Abstract
Protein transport across membranes is a fundamental and essential cellular activity in all organisms. In bacteria, protein export across the cytoplasmic membrane, driven by dynamic interplays between the protein-conducting SecYEG channel (Sec translocon) and the SecA ATPase, is enhanced by the proton motive force (PMF) and a membrane-integrated Sec component, SecDF. However, the structure and function of SecDF have remained unclear. We solved the first crystal structure of SecDF, consisting of a pseudo-symmetrical 12-helix transmembrane domain and two protruding periplasmic domains. Based on the structural features, we proposed that SecDF functions as a membrane-integrated chaperone, which drives protein movement without using the major energetic currency, ATP, but with remarkable cycles of conformational changes, powered by the proton gradient across the membrane. By a series of biochemical and biophysical approaches, several functionally important residues in the transmembrane region have been identified and our model of the SecDF function has been verified.
Keywords: Sec translocon, SecDF, protein transport
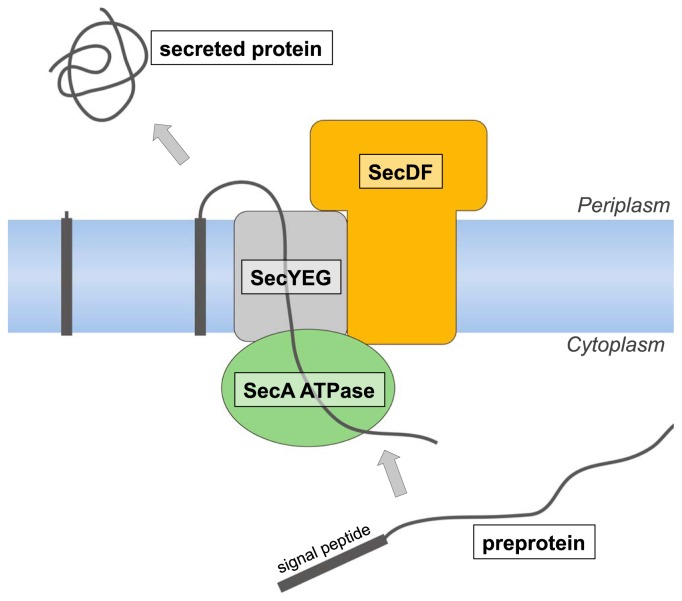
Newly synthesized preproteins that contain a N-terminal cleavable signal peptide for secretion are targeted to the membrane and then translocated across it 1,2. The Sec translocon provides a channel-like pathway for protein translocation across membranes, which normally prevent even the permeation of small ions. The bacterial Sec translocon, consisting of three membrane proteins, SecY, SecE and SecG, is embedded within the cytoplasmic membrane and forms a protein-conducting channel to export proteins from the cytosol to the periplasmic space. In eukaryotic cells, the corresponding process across the ER membrane is achieved by the SecYEG-homolog Sec61αγβ machinery. To elucidate the molecular mechanism of the Sec translocon machinery, structural and functional studies of Sec proteins have been performed. Although there is an hourglass-shaped pathway in the center of SecY for protein transport3,4, the Sec translocon does not function by itself. SecYEG transiently interacts with SecA ATPase and SecDF (Fig. 1). All of the Sec proteins are considered to function through large conformational changes during protein translocation via the translocon. Synthesized preproteins, which are prevented from folding in the cytosol by a chaperone such as SecB, are directed to the membrane-bound SecA. Conformational changes of SecA, accompanied by ATP hydrolysis cycles, then achieve the insertion of the preprotein into the Sec translocon5,6. Using repeated ATP-induced conformational changes, SecA drives the stepwise protein secretion7. Although the protein translocation is enhanced by the PMF8–10 and SecDF11–17, the structure and role of SecDF have remained unclear. In this review, we describe the first 3D structure of SecDF and propose a new working model of the PMF-driven protein translocation by SecDF, based on its structural features and subsequent functional analyses18–20.
Figure 1.
Protein translocation via the Sec translocon complex.
Role of SecDF
The functional importance of SecDF in protein translocation as well as membrane protein biogenesis was previously shown in vivo. A SecDF-deficient Escherichia coli strain is severely defective in protein export at any temperature and cold sensitive for growth. SecDF is proposed to facilitate the preprotein release to the periplasmic side of the membrane in the later stage of translocation. Since the exact role of SecDF in protein translocation has not yet been defined, we examined whether SecDF functions in the steps subsequent to the initiation of translocation, using an in vitro reaction system21. As a result, the initiation of translocation required SecA ATPase, whereas the later steps occurred even in the absence of ATP, and were dependent on the presence of both SecDF and PMF. These results show that SecDF is involved in the PMF-driven progression and completion of protein translocation.
Architecture of SecDF (F form and I form)
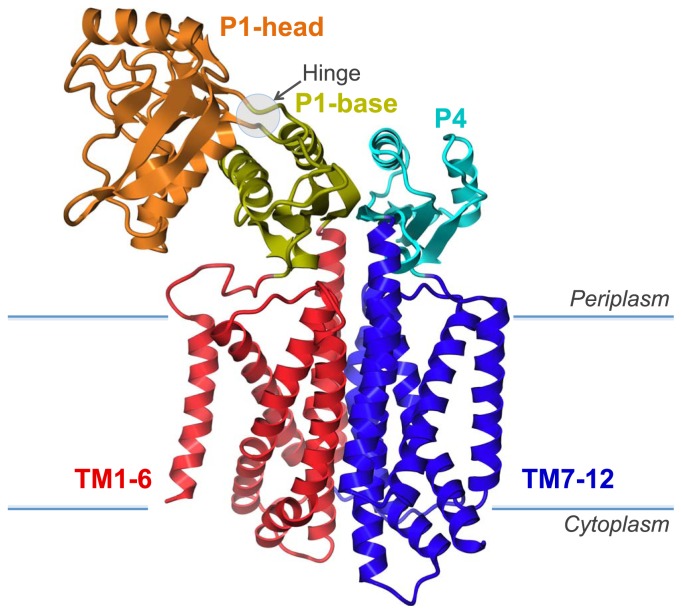
The crystal structure of full-length SecDF, consisting of a 12-helix transmembrane domain and two protruding periplasmic domains, the P1 and P4 domains, was determined at 3.3Å resolution by a single-wavelength anomalous dispersion analysis (Fig. 2; PDB ID 3AQP). TM1–6 and TM7–12 are assembled pseudo-symmetrically. The periplasmic surface of the TM regions is covered by an anti-parallel β-sheet, which is formed by eight-strands of the P1 and P4 domains. The P1 domain is divided into head and base subdomains. The P4 domain is structurally homologous to the P1 base subdomain, and both are assembled in a pseudosymmetrical manner similar to that of the TM region. The characteristic P1 head domain protrudes into the periplasmic space, while the two loops connecting the head and base subdomains form a constricted region.
Figure 2.
Crystal structure of SecDF. SecDF consists of a pseudosymmetrical 12-helix transmembrane domain (TM) and two protruding periplasmic domains (P1 and P4).
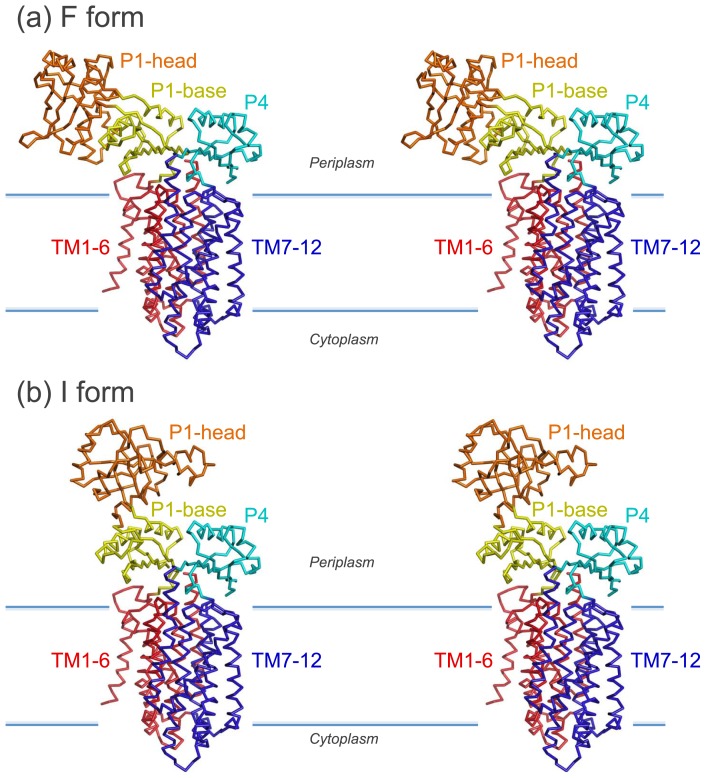
A comparison of the full-length crystal structure (called the F form; Fig. 3a) with the model of the SecDF structure, which was built by superimposing the base subdomain of the isolated P1 structure (PDB ID 3AQO) onto that of the full-length SecDF (called the I form; Fig. 3b), revealed a significant difference in the head-to-base orientation of the P1 domain. This conformational change is due to the ~120° rigid-body rotation of the head domain, which is attributed to a hinge motion at the connecting loops between the head and base domains.
Figure 3.
Conformational transition of SecDF. Stereo-views of the F form (a) and the I form (b).
In vivo and in vitro analyses using E. coli SecDF revealed that the head domain is functionally essential and that the conformational flexibility of the P1 domain is required for effective protein translocation. In addition, biochemical experiments suggested that the flexible P1 domain directly interacts with unfolded proteins, such as preproteins. Taken together, the conformational changes induced by preprotein-binding to the P1 domain could contribute to the enhancement of protein translocation.
SecDF driven by proton flow
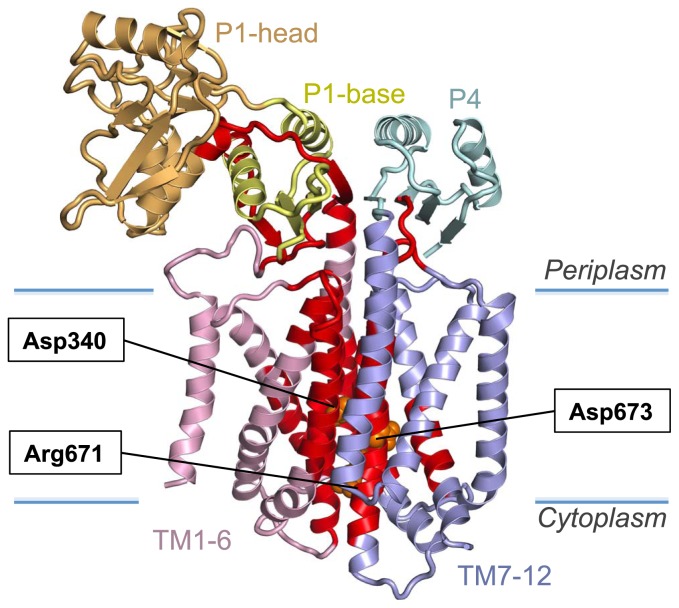
The TM arrangement of SecDF is similar to that of a member of the RND superfamily22, Multi-Drug Efflux Transporter AcrB23. Homotrimeric AcrB extrudes a variety of drugs by utilizing the proton gradient across the membrane, whereas SecDF functions as a monomer associated with SecYEG. We propose that the conserved Asp and Arg residues at the transmembrane interface between the TM1-6 and TM7-12 bundles (Fig. 4), in a similar manner to the conserved charged residues of AcrB24, play essential roles in the movements of protons and preproteins. The mutants in which the charged residues in E. coli SecDF were replaced by uncharged residues lacked the SecDF activity. This observation is consistent with the hypothesis that the charged residues in the TM region of SecDF participate in proton transport. Furthermore, function analysis of Vibrio alginolyticus SecDF demonstrated that a cation gradient across the membrane was required for the SecDF activity.
Figure 4.
Conserved residues of SecDF. The conserved regions of SecDF are shown in red. Functionally important, conserved residues are colored orange in a sphere representation.
To verify that SecDF conducts protons, we performed biophysical analyses. The SecDF-dependent channel activities were detected by patch clamp experiments. The ion-conduction was regulated by a proton gradient and by a binding of an unfolded protein to SecDF. We also confirmed SecDF-dependent proton import using a proton-specific fluorescence probe. The highly conserved SecDF regions25 are clustered from the center of the TM to the periplasmic base region underneath the head (Fig. 4). SecDF mutants lacking the flexible P1-head domain were defective in not only the enhancement of protein translocation but also the ion-conductance signals in the patch clamp assay. The hinge motion of the P1 domain is likely to be functionally related to the proton flow through the TM region of SecDF. Consequently, we conclude that the proton gradient across the membrane is the driving force for SecDF function.
A working model of SecDF-enhanced protein translocation
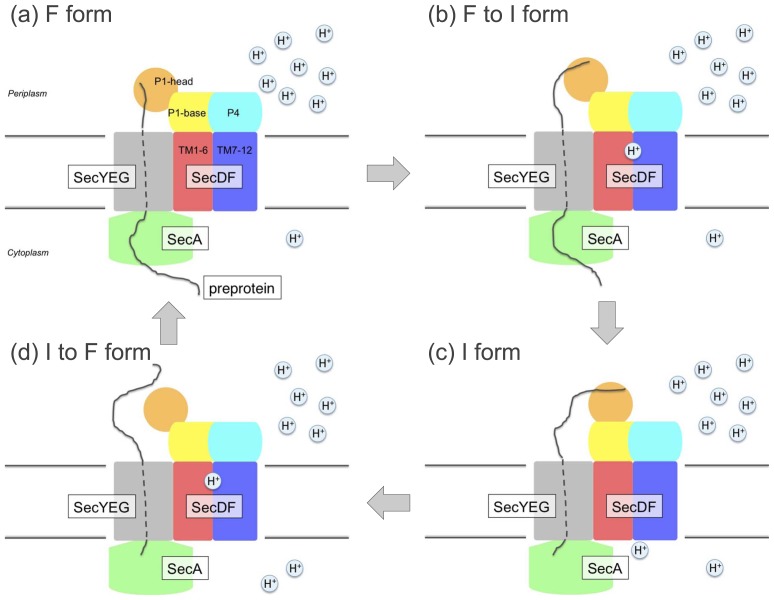
Based on its structural features and subsequent functional analyses, we verified the following SecDF functions: (1) The driving force for the achievement of ATP-independent protein translocation by SecDF is the PMF. (2) The TM region of SecDF participates in proton transport and contains several essential hydrophilic residues. (3) The P1 domain of SecDF, which interacts with an unfolded protein, undergoes functionally important structural alterations. Taken together, we propose a working model of SecDF-mediated enhancement of protein translocation (Fig. 5). SecYEG could reside just beneath the protruding head domain of SecDF (Fig. 5a), enabling it to capture a translocating preprotein via the translocon channel. The protein-capturing F form could then change to the I state, preventing the backward movement26 of the substrate and driving the forward movement (Fig. 5b). Thus, SecDF can actively assist with protein translocation. The conversion from the I to F form could occur after the bound protein is released from the I form (Fig. 5c, d). The reformed F form can bind to the more C-terminal part of the substrate. By repeating this conformational transition cycle coupled with proton flow, SecDF can enhance protein transport.
Figure 5.
Working model of PMF-driven protein translocation by SecDF. (a), F form, preprotein-capturing state. (b), F form to I form, preprotein-holding state (c), I form, preprotein-releasing state, (d) I form returned to F form.
Concluding remarks
In this study, we elucidated the mechanism of protein translocation enhancement by SecDF. However, further studies will be needed to define the exact proton pathway in SecDF and to reveal the mechanism of energy transduction, by which the proton flow-induced conformational transition in the TM region is converted into the motion of the P1 head, which presumably occurs within the conserved region of SecDF. Although the crystal structural information for all of the Sec proteins, Sec translocon, SecA ATPase and SecDF, is now available, it is unclear how the Sec proteins interact with each other and form the Sec translocon complex. Visualization of the dynamic Sec machinery will require further structural-based mechanistic studies as well as the full complex structure of the Sec machinery.
Acknowledgments
The original study was reported in collaboration with H. Mori, Y. Echizen, R. Ishitani, S. Fukai, T. Tanaka, A. Perederina, D.G. Vassylyev, T. Kohno, A. D. Maturana and K. Ito. This review was supported by JSPS and MEXT KAKENHI (22020008, 21770135, 22121503 and 20227003).
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.du Plessis DJ, Nouwen N, Driessen AJ. The Sec translocase. Biochim Biophys Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 4.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 7.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 8.Driessen AJ, Wickner W. Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc Natl Acad Sci USA. 1991;88:2471–2475. doi: 10.1073/pnas.88.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiozuka K, Tani K, Mizushima S, Tokuda H. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in. Escherichia coli J Biol Chem. 1990;265:18843–18847. [PubMed] [Google Scholar]
- 10.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci USA. 1996;93:5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagara K, Matsuyama S, Mizushima S. SecF stabilizes SecD and SecY, components of the protein translocation machinery of the Escherichia coli cytoplasmic membrane. J Bacteriol. 1994;176:4111–4116. doi: 10.1128/jb.176.13.4111-4116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hand NJ, Klein R, Laskewitz A, Pohlschroder M. Archaeal and bacterial SecD and SecF homologs exhibit striking structural and functional conservation. J Bacteriol. 2006;188:1251–1259. doi: 10.1128/JB.188.4.1251-1259.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 15.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:265–270. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arkowitz RA, Wickner W. SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J. 1994;13:954–963. doi: 10.1002/j.1460-2075.1994.tb06340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukazaki T, Mori H, Fukai S, Numata T, Perederina A, Adachi H, Matsumura H, Takano K, Murakami S, Inoue T, Mori Y, Sasaki T, Vassylyev DG, Nureki O, Ito K. Purification, crystallization and preliminary X-ray diffraction of SecDF, a translocon-associated membrane protein, from Thermus thermophilus. Acta Crystallogr F. 2006;62:376–380. doi: 10.1107/S1744309106007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Echizen Y, Tsukazaki T, Dohmae N, Ishitani R, Nureki O. Crystallization and preliminary X-ray diffraction of the first periplasmic domain of SecDF, a translocon-associated membrane protein, from Thermus thermophilus. Acta Crystallogr F. 2011;67:1367–1370. doi: 10.1107/S1744309111031885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:30862–30868. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 22.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 23.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 24.Seeger MA, von Ballmoos C, Verrey F, Pos KM. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry. 2009;48:5801–5812. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 25.Eichler J, Wickner W. The SecA subunit of Escherichia coli preprotein translocase is exposed to the periplasm. J Bacteriol. 1998;180:5776–5779. doi: 10.1128/jb.180.21.5776-5779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiebel E, Driessen AJ, Hartl FU, Wickner W. ΔμH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]