Abstract
The bacterial family Enterobacteriaceae is notable for its well studied human pathogens, including Salmonella, Yersinia, Shigella, and Escherichia spp. However, it also contains several plant pathogens. We report the genome sequence of a plant pathogenic enterobacterium, Erwinia carotovora subsp. atroseptica (Eca) strain SCRI1043, the causative agent of soft rot and blackleg potato diseases. Approximately 33% of Eca genes are not shared with sequenced enterobacterial human pathogens, including some predicted to facilitate unexpected metabolic traits, such as nitrogen fixation and opine catabolism. This proportion of genes also contains an overrepresentation of pathogenicity determinants, including possible horizontally acquired gene clusters for putative type IV secretion and polyketide phytotoxin synthesis. To investigate whether these gene clusters play a role in the disease process, an arrayed set of insertional mutants was generated, and mutations were identified. Plant bioassays showed that these mutants were significantly reduced in virulence, demonstrating both the presence of novel pathogenicity determinants in Eca, and the impact of functional genomics in expanding our understanding of phytopathogenicity in the Enterobacteriaceae.
The bacterial family Enterobacteriaceae includes many well studied human pathogens within genera such as Salmonella, Yersinia, and Shigella, as well as the model commensal and pathogenic species Escherichia coli, and the insect pathogen Photorhabdus luminescens. More genome sequences have been determined from this family than from any other (1–6). However, the Enterobacteriaceae also include several significant plant pathogens, and here we present the complete genome sequence of such a pathogen, Erwinia carotovora subsp. atroseptica (Eca).
Eca, also named Pectobacterium carotovorum subsp. atrosepticum (7), and recently, Pectobacterium atrosepticum (8), is a commercially important pathogen restricted to potato in temperate regions, where it causes blackleg in the field and soft rot of tubers after harvest. In contrast, the related soft rot bacteria Erwinia carotovora subsp. carotovora (Ecc) and Erwinia chrysanthemi (Ech) have broader host ranges, including potato, and cause disease in warmer climates (9). The soft rot erwiniae also live as epiphytes and endophytes on plants or saprophytes in soil and ground water (9, 10), although little is known about these nonpathogenic lifestyles. Moreover, the molecular bases of differences in host range and the requirements for establishing disease are largely unknown. Although soft rot pathogenesis relies primarily on the prolific production of extracellular plant cell wall-degrading enzymes (PCWDEs) that cause extensive tissue maceration (9, 11), discoveries in recent years suggest the process may be far more subtle and complex than previously thought (12).
Genome sequencing of the phytopathogens Pseudomonas syringae pv. tomato (13), Ralstonia solanacearum (14), Xanthomonas campestris pv. campestris and Xanthomonas axonopodis pv. citri (15), Xylella fastidiosa (16, 17), and Agrobacterium tumefaciens (18, 19), has yielded a wealth of information on novel and shared candidate phytopathogenicity determinants. Here, we describe the complete genome sequence of Eca strain SCRI1043 (Eca1043) and assess its similarity to other members of the Enterobacteriaceae and to other plant pathogenic bacteria. We describe a wide array of genes previously unknown in Eca and, in some cases, in the Enterobacteriaceae, which are potentially involved in pathogenicity and metabolism. In a functional genomic investigation, we provide evidence that genes in two identified clusters have a role in disease development.
Materials and Methods
Genome Sequencing and Annotation. The strain Eca SCRI1043 (American Type Culture Collection catalog no. BAA-672) was isolated from a potato stem with blackleg disease symptoms in 1985 in Perthshire, Scotland (the sample used to provide DNA for sequencing was frozen shortly after isolation). This strain has been used in epidemiological and molecular studies for many years and is amenable to genetic manipulation (20, 21). The initial shotgun was generated from 54,600 paired-end sequences from two libraries in pUC18 with insert sizes of 2.2–2.5 and 2.5–4.0 kb, and 25,800 paired-end reads from a 4.5- to 5.5-kb insert library in pMAQ1b by using dye terminator chemistry on ABI3700 automated sequencers, giving 8.2-fold coverage. Paired-end sequences from 500 clones from a 12- to 23-kb library in pBACe3.6 were used to generate a scaffold with 1.8-fold clone coverage. All identified repeats were bridged by using read pairs or end-sequenced PCR products.
The initial shotgun sequences were assembled by using phrap (www.phrap.org), and gap closure and finishing used GAP4 (http://staden.sourceforge.net) in combination with directed sequencing (primer walks and subcloning and shotgun sequencing of bacterial artificial chromosome clones or PCR products that spanned sequence gaps). The final sequence was composed of 106,500 reads, giving an average 10.2-fold coverage. The finished sequence was compared with DNA sequences in public databases by using fasta, blastx, and blastn (22). Potential coding sequences (CDSs) were predicted by a combination of orpheus (23) and glimmer (24) and manual curation, and were searched for pfam (25) domains, and compared with the protein databases by using blastp. artemis (26) was used to collate data and facilitate annotation. Stable noncoding RNAs were identified by comparison with the rfam database (27). Metabolic pathways were examined by using the kegg database (www.genome.ad.jp/kegg).
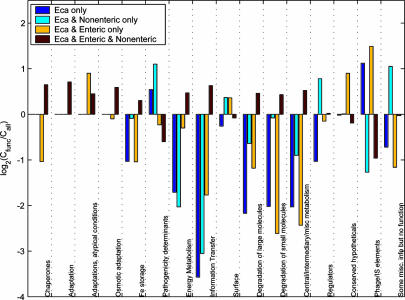
Comparative Genomics. Each CDS from the Eca1043 genome was used as the query in a fasta protein search against the complete set of CDSs from each of 64 fully annotated genomes obtained from the National Center for Biotechnology Information web site (ftp.ncbi.nih.gov/genomes/Bacteria; Fig. 1, and Table 2, which is published as supporting information on the PNAS web site). Where the resulting top hit (ranked by score; hits with equivalent scores treated equally) was aligned with the query over at least 80% of its length to >30% identity, it was used as the query in a search against the Eca genome. Where the original CDS from Eca1043 was found to be the top match to this second query, the pair of sequences was termed a reciprocal best hit (RBH). For each genome, results for chromosomal and extrachromosomal sequences were obtained independently, and were then concatenated for further analyses. The number of CDSs in each of four groups as follows: (i) Eca only, (ii) shared with nonenterobacteria only, (iii) shared only with other enterobacteria, and (iv) shared with enterobacteria and nonenterobacteria, was counted for each functional class (Cfunc; see EMBL/GenBank accession no. BX950851 for feature class qualifiers) and for all CDSs, normalized to each functional class (Call). The value log2(Cfunc/Call) was plotted to illustrate over- or under-representation of each group (Fig. 2).
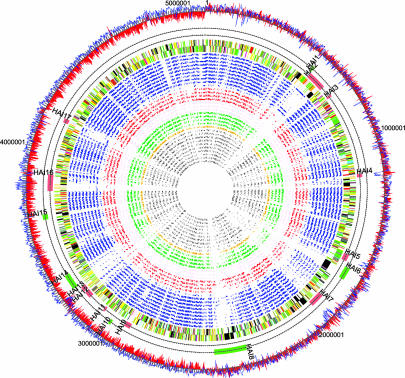
Fig. 1.
Comparison of the Eca genome sequence with other bacterial genomes. Inner to outer tracks show the locations of RBHs found by reciprocal fasta of Eca CDSs against those from 32 bacterial genomes (circular plot): Gram-positive (gray); Shewanella oneidensis (ochre); nonenteric animal pathogens (green); plant-associated bacteria (brown); nonenteric plant pathogens (red); and enterobacteria (blue) (Table 2). The locations of CDSs on the Eca genome colored by functional class (see legend to Fig. 5). Two tracks indicating HAIs listed in Table 1. Shown are islands with evidence of recent acquisition (red bars) and possible islands based on reciprocal FASTA analysis (green bars). A plot of G+C skew (red) and percent G+C content (blue).
Fig. 2.
Ratio of observed to expected CDSs by functional class and bacterial group distribution. The log2(Cfunc/Call) ratio of CDSs from Eca strain SCRI1043 shared with no other species (i, dark blue), nonenteric only (ii, cyan) enteric only (iii, yellow), or enteric and nonenteric species (iv, red), by functional class.
Transposon Mutation Grid. Donor (E. coli S17–1[λpir] carrying the Tn5 cassette [mTn5-gusA-pgfp21] in the pUT vector; ref. 28) and recipient (Eca1043) strains were mated and transconjugants recovered on M9Kan. A total of 9,216 transconjugants were grown in 24 384-well microtiter plates (M9Kan broth plus 4% glycerol), constituting the mutation grid. Clones from each of the 24 384-well plates were pooled and DNA extracted for PCR screening by using primers designed to anneal to the gene of interest and the end of the transposon cassette. Having identified a plate pool containing a mutant, PCR from each clone within that plate was conducted by using a 384-well PCR machine.
Primers used to identify mutants were: VIRB4F 5′-CATTACCTCAGTTATGGCTATTACAGGGC-3′ (virB4); PKSF3 5′-CGGATCGGTGGTTCCGTC-3′ (cfa6); PKSR1 5′-GGAGCACTATGCTTCCTGCAC-3′ (cfa7); and Tn5R 5′-TATCCTCCTTAGCTAGTCAGG-3′ (mTn5-gusA-pgfp).
The purified PCR products were sequenced in both directions to confirm the location of the transposon. The mutations were transduced into a clean genetic background before analysis (20).
Pathogenicity Test. Eca1043-susceptible potato plants cv. Estima were grown in compost to a height of ≈20 cm. Eca cells were grown to log phase (overnight) in LB broth, and cells were harvested by centrifugation at 4,000 × g for 10 min, washed in PBS, and resuspended in the same volume of PBS. Two dilutions (104 and 108 cells per ml–1) in PBS were prepared from these suspensions, and potato plant stems were inoculated under the second fully expanded leaf by using a micropipette tip with 10 μl of bacterial suspension (equivalent to 102 and 106 cells per inoculation site). The inoculation site was then covered with parafilm to avoid desiccation. At least 12 replicas for each strain and each inoculum level were used. The inoculated plants were kept in the glasshouse under controlled temperature conditions (≈22°C) and high humidity and the presence and length (millimeters) of necrotic lesions was recorded daily for 14 days. genstat for Windows, version 6.1.0.200 (Lawes Agricultural Trust), was used to determine the least significant difference (LSD; P < 0.05) defined as the smallest difference between any two means that can be described as being significantly different. LSD was derived from the residual mean square of the analysis of variance from means averaged across all days.
Results and Discussion
Genome Composition. The genome of Eca1043 is a single circular chromosome of 5,064,019 bp with a G + C content of 50.97%, 4,491 predicted CDSs, seven rRNA operons, 76 tRNAs, and 25 predicted stable noncoding RNAs. There are 183 bacteriophage genes, including two complete prophages with similarity to phage sequences in other enterobacteria and, in one case, in other plant pathogens (Table 1). Fifty-two of the 4,491 CDSs are pseudogenes (including 22 phage or transposase genes). This figure is similar to the established enterobacteria such as E. coli CFT073 (62 CDSs) and Salmonella enterica serovar Typhimurium (39 CDSs), but is relatively low compared with the recently diverged enterobacteria such as S. enterica serovar Typhi (204 CDSs) (3) and Yersinia pestis (149 CDSs) (4). The positions of 11 putative horizontally acquired genomic islands (HAIs) having some of the hallmarks of recent gene transfer events, such as atypical percent G+C, location at or near tRNA genes, and the presence of pathogenicity and/or phage/plasmid genes, were determined (Fig. 1 and Table 1).
Table 1. Putative HAIs and their phenotypic traits.
| CDS identifiers | Island no. | Acquisition evidence | Putative phenotype(s) |
|---|---|---|---|
| ECA0499-0510 | HAI1 | fasta analysis* | Capsular polysaccharide biosynthesis |
| ECA0516-0614 | HAI2 | Integrase and tRNA | Polyketide phytotoxin biosynthesis (cfa) |
| Agglutination/adhesion | |||
| ECA0665-0678 | HAI3 | Phage genes | |
| ECA1054-1067 | HAI4 | Integrase | |
| Low-percent GC region | |||
| ECA1416-1443 | HAI5 | Low-percent GC region | Exopolysaccharide and O-antigen biosynthesis |
| ECA1466-1488 | HAI6 | fasta analysis* | Nonribosomal peptide phytotoxin |
| ECA1598-1679 | HAI7 | Integrase and tRNA | Type IV secretion (virB) |
| Low-percent GC region | Integrated plasmid | ||
| Agglutination/adhesion | |||
| ECA2045-2182 | HAI8 | fasta analysis* | Type III secretion (hrp) |
| Agglutination/adhesion (hecAB) | |||
| ECA2598-2637 | HAI9 | Phage genes | P2 family prophage |
| ECA2694-2705 | HAI10 | fasta analysis* | Phenazine antibiotic biosynthesis (ehp) |
| ECA2750-2759 | HAI11 | Phage genes | |
| Integrase and tRNA | |||
| ECA2850-2879 | HAI12 | Integrase and tRNA | Rhs and its accessory element VgrG |
| ECA2889-2921 | HAI13 | Integrase and tRNA | Putative integrated plasmid |
| Low-percent GC region | |||
| ECA2936-3000 | HAI14 | fasta analysis* | Nitrogen fixation (nif) |
| ECA3262-3270 | HAI15 | fasta analysis* | Agglutination/adhesion (aggA) |
| ECA3378-3460 | HAI16 | Integrase and tRNA | |
| ECA3695-3742 | HAI17 | Phage genes | Prophage |
Absence in other enteric bacteria determined by reciprocal fasta analysis.
Comparative Genomics and Evolution. Reciprocal fasta analysis of the EcaCDS set was performed against 64 published bacterial genome sequences, including 15 enterobacteria, and all available plant pathogenic and plant-associated bacteria (Fig. 1, Table 2, and for a linear version of the diagram see Fig. 5, which is published as supporting information on the PNAS web site). The largest numbers of RBHs were with other enterobacteria, and 2,982 (66.4%) CDSs have RBHs in at least one member of this family. This analysis helped to confirm the locations of the 11 HAIs (see above). Moreover, from regions of the genome lacking RBHs to other enterobacterial genomes, six additional candidate HAIs, possibly representing older acquisition events, were identified (Fig. 1 and Table 1).
The full set of RBHs was used to determine CDSs in Eca1043 that were shared with (i) no other genomes, (ii) nonenterobacterial genomes only, (iii) enterobacterial genomes only, or (iv) both enterobacterial and nonenterobacterial genomes. The distribution of CDSs among groups i–iv by major functional class, relative to the overall distribution of CDSs across the groups, is shown in Fig. 2. As expected, CDSs involved in core bacterial functions, such as energy metabolism, had a greater proportion of RBHs in group iv. The distribution of pathogenicity determinants was of particular interest and this analysis focused on CDSs with a probable direct involvement in pathogenesis, e.g., those coding for toxins, type III secretion system (T3SS) proteins, PCWDEs, etc. (148 CDSs; Table 3, which is published as supporting information on the PNAS web site). CDSs encoding pathogenicity determinants comprised a greater proportion of RBHs in groups i and ii. Because many of these CDSs lie within predicted HAIs, this finding may indicate the impact of horizontal gene transfer on the evolution of Eca as a plant pathogen.
To investigate further the possible acquisition of pathogenicity determinants from nonenterobacterial genomes (represented by group ii) CDSs from Eca making RBHs only with nonenterobacterial genomes were identified (Fig. 6, which is published as supporting information on the PNAS web site). This finding revealed a number of interesting relationships, most striking of which were the RBHs for putative pathogenicity determinants shared between Eca and the phytopathogen P. syringae pv. tomato DC3000 (referred to as P. syringae throughout the text). These relationships correspond to two P. syringae pathogenicity gene clusters: the hrp (T3SS) gene cluster (HAI8) (also shared with Pseudomonas aeruginosa), and the cfa gene cluster (within HAI2) involved in phytotoxin synthesis (29). Several organisms, including the plant-associated Pseudomonas putida and three animal pathogenic Vibrio species, share genes from a cluster (HAI15) that includes aggA, which is involved in adherence to plant roots in P. putida (30). Other RBHs include avrXca, which is involved in avirulence in X. campestris pv. raphani (31), and VirB/type IV secretion genes (HAI7) in A. tumefaciens (32); the latter also being present in X. axonopodis pv. citri and Sinorhizobium meliloti. Finally, plant pathogens, plant-associated bacteria, and more distantly related bacteria, including Bacillus and Streptomyces spp., make RBHs to CDSs encoding several PCWDEs. These data provide evidence for horizontal transfer of putative pathogenicity determinants (HAIs 2, 7, 8, and 15) during the evolution of Eca1043.
Survival in Diverse Environments. The gene content of Eca1043 suggests that all major metabolic pathways are intact, and that it has the ability to use a range of different nutrients for adaptation to diverse environments. There are 36 putative methyl-accepting chemotaxis protein genes, more than in other sequenced enterobacterial genomes, e.g., 11 in S. enterica serovar Typhi and 10 in Yersinia enterocolitica, or in most other sequenced phytopathogens, e.g., 20 in X. campestris pv. campestris, 22 in X. axonopodis pv. citri, and 20 in R. solanacearum (www.genome.ad.jp/kegg). Similarly, 80 (2% of CDSs) putative ABC transport uptake systems were identified, compared with only 44 (1% of CDSs) in E. coli K12. The genome also contains 336 (7% of CDSs) putative regulators, compared with 178 (4% of CDSs) in E. coli K12 (1). Many regulators may have been acquired through horizontal gene transfer (Fig. 2), and a number are also directly implicated in virulence (Table 3). Interestingly, other phytopathogens also possess large numbers of putative regulators (8% and 12% of CDSs in R. solanacearum and P. syringae, respectively), which may reflect similarities in lifestyle. These findings suggest that Eca1043 is able to respond to a wide range of nutrient sources, or other environmental cues, in keeping with its variety of lifestyles and habitats.
Eca1043 appears to have unexpected metabolic traits of potential advantage for survival in the soil or rhizosphere, including the potential to fix nitrogen (nif genes, HAI14) and catabolize opines (e.g., occQMPJ). Such traits have not been recognized in the soft rot erwiniae, and are absent in other sequenced enterobacterial genomes, but are important in other bacteria for colonization of plants (33–35). In addition, HAI10 carries genes similar to those for phenazine antibiotic production in the closely related plant pathogen Pantoea agglomerans (ehp genes) (36), which may provide a competitive advantage in soil or on the host.
Pathogenicity Determinants. The Eca1043 genome carries many factors previously implicated in the pathogenesis of soft rot erwiniae (12), but many more putative pathogenicity genes novel to this group of pathogens have now been revealed. A total of 393 CDSs with a possible involvement in pathogenicity, including the 148 CDSs used in the comparative genomic analyses and other CDSs related to processes such as adhesion, iron acquisition, and motility, are listed in Table 3, and key findings are summarized below.
The main virulence determinants in Eca are PCWDEs that cause extensive tissue maceration during the latter stages of infection (11). Eca1043 has 20 putative pectinase genes, 11 of which were previously unknown in Eca. Two of these genes are most similar to genes from A. tumefaciens and Bacillus spp. Seven CDSs may encode other PCWDEs, including cellulases, four of which are novel to the erwiniae. In addition to known Ecc regulators that control the PCWDEs and other virulence factors (Table 3), the genome contains other candidate virulence regulators. The LuxR homologue ECA1561 encodes a putative novel cognate binding protein of N-acyl homoserine lactones. Homologues of some of the previously identified quorum sensing-related genes are present in the genome, including expI, encoding the N-acyl homoserine lactone synthetase, and the contiguous putative regulator expR. Strain SCRI1043 does not make a carbapenem antibiotic (G.P.C.S., unpublished work) and the corresponding biosynthetic genes are absent. It therefore seems unlikely that ECA1561 encodes a functional homologue of CarR, the LuxR-type regulator of carbapenem biosynthesis previously identified in some Ecc strains. The function of the cryptic ECA1561 is currently unknown but one possibility would be a role as a “missing link” in the quorum-sensing control of PCWDEs (11).
In Ech, PCWDE production and full pathogenicity are linked to production of the siderophores chrysobactin and achromobactin, which scavenge iron from the iron-poor environment of the host (37, 38). Although Ecc strains produce chrysobactin and aerobactin siderophores, they have no obvious role in potato disease, suggesting that other iron acquisition systems may be involved (39). Little is known about iron acquisition in Eca. However, Eca1043 has genes for the achromobactin uptake and transport system (cbrABCD) and enterobactin synthesis (entAFBECD). The latter has not previously been identified in the erwiniae but is a well characterized siderophore in E. coli and Salmonella spp (40). Two additional putative siderophore-independent systems similar to ferric citrate uptake in E. coli (fecIRABCD), and to the HasA heme-binding protein and its cognate secretion system in Serratia marcesens (41), are also identified in the erwiniae. Genes involved in hemin storage (hmsHFRS) are also present, although a second operon containing hmsT present in Y. pestis is absent in Eca1043 (42).
Several Ech genes involved in defense against the plant oxidative burst or antimicrobial peptides are present in Eca1043 (12). However, there are additional putative defenses against active oxygen species: catalase peroxidase (katB), paraquat-inducible genes (pqiAB), a broad specificity flavoprotein reductase (similar to P. putida xenA; ref. 43), and peroxide resistance genes (similar to X. campestris pv. phaseoli ahpC, ohr, and ohrR; refs. 44 and 45).
Eca1043 is the only enterobacterium sequenced to date that possesses all six known Gram-negative protein secretion systems. To our knowledge, the only other bacterium identified with representatives of all these systems is another plant pathogen, X. axonopodis pv. citri (15). The known type I and II systems, which secrete PCWDEs and other pathogenicity determinants (11), are present together with types III, IV, and V (autotransporter), and two-partner secretion and additional putative type I systems (Table 3). Also revealed in the genome are the twin arginine-targeting pathway tat genes, which offer an alternative route to sec-dependant export for folded proteins to reach the periplasm. Such proteins include virulence factors in some species (46, 47), although no Eca proteins have yet been identified that use this system.
A complete T3SS, recently identified in Eca SCRI1039 (48), and carrying putative effector, helper, and avirulence genes similar to those in Erwinia amylovora, Ecc, and Ech (12), is also present in Eca1043 (HAI8), but carries eight additional CDSs absent in these closely related pathogens. Elsewhere in the genome there are T3SS-associated genes resembling srfABC in Salmonella spp. SrfC is similar to HopPtoL in P. syringae, encoding putative T3SS-delivered effector proteins (49, 50). In contrast to the broad host range phytopathogens P. syringae and R. solanacearum (13, 14, 51), large numbers of candidate type III (Hrp) effectors have not so far been identified in Eca1043. If confirmed, this finding may reflect the narrow host range of Eca and/or its lack of a “race structure;” i.e., strains that are incompatible on specific potato host genotypes have not been found.
A surprise finding was a gene cluster within HAI7 that resembles both a plasmid conjugation system in E. coli and the pathogenicity-related type IV secretion system (T4SS) locus in A. tumefaciens (32) Although this may simply be an integrated plasmid, and not directly related to pathogenicity, recent discoveries show that conjugation machinery can also deliver virulence proteins (52), and that similar loci are present in two other phytopathogens (15, 16). We thus investigated its possible role in virulence (see below).
The Eca1043 genome has a number of genes associated with agglutination and cell attachment, similar to those in both plant and animal pathogens and plant-associated bacteria. These genes include two putative two-partner secretion systems (e.g., hecAB and HAI8) found in all sequenced necrogenic plant pathogens, which may offer a common mechanism of host attachment (13, 53, 54). Another cluster (HAI15) includes genes encoding a large hemagglutinin-like protein (4,588 aa) and a putative adhesion protein (aggA; ref. 55). Other CDSs (e.g., within HAI2) appear to encode type IV pili and other fimbrial proteins similar to those in other enterobacteria and plant-associated bacteria, which in plant pathogens such as R. solanacearum are involved in adhesion to the host plant (56). Eca1043 also has CDSs similar to polysaccharide biosynthesis genes (HAI1) that more closely resemble those of distantly related (often plant-associated) bacteria than those of other enterobacteria. Flagellar motility is also recognized as contributing to the pathogenicity of Eca (57) and a large number of genes similar to those in other enterobacteria are devoted to this function.
Additional CDSs potentially encoding secreted pathogenicity proteins include those that resemble eukaryotic genes (putative lipases and a carbonic anhydrase), an avirulence gene from X. campestris pv. raphani (avrXca; ref. 31), and a necrosis-inducing gene from Fusarium oxysporum (nep1; ref. 58). The latter two are required for full virulence of Eca1043 (G.P.C.S., unpublished work).
The largest genes in the Eca1043 genome (HAI6) appear to encode a nonribosomal peptide (NRP) synthetase system. These genes are similar to that producing the P. syringae pv. syringae phytotoxin syringomycin (syrE), which induces necrosis by forming transmembrane pores and causing electrolyte leakage (29). Interestingly, these genes are not present in P. syringae pv. tomato DC3000 (13), and NRP phytotoxins have not previously been identified in enterobacterial plant pathogens. However, similar genes have been found in R. solanacearum (14) and X. axonopodis pv. citri (15).
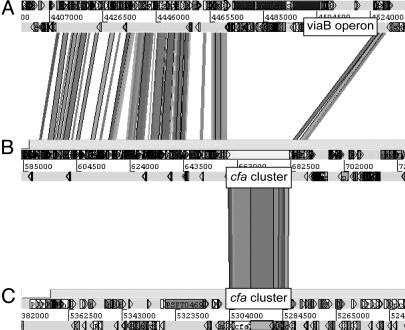
HAI2 shows a high level of conservation of both sequence and gene order to the SPI-7 pathogenicity island in S. enterica serovar Typhi (Fig. 3). The presence of similar islands in distantly related bacteria, such as P. aeruginosa and X. axonopodis pv. citri, has led to the suggestion that this island is a mobile element (59). However, the viaB operon in SPI-7, encoding the S. enterica serovar Typhi Vi exopolysaccharide pathogenicity determinant, is absent in Eca1043 (Fig. 3). This finding is perhaps unsurprising because SPI-7 has a mosaic structure across S. enterica isolates and may have arisen as a result of several independent insertion events (59). Interestingly, at an equivalent location to the viaB operon in SPI-7, HAI2 contains genes highly similar to the cfa gene cluster in P. syringae (Fig. 3). The cfa genes in P. syringae encode synthetases of the polyketide component (coronafacic acid) of the coronatine phytotoxin, a molecular mimic in plant-signaling pathways (29). However, genes encoding synthetases of the second component of coronatine, coronamic acid, are absent. The Eca1043 cfa-like cluster and S. enterica viaB operon could be “islands within an island,” and their similar locations in SPI-7-like loci may indicate a recombination “hot spot.” Polyketide phytotoxins have not previously been identified in enterobacterial plant pathogens but are important pathogenicity determinants in P. syringae. This cluster was thus investigated for its role in virulence (see below).
Fig. 3.
Screen shot adapted from Artemis Comparison Tool (www.sanger.ac.uk/Software/ACT). (A–C) RBHs between the SPI-7-like regions of S. enterica serovar Typhi (A) and Eca SCRI1043 (B), and the genomic region containing the cfa gene cluster of P. syringae pv. tomato (C). The locations of the cfa gene cluster and the viaB operon are indicated. Shaded lines between the genomes represent RBHs.
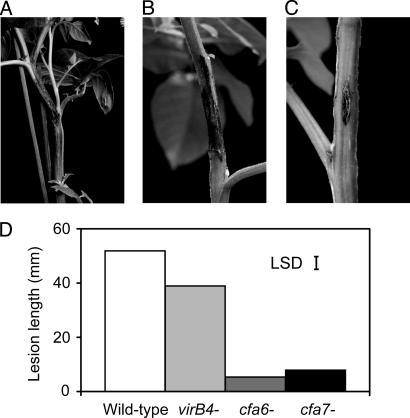
Functional Genomics. As two major findings from the Eca1043 genome sequence and comparative genomics study, the putative T4SS and cfa-like polyketide phytotoxin clusters were selected to investigate their role in virulence. For this purpose, an arrayed set of transposon insertion mutants (mutation grid) was constructed. By PCR screening of pooled clones from the grid, mutants carrying insertions within these clusters (ECA1620, virB4, T4SS, and ECA0602–0603, cfa6, and cfa7) were isolated, and were inoculated into potato stems. The virB4 mutant showed a statistically significant reduction in virulence (P < 0.05), whereas virulence was greatly reduced in the cfa6 and cfa7 mutants (P < 0.05; Fig. 4). Although only a preliminary indication in the absence of other experiments, such as measuring bacterial growth, the reduced virulence in the virB4 mutant suggests that the putative T4SS does appear to be more than simply integrated conjugation machinery and may have been recruited for delivery of virulence factors. Moreover, although only one of the two gene clusters required for coronatine synthesis in P. syringae is present in Eca1043, this cluster appears to play a major role in disease development, possibly through the production of an alternative polyketide phytotoxin.
Fig. 4.
Mutations in putative type IV secretion and polyketide phytotoxin synthesis genes affect disease development. Lesion development on potato plant stem over a 14-day period after inoculation of Eca strain SCRI1043 wild-type and mutant strains at 104 cells per inoculation site. (A) Wild type. (B) Mutant Eca1043 virB4::Tn5. (C) Mutant Eca1043 cfa6::Tn5. (D) Bar graph showing length of rot (millimeters) for wild-type and mutants Eca1043virB4::Tn5, Eca1043cfa6::Tn5, and Eca1043cfa7::Tn5. LSD = LSD of means across all days (value 4.82).
Conclusions. Here, we report the complete genome sequence of a plant pathogen from the Enterobacteriaceae, a family noted for its human pathogens. The sequence has revealed a metabolic plasticity that fits a lifestyle adapted to different environments in and around the plant host. It contains all of the known Gram-negative protein targeting systems and many PCWDEs, which is consistent with a lifestyle as a “rotter,” whereas factors such as the T3SS imply more subtle interactions with its host. However, the genome has also revealed putative pathogenicity determinants novel to Eca1043, to other soft rot erwiniae and, in many cases, to the enterobacteria. Many of these determinants lie within putative HAIs and resemble genes in other plant pathogens, suggesting horizontal exchange of genomic islands during the evolution of plant pathogens. Finally, we carried out functional analyses on two gene clusters in what appear to be recently acquired islands in Eca1043, providing evidence that a T4SS and a polyketide phytotoxin act as virulence determinants in nontumorogenic and non-Pseudomonas phytopathogens, respectively. A wider functional genomics program used to identify all components contributing to the pathogenic lifestyle of this important bacterial plant pathogen is warranted for future study.
Supplementary Material
Acknowledgments
We thank the core sequencing and informatics teams at the Sanger Institute for their assistance and the Wellcome Trust for its support of the Sanger Institute Pathogen Sequencing Unit. This work was supported by the Scottish Executive Environment and Rural Affairs Department and the Biotechnology and Biological Sciences Research Council. M.C.H. was funded by a North Atlantic Treaty Organization studentship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Eca, Erwinia carotovora subsp. atroseptica; Ecc, Erwinia carotovora subsp. carotovora; Ech, Erwinia chrysanthemi; CDS, coding sequence; HAI, horizontally acquired genomic island; RBH, reciprocal best hit; PCWDE, plant cell wall-degrading enzyme; T3SS, type III secretion system; T4SS, type IV secretion system; LSD, least significant difference.
Data deposition: The sequence reported in this paper has been deposited in the EMBL/GenBank database (accession no. BX950851).
References
- 1.Blattner, F. R., Plunkett G., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., ColladoVides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 2.Perna, N. T., Plunkett, G., Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A, et al. (2001) Nature 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 3.Parkhill, J., Dougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., Churcher, C., Mungall, K. L., Bentley, S. D., Holden, M. T., et al. (2001) Nature 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 4.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentise, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523–527. [DOI] [PubMed] [Google Scholar]
- 5.Welch, R. A., Burland, V., Plunkett, G., Redford, P., Roesch, P., Rasko, D., Buckles, E. L., Liou, S. R., Boutin, A., Hackett, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., Bocs, S., Boursaux-Eude, C., Chandler, M., Charles, J. F., et al. (2003) Nat. Biotechnol. 21, 1307–1313. [DOI] [PubMed] [Google Scholar]
- 7.Hauben, L., Moore, E. R., Vauterin, L., Steenackers, M., Mergaert, J., Verdonck, L. & Swings, J. (1998) Syst. Appl. Microbiol. 21, 384–397. [DOI] [PubMed] [Google Scholar]
- 8.Gardan, L., Gouy, C., Christen, R. & Samson, R. (2003) Int. J. Syst. Evol. Microbiol. 53, 381–391. [DOI] [PubMed] [Google Scholar]
- 9.Perombelon, M. C. M. (2002) Plant Pathol. 51, 1–12. [Google Scholar]
- 10.Perombelon, M. C. M. & Kelman, A. (1980) Annu. Rev. Phytopathol. 18, 361–387. [Google Scholar]
- 11.Py, B., Barras, F., Harris, S., Robson, N. & Salmond, G. P. C. (1998) Methods Microbiol. 27, 157–168. [Google Scholar]
- 12.Toth, I. K., Bell, K. S., Holeva, M. C. & Birch, P. R. J. (2003) Mol. Plant Pathol. 4, 17–30. [DOI] [PubMed] [Google Scholar]
- 13.Buell, C. R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I. T., Gwinn, M. L., Dodson, R. J., Deboy, R. T., Durkin, A. S., Kolonay, J. F., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., Billault, A., Brottier, P., Camus, J. C., Cattolico, L., et al. (2002) Nature 415, 497–502. [DOI] [PubMed] [Google Scholar]
- 15.da Silva, A. C. R., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M. C., et al. (2002) Nature 417, 459–463. [DOI] [PubMed] [Google Scholar]
- 16.Simpson, A. J., Reinach, F. C., Arruda, P., Abreu, F. A., Acencio, M., Alvarenga, R., Alves, L. M., Araya, J. E., Baia, G. S., Baptista, C. S., et al. (2000) Nature 406, 151–157. [DOI] [PubMed] [Google Scholar]
- 17.Van Sluys, M. A., de Oliveira, M. C., Monteiro-Vitorello, C. B., Miyaki, C. Y., Furlan, L. R., Camargo, L. E., de Silva, A. C., Moon, D. H., Takita, M. A., Lemos, E. G., et al. (2003) J. Bacteriol. 185, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodner, B., Hinkle, G., Gattung, S., Miller, N., Blanchard, M., Qurollo, B., Goldman, B. S., Cao, Y., Askenazi, M., Halling, C., et al. (2001) Science 294, 2323–2328. [DOI] [PubMed] [Google Scholar]
- 19.Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., Zhou, Y., Chen, L., Wood, G. E., Almeira, N. F., et al. (2001) Science 294, 2317–2323. [DOI] [PubMed] [Google Scholar]
- 20.Toth, I. K., Mulholland, V., Cooper, V., Bentley, S., Shih, Y.-L., Perombelon, M. C. M. & Salmond, G. P. C. (1997) Microbiology 143, 2433–2438. [DOI] [PubMed] [Google Scholar]
- 21.Hinton, J. C. D., Perombelon, M. C. M. & Salmond, G. P. C. (1985) J. Bacteriol. 161, 786–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 23.Frishman, D., Mironov, A., Mewes, H. W. & Gelfand, M. (1998) Nucleic Acids Res. 26, 2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M., Sonnhammer, E. L., et al. (2002) Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A. & Eddy, S. R. (2003) Nucleic Acids Res. 31, 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi, C., Lambrecht, M., Vanderleyden, J. & Michiels, J. (1999) J. Microbiol. Methods 35, 85–92. [DOI] [PubMed] [Google Scholar]
- 29.Bender, C. L., Alarcon-Chaidez, F. & Gross, D.C. (1999) Microbiol. Mol. Biol. Rev. 63, 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buell, C. R. & Anderson, A. J. (1992) Mol. Plant–Microbe Interact. 5, 154–162. [DOI] [PubMed] [Google Scholar]
- 31.Parker, J. E., Barber, C. E., Fan, M. J. & Daniels, M. J. (1993) Mol. Plant–Microbe Interact. 6, 216–224. [DOI] [PubMed] [Google Scholar]
- 32.Christie, P. J. (2001) Mol. Microbiol. 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz, R. A., Klopprogge, K. & Grabbe, R. (2002) J. Mol. Microbiol. Biotechnol. 4, 235–242. [PubMed] [Google Scholar]
- 34.Steibl, H. D., Siddavattam, D. & Klingmuller, W. (1995) Plasmid 34, 223–228. [DOI] [PubMed] [Google Scholar]
- 35.Valdivia, R. H., Wang, L. & Winans, S. C. (1991) J. Bacteriol. 173, 6398–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giddens, S. R., Feng, Y. J. & Mahanty, H. K. (2002) Mol. Microbiol. 45, 769–783. [DOI] [PubMed] [Google Scholar]
- 37.Expert, D. (1999) Annu. Rev. Phytopathol. 37, 307–334. [DOI] [PubMed] [Google Scholar]
- 38.Franza, T., Michaud-Soret, I., Piquerel, P. & Expert, D. (2002) Mol. Plant–Microbe Interact. 15, 1181–1191. [DOI] [PubMed] [Google Scholar]
- 39.Bull, C. T., Carnegie, S. R. & Loper, J. E. (1996) Phytopathology 86, 260–266. [Google Scholar]
- 40.Raymond, K. N., Dertz, E. A. & Kim, S. S. (2003) Proc. Natl. Acad. Sci. USA 100, 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letoffe, S., Ghigo, J. M. & Wandersman, C. (1994) J. Bacteriol. 176, 5372–5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones, H. A., Lillard, J. W. & Perry, R. D. (1999) Microbiology 145, 2117–2128. [DOI] [PubMed] [Google Scholar]
- 43.Blehert, D. S., Fox, B. G. & Chambliss, G. H. (1999) J. Bacteriol. 181, 6254–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panmanee, W., Vattanaviboon, P., Eiamphungporn, W., Whangsuk, W., Sallabhan, R. & Mongkolsuk, R. (2002) Mol. Microbiol. 45, 1647–1654. [DOI] [PubMed] [Google Scholar]
- 45.Vattanaviboon, P., Whangsuk, W., Panmanee, W., Klomsiri, C., Dharmsthiti, S. & Mongkolsuk, R. (2002) Biochem. Biophys. Res. Commun. 299, 177–182. [DOI] [PubMed] [Google Scholar]
- 46.Dilks, K., Rose, R.W., Hartmann, E. & Pohlschroder, M. (2003) J. Bacteriol. 185, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding, Z. Y. & Christie, P. J. (2003) J. Bacteriol. 185, 760–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell, K. S., Avrova, A. O., Holeva, M. C., De Jong, W., Toth, I. K., Waugh, R., Bryan, G. & Birch, P. R. J. (2002) Microbiology 148, 1367–1378. [DOI] [PubMed] [Google Scholar]
- 49.Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., Schechter, L. M., Janes, M. D., Buell, C. R., Tang, X., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worley, M. J., Ching, K. H. L. & Heffron, F. (2000) Mol. Microbiol. 36, 749–761. [DOI] [PubMed] [Google Scholar]
- 51.Zwiesler-Vollick, J., Plovanich-Jones, A. E., Nomura, K., Bandyopadhyay, S., Joardar, V., Kunkel, B. N. & He, S. Y. (2002) Mol. Microbiol. 45, 1207–1218. [DOI] [PubMed] [Google Scholar]
- 52.Seubert, A., Hiestand, R., de la Cruz, F. & Dehio, C. (2003) Mol. Microbiol. 49, 1253–1266. [DOI] [PubMed] [Google Scholar]
- 53.Rojas, C. M., Ham, J. H., Deng, W. L., Doyle, J. J. & Collmer, A. (2002) Proc. Natl. Acad. Sci. USA 99, 13142–13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Sluys, M. A., Monteiro-Vitorello, C. B., Camargo, L. E., Menck, C. F., de Silva, A. C., Ferro, J. A., Oliveiro, M. C., Setubal, J. C., Kitajima, J. P., Simpson, A. J., et al. (2002) Annu. Rev. Phytopathol. 40, 169–189. [DOI] [PubMed] [Google Scholar]
- 55.Buell, C. R., Whetton, R., Tari, P. & Anderson, A. J. (1993) Can. J. Microbiol. 39, 787–794. [DOI] [PubMed] [Google Scholar]
- 56.Kang, Y. W., Liu, H. L., Genin, S., Schell, M. A. & Denny, T. P. (2002) Mol. Microbiol. 46, 427–437. [DOI] [PubMed] [Google Scholar]
- 57.Mulholland, V., Hinton, J., Sidebottom, J. M., Toth, I. K., Hyman, L. J., Perombelon, M. C. M., Reeves, P. J. & Salmond, G. P. C., et al. (1993) Mol. Microbiol. 9, 343–356. [DOI] [PubMed] [Google Scholar]
- 58.Bailey, B. A. (1995) Phytopathology 85, 1250–1255. [Google Scholar]
- 59.Pickard, D., Wain, J., Baker, S., Line, A., Chohan, S., Fookes, M., Barron, A., Goara, P. O., Chabalgoity, J. A., Thanky, N., et al. (2003) J. Bacteriol. 185, 5055–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.