myeloma patients. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:679–688. doi: 10.1007/s13277-012-0362-y. [DOI] [PubMed] [Google Scholar]
- 151.Rapraeger AC. Synstatin: a selective inhibitor of the syndecan-1-coupled IGF1R-alphavbeta3 integrin complex in tumorigenesis and angiogenesis. The FEBS journal. 2013;280:2207–2215. doi: 10.1111/febs.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cassinelli G, Lanzi C, Tortoreto M, Cominetti D, Petrangolini G, Favini E, Zaffaroni N, Pisano C, Penco S, Vlodavsky I, Zunino F. Antitumor efficacy of the heparanase inhibitor SST0001 alone and in combination with antiangiogenic agents in the treatment of human pediatric sarcoma models. Biochemical pharmacology. 2013;85:1424–1432. doi: 10.1016/j.bcp.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 153.Pisano C, Vlodavsky I, Ilan N, Zunino F. The potential of heparanase as a therapeutic target in cancer. Biochemical pharmacology. 2014;89:12–19. doi: 10.1016/j.bcp.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ramani VC, Purushothaman A, Stewart MD, Thompson CA, Vlodavsky I, Au JL, Sanderson RD. The heparanase/syndecan-1 axis in cancer: mechanisms and therapies. The FEBS journal. 2013;280:2294–2306. doi: 10.1111/febs.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gialeli C, Theocharis AD, Kletsas D, Tzanakakis GN, Karamanos NK. Expression of matrix macromolecules and functional properties of EGF-responsive colon cancer cells are inhibited by panitumumab. Investigational new drugs. 2013;31:516–524. doi: 10.1007/s10637-012-9875-x. [DOI] [PubMed] [Google Scholar]
- 156.Malavaki CJ, Roussidis AE, Gialeli C, Kletsas D, Tsegenidis T, Theocharis AD, Tzanakakis GN, Karamanos NK. Imatinib as a key inhibitor of the platelet-derived growth factor receptor mediated expression of cell surface heparan sulfate proteoglycans and functional properties of breast cancer cells. The FEBS journal. 2013;280:2477–2489. doi: 10.1111/febs.12163. [DOI] [PubMed] [Google Scholar]
- 157.Gotte M. The FEBS journal. 2014 SAME MRS. [Google Scholar]
- 158.Garusi E, Rossi S, Perris R. Antithetic roles of proteoglycans in cancer. Cellular and molecular life sciences : CMLS. 2012;69:553–579. doi: 10.1007/s00018-011-0816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang J, Price MA, Neudauer CL, Wilson C, Ferrone S, Xia H, Iida J, Simpson MA, McCarthy JB. Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. The Journal of cell biology. 2004;165:881–891. doi: 10.1083/jcb.200403174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, Overall CM, McCarthy JB. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A) The Biochemical journal. 2007;403:553–563. doi: 10.1042/BJ20061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Iida J, Pei D, Kang T, Simpson MA, Herlyn M, Furcht LT, McCarthy JB. Melanoma chondroitin sulfate proteoglycan regulates matrix metalloproteinase-dependent human melanoma invasion into type I collagen. The Journal of biological chemistry. 2001;276:18786–18794. doi: 10.1074/jbc.M010053200. [DOI] [PubMed] [Google Scholar]
- 162.Cattaruzza S, Nicolosi PA, Braghetta P, Pazzaglia L, Benassi MS, Picci P, Lacrima K, Zanocco D, Rizzo E, Stallcup WB, Colombatti A, Perris R. NG2/CSPG4-collagen type VI interplays putatively involved in the microenvironmental control of tumour engraftment and local expansion. Journal of molecular cell biology. 2013;5:176–193. doi: 10.1093/jmcb/mjt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang X, Katayama A, Wang Y, Yu L, Favoino E, Sakakura K, Favole A, Tsuchikawa T, Silver S, Watkins SC, Kageshita T, Ferrone S. Functional characterization of an scFv-Fc antibody that immunotherapeutically targets the common cancer cell surface proteoglycan CSPG4. Cancer research. 2011;71:7410–7422. doi: 10.1158/0008-5472.CAN-10-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Poli A, Wang J, Domingues O, Planaguma J, Yan T, Rygh CB, Skaftnesmo KO, Thorsen F, McCormack E, Hentges F, Pedersen PH, Zimmer J, Enger PO, Chekenya M. Targeting glioblastoma with NK cells and mAb against NG2/CSPG4 prolongs animal survival. Oncotarget. 2013;4:1527–1546. doi: 10.18632/oncotarget.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rivera Z, Ferrone S, Wang X, Jube S, Yang H, Pass HI, Kanodia S, Gaudino G, Carbone M. CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5352–5363. doi: 10.1158/1078-0432.CCR-12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, McCarthy JB, Brufsky A, Chivukula M, Khoury T, Hsu DS, Barry WT, Lyerly HK, Clay TM, Ferrone S. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. Journal of the National Cancer Institute. 2010;102:1496–1512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Geldres C, Savoldo B, Hoyos V, Caruana I, Zhang M, Yvon E, Del Vecchio M, Creighton CJ, Ittmann M, Ferrone S, Dotti G. T lymphocytes redirected against the chondroitin sulfate proteoglycan-4 control the growth of multiple solid tumors both in vitro and in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:962–971. doi: 10.1158/1078-0432.CCR-13-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Korpetinou A, Skandalis SS, Labropoulou VT, Smirlaki G, Noulas A, Karamanos NK, Theocharis AD. Serglycin: At the Crossroad of Inflammation and Malignancy. Frontiers in oncology. 2014;3:327. doi: 10.3389/fonc.2013.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.He L, Zhou X, Qu C, Tang Y, Zhang Q, Hong J. Serglycin (SRGN) overexpression predicts poor prognosis in hepatocellular carcinoma patients. Medical oncology. 2013;30:707. doi: 10.1007/s12032-013-0707-4. [DOI] [PubMed] [Google Scholar]
- 170.Korpetinou A, Skandalis SS, Moustakas A, Happonen KE, Tveit H, Prydz K, Labropoulou VT, Giannopoulou E, Kalofonos HP, Blom AM, Karamanos NK, Theocharis AD. Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PloS one. 2013;8:e78157. doi: 10.1371/journal.pone.0078157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li XJ, Ong CK, Cao Y, Xiang YQ, Shao JY, Ooi A, Peng LX, Lu WH, Zhang Z, Petillo D, Qin L, Bao YN, Zheng FJ, Chia CS, Iyer NG, Kang TB, Zeng YX, Soo KC, Trent JM, Teh BT, Qian CN. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer research. 2011;71:3162–3172. doi: 10.1158/0008-5472.CAN-10-3557. [DOI] [PubMed] [Google Scholar]
- 172.Purushothaman A, Toole BP. Serglycin proteoglycan is required for multiple myeloma cell adhesion, in vivo growth, and vascularization. The Journal of biological chemistry. 2014;289:5499–5509. doi: 10.1074/jbc.M113.532143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Skliris A, Labropoulou VT, Papachristou DJ, Aletras A, Karamanos NK, Theocharis AD. Cell-surface serglycin promotes adhesion of myeloma cells to collagen type I and affects the expression of matrix metalloproteinases. The FEBS journal. 2013;280:2342–2352. doi: 10.1111/febs.12179. [DOI] [PubMed] [Google Scholar]
- 174.Theocharis AD, Seidel C, Borset M, Dobra K, Baykov V, Labropoulou V, Kanakis I, Dalas E, Karamanos NK, Sundan A, Hjerpe A. Serglycin constitutively secreted by myeloma plasma cells is a potent inhibitor of bone mineralization in vitro. The Journal of biological chemistry. 2006;281:35116–35128. doi: 10.1074/jbc.M601061200. [DOI] [PubMed] [Google Scholar]
- 175.Korpetinou A, Milia-Argeiti E, Labropoulou V, Theocharis A. 7.6 Serglycin: a novel player in the terrain of neoplasia in. In: Karamanos N, editor. Extracellular Matrix: Pathobiology and Signaling. DE GRUYTER; 2012. pp. 677–688. [Google Scholar]
- 176.Abrink M, Grujic M, Pejler G. Serglycin is essential for maturation of mast cell secretory granule. The Journal of biological chemistry. 2004;279:40897–40905. doi: 10.1074/jbc.M405856200. [DOI] [PubMed] [Google Scholar]
- 177.Braga T, Grujic M, Lukinius A, Hellman L, Abrink M, Pejler G. Serglycin proteoglycan is required for secretory granule integrity in mucosal mast cells. The Biochemical journal. 2007;403:49–57. doi: 10.1042/BJ20061257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Skliris A, Happonen KE, Terpos E, Labropoulou V, Borset M, Heinegard D, Blom AM, Theocharis AD. Serglycin inhibits the classical and lectin pathways of complement via its glycosaminoglycan chains: implications for multiple myeloma. European journal of immunology. 2011;41:437–449. doi: 10.1002/eji.201040429. [DOI] [PubMed] [Google Scholar]
- 179.Zernichow L, Dalen KT, Prydz K, Winberg JO, Kolset SO. Secretion of proteases in serglycin transfected Madin-Darby canine kidney cells. The FEBS journal. 2006;273:536–547. doi: 10.1111/j.1742-4658.2005.05085.x. [DOI] [PubMed] [Google Scholar]
- 180.Theocharis AD, Tsolakis I, Tzanakakis GN, Karamanos NK. Chondroitin sulfate as a key molecule in the development of atherosclerosis and cancer progression. Advances in pharmacology. 2006;53:281–295. doi: 10.1016/S1054-3589(05)53013-8. [DOI] [PubMed] [Google Scholar]
- 181.Cooney CA, Jousheghany F, Yao-Borengasser A, Phanavanh B, Gomes T, Kieber-Emmons AM, Siegel ER, Suva LJ, Ferrone S, Kieber-Emmons T, Monzavi-Karbassi B. Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast cancer research : BCR. 2011;13:R58. doi: 10.1186/bcr2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hadler-Olsen E, Fadnes B, Sylte I, Uhlin-Hansen L, Winberg JO. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011;278:28–45. doi: 10.1111/j.1742-4658.2010.07920.x. [DOI] [PubMed] [Google Scholar]
- 183.Murphy G, Nagase H. Localizing matrix metalloproteinase activities in the pericellular environment. FEBS J. 2011;278:2–15. doi: 10.1111/j.1742-4658.2010.07918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu WH, Woessner JF., Jr Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7) J Biol Chem. 2000;275:4183–4191. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- 185.Tocchi A, Parks WC. Functional interactions between matrix metalloproteinases and glycosaminoglycans. FEBS J. 2013;280:2332–2341. doi: 10.1111/febs.12198. [DOI] [PubMed] [Google Scholar]
- 186.Ra HJ, Harju-Baker S, Zhang F, Linhardt RJ, Wilson CL, Parks WC. Control of promatrilysin (MMP7) activation and substrate-specific activity by sulfated glycosaminoglycans. J Biol Chem. 2009;284:27924–27932. doi: 10.1074/jbc.M109.035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ryu HY, Lee J, Yang S, Park H, Choi S, Jung KC, Lee ST, Seong JK, Han IO, Oh ES. Syndecan-2 functions as a docking receptor for pro-matrix metalloproteinase-7 in human colon cancer cells. J Biol Chem. 2009;284:35692–35701. doi: 10.1074/jbc.M109.054254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Koyama Y, Naruo H, Yoshitomi Y, Munesue S, Kiyono S, Kusano Y, Hashimoto K, Yokoi T, Nakanishi H, Shimizu S, Okayama M, Oguri K. Matrix metalloproteinase-9 associated with heparan sulphate chains of GPI-anchored cell surface proteoglycans mediates motility of murine colon adenocarcinoma cells. J Biochem. 2008;143:581–592. doi: 10.1093/jb/mvn006. [DOI] [PubMed] [Google Scholar]
- 189.Gao G, Plaas A, Thompson VP, Jin S, Zuo F, Sandy JD. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J Biol Chem. 2004;279:10042–10051. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- 190.Echtermeyer F, Bertrand J, Dreier R, Meinecke I, Neugebauer K, Fuerst M, Lee YJ, Song YW, Herzog C, Theilmeier G, Pap T. Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nature medicine. 2009;15:1072–1076. doi: 10.1038/nm.1998. [DOI] [PubMed] [Google Scholar]
- 191.Munesue S, Yoshitomi Y, Kusano Y, Koyama Y, Nishiyama A, Nakanishi H, Miyazaki K, Ishimaru T, Miyaura S, Okayama M, Oguri K. A novel function of syndecan-2, suppression of matrix metalloproteinase-2 activation, which causes suppression of metastasis. J Biol Chem. 2007;282:28164–28174. doi: 10.1074/jbc.M609812200. [DOI] [PubMed] [Google Scholar]
- 192.Sorensen HP, Vives RR, Manetopoulos C, Albrechtsen R, Lydolph MC, Jacobsen J, Couchman JR, Wewer UM. Heparan sulfate regulates ADAM12 through a molecular switch mechanism. J Biol Chem. 2008;283:31920–31932. doi: 10.1074/jbc.M804113200. [DOI] [PubMed] [Google Scholar]
- 193.Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci U S A. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Malla N, Berg E, Theocharis AD, Svineng G, Uhlin-Hansen L, Winberg JO. In vitro reconstitution of complexes between pro-matrix metalloproteinase-9 and the proteoglycans serglycin and versican. The FEBS journal. 2013;280:2870–2887. doi: 10.1111/febs.12291. [DOI] [PubMed] [Google Scholar]
- 195.Winberg JO, Kolset SO, Berg E, Uhlin-Hansen L. Macrophages secrete matrix metalloproteinase 9 covalently linked to the core protein of chondroitin sulphate proteoglycans. J Mol Biol. 2000;304:669–680. doi: 10.1006/jmbi.2000.4235. [DOI] [PubMed] [Google Scholar]
- 196.Winberg JO, Berg E, Kolset SO, Uhlin-Hansen L. Calcium-induced activation and truncation of promatrix metalloproteinase-9 linked to the core protein of chondroitin sulfate proteoglycans. European journal of biochemistry / FEBS. 2003;270:3996–4007. doi: 10.1046/j.1432-1033.2003.03788.x. [DOI] [PubMed] [Google Scholar]
- 197.Labropoulou VT, Theocharis AD, Symeonidis A, Skandalis SS, Karamanos NK, Kalofonos HP. Pathophysiology and pharmacological targeting of tumor-induced bone disease: current status and emerging therapeutic interventions. Current medicinal chemistry. 2011;18:1584–1598. doi: 10.2174/092986711795471275. [DOI] [PubMed] [Google Scholar]
- 198.Zhang L, Yang M, Yang D, Cavey G, Davidson P, Gibson G. Molecular interactions of MMP-13 C-terminal domain with chondrocyte proteins. Connect Tissue Res. 2010;51:230–239. doi: 10.3109/03008200903288902. [DOI] [PubMed] [Google Scholar]
- 199.Pejler G, Sadler JE. Mechanism by which heparin proteoglycan modulates mast cell chymase activity. Biochemistry. 1999;38:12187–12195. doi: 10.1021/bi991046b. [DOI] [PubMed] [Google Scholar]
- 200.Lindstedt L, Lee M, Kovanen PT. Chymase bound to heparin is resistant to its natural inhibitors and capable of proteolyzing high density lipoproteins in aortic intimal fluid. Atherosclerosis. 2001;155:87–97. doi: 10.1016/s0021-9150(00)00544-x. [DOI] [PubMed] [Google Scholar]
- 201.Pejler G, Berg L. Regulation of rat mast cell protease 1 activity. Protease inhibition is prevented by heparin proteoglycan. European journal of biochemistry / FEBS. 1995;233:192–199. doi: 10.1111/j.1432-1033.1995.192_1.x. [DOI] [PubMed] [Google Scholar]
- 202.Hallgren J, Spillmann D, Pejler G. Structural requirements and mechanism for heparin-induced activation of a recombinant mouse mast cell tryptase, mouse mast cell protease-6: formation of active tryptase monomers in the presence of low molecular weight heparin. The Journal of biological chemistry. 2001;276:42774–42781. doi: 10.1074/jbc.M105531200. [DOI] [PubMed] [Google Scholar]
- 203.Sakai K, Ren S, Schwartz LB. A novel heparin-dependent processing pathway for human tryptase. Autocatalysis followed by activation with dipeptidyl peptidase I. The Journal of clinical investigation. 1996;97:988–995. doi: 10.1172/JCI118523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li Z, Hou WS, Bromme D. Collagenolytic activity of cathepsin K is specifically modulated by cartilage-resident chondroitin sulfates. Biochemistry. 2000;39:529–536. doi: 10.1021/bi992251u. [DOI] [PubMed] [Google Scholar]
- 205.Li Z, Hou WS, Escalante-Torres CR, Gelb BD, Bromme D. Collagenase activity of cathepsin K depends on complex formation with chondroitin sulfate. The Journal of biological chemistry. 2002;277:28669–28676. doi: 10.1074/jbc.M204004200. [DOI] [PubMed] [Google Scholar]
- 206.Li Z, Yasuda Y, Li W, Bogyo M, Katz N, Gordon RE, Fields GB, Bromme D. Regulation of collagenase activities of human cathepsins by glycosaminoglycans. The Journal of biological chemistry. 2004;279:5470–5479. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- 207.Cherney MM, Lecaille F, Kienitz M, Nallaseth FS, Li Z, James MN, Bromme D. Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions. The Journal of biological chemistry. 2011;286:8988–8998. doi: 10.1074/jbc.M110.126706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nallaseth FS, Lecaille F, Li Z, Bromme D. The role of basic amino acid surface clusters on the collagenase activity of cathepsin K. Biochemistry. 2013;52:7742–7752. doi: 10.1021/bi401051j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sage J, Mallevre F, Barbarin-Costes F, Samsonov SA, Gehrcke JP, Pisabarro MT, Perrier E, Schnebert S, Roget A, Livache T, Nizard C, Lalmanach G, Lecaille F. Binding of chondroitin 4-sulfate to cathepsin S regulates its enzymatic activity. Biochemistry. 2013;52:6487–6498. doi: 10.1021/bi400925g. [DOI] [PubMed] [Google Scholar]
- 210.Almeida PC, Nantes IL, Chagas JR, Rizzi CC, Faljoni-Alario A, Carmona E, Juliano L, Nader HB, Tersariol IL. Cathepsin B activity regulation. Heparin-like glycosaminogylcans protect human cathepsin B from alkaline pH-induced inactivation. The Journal of biological chemistry. 2001;276:944–951. doi: 10.1074/jbc.M003820200. [DOI] [PubMed] [Google Scholar]
- 211.Caglic D, Pungercar JR, Pejler G, Turk V, Turk B. Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. The Journal of biological chemistry. 2007;282:33076–33085. doi: 10.1074/jbc.M705761200. [DOI] [PubMed] [Google Scholar]
- 212.Nascimento FD, Rizzi CC, Nantes IL, Stefe I, Turk B, Carmona AK, Nader HB, Juliano L, Tersariol IL. Cathepsin X binds to cell surface heparan sulfate proteoglycans. Archives of biochemistry and biophysics. 2005;436:323–332. doi: 10.1016/j.abb.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 213.Yasuda Y, Li Z, Greenbaum D, Bogyo M, Weber E, Bromme D. Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages. The Journal of biological chemistry. 2004;279:36761–36770. doi: 10.1074/jbc.M403986200. [DOI] [PubMed] [Google Scholar]
- 214.Beckman M, Freeman C, Parish CR, Small DH. Activation of cathepsin D by glycosaminoglycans. The FEBS journal. 2009;276:7343–7352. doi: 10.1111/j.1742-4658.2009.07444.x. [DOI] [PubMed] [Google Scholar]
- 215.Higgins WJ, Fox DM, Kowalski PS, Nielsen JE, Worrall DM. Heparin enhances serpin inhibition of the cysteine protease cathepsin L. The Journal of biological chemistry. 2010;285:3722–3729. doi: 10.1074/jbc.M109.037358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Casu B, Naggi A, Torri G. Heparin-derived heparan sulfate mimics to modulate heparan sulfate-protein interaction in inflammation and cancer. Matrix Biol. 2010;29:442–452. doi: 10.1016/j.matbio.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Selent J, Kaleta J, Li Z, Lalmanach G, Bromme D. Selective inhibition of the collagenase activity of cathepsin K. The Journal of biological chemistry. 2007;282:16492–16501. doi: 10.1074/jbc.M700242200. [DOI] [PubMed] [Google Scholar]
- 218.Alter SC, Metcalfe DD, Bradford TR, Schwartz LB. Regulation of human mast cell tryptase. Effects of enzyme concentration, ionic strength and the structure and negative charge density of polysaccharides. Biochem J. 1987;248:821–827. doi: 10.1042/bj2480821. [DOI] [PMC free article] [PubMed] [Google Scholar]