Summary
Adhesion G-protein-coupled receptors (aGPCRs) play critical roles in diverse neurobiological processes including brain development, synaptogenesis, and myelination. aGPCRs have large alternatively spliced extracellular regions (ECRs) that likely mediate intercellular signaling; however, the precise roles of ECRs remain unclear. The aGPCR GPR56/ADGRG1 regulates both oligodendrocyte and cortical development. Accordingly, human GPR56 mutations cause myelination defects and brain malformations. Here, we determined the crystal structure of the GPR56 ECR, the first structure of any complete aGPCR ECR, in complex with an inverse-agonist monobody, revealing a GPCR-Autoproteolysis-Inducing domain and a previously unidentified domain that we term Pentraxin/Laminin/neurexin/sex-hormone-binding-globulin-Like (PLL). Strikingly, PLL domain deletion caused increased signaling and characterizes a GPR56 splice variant. Finally, we show that an evolutionarily conserved residue in the PLL domain is critical for oligodendrocyte development in vivo. Thus, our results suggest that the GPR56 ECR has unique and multifaceted regulatory functions, providing novel insights into aGPCR roles in neurobiology.
Keywords: oligodendrocyte development, adhesion GPCR, monobody, X-ray crystallography, protein engineering
Introduction
Brain development requires precise coordination of numerous key processes that are individually complex. For instance, to ensure rapid action potential propagation in the vertebrate nervous system, many axons must be insulated by myelin, a multilamellar lipid-rich membrane (Nave and Trapp, 2008). In the central nervous system (CNS), oligodendrocytes govern myelination by extending and iteratively wrapping their plasma membranes around axon segments (Snaidero et al., 2014). Loss of myelin leads to severe neurological disorders such as multiple sclerosis (Noseworthy et al., 2000, Olmos-Serrano et al., 2016). The interplay between the control of myelination and cortical development is poorly understood
Genetic studies have revealed that GPR56/ADGRG1, a cell-surface G protein-coupled receptor (GPCR), plays important roles in both oligodendrocyte and cortex development, potentially providing a molecular link between these processes. (Piao et al., 2004, Li et al., 2008, Koirala et al., 2009, Bae et al., 2014, Ackerman et al., 2015, Giera et al., 2015). Mutations in GPR56 cause a human brain malformation called bilateral frontoparietal polymicrogyria (BFPP) that is characterized by disorganized cortex lamination and patterning, especially in the frontal cortex, the region responsible for many human-specific functions (Piao et al., 2004). In addition to cortex malformation, the brains of BFPP patients exhibit myelination abnormalities, such as a reduced white matter volume, indicative of myelinated axon defects (Piao et al., 2004, Bahi-Buisson et al., 2010). Furthermore, recent studies have revealed that GPR56 has a critical role in the regulation of oligodendrocyte development in both zebrafish (Ackerman et al., 2015) and mouse (Giera et al., 2015). Altogether, these studies have established GPR56 as a key molecule with multiple functions in CNS development.
GPR56 belongs to the adhesion G protein-coupled receptor (aGPCR) family, a large family of chimeric proteins that have both adhesion and signaling functions (Langenhan et al., 2013, Hamann et al., 2015). aGPCRs are cell-surface molecules that are believed to mediate intercellular communication via cell-cell and cell-matrix interactions. Many aGPCRs have critical roles in nervous system function including peripheral nervous system myelination by Schwann cells (Monk et al., 2009, Mogha et al., 2013), CNS angiogenesis (Nishimori et al., 1997, Kuhnert et al., 2010), and excitatory synapse formation (O'Sullivan et al., 2012). As in the canonical GPCR families, aGPCRs have a seven-pass transmembrane helix bundle (7TM) that, for many aGPCRs, can be activated to initiate a signaling cascade via interactions with cytosolic G proteins. Unlike the canonical GPCR families, aGPCRs also have large and diverse extracellular regions (ECRs), mainly composed of domains generally involved in adhesion-related functions (Langenhan et al., 2013). Although this architecture is suggestive of functional importance of the ECRs, their biological roles are incompletely understood.
aGPCRs are characterized by the presence of an extracellular GPCR-Autoproteolysis-INducing (GAIN) domain located immediately N-terminal to the 7TM (Arac et al., 2012). During aGPCR maturation, autoproteolysis occurs within the GAIN domain (Lin et al., 2004), cleaving the receptor into two fragments: (1) an N-terminal fragment (NTF) comprising various extracellular adhesion domains and the majority of the GAIN domain; (2) a membrane-bound C-terminal fragment (CTF) comprising the C-terminal β-strand of the GAIN domain, termed the ‘Stachel peptide’ (Liebscher et al., 2014), (also called ‘tethered agonist’ (Stoveken et al., 2015), or ‘stalk’ (Kishore et al., 2015)), the 7TM, and the intracellular region (Figure 1A). After autoproteolysis, the NTF and CTF remain associated to form the mature, plasma membrane-localized receptor (Paavola et al., 2011, Arac et al., 2012). To date, two non-mutually exclusive models have been proposed for ECR-regulated aGPCR activation. According to the ‘shedding’ model, ligand binding to the adhesion domains in the ECR may induce dissociation of the NTF from the membrane-anchored CTF, termed ‘shedding’. After shedding, the Stachel peptide on the CTF is freed from the GAIN domain and functions as a tethered agonist to activate the 7TM (Liebscher et al., 2014, White et al., 2014, Stoveken et al., 2015, Demberg et al., 2015, Hamann et al., 2015, Scholz et al., 2015, Petersen et al., 2015). An alternative model suggests that aGPCR extracellular domains govern receptor activity by directly interacting with the 7TM, perhaps in a ligand-dependent fashion (Paavola et al., 2011, Kishore et al., 2015). However, without detailed characterization of aGPCR ECRs, the validities of these models have been difficult to verify.
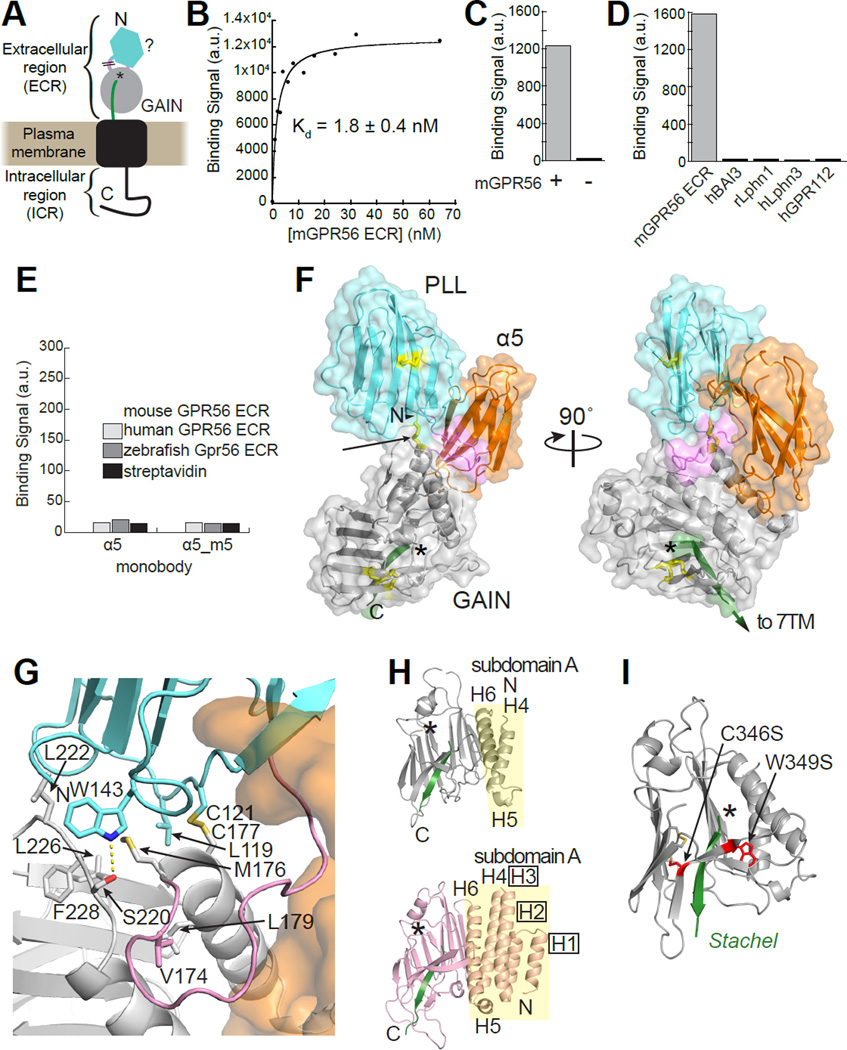
Figure 1. Crystal structure of GPR56 extracellular region in complex with high-affinity and specific monobody.
(A) Schematic of predicted GPR56 domain structure including ECR composed of unidentified N-terminal domain (cyan), linker (pink), and cleaved GAIN domain (NTF, gray; Stachel, green; autoproteolysis site, *). Though the terms ‘Extracellular Domain’ or ‘Ectodomain’ (both abbreviated ECD) are conventional, we have chosen to refer to the extracellular part of GPR56 as ‘extracellular region’ (ECR) to avoid confusion given that the ECR is composed to two protein domains (Rossmann and Argos, 1981). \\ represents unclear domain/linker boundary. (B) Binding titration of purified mouse GPR56 ECR to yeast-displayed monobody α5. Bound GPR56 was quantified using flow cytometry. (C) Binding signal of purified α5 (25 nM) to HEK293T cells overexpressing full-length mouse GPR56 (+) and control cells (−) detected by flow cytometry. (D) Binding signals of different purified aGPCR extracellular fragments at 250 nM (BAI3, ADGRB3; Lphn1, ADGRL1; Lphn3, ADRGL3; GPR112, ADGRG4; m, mouse; h, human; r, rat) to yeast-displayed α5. (E) Binding signal of α5 and α5_m5 to purified GPR56 ECR-coated M280 beads (see Figure S2B). (F) The crystal structure of GPR56 ECR in complex with α5 (orange). Cys residues involved in a disulfide bond are colored yellow, with the interdomain disulfide bond (C121-C177) indicated by the arrow. The linker and Stachel are colored pink and green, respectively and the asterisk indicates the autoproteolysis site. (G) Close-up view of the binding interface between the PLL domain, PLL-GAIN linker, and GAIN domain. Residues at the binding interface are shown as sticks. The PLL domain, PLL-GAIN linker, and GAIN domain are colored cyan, pink, and gray, respectively. α5 is shown as a transparent orange surface. Polar contacts are indicated by yellow dashes. (H) Crystal structures of autoproteolyzed GAIN domains of GPR56 (top) and Lphn1 (PDB: 4DLQ; bottom) in identical orientations. The α-helices in subdomain A (yellow background) are labeled, and the boxed labels indicate α-helices present in Lphn1 but not GPR56. (I) Human disease-causing GPR56 mutations (red) mapped to the GAIN domain. See also Figures S1–S2.
Human and mouse GPR56 are 693 and 687 residues long, respectively, each including a ~377-residue ECR. The localization of six BFPP mutations to the GPR56 ECR suggests a critical role for the ECR in GPR56 function (Singer et al., 2013, Fujii et al., 2014). The ECR comprises an N-terminal domain with no previously defined motifs and a GAIN domain (Figure 1A) (Arac et al., 2012). Truncation-based studies suggest that the N-terminal domain mediates the interaction of GPR56 with two known natural extracellular ligands, collagen III and tissue transglutaminase 2 (TG2), and that these interactions may regulate GPR56 function (Luo et al., 2011, Yang et al., 2014). However, sequence-based bioinformatics analyses have failed to determine the identity of the N-terminal domain as well as the domain boundaries of the N-terminal domain and GAIN domain.
Alternative splicing (AS) has been observed in the coding and non-coding regions of GPR56 transcripts (Kim et al., 2010, Bae et al., 2014). AS in non-coding upstream elements of GPR56 regulates human-specific cerebral cortical patterning, leading to the suggestion that rapid evolution of GPR56 AS might have influenced cortex evolution of gyrencephalic brains, such as the human brain (Bae et al., 2014). Intriguingly, the coding region of GPR56 also undergoes AS to generate four variants in humans, two of which include large deletions in the ECR. (Kim et al., 2010). Considering the established pathophysiological importance of AS in the coding regions of many proteins, especially in the brain (Chen and Manley, 2009, Kalsotra and Cooper, 2011, Irimia et al., 2014), it is likely that these four GPR56 splice variants have distinct and important roles. However, due in large part to the absence of three-dimensional structural information, the effect of AS on ECR architecture is unknown and isoform-specific roles of GPR56 remain elusive.
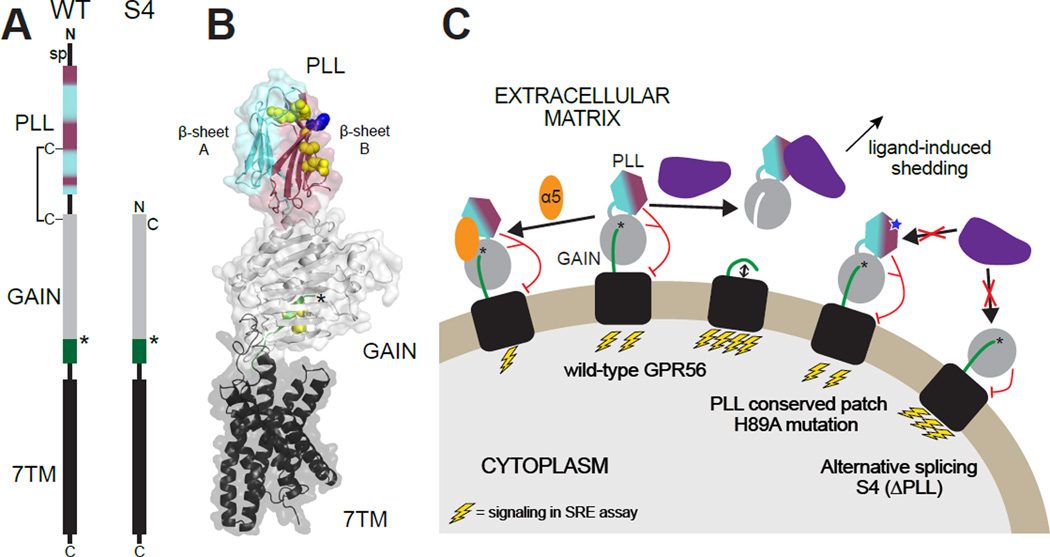
In this study, we set out to determine the 3D structure of the entire ECR of GPR56 at atomic resolution. To this end, we engineered monobodies that recognize the ECR of GPR56. Monobodies are synthetic binding proteins based on the human fibronectin type-III (FN3) scaffold (Koide et al., 1998), which have recently emerged as powerful tools to facilitate structure determination as ‘crystallization chaperones.’ Monobodies can also act as agonists or antagonists, further underscoring their utility in probing the function of a given protein (Wojcik et al., 2010, Sha et al., 2013, Stockbridge et al., 2015). In this study, we determined the crystal structure of the ECR of GPR56 in complex with a monobody; excitingly, this represents the first crystal structure of a full ECR for any aGPCR. The structure revealed the identity and boundaries of two extracellular domains: a previously unidentified N-terminal domain with low homology to all known folds, and a short but functional GAIN domain. Notably, we discovered that the entire newly defined N-terminal domain was deleted in GPR56 splice variant 4 (S4), but not the other variants, and deletion of this domain increased basal activity of the receptor. Finally, we identified a highly conserved, surface-exposed patch on the N-terminal domain, mutation of which abolished GPR56 function in vivo. Together these results elucidate the multifaceted manner by which the ECR regulates GPR56 function and broadens our understanding of aGPCR biology and oligodendrocyte development.
Results
A specific and high-affinity monobody directed to the extracellular region of GPR56
Using a baculovirus expression system as previously described (Arac et al., 2012), recombinant mouse GPR56 ECR was purified from High Five insect cells. The protein was folded, monomeric, and properly underwent autoproteolysis within the GAIN domain (Figure S1A–D). From combinatorial phage-display libraries (Wojcik et al., 2010, Koide et al., 2012), monobody clones that bound to mouse GPR56 ECR were enriched. After gene shuffling and additional library sorting using yeast surface display (Koide et al., 2012), a total of 13 monobodies with different degrees of affinity for the GPR56 ECR were identified (Figure S2A). The clone with the highest affinity, termed Mb(mGPR56_α5), was chosen for further analyses (Figure S2A). This clone will be abbreviated as ‘α5’ hereafter.
α5 bound mouse GPR56 ECR purified from insect cells with an apparent dissociation constant (KD)=1.8±0.4 nM in the yeast surface display format, as expected from the design of monobody selection (Figure 1B, S2B). Importantly, the apparent KD values of monobodies determined in this manner are consistent with those from more conventional biophysical measurements of purified monobodies such as surface plasmon resonance (Koide et al., 2012, Sha et al., 2013). Purified α5 also bound to full-length mouse GPR56 expressed on the surface of HEK293 cells with apparent KD=17±2 nM, indicating that this monobody recognizes the ECR in the context of full-length GPR56 (Figure 1C, S2B–C). To determine the α5 residues responsible for interacting with GPR56, mutations were made in several regions of α5. Variants harboring mutations in the so-called CD or FG variable loop of α5 (termed α5_m2, α5_m4 or α5_m5; Figure S2D) independently decreased affinity by >100-fold, suggesting that both of these loops interact with GPR56. To assess the specificity of α5, we tested its ability to bind to other aGPCR extracellular fragments that contain GAIN domains. No binding was detected between α5 and any of these fragments including GPR112/ADGRG4, an aGPCR in the same subfamily as GPR56 (Figure 1D). Furthermore, no binding was detected between α5 and human or zebrafish GPR56 ECR (Figure 1E). Finally, differential scanning fluorimetry showed α5 increased the thermostability of the GPR56 ECR (Figure S2E). Together, these results show that α5 has high affinity and specificity for mouse GPR56 ECR.
The structure of the α5-GPR56 ECR complex reveals two domains with an interdomain disulfide bond
We first attempted to crystallize the GPR56 ECR alone using standard techniques but obtained only crystals that diffracted poorly (>8Å resolution; data not shown). Thus, we instead used α5 as a crystallization chaperone, which yielded high-quality crystals (Figure S1E–F). We determined the structure of the GPR56 ECR-α5 complex at 2.5 Å resolution with experimental phases obtained from iodine single wavelength anomalous diffraction data (Figure 1F and Table 1).
Table 1.
Data collection and refinement statistics.
| Protein | GPR56 ECR-α5 Native dataset |
GPR56 ECR-α5 Iodide SAD |
|---|---|---|
| Integration Package | HKL2000 | HKL2000 |
| Wavelength (Å) | 1.033 | 1.771 |
| Space group | P 65 | P 65 |
| Cell dimensions | ||
| a, b, c (Å) | 120.34, 120.34, 72.85 | 121.23, 121.23, 72.67 |
| c (°) | 90, 120, 120 | 90, 120, 120 |
| Resolution (Å) | 46.39 – 2.45 (2.49 – 2.45) |
46.58 – 3.00 (3.05 – 3.00) |
| Rsym or Rmerge | 0.035 (0.421) | 0.057 (0.639) |
| CC1/2 | 0.923 (0.663) | 0.934(0.691) |
| I / σI | 19.75 (1.5) | 13.9 (1.4) |
| Completeness (%) | 99.9 (99.5) | 89.2 (80.3) |
| Redundancy | 5.7 (5.7) | 11.2 (10.9) |
| Number measured reflections | 631,603 | 451,228 |
| Number unique reflections | 22,286 | 12,201 |
| Refinement Statistics | ||
| Rwork/Rfree | 0.215/0.263 | |
| Number of atoms | ||
| Protein | 3576 | |
| Water | 24 | |
| Other | 98 | |
| Average B-factors (Å2) | ||
| Protein | 79.5 | |
| Water | 60.0 | |
| Other | 101.8 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 0.611 | |
| Ramachandran plot statistics (%) | ||
| Most favorable | 96.0 | |
| Allowed | 4.0 | |
| Disallowed | 0.0 |
This structure revealed two domains in the GPR56 ECR with an overall dimension of 84Å × 54Å × 42Å: a previously unidentified domain with a β-sandwich architecture at the N-terminus (P28-S160) and, as predicted from the sequence, a GAIN domain at the C-terminus (M176-S391). The 15-residue linker between the two GPR56 domains is ordered in the crystal, despite its lack of defined secondary structure. We found an interdomain disulfide bond linking the two domains formed by cysteine residues C121 and C177, both of which are highly conserved among GPR56 orthologs (Alignment S1). This disulfide bond may restrict the movements of the two domains with respect to each other as observed in other proteins with an interdomain disulfide bond (Bustanji and Samori, 2002). We observe an interface between the N-terminal domain and GAIN composed of mostly conserved and hydrophobic residues (L119, W143, M176, and F228). At this interface, we observe a buried surface area of ~680Å2 (~300Å2 between the two domains and an additional ~380Å2 contributed by the linker; Figure 1G). It is likely that the interdomain disulfide bond and the hydrophobic residues are sufficient to stabilize this conformation of the ECR.
α5 interacts with the N-terminal domain and GAIN domain simultaneously via its CD and FG variable loops, respectively, which are located at opposite ends of the scaffold (Figure S2F–G). This is consistent with the α5 mutagenesis data (Figure S2D). At the GPR56 ECR-α5 interface, we observe a buried surface area of ~1620Å2 (N-terminal domain-α5, linker-α5, and GAIN-α5 contributing ~260Å2, ~300Å2, and ~1060Å2, respectively). The fact that we were able to readily generate a monobody with high affinity suggests that α5 binds to a highly populated conformational species, rather than a high-energy, rare species that would require an extraordinarily high-affinity monobody to capture (Koide, 2009). Thus, it is likely that the conformation of GPR56 ECR captured in our crystal structure represents a dominant conformational state.
Superimposition of the GPR56 GAIN domain structure with the previously determined crystal structures of the GAIN domains of aGPCRs Latrophilin 1 (Lphn1/ADGRL1) and brain angiogenesis inhibitor 3 (BAI3/ADGRB3) shows that, intriguingly, subdomain A of GPR56 contains only three helices and is much smaller than subdomains A of Lphn1 and BAI3 that each have six helices (Figure 1H) (Arac et al., 2012). On the other hand, the conformation of subdomain B, which contains the autoproteolysis site and the Stachel, is highly conserved among the three aGPCRs, particularly around the Stachel (Arac et al., 2012) (Figure S1G–I). Analysis of the 2Fo-Fc electron density map confirmed that the GPR56 ECR underwent autoproteolysis (Figure S1G). Mass spectrometry showed that the GPR56 GAIN domain alone was sufficient to mediate autoproteolysis (Figure S1C), consistent with previous observations for Lphn1 and BAI3 (Arac et al., 2012). Two BFPP mutations now unambiguously mapped to the GAIN domain: C346S, which eliminates a conserved disulfide bond, and W349S, which mutates a conserved hydrophobic core residue, are likely to cause global folding problems of the GAIN domain, consistent with previous findings that these mutants undergo little to no autoproteolysis (Jin et al., 2007, Chiang et al., 2011) (Figure 1I). Together, these results show that even though the GAIN domain of GPR56 is unexpectedly smaller than other known GAIN domains (Arac et al., 2012), it retains autoproteolytic activity.
The N-terminal domain of GPR56 has a previously unidentified fold
The crystal structure of the GPR56 ECR reveals a 133 amino acid, 12-stranded β-sandwich domain at the N-terminus of mature GPR56 (residues P28-S160) (Figure 1F). We denote the two β-sheets as β-sheet A (strands 2–5, 9 and 12) and β-sheet B (strands 1, 6–8, 10 and 11). Using the DALI (Holm and Rosenstrom, 2010) and HorA (Kim et al., 2009) servers, we found that this domain has weak homology to the pentraxin (PTX) and laminin/neurexin/sex hormone-binding globulin (LNS) domain families, but no strong homology to any known fold (top DALI hit: LNS, Z-score=6.5; top HorA hit: PTX, combined score=4.48). Superimposition of the GPR56 N-terminal domain with LNS or PTX domains yields a high backbone rmsd (5.7 Å and 4.7 Å, respectively), whereas superimposition of LNS domain with PTX domain yields a lower backbone rmsd (~3.2 Å), suggesting that the GPR56 N-terminal domain has diverged more from both PTX and LNS domains than the PTX and LNS domains have from each other (Figure 2A). Though the N-terminal domain of GPR56 has low sequence identity with PTX and LNS domains (18% and 19%, respectively for the family member with the highest identity), we found that it has a conserved motif (HΦC91xxWxxxxG) that we identified among canonical PTX domains (Alignments S2, S3). Thus we termed this GPR56 domain as the PTX/LNS-Like (PLL) domain.
Figure 2. The PLL domain of GPR56 is a previously unidentified domain that likely diverged from the pentraxin and LNS folds.
(A) The PLL domain of GPR56, the PTX domain of C-reactive protein (PDB ID: 3PVN), and the LNS domain of Neurexin-1 beta (PDB ID: 3QCW) in similar orientation. β-Strands are numbered from N to C terminus, and equivalent β-strands are colored in the same manner. Cys residues involved in a disulfide bond are colored yellow. (B) Schematic of β-strand connectivity comprising the two β-sheets of each domain. Wavy arrows represent loops with geometry similar to a β-strand. (C) Human disease mutations (red) mapped to the PLL domain.
The connectivity of the β-strands in the PLL domain of GPR56 is substantially different from the completely conserved connectivity within the PTX and LNS families (Figure 2A–B). Interestingly, the majority of the changes in β-strand connectivity map to β-sheet A. All PTX and LNS domains have completely antiparallel β-sheets, whereas β-sheet A of the PLL domain of GPR56 is a mixed β-sheet with β2 and β4 strands parallel to each other (Figure 2A–B). In contrast, the other β-sheet, β-sheet B, is antiparallel as in the PTX and LNS domains, and contains all six PLL domain-localized BFPP mutations (Figure 2A–C). Additionally, the locations of β2, β11, and β12 strands with respect to the other β-strands sets the PLL domain apart from the known PTX and LNS folds (Figure 2B). Thus, the PLL domain of GPR56 has a unique fold that likely diverged from the PTX and LNS domain folds.
The PLL domain is deleted in GPR56 splice variant 4
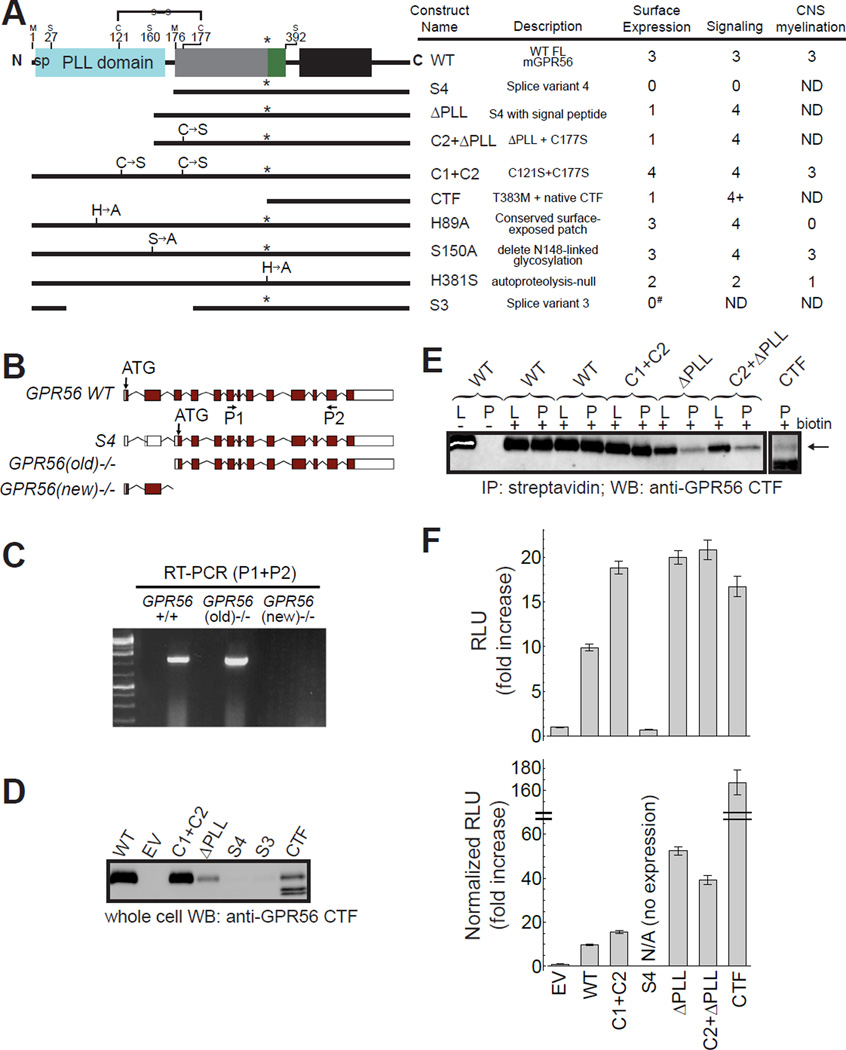
Although AS of upstream regulatory elements of GPR56 controls regional cerebral cortical patterning (Bae et al., 2014), the role of AS of GPR56’s coding sequence is unknown. AS in the coding region of genes is an important mechanism to greatly expand the functional and regulatory capacity of metazoan genomes and its regulatory role in brain function has been repeatedly demonstrated (Braunschweig et al., 2013, Irimia et al., 2014, Anderson et al., 2015). For instance, recent studies suggest a specific expression pattern for hundreds of alternatively spliced isoforms of neurexins, key proteins that organize synapse architecture and encode cellular identity and diversity (Fuccillo et al., 2015). The coding sequence of human GPR56 consists of 13 exons and the ECR is encoded by exons 2–9 (Figure 3A–B). AS occurs in exons 2, 3, and 10, resulting in a total of five variants (WT and S1–S4), of which only S3 and S4 result in substantial changes to the ECR (Kim et al., 2010). S3 has a large deletion encompassing the 3’ end of exon 2 and all of exon 3 (ΔR38-Q207) including the C-terminal portion of the PLL domain and a small N-terminal portion of the GAIN domain, likely resulting in a hybrid domain with unknown structure (Figure 3A). In S4, 43 nucleotides at the 5’ end of exon 2 are deleted, resulting in a frameshift and therefore a new translation start site at M176 (Figure 3A–C). AS in the ECR-coding region of human and mouse GPR56 is identical, specifically the formation of S3 and S4. Strikingly, the crystal structure of the GPR56 ECR revealed that M176 corresponds exactly to the first residue of the GAIN domain. Therefore, S4 lacks the N-terminal 175 residues including the signal peptide sequence (M1-G26), PLL domain (S27-S160), and PLL-GAIN linker (F161-D175). These observations suggest that by regulating AS, a cell may generate GPR56 with or without a PLL domain in the ECR, which could diversify functionality.
Figure 3. Precise deletion of the PLL domain, as in GPR56 splice variant 4, leads to increased basal activity.
(A) Domain architecture schematics and function-metrics of important GPR56 constructs (data compiled from D–F, Figure 5 and Table S1). Residue numbers for domain boundaries based on the crystal structure and the interdomain disulfide bond are shown. Function metrics used are: (0) none, (1) very little, (2) less than WT, (3) comparable to WT, (4) more than WT, (ND) not determined. #see also Figure S3. (B) Expected transcripts in two knockout mouse alleles. The starting ATG for WT GPR56 is in exon 2. The S4 variant has its starting ATG in exon 4. The targeting strategy for GPR56(old)−/− mice was to delete exons 2 and 3, which preserved the S4 variant, whereas the GPR56(new)−/− allele deletes exon 4–6, causing a frameshift that leads to a deletion of all splicing variants of GPR56. (C) RT-PCR showing the presence of the S4 transcript in GPR56(old)−/− but absence in GPR56(new)−/− mouse brains. (D–E) In order to quantify cell-surface expression of GPR56 mutant constructs with decreased affinity for α5, IP-western blot was performed. (D) Western blot of whole cell lysates of cells expressing different GPR56 constructs. (E) Western blot of lysate (L) and lysate subject to streptavidin pull-down (P) of HEK293T cells transfected with WT and mutant GPR56 constructs. (F) Basal activity of mutant GPR56 constructs as measured by the SRE-luciferase reporter assay. Top: Basal activity of mutant constructs. Bottom: Basal activity of mutant constructs normalized for cell-surface expression using band densities from E. Data are presented as mean ± S.E.M.; n = 3. sp, signal peptide. See also Figures S3–S4.
PLL domain deletion increases GPR56 basal activity
In order to test the role of the PLL domain in GPR56 G protein signaling, we generated various constructs and assayed their cell surface expression and signaling capability. These constructs include splice variant 4 (termed ‘S4’, deleting residues M1-D175 including the signal peptide), and splice variant 4 with the signal peptide (effectively a deletion of the PLL domain and PLL-GAIN linker, termed ‘ΔPLL’, deleting residues G26-D175). Due to the interdomain disulfide bond, PLL domain deletion in both constructs generates a free cysteine that may mediate nonspecific interactions. Therefore, we also generated a construct with the C177S mutation on ΔPLL (termed ‘C2+ΔPLL’). Additionally, we generated a construct corresponding to full-length GPR56 without the interdomain disulfide bond (termed ‘C1+C2’ corresponding to C121S+C177S) and one that corresponds to the cleaved C-terminal fragment (CTF, including the Stachel and 7TM) that has been reported to have dramatically increased basal activity (Figure 3A, Table S1) (Stoveken et al., 2015). Proper cell-surface expression and trafficking of these constructs in HEK293T cells was quantified using a cell-surface biotinylation assay followed by streptavidin pull-down and western blot using an antibody directed to the C-terminus of GPR56 (Paavola et al., 2011, Stoveken et al., 2015). We note that we did not attach an N-terminal epitope tag such as FLAG to these constructs, because modifications near the N-terminus led to dramatically reduced cell-surface expression and signaling (Figure S3).
Human and mouse GPR56 are both reported to activate Gα13, which is upstream of RhoA and serum response element (SRE) (Luo et al., 2011, Stoveken et al., 2015) (Figure S4A). Interestingly, we found mouse GPR56 weakly coupled to an additional G protein, Gαq, but not Gαi/o, which can couple to human GPR56 (Figure S4B–D) (Stoveken et al., 2015), likely illustrating different roles for GPR56 across species. We used an SRE-luciferase assay to measure Gα13 G protein signaling of HEK293T cells overexpressing WT or mutant GPR56 constructs. Overexpression of full-length GPR56 in HEK293T cells resulted in higher luciferase activity when compared to cells transfected with an empty vector, showing that the basal activity of overexpressed GPR56 can be detected in this assay. We detected much lower basal activity for S4 expressed in HEK293T cells relative to cells expressing WT GPR56, as reported (Kim et al., 2010), but we found that S4 had undetectable expression in this system (Figure 3D), rendering it difficult to evaluate the functional state of S4. In contrast, ΔPLL, which differs from S4 only by the presence of the N-terminal secretion signal sequence, showed detectable surface expression in HEK293T cells (Figure 3D–E); thus, we used this construct to represent the function of S4 in HEK293T-based signaling assays. Importantly, the majority of GPCRs do not contain an N-terminal secretion signal peptide, yet are still properly trafficked to the plasma membrane (Schulein et al., 2012), suggesting that despite the requirement of a signal peptide in HEK293T cell culture (as in ΔPLL), GPR56 S4 may be properly trafficked in vivo, consistent with its detectable expression in mouse brain (Figure 3C).
Strikingly, the ΔPLL construct had higher basal activity than WT GPR56 (~2 fold without normalization for surface expression and ~5 fold with normalization). This suggests that deletion of the PLL domain in S4 would result in increased basal G protein signaling as long as S4 is properly trafficked to the plasma membrane. C2+eS4, which removes the unpaired Cys residue in the GAIN domain after the deletion of the PLL domain, produced similar results (~2 fold and ~4 fold increase in basal activity without and with normalization for surface expression, respectively), indicating that the elevated activity is not caused by potentially anomalous conjugation involving this unpaired sulfhydryl group (Figure 3F). Moreover, eliminating the disulfide bond between the PLL and GAIN domains in full-length GPR56 also increased GPR56 basal activity (Figure 3F), suggesting the importance of restricting the flexibility within the ECR in keeping the receptor in the basal state. We generated several additional GPR56 ECR mutants and assayed their basal activity using the same SRE-luciferase reporter (Table S1). Altogether, these results suggest that the ECR regulates GPR56 signaling via complex mechanisms and that AS of the coding region modulates GPR56 function.
The two β-sheets of the PLL domain experienced different evolutionary pressures
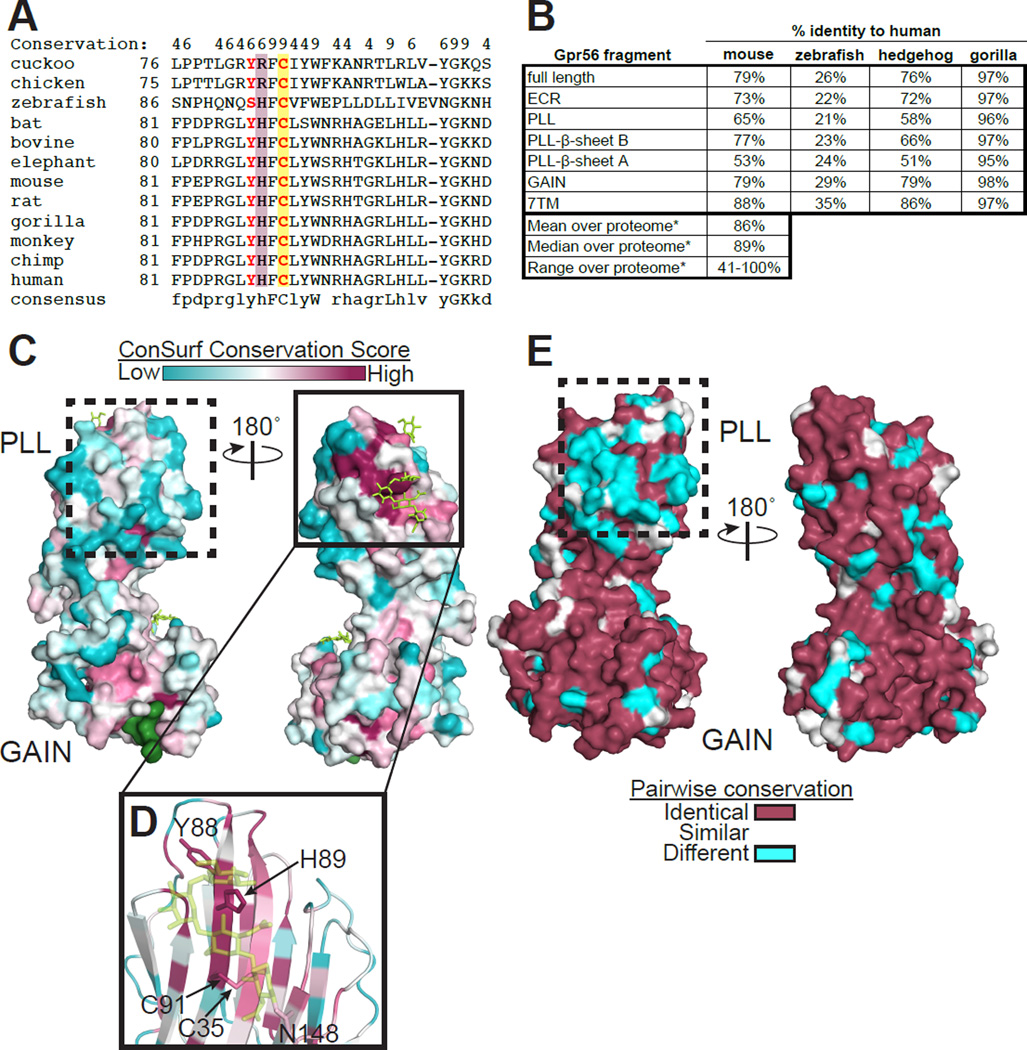
In order to characterize the evolution of the PLL domain, GPR56 protein sequences from various species were aligned (Figure 4A). The PLL domain of GPR56 has lower sequence identity (65% between mouse and human) when compared with those for the GAIN (79%) and 7TM domains (88%), suggesting the individual domains of GPR56 have evolved to different degrees across species (Figure 4B). Furthermore, these analyses revealed that even the two β-sheets of the PLL domain have strikingly different sequence conservation across species: β-sheet B had 77% sequence identity between mouse and human, while β-sheet A had only 53%. This distinctly lower conservation suggests that β-sheet A has evolved rapidly since the divergence of mouse, human, and hedgehog from their common ancestor about 105 million years ago (Figure S5A, Alignment S4).
Figure 4. The two β-sheets of the PLL domain experienced different evolutionary pressures.
(A) Sequence alignment of a segment of the PLL domain from 12 species of GPR56. Conservation scores greater than 3 are shown at the top, with 9 representing the highest conservation. Residues found mutated in BFPP patients (Y88 and C91) are in red. C91, participating in the intra-PLL domain disulfide bond, is highlighted in yellow. H89 is highlighted in maroon. (B) Sequence identities for different fragments of GPR56 between human and the indicated species. *Taken from Makalowski and Boguski (1998). (C) Conservation score of each residue is mapped on the GPR56 ECR structure. Stachel and N-linked glycans are shown as green surface and yellow sticks, respectively. Note that the conserved patch on β-sheet B (solid box, panel D) and the non-conserved patch on β-sheet A (dashed boxes in C and E) are the most and least conserved patches in the entire ECR, respectively. (D) Close-up of the most conserved surface-exposed patch. The sidechains of the residues with the highest conservation score shown as sticks. (E) Pairwise surface conservation between human and mouse GPR56. See also Figure S5.
Functional sites (i.e. natural ligand-binding sites) are often highly conserved patches on protein surfaces. Thus, we performed surface conservation analysis of 102 GPR56 protein sequences from diverse organisms using the ConSurf server (Celniker et al., 2013) to identify any putative functional sites on the GPR56 ECR. When the conservation score for each residue was mapped onto the GPR56 ECR structure (Figure 4C), the largest and most obvious conserved patch on the entire ECR was formed by a group of residues (G86, Y88, H89, and G106) located on β-sheet B of the PLL domain (Figure 4D). The residues involved in the conserved intra-PLL domain disulfide bond between strands 1 and 6 (C35 and C91) also contribute to this conserved patch. Finally, a glycan with a conserved N-linked glycosylation motif (N148-X-S150) sits adjacent to this patch. Notably, the side chain of H89 points out of β-sheet B into the solvent. Pairwise surface conservation analysis between human GPR56 and protein sequences from various organisms (gorilla, mouse, hedgehog, and zebrafish) shows that this patch is indeed highly conserved, even in zebrafish, the earliest known organism with GPR56 in its genome (Figure 4E, S5A–B). Thus, we speculated that this conserved patch, particularly H89, has an essential role in GPR56 function (see below for in vitro and in vivo analysis).
A residue in the conserved patch of the PLL domain is critical for oligodendrocyte development in zebrafish
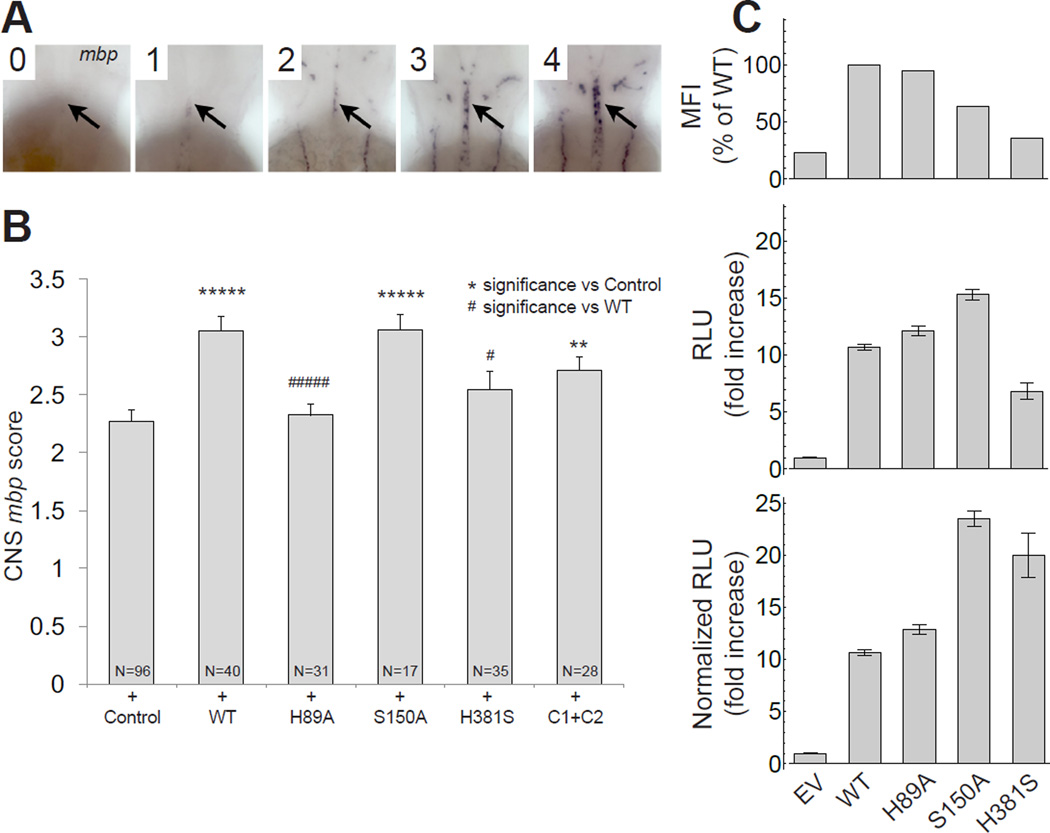
We hypothesized that the residues comprising the aforementioned conserved patch of the PLL domain are involved in an evolutionarily conserved function of GPR56. To dissect this in vivo, we tested GPR56 point mutants using zebrafish. Briefly, in zebrafish, Gpr56 promotes oligodendrocyte proliferation in the CNS, such that loss of this aGPCR results in reduced numbers of mature oligodendrocytes and myelinated axons (Ackerman et al., 2015). Gpr56 activity in zebrafish can be readily measured by assessing the expression of myelin basic protein (mbp), which encodes a structural component of the myelin sheath. Importantly, transient expression of mouse GPR56 mRNA increases mbp expression above WT levels as reported previously (Ackerman et al., 2015) and confirmed in the present study (Figure 5A–B).
Figure 5. The conserved patch of the PLL domain is required for GPR56 function in vivo.
(A) Injection of WT and mutant mouse GPR56 mRNAs generate embryos with varied levels of CNS myelin basic protein (mbp) expression (black arrow, hindbrain) at 65 hours post-fertilization (hpf). Embryos were given the following scores to signify (0) none, (1) weak, (2) modest, (3) WT, and (4) excess CNS mbp expression. (B) Average CNS mbp score (± S.E.M.) for phenol red (+ Control), WT, H89A, H381S, S150A, and C121S+C177S (C1+C2) injected embryos (from left to right). Injection of WT GPR56 causes an increase in CNS mbp score compared to control-injected embryos (p<7.56×10−6). H381S and H89A abolish the effect of GPR56 overexpression on CNS mbp expression (no significant difference from control injected, significantly less than WT injected: H381S, p<.02; H89A, p<2.68×10−5). S150A and C1+C2 do not affect GPR56-induced CNS mbp overexpression (v. control injected: S150A, p<3.78×10−5; C1+C2, p<.005). (C) Cell surface expression and basal activity of mutant GPR56 constructs as measured by the SRE-luciferase reporter assay. Top: Cell-surface expression of untagged WT and mutant GPR56 constructs with affinity for α5 comparable to WT measured by flow cytometry. Middle: Basal activity of mutant constructs. Bottom: Basal activity of mutant constructs normalized for cell-surface expression using MFI from Top. Data are presented as mean ± S.E.M.; n = 3.
We tested the following mouse GPR56 point mutations in this assay: H89A, S150A, H381S, and C121S+C177S (C1+C2), described in Figure 3A. Strikingly, injection of mRNA encoding the mouse GPR56 H89A mutant failed to enhance mbp expression, suggesting an essential role of this evolutionarily conserved residue in CNS myelination (Figure 5B). The H381S mutant also failed to enhance mbp expression, suggesting a possible role for receptor autoproteolysis in GPR56-dependant oligodendrocyte development, consistent with previous studies that implicate autoproteolysis in GPR56 function (Luo et al., 2014, Kishore et al., 2015). On the other hand, S150A and C1+C2 resulted in a significant increase in mbp expression, similar to injection of WT GPR56 (Figure 5B). To ensure the in vivo effects of these mutants were not simply due to mutation-dependent cell-surface expression, we quantified surface expression and SRE signaling in HEK293T cells. We found H89A had no effect on surface expression or basal activity as compared to WT GPR56, suggesting any differences between WT and H89A phenotypes in vivo are not due to cell-surface expression or basal activity (Figure 5C). Altogether, these results reveal that the conserved patch of the PLL domain mediates an essential function in CNS myelination.
α5 monobody is an allosteric inverse-agonist for GPR56
We hypothesized that since α5 interacts with the GAIN and PLL domains, both shown to regulate signaling, α5 may itself modulate GPR56 basal activity. We found that addition of α5 to the SRE-luciferase signaling assay causes a ~25% decrease in GPR56 basal activity with an IC50=65 ± 14 nM (Figure 6A–B), the same order of magnitude as the measured affinity of purified α5 to GPR56-expressing HEK293T cells (KD=17±2nM, Figure S2C). The addition of an unrelated, non-binding monobody, or α5_m5 (double Tyr to Ala mutant in the FG loop with >100-fold decreased affinity for GPR56 [Figures 1E and S2D]), produced no significant effect on GPR56 basal activity (Figure 6A). In addition, α5 had no effect on the background luminescence of HEK293T cells transfected with an empty vector (Figure 6A), demonstrating the specificity of α5 in this assay. The binding site of α5 is poorly conserved among mouse, human, and zebrafish GPR56 (Figure 6C), consistent with the observation that α5 does not detectibly interact with human or zebrafish GPR56 (Figure 1E). Furthermore, as expected from the crystal structure, α5 binds to full-length mouse GPR56 and purified soluble mouse GPR56 ECR, but it does not detectibly interact with N-terminally truncated mouse GPR56 constructs expressed in HEK293T cells including ΔPLL (Figure S3). Thus, the entire ECR is necessary for α5 binding, indicating that any effect on GPR56 activity mediated by α5 is due to its interaction with the ECR and not the 7TM, the canonical site for GPCR ligand interaction. Therefore, α5 represents an ‘allosteric inverse-agonist’ for GPR56 (Christopoulos, 2014).
Figure 6. Monobody α5 is an allosteric inverse-agonist for GPR56.
(A) Effect of 1 µΜ monobody on GPR56 activation as measured by SRE-luciferase assay in HEK293T cells. Data are presented as mean ± S.E.M.; n = 3; *, p<0.01 compared to Mock by two-tailed Student’s t-test; NS, not significant compared to Mock. (B) SRE-luciferase activity in HEK293T cells is plotted as a function of α5 concentration. Line represents the best fit of the 1:1 binding model for calculation of IC50. (C) Stereo image of the interface between α5 and GPR56 ECR. ECR is colored by conservation score, as in Figure 4C. α5 residues important for GPR56 binding (Figure S2D) shown as sticks.
Discussion
CNS myelination likely requires GPR56 activation by a PLL-binding ligand
Our study yielded the first crystal structure of the full ECR of an aGPCR and provides a functional framework to understand the molecular mechanisms by which aGPCR ECRs govern receptor function. The crystal structure revealed a previously unidentified PTX/LNS-Like (PLL) domain at the N-terminus of the GPR56 ECR (Figure 2). Both PTX and LNS domains predominantly occur in secreted proteins and in ECRs of cell-surface proteins. LNS domains in particular are mostly known for their adhesion properties, especially in the brain, and exist in adhesion molecules such as agrins, laminins, and neurexins to mediate cell-cell and cell-extracellular matrix interactions (Rudenko et al., 2001, Arac et al., 2007, Domogatskaya et al., 2012). Thus, the remote similarity of the PLL domain to PTX and LNS domains supports an adhesion-related role for the PLL domain in GPR56 as was previously suggested (Koirala et al., 2009) (Figure 2A–B, Alignments S2 and S3). Future studies focusing on biochemical and structural characterization of ECR-ligand interactions will provide more insight into the pathophysiological role of GPR56.
The suggested ‘shedding’ mechanism for aGPCR activation involves engagement of an aGPCR ECR by an extracellular ligand, which results in separation of the NTF from the CTF and initiation of G protein signaling (Stoveken et al., 2015, Liebscher et al., 2014). On the other hand, recent studies suggest that the non-shed NTF of GPR56 may negatively regulate signaling by interacting with the 7TM directly (Kishore et al., 2015). Importantly, ligand binding may activate GPR56 through each of these mechanisms individually or perhaps in concert (Figure S6). It is widely accepted that GPR56 signals through SRE via RhoA and Gα13 (Iguchi et al., 2008, Luo et al., 2014, Stoveken et al., 2015) and it has been shown that transient expression of constitutively active RhoA can suppress myelination defects in gpr56 mutant zebrafish (Ackerman et al., 2015). We discovered a surface-exposed conserved patch on the PLL domain that is necessary to promote CNS myelination in vivo, but that does not affect basal activity in vitro (Figures 4 and 5). Thus, as is common for highly conserved patches, we speculate that this patch on the PLL domain directly engages a GPR56 ligand such as collagen III, TG2, or an as yet unidentified ligand. Indeed, truncation-based analyses suggest that the regions of GPR56 responsible for binding TG2 and collagen III are within residues 108–177 and 27–160, respectively, both of which map to the PLL domain (Yang et al., 2011, Luo et al., 2012). Together, these observations are consistent with the hypothesis that CNS myelination is dependent on GPR56 activation induced by a PLL domain-binding ligand (Figure S6).
Alternative splicing restricts PLL domain expression
The crystal structure of the GPR56 ECR defined the boundaries of the PLL and GAIN domains, and, remarkably, showed that S4, a GPR56 splice variant present in both human and mouse, encodes an isoform that lacks the entire PLL domain, precisely starting with the first residue of the GAIN domain (Figures 3A–B and 7A). Intriguingly, in the originally described GPR56 knock-out mouse, which presents with phenotypes of cortical neuronal ectopia and impaired oligodendrocyte development, the expression of the full-length GPR56, but not the S4 transcript, is disrupted (Figure 3B) (Li et al., 2008, Giera et al., 2015). This demonstrates that GPR56 S4 expression alone is not sufficient for normal CNS development and suggests that the PLL domain has a critical role in both cortical development and oligodendrocyte development. We speculate that observations of a recently published null knock-out mouse with all GPR56 isoforms deleted will reveal distinct, and perhaps more severe phenotypes (Giera et al., 2015).
Figure 7. Working model of mechanisms underlying GPR56 function.
(A) Schematic of GPR56 domain structure comparing WT and S4. In all panels, the PLL domain is colored cyan and maroon, corresponding to β-sheets A and B, respectively. (B) Scale model of full-length GPR56 based on the crystal structure of the ECR and a model of the 7TM (generated based on GCGR structure, PDB ID: 46LR). An arbitrary orientation of the ECR with respect to the 7TM is chosen. Residues mutated in BFPP are shown as yellow spheres. H89 is shown as blue spheres. (C) A working model of aGPCR signaling involves ligand-induced activation. In this model, full-length GPR56 is activated when a natural ligand binds to the conserved patch on the PLL domain including H89, causing conformational changes, perhaps including shedding. Introducing the H89A mutation (blue star) to the conserved patch of the PLL domain or deleting the PLL domain completely (as in S4) would result in abrogation of ligand binding and therefore no ligand-induced activation. Binding of α5 likely stabilizes the ECR, causing decreased signaling. sp, signal peptide. See Figure S6 for further possibilities.
Taken together, our results support a model in which domains in the ECR directly or indirectly regulate the distinct but interrelated functions of GPR56: a possible adhesion function mediated by the PLL domain and a G protein signaling function mediated by the 7TM domain, (Figure 7, S6). With regard to mechanism, we show that ECR modification leads to altered basal activity by allosterically altering NTF shedding propensity and/or altering the conformational states sampled by the 7TM (e.g., absence of the PLL domain leads to increased basal activity) (Figure S6). Moreover, by fine-tuning the expression levels of different GPR56 isoforms, cells may regulate their response to diverse extracellular ligands as well as their basal level of G protein signaling (Figure 7).
aGPCR inhibition by an ECR-directed synthetic allosteric inverse-agonist
The α5 monobody represents, to our knowledge, the first synthetic allosteric inverse-agonist that interacts with the GPR56 ECR with high affinity and specificity (Figures 1B–F, 6A–C, S2, S3). Agonistic antibodies directed to GPR56 have been reported but their mechanisms of action are not fully understood (Ohta et al., 2015, Yang et al., 2015). The lack of well-characterized agonists and antagonists has hampered mechanistic studies of GPR56 and other aGPCRs. Our success in generating a modulator of GPR56 suggests that like the canonical GPCRs, drugging aGPCRs, including GPR56, is a realistic possibility. Such synthetic modulators will help advance mechanistic analyses of aGPCRs. Our finding that α5 alters basal activity by binding to the ECR is an encouraging proof of concept for developing highly selective modulators of aGPCRs. A major challenge in GPCR-targeted drug design is the high conservation of 7TM, which demands high specificity of drugs so as to minimize undesirable side-effects (Schlyer and Horuk, 2006). As aGPCR ECRs are much more diverse than 7TMs, the pursuit of aGPCR ECR-targeted (i.e., allosteric) synthetic ligands, such as monobodies or antibodies, will likely result in highly specific reagents. Furthermore, as the therapeutic potential of allosteric GPCR modulators that exhibit moderate effects has been demonstrated (Wootten et al., 2013, Christopoulos, 2014, Wootten et al., 2016), this work validates the aGPCR ECR as a ‘druggable’ target.
Experimental Procedures
Monobody generation
Biotinylated mouse GPR56 ECR was used as a target for phage-display selection from a ‘side and loop’ monobody library as previously described (Koide et al., 2012). The naïve library contained ~109 different clones. Four rounds of selection were performed at target concentrations of 1) 100 nM (tetramerized), 2) 100 nM (monomeric), 3) 50 nM (monomeric), 4) 50 nM (monomeric). A yeast display library containing ~106 different clones was constructed from the output of phage display selection. A single round of positive sorting of the yeast display library was done using fluorescent-activated cell sorting (FACS) using dye-labeled GPR56 ECR to stain yeast. Binding assay for testing the affinity and specificity of individual monobody clones was performed using yeast surface display as described previously (Sha et al., 2013).
X-ray crystallography data collection
Purified mouse GPR56 ECR was mixed with purified monobody α5 and the complex was purified by size-exclusion chromatography (Superdex 200 10/300 GL; GE Healthcare) and concentrated to ~22 mg/mL protein complex. Crystals grew to >0.1mm in 80 mM sodium acetate pH 4.6, 19.5% glycerol, 16.9% PEG 600, 7.6% PEG 1000. To obtain phase information, some crystals were treated with KI3 using vaporizing iodine labeling (Miyatake et al., 2006). Native and iodinated diffraction data were collected to 2.45 and 3.00Å, respectively at the Advanced Photon Source, beamline 23-ID-B.
SRE-luciferase assay
HEK293T cells were transfected with Gpr56 (WT or mutant) and dualLuc-SRE using FUGENE6. After 24 hours, media was replaced with DMEM + 0% FBS. For monobody treatment, monobody was added to cells 6.5 hours after the start of serum starvation. After 12 hours total of serum starvation, media was aspirated. Cells were lysed using the Dual-Glo® Luciferase Assay System from Promega.
Zebrafish embryo synthetic mRNA injections
The full-length WT mouse GPR56 cDNA clone (openbiosystems clone ID: 3709247) and all mutant derivatives of GPR56 (H89A, S150A, H381S, C121S+C177S) were linearized with NotI, transcribed using the mMESSAGE mMACHINE® SP6 ULTRA kit (Ambion), combined with phenol-red dye and injected at a final concentration of 50 pg in 2 nl. To control for adverse side-effects resulting from mechanical stress during injection, we also injected zebrafish embryos with an equal volume of phenol-red diluted 1:5 in water.
Supplementary Material
Acknowledgments
We thank Engin Özkan and the staff at the Advanced Photon Source at Argonne National Labs, specifically Craig Ogata at GM/CA, for their help with x-ray crystallography. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We also thank Navraj Pannu for his assistance in implementing the CRANK2 software package for experimental phasing. Yue Lu and Olha Nazarko provided technical assistance. BirA was a kind gift from the G. Montelione lab. We thank the A. Kossiakoff lab for the use of their luminescence plate reader and the T. Sosnick lab for the use of their CD spectrometer. Supported by Brain Research Foundation (D.A.), Big Ideas Generator (D.A.), and NIH grants U54-GM087519 (S.K.), R01-GM120322 (D.A.), F30-GM116455 (G.S.S.), F31-NS087801 (S.D.A.), R01-NS079445 (K.R.M.), and T32GM007183.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The coordinates and diffraction data have been deposited in the RCSB PDB (PDB: 5KVM).
Author Contributions
G.S.S., S.K. and D.A. designed all experiments, interpreted results and wrote the manuscript. G.S.S. performed all experiments involving aGPCR ECR cloning, purification, MAALS, and crystallography (with assistance from C.D.); monobody engineering, purification, and characterization (with guidance from A.K.); mammalian cell expression; SRE signaling; bioinformatics; and mass spectrometry. A.K. and S.K. provided the monobody libraries; S.D.A. and K.R.M. designed and performed zebrafish experiments. R.L. and X.P. designed and performed RT-PCR experiments. H.M.S. and G.G.T. designed and performed direct G protein coupling experiments (with assistance from K.L.). All authors discussed the results and provided comments and revisions on the manuscript.
References
- Ackerman SD, Garcia C, Piao X, Gutmann DH, Monk KR. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Galpha12/13 and RhoA. Nat Commun. 2015;6:6122. doi: 10.1038/ncomms7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Aoto J, Tabuchi K, Foldy C, Covy J, Yee AX, Wu D, Lee SJ, Chen L, Malenka RC, Sudhof TC. beta-Neurexins Control Neural Circuits by Regulating Synaptic Endocannabinoid Signaling. Cell. 2015;162:593–606. doi: 10.1016/j.cell.2015.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC, Brunger AT. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac D, Boucard AA, Ozkan E, Strop P, Newell E, Sudhof TC, Brunger AT. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Bae BI, Tietjen I, Atabay KD, Evrony GD, Johnson MB, Asare E, Wang PP, Murayama AY, Im K, Lisgo SN, Overman L, Sestan N, Chang BS, Barkovich AJ, Grant PE, Topcu M, Politsky J, Okano H, Piao X, Walsh CA. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi-Buisson N, Poirier K, Boddaert N, Fallet-Bianco C, Specchio N, Bertini E, Caglayan O, Lascelles K, Elie C, Rambaud J, Baulac M, An I, Dias P, Des Portes V, Moutard ML, Soufflet C, El Maleh M, Beldjord C, Villard L, Chelly J. GPR56-related bilateral frontoparietal polymicrogyria: further evidence for an overlap with the cobblestone complex. Brain. 2010;133:3194–3209. doi: 10.1093/brain/awq259. [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic Integration of Splicing within Gene Regulatory Pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustanji Y, Samori B. The mechanical properties of human angiostatin can be modulated by means of its disulfide bonds: A single-molecule force-spectroscopy study. Angewandte Chemie-International Edition. 2002;41:1546–1548. doi: 10.1002/1521-3773(20020503)41:9<1546::aid-anie1546>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Israel Journal of Chemistry. 2013;53:199–206. [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang NY, Hsiao CC, Huang YS, Chen HY, Hsieh IJ, Chang GW, Lin HH. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J Biol Chem. 2011;286:14215–14225. doi: 10.1074/jbc.M110.183830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Mol Pharmacol. 2014;86:463–478. doi: 10.1124/mol.114.094342. [DOI] [PubMed] [Google Scholar]
- Demberg LM, Rothemund S, Schoneberg T, Liebscher I. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem Biophys Res Commun. 2015;464:743–747. doi: 10.1016/j.bbrc.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu Rev Cell Dev Biol. 2012;28:523–553. doi: 10.1146/annurev-cellbio-101011-155750. [DOI] [PubMed] [Google Scholar]
- Fuccillo MV, Foldy C, Gokce O, Rothwell PE, Sun GL, Malenka RC, Sudhof TC. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Ishikawa N, Kobayashi Y, Kobayashi M, Kato M. Compound heterozygosity in GPR56 with bilateral frontoparietal polymicrogyria. Brain Dev. 2014;36:528–531. doi: 10.1016/j.braindev.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Giera S, Deng Y, Luo R, Ackerman SD, Mogha A, Monk KR, Ying Y, Jeong SJ, Makinodan M, Bialas AR, Chang BS, Stevens B, Corfas G, Piao X. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat Commun. 2015;6:6121. doi: 10.1038/ncomms7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann J, Aust G, Arac D, Engel FB, Formstone C, Fredriksson R, Hall RA, Harty BL, Kirchhoff C, Knapp B, Krishnan A, Liebscher I, Lin HH, Martinelli DC, Monk KR, Peeters MC, Piao X, Promel S, Schoneberg T, Schwartz TW, Singer K, Stacey M, Ushkaryov YA, Vallon M, Wolfrum U, Wright MW, Xu L, Langenhan T, Schioth HB. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallieres M, Tapial J, Raj B, O'hanlon D, Barrios-Rodiles M, Sternberg MJ, Cordes SP, Roth FP, Wrana JL, Geschwind DH, Blencowe BJ. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Tietjen I, Bu L, Liu-Yesucevitz L, Gaur SK, Walsh CA, Piao X. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16:1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Cheng H, Grishin NV. HorA web server to infer homology between proteins using sequence and structural similarity. Nucleic Acids Research. 2009;37:W532–W538. doi: 10.1093/nar/gkp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Han JM, Park CR, Shin KJ, Ahn C, Seong JY, Hwang JI. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J Cancer Res Clin Oncol. 2010;136:47–53. doi: 10.1007/s00432-009-0635-z. [DOI] [PubMed] [Google Scholar]
- Kishore A, Purcell RH, Nassiri-Toosi Z, Hall RA. Stalk-dependent and stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1) J Biol Chem. 2015;291:3385–3394. doi: 10.1074/jbc.M115.689349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. Teaching an old scaffold new tricks: monobodies constructed using alternative surfaces of the FN3 scaffold. J Mol Biol. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol. 2009;19:449–457. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Jin Z, Piao X, Corfas G. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci. 2009;29:7439–7449. doi: 10.1523/JNEUROSCI.1182-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F, Mancuso MR, Shamloo A, Wang HT, Choksi V, Florek M, Su H, Fruttiger M, Young WL, Heilshorn SC, Kuo CJ. Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science. 2010;330:985–989. doi: 10.1126/science.1196554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan T, Aust G, Hamann J. Sticky signaling--adhesion class G protein-coupled receptors take the stage. Sci Signal. 2013;6:re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- Li S, Jin Z, Koirala S, Bu L, Xu L, Hynes RO, Walsh CA, Corfas G, Piao X. GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28:5817–5826. doi: 10.1523/JNEUROSCI.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebscher I, Schon J, Petersen SC, Fischer L, Auerbach N, Demberg LM, Mogha A, Coster M, Simon KU, Rothemund S, Monk KR, Schoneberg T. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014;9:2018–2026. doi: 10.1016/j.celrep.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Jeong SJ, Yang A, Wen M, Saslowsky DE, Lencer WI, Arac D, Piao X. Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PLoS One. 2014;9:e100043. doi: 10.1371/journal.pone.0100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Jin Z, Deng Y, Strokes N, Piao X. Disease-associated mutations prevent GPR56-collagen III interaction. PLoS One. 2012;7:e29818. doi: 10.1371/journal.pone.0029818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci U S A. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake H, Hasegawa T, Yamano A. New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL) Acta Crystallogr D Biol Crystallogr. 2006;62:280–289. doi: 10.1107/S0907444905041909. [DOI] [PubMed] [Google Scholar]
- Mogha A, Benesh AE, Patra C, Engel FB, Schoneberg T, Liebscher I, Monk KR. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J Neurosci. 2013;33:17976–17985. doi: 10.1523/JNEUROSCI.1809-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, Moens CB, Talbot WS. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–1405. doi: 10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K, Yoshida S, Ono M, Kuwano M, Nakamura Y, Tokino T. A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene. 1997;15:2145–2150. doi: 10.1038/sj.onc.1201542. [DOI] [PubMed] [Google Scholar]
- Noseworthy JH, Lucchinetti C, Rodriguez M, Weinshenker BG. Multiple sclerosis. N Engl J Med. 2000;343:938–952. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- O'sullivan ML, De Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, 3rd, Ghosh A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Sakaguchi S, Kobayashi Y, Mizuno N, Tago K, Itoh H. Agonistic antibodies reveal the function of GPR56 in human glioma U87-MG cells. Biol Pharm Bull. 2015;38:594–600. doi: 10.1248/bpb.b14-00752. [DOI] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, Pletikos M, Jankovic-Rapan L, Cramer NP, Galdzicki Z, Goodliffe J, Peters A, Sethares C, Delalle I, Golden JA, Haydar TF, Sestan N. Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination. Neuron. 2016;89:1208–1222. doi: 10.1016/j.neuron.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. The GPR56 N-terminus controls receptor signaling activity. The Journal of biological chemistry. 2011;286:28914–28921. doi: 10.1074/jbc.M111.247973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SC, Luo R, Liebscher I, Giera S, Jeong SJ, Mogha A, Ghidinelli M, Feltri ML, Schoneberg T, Piao X, Monk KR. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron. 2015;85:755–769. doi: 10.1016/j.neuron.2014.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, Ross ME, Michaud JL, Descarie JC, Barkovich AJ, Walsh CA. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Argos P. Protein folding. Annu Rev Biochem. 1981;50:497–532. doi: 10.1146/annurev.bi.50.070181.002433. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Hohenester E, Muller YA. LG/LNS domains: multiple functions -- one business end? Trends Biochem Sci. 2001;26:363–368. doi: 10.1016/s0968-0004(01)01832-1. [DOI] [PubMed] [Google Scholar]
- Schlyer S, Horuk R. I want a new drug: G-protein-coupled receptors in drug development. Drug Discov Today. 2006;11:481–493. doi: 10.1016/j.drudis.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Scholz N, Gehring J, Guan C, Ljaschenko D, Fischer R, Lakshmanan V, Kittel RJ, Langenhan T. The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep. 2015;11:866–874. doi: 10.1016/j.celrep.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Schulein R, Westendorf C, Krause G, Rosenthal W. Functional significance of cleavable signal peptides of G protein-coupled receptors. Eur J Cell Biol. 2012;91:294–299. doi: 10.1016/j.ejcb.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Sha F, Gencer EB, Georgeon S, Koide A, Yasui N, Koide S, Hantschel O. Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains. Proc Natl Acad Sci U S A. 2013;110:14924–14929. doi: 10.1073/pnas.1303640110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer K, Luo R, Jeong SJ, Piao X. GPR56 and the developing cerebral cortex: cells, matrix, and neuronal migration. Mol Neurobiol. 2013;47:186–196. doi: 10.1007/s12035-012-8343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaidero N, Mobius W, Czopka T, Hekking LH, Mathisen C, Verkleij D, Goebbels S, Edgar J, Merkler D, Lyons DA, Nave KA, Simons M. Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell. 2014;156:277–290. doi: 10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Kolmakova-Partensky L, Shane T, Koide A, Koide S, Miller C, Newstead S. Crystal structures of a double-barrelled fluoride ion channel. Nature. 2015;525:548–551. doi: 10.1038/nature14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoveken HM, Hajduczok AG, Xu L, Tall GG. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc Natl Acad Sci U S A. 2015;112:6194–6199. doi: 10.1073/pnas.1421785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Wrann CD, Rao RR, Nair SK, Jedrychowski MP, You JS, Martinez-Redondo V, Gygi SP, Ruas JL, Hornberger TA, Wu Z, Glass DJ, Piao X, Spiegelman BM. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci U S A. 2014;111:15756–15761. doi: 10.1073/pnas.1417898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik J, Hantschel O, Grebien F, Kaupe I, Bennett KL, Barkinge J, Jones RB, Koide A, Superti-Furga G, Koide S. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat Struct Mol Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D, Christopoulos A, Sexton PM. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat Rev Drug Discov. 2013;12:630–644. doi: 10.1038/nrd4052. [DOI] [PubMed] [Google Scholar]
- Wootten D, Miller LJ, Koole C, Christopoulos A, Sexton PM. Allostery and Biased Agonism at Class B G Protein-Coupled Receptors. Chemical Reviews. 2016 doi: 10.1021/acs.chemrev.6b00049. [DOI] [PubMed] [Google Scholar]
- Yang L, Chen G, Mohanty S, Scott G, Fazal F, Rahman A, Begum S, Hynes RO, Xu L. GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res. 2011;71:5558–5568. doi: 10.1158/0008-5472.CAN-10-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Friedland S, Corson N, Xu L. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res. 2014;74:1022–1031. doi: 10.1158/0008-5472.CAN-13-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TY, Chiang NY, Tseng WY, Pan HL, Peng YM, Shen JJ, Wu KA, Kuo ML, Chang GW, Lin HH. Expression and immunoaffinity purification of recombinant soluble human GPR56 protein for the analysis of GPR56 receptor shedding by ELISA. Protein Expr Purif. 2015;109:85–92. doi: 10.1016/j.pep.2014.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.