Abstract
Respiratory syncytial virus (RSV) is one of the major pathogens responsible for lower respiratory tract infections (LRTI) in young children, the elderly, and the immunosuppressed. Currently, there are no antiviral drugs or vaccines available that effectively target RSV infections, proving a significant challenge in regards to prevention and treatment. An in-depth understanding of the host-virus interactions that underlie assembly and budding would inform new targets for antiviral development.
Current research suggests that the polymerised form of actin, the filamentous or F-actin, plays a role in RSV assembly and budding. Treatment with cytochalasin D, which disrupts F-actin, has been shown to inhibit virus release. In addition, the actin cytoskeleton has been shown to interact with the RSV matrix (M) protein, which plays a central role in RSV assembly. For this reason, the interaction between these two components is hypothesised to facilitate the movement of viral components in the cytoplasm and to the budding site. Despite increases in our knowledge of RSV assembly and budding, M-actin interactions are not well understood. In this review, we discuss the current literature on the role of actin cytoskeleton during assembly and budding of RSV with the aim to integrate disparate studies to build a hypothetical model of the various molecular interactions between actin and RSV M protein that facilitate RSV assembly and budding.
Keywords: Cytoskeleton, Actin, Microfilaments, Molecular motors, Virus assembly, Virus budding, Matrix protein
Background
Respiratory syncytial virus (RSV) is the major cause of lower respiratory tract disease in infants and young children [1] and a major viral agent responsible for respiratory tract disease in immunosuppressed individuals and the elderly [1]. In 2009, the World Health Organisation estimated that globally there are 64 million annual RSV cases leading to 160,000 deaths (http://apps.who.int/vaccine_research/diseases/ari/en/index2.html). Antiviral drugs and vaccines that effectively target RSV infections are currently unavailable, representing a significant challenge in regards to RSV disease prevention and treatment [2]. Virus infection is initiated via inoculation of the nose or eyes which can occur through direct contact or inhalation of airborne infectious particles. This results in viral replication in the nasopharynx over 4–5 days, the virus occasionally spreads to the lower respiratory tract over the next few days [3].
RSV is a member of the Pneumoviridae family, which consists of two genera, Metapneumovirius and Orthopneumovirus, with RSV belonging to the latter [4]. The virion is pleomorphic, being either spherical or filamentous [5]. Similar to the closely related Paramyxoviruses, the negative sense RNA genome of RSV is tightly encapsidated within the nucleocapsid, which is composed of nucleocapsid protein N, the RNA polymerase L and its cofactor phosphoprotein P, as well as the M2-1 protein; external to the nucleocapsid is a layer of matrix (M) protein which acts as a bridge between the nucleocapsid and the lipid bilayer envelope. Embedded in the envelope are the fusion (F), large (G) and small hydrophobic (SH) glycoproteins. M2-2 and two non-structural proteins NS1 and NS2 are not found in the virion in any significant amount but have important roles in the RSV replication cycle [5–12]. Figure 1 shows the genome organisation of prototypic members of the Paramyxovirus and Pneumovirus families.
Fig. 1.
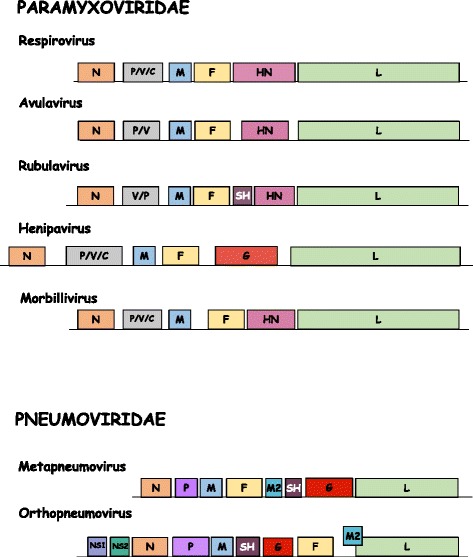
Genome organisation of Paramyxovirus and Pneumovirus genera. The nucleoprotein (N), phosphoprotein (P), matrix (M) protein, fusion (F) protein, and RNA-dependent RNA polymerase (L), are conserved in this order among the viruses belonging to both families. The attachment protein (H, HN, or G), differs amongst the viruses depending on haemagglutinin (H) presence, haemagglutinin-neuraminidase (HN) activity, or neither H present or HN activity (G). Only three genera contain the small hydrophobic SH protein, while both genera belonging to the Pneumovirinae subfamily (Pneumovirus and Metapneumovirus) also possess M2, NS1, and NS2 genes. However, gene positions vary between the Pneumovirus and Metapneumovirus genera
The mechanisms that drive assembly and budding differ among the viruses and thus, no particular mechanism can be assumed for all members. However, in all cases, the M protein drives coordinated interplay between viral structural components to facilitate assembly of infectious progeny. For this reason, M protein is considered as the key organizer of virus assembly [13]. Not only does the M protein interact with viral glycoproteins and the nucleocapsid, it also interacts with the cytoskeleton in infected cells [14], and bundles of actin microfilaments in epithelial cells and fibroblasts are altered during infection. Additionally, similar to members of Paramyxoviridae family, RSV has been shown to require actin or tubulin or both for effective transcriptase activity in vitro [15, 16] underscoring the importance of host cell cytoskeleton in Orthopneumovirus lifecycle.
Despite at least two decades of sporadic research investigating cytoskeleton involvement in RSV infection, the role of cytoskeleton in RSV assembly and/or budding is still unclear. In this review we bring together our current knowledge and understanding of the role of the cytoskeleton in RSV infection, drawing on literature on such interactions in other related viruses; the aim being to integrate diverse studies in order to form a rational informed model of RSV-cytoskeleton interactions.
Virus life cycle
The infectious cycle of RSV follows a similar route to the paramyxovirus lifecycle, however, with several differences, which are detailed in following sections. The paramyxovirus life cycle begins upon attachment of the virus to the host cell and fusion of the viral membrane to the host cell membrane facilitated by the two major glycoproteins, the attachment protein (H, HN, or G depending on the virus; see Fig. 1) and the F protein (Fig. 1). Membrane fusion follows and precedes the release of the viral genome into the cytoplasm [17]. Upon release, the viral genome undergoes primary transcription, followed by translation and replication to form new genomes. The newly synthesised viral genomes are associated with the N protein and wrapped around a nucleocapsid core containing L and P proteins [18]. This process forms the ribonucleoprotein complex (RNPs), which are then transported to the plasma membrane where interaction with the membrane glycoprotein complex takes place, facilitated by the M protein. At the plasma membrane, the RNPs, the M protein, and the glycoproteins form the paramyxovirus particles, which are then released through membrane scission [13].
RSV entry
Of the three RSV envelope glycoproteins, only F is essential for attachment and entry although G deleted strains are highly attenuated in vivo [19]. Glycosaminoglycans (GAGs) are cell surface receptors implicated in RSV attachment in cell culture, as well for many other viruses [20, 21]. Interestingly, nucleolin has been shown to function as an entry receptor for RSV via F glycoprotein; non-permissive cells become susceptible to RSV infection when transfected to express human nucleolin [22]. As nucleolin is ubiquitously expressed, there is probably another receptor that determines tropism. Of direct relevance to this review is the fact that cell surface nucleolin mediates internalisation of ligands via its interaction with the actin cytoskeleton [23]. Entry of RSV genome has been described as F mediated fusion of viral envelope and host cell membrane (Fig. 2), though some evidence implicates clathrin mediated endocytosis in the entry process, independent of endosomal acidification [24, 25].
Fig. 2.
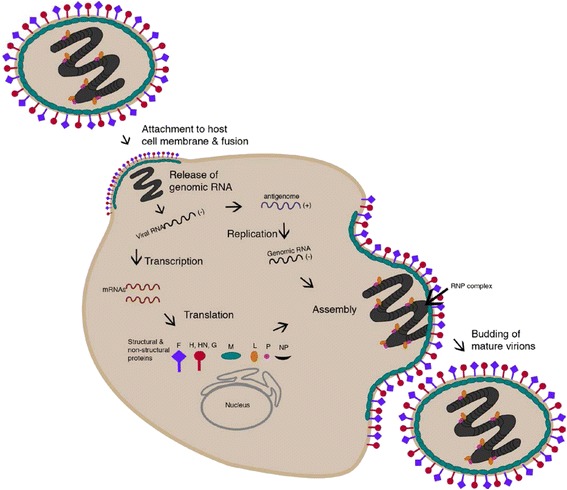
Paramyxovirus Lifecycle. Attachment to the host cell membrane initiates the viral lifecycle and entry occurs by the release of the viral genome into the cytoplasm. The negative-sense RNA genome undergoes primary transcription to produce mRNAs. The viral genome is replicated in a two-step process catalyzed by the viral polymerase (composed of phosphoprotein (P) and large (L) protein components) where antigenome intermediates are produced from genomic templates before the production of negative-sense genomes. The helical nucleocapsid is formed by association of the newly synthesised nucleoprotein (NP) with the nascent genomic RNA. This newly formed structure then interacts with the RNA-dependent RNA polymerase complex (RNPs). RNPs are transported to the plasma membrane by M protein where they interact with surface glycoproteins for assembly, before membrane scission and release of virus particles. The figure was generated using Adobe Illustrator
RSV transcription and translation
The minimum protein requirement for RSV transcription is N, P and L. In infected cells, these proteins are found in cytoplasmic inclusions, which also contain viral RNA of both polarities and are presumed to be sites of transcription [26]; recent studies suggest that inclusions are not structurally or functionally homogenous [27] being implicated in innate response to RSV infection. Transcription results in 10 capped, methylated and polyadenylated mRNAs which are translated by host cell machinery. Each gene is flanked by a gene-start (GS) and gene-end (GE) sequence [28, 29]. A promoter at the 3’ end of the genome acts as the only attachment site for the polymerase complex [5]. Genes are then transcribed sequentially from 3’ to 5’ end. However, due to polymerase drop off at intergenic regions, there is a gradient of expression, with genes at the 3’ end expressed at higher levels than genes at the 5’ end. Hence, proteins that the virus needs early and in greater numbers such as the interferon antagonists, non-structural proteins NS1 and NS2, are produced much more than proteins needed in small supply such as the polymerase L protein. The end of the M2 gene and start of the L gene overlap by 68 nucleotides. The polymerase complex overcomes this difficulty by backtracking to the start of the L gene after completing transcription of M2 [30]. This means that to complete transcription of L, the polymerase complex must “read through” the GE sequence of M2, something that it does not do in 90 % of cases [5]. The benefit of this added complexity, if any, is uncertain. The M2 gene also has the distinction of coding an additional protein via a second, slightly overlapping open reading frame. The M2-2 protein thus produced, has a non-structural function and is dispensable for RSV replication [31].
RSV replication
Replication of new negative sense genomes requires the production of a complete positive sense anti-genome as an intermediate. This is accomplished with the polymerase complex used for mRNA transcription but operating in “read through” mode where GS and GE signals are not recognised. The switch from transcription mode to replication mode is of great interest to researchers but little is known of this mechanism aside from M2-2 being implicated. In cells infected with RSV without M2-2, the level of viral mRNA was increased while genomic RNA was decreased [31].
RSV assembly
Glycoproteins are translated and traffic through the secretory pathway to the apical surface, while internal virion proteins remain in the cytoplasm. Generation of nascent RSV genomic RNA probably occurs in cytoplasmic inclusions. N and P form RNPs that associate with the M protein, then traffic to the apical cell surface where they meet the glycoproteins. At the cell surface, viral proteins assemble into viral filaments that contain both viral structural proteins and viral genomic RNA. These filaments are thought to assemble into infectious virion particles prior to separation from the cell surface, since they can contribute to cell-cell spread of the virus and morphologically resemble the filamentous form of virions seen in electron micrograph studies of virus produced in polarized cells [5].
Although significant advances have been made in our understanding of RSV assembly and elucidation of the interactions between M and other viral proteins, our current knowledge on how the RNPs and the envelope glycoprotein complexes move from their cytoplasmic location to the plasma membrane is still limited. Literature from work with other viruses suggests a possible role for the cytoskeleton in this movement. Specifically, proteins associated with the actin network are implicated in RSV budding [32–34].
RSV budding
As mentioned previously, RSV infection is restricted to the superficial cells of the respiratory epithelium. In keeping with this, budding in polarised epithelial cells occurs at the apical membrane [35, 36] and preferentially from lipid rafts [37, 38]. Viral budding occurs in a Vps-4, ESCRT pathway independent manner resulting in pleomorphic particles that can be spherical or filamentous [39]. Recent literature suggests that RSV proteins are the main drivers of the formation of filamentous virus, with cytoskeletal proteins playing a supporting role [40].
It is likely that most, if not all, viruses utilise the cytoskeleton in some way. The most compelling argument to support this is that while salts and gases are small enough to diffuse through the cytoplasm, larger entities such as mature virions and large subviral particles must utilise the cytoskeleton transport system for active transport [41]. Support for this idea comes from work that examines the viscosity and diffusion properties of the cytoplasm. It was found that the diffusion of hydrophilic, electroneutral beads of 80 nm in radius was between 500 and 1000 fold lower in the cytoplasm than in aqueous solution [42]. Data from the same study has been used to estimate that vaccinia virus would take 5 h to diffuse a mere 10 μm in the cytosol [43]; however vaccinia virus movement from the juxtanuclear region to the periphery is significantly faster (>2 μm/s).
In the following sections we provide a brief overview of the cytoskeleton, followed by examples of its exploitation by chosen viruses; finally, we present current literature on the role of cytoskeleton in RSV assembly and budding and possible interactions with M protein.
The cytoskeleton
The cytoskeleton is composed of three different types of protein filaments which in order of smallest to largest are (filamentous) actin, intermediate filaments and microtubules. Apart from being the scaffolding that gives the cell its shape and strength, the cytoskeleton has other important functions. Not only does the cytoskeleton play a role in positioning and holding organelles in place, it also allows for cell locomotion. In addition, events such as cytokinesis and the movement of chromosomes during anaphase rely on the cytoskeleton and its associated molecular motors [44].
Actin
Actin is the most abundant cytoskeletal protein in eukaryotes and typically accounts for 5 to 10 % of total protein [44]. Actin monomers (G-actin) assemble into filaments (F-actin) with a distinct polarity; they have a fast growing barbed end and a slow growing pointed end [45]. The implication of this polarity is that when used as a track for intracellular transport, motor proteins only carry cargo in a specific direction. Actin filaments can be arranged into a variety of linear bundles and two-dimensional arrays, or three-dimensional gels [46]. Numerous actin binding proteins regulate the polymerisation and organisation of F-actin and it is these proteins that are sometimes the target of viral manipulation [32]. Although actin is present throughout the cytoplasm it is most abundant at the periphery of the cell which is where polar actin networks such as filopodia, lamellipodia and pseudopodia reside [47]. The peripheral actin network is sometimes referred to as the actin cortex. The actin cortex is generally arranged with plus ends facing outward and is the key player in cell motility which is based on filament assembly and the action of myosins [48]. The actin cortex has been proposed to be an obstacle to viral entry [49] and this might be why some viruses use the endocytic pathway which easily crosses the actin cortex [46]. Regulated binding to actin filaments can control the localisation of viral proteins such as the nucleoprotein of influenza virus [50].
Microtubules
Microtubules are the largest of the cytoskeleton filaments and like actin filaments they display a distinct polarity as well as a fast growing end and a slow growing end. Microtubules are composed of dimers of α-tubulin and β-tubulin arranged head to tail lengthwise and 13 side by side around a hollow core [44]. Most of the microtubules in a cell radiate outward from a microtubule organising centre (MTOC), also called a centrosome which is usually located near the nucleus. Microtubules extend from the MTOC with plus ends facing outward. Different cell types such as polarised epithelial cells and neurons can have specialised microtubule networks. Microtubules in axons are longitudinally arranged with plus ends pointing toward the axon terminal and minus end pointing toward the soma [51] while in polarised epithelial cells microtubules are arranged with plus ends pointing toward basal membrane and minus ends pointing toward the apical membrane [46]. The main role of microtubules in viral infection is to provide the tracks that the motor proteins kinesin and dynein use for long range transport of viral proteins, nucleocapsids and enveloped virions. This role has been described previously in many reviews [41, 52, 53], and will be described briefly in this review.
Intermediate filaments
Intermediate filaments have several important differences to F-actin and microtubules. They exhibit no treadmilling behaviour and have no polarity [44]. The general role of intermediate filaments is more of a structural support role and to provide scaffolding for the localisation of cellular processes, especially the nucleus, around which intermediate filaments form a ring. They are composed of over 65 different proteins enabling intermediate filaments to be turned to a variety of different purposes [44]. Intermediate filaments are known to be disassembled or rearranged by viral proteases [54, 55]. This may serve the virus by removing a barrier to free diffusion or it may weaken the cell enough to enhance cell lysis and release of viral progeny.
Molecular motors
Motor proteins use ATP hydrolysis to power their movement along cytoskeleton filaments. If the motor protein is fixed, it moves the filament and if the motor protein is not fixed, it moves cargo along the filament. Myosins transport cargo along actin filaments, whereas kinesins and dynein move along microtubules. There are 18 classes of myosin, grouped according to homologous sequences in the tail domain which determines cargo specificity. The most well-known and best studied is myosin II, the myosin responsible for muscle contraction. Most classes of myosin are plus end directed motors with the exception of myosin VI and possibly IXb, which are minus end directed [46]. Kinesins and dyneins transport membrane vesicles, macromolecules, organelles and viral particles/proteins toward the plus end and minus end, respectively, of microtubules.
The cytoskeleton in virus assembly
The host cell cytoskeleton plays an important role in assembly and budding of several viruses across different families.
Vaccinia virus
Vaccinia virus is a large double stranded DNA virus that exploits actin and tubulin cytoskeletons to ensure that its components are at the right place at the right time for optimal infectious virus production. After entry, viral cores use microtubules to move centrally where viral gene expression and formation of intracellular mature virus (IMV) occur [56]. The IMVs now use microtubules to move from viral factories to areas near the MTOC where they acquire an envelope from the trans-Golgi network or early endosomes and become intracellular enveloped virions (IEV) [57]. IEV is the intermediate between IMV and cell-associated enveloped virions (CEV). After acquiring the envelope, IEVs again use microtubules but this time in the opposite direction as they travel toward the periphery where they fuse with the plasma membrane and are exposed on the outside of the plasma membrane as CEV [58]. The next stage of the vaccinia lifecycle illustrates an impressive subversion of the actin cytoskeleton by the virus. CEV are able to induce the polymerisation of actin, creating protrusions known as actin tails that facilitate cell to cell spread by pushing the exposed CEV into adjacent cells [59].
Poliovirus
The positive sense RNA virus Poliovirus exploits the actin cytoskeleton during infection; after binding, poliovirus is internalized through an actin dependent pathway followed by rapid release of the RNA [60]. Interestingly, poliovirus was shown to move rapidly in the cytoplasm with unusually high speeds, independent of microtubules, but both ATP- and actin-dependent, suggesting an active transport mechanism. However, the viral trajectory and speed could not be accounted for by transport along microfilaments and a model wherein poliovirus induces changes in the activity of motor proteins was proposed [61].
Herpesvirus
Neurotropic viruses such as herpes simplex viruses (HSV) need to travel long distances within neurons. HSV takes control over the cytoskeleton early in infection by remodelling the cortical actin cytoskeleton [62]. The purpose of this remodelling may be to prepare the cell for efficient exit of virus later in infection [63]. Viral capsids interact directly with dynein [64], in order to travel the long distance to reach the nucleus. After entering the nucleus, HSV genomes are transcribed, replicated and packaged into new capsids. HSV induces the formation of actin in the nucleus even in cells where there is normally no nuclear actin [65]. Newly formed capsids recruit myosin for active transport along these actin filaments to the periphery of the nucleus [66]. Several HSV proteins have been shown to induce conformational changes in the nuclear lamina that may facilitate nuclear egress [67, 68]. Virions then travel back down the axon to the site of exit via microtubule transport through the recruitment of kinesin [52].
Cytoskeleton in paramyxovirus, pneumovirus life cycle
Entry
The host actin cytoskeleton plays an important role in the life cycle of paramyxoviruses [15, 69, 70]. The actin network creates a physical barrier against the invading virus, which must be overcome to enable productive infection [71]. This is made possible by the remodelling of the actin skeleton that is stimulated during paramyxovirus (and RSV) infection. Bundles of microfilaments are established through the formation of stress fibres [71] regulated by the Rho-family of GTPases, which play a role in controlling signal transduction pathways that generate changes in the actin cytoskeleton [71, 72] and are involved in influencing the assembly or disassembly of F-actin [73].
Assembly
Several paramyxoviruses, including Sendai (SeV) and measles virus (MeV), utilise vesicles that contain key regulators of microtubule transport such as Rab11A. The apical recycling endosome (ARE) pathway, that Rab11 endosomes are associated with, is important for the transport of proteins in polarized cells [13, 39, 74]. Although this notion suggests that Rab11A is required only in polarized cells, it has been demonstrated that the requirement is for both polarized and non-polarised cells with regards to SeV assembly [39]. In MeV infection, Rab11A is required only for virus production in polarized cells and not for RNP transport [75]. The presence of a dominant-negative form of the Rab11-interacting protein myosin Vb (MVb), results in reduced RSV assembly at the apical membrane. Of interest is Rab11-FIP2 (FIP2), another Rab11 interacting protein that may be involved in formation of filamentous virions [39]. Parainfluenza virus 3 (hPIV3) requires microfilaments for replication; however, its assembly is dependent on microtubules. This is in contrast to SeV and MeV where assembly depends on microfilaments as well as microtubules [75].
Budding
Cytoskeletal components are also involved in the budding of parvirus particles [13, 76, 77]. Actin involvement in budding was first discovered through studies undertaken on SeV and MeV particles, where large quantities of actin were detected in purified preparations of the two viruses. Studies on SeV have suggested that expression of M or F allows for the production of virus like particles (VLPs) that contain actin; both M and F proteins are required for budding. Significantly, sequences within M and F proteins resemble actin-binding domains [78, 79]. Mutations in the actin binding domains of SeV F result in significant reduction in VLP production suggesting that SeV F protein binding to actin is required for budding to take place [13].
Cytochalasin D, a specific inhibitor of actin polymerisation, inhibits release and reduces infectivity as well as entry of paramyxoviruses [71, 75]. As the microfilament network plays a role in the trafficking of RNP complexes, it is not surprising that virus release is affected by such drugs [13, 71]. However, treatment of hPIV3 infected A549 alveolar carcinoma lung cells with cytochalasin D had no effect on virus release. In contrast, microtubule depolymerisation drugs seemed to have more of a inhibitory effect on hPIV3 virus release [75]. Therefore, it is evident that each virus utilizes the cytoskeleton differently, however, the cytoskeleton does play an important role in replication, assembly, transportation, and budding of paramyxoviruses (Table 1).
Table 1.
Requirement of cytoskeletal components in paramyxovirus replication and assembly
| Virus | Cytoskeletal component | Virus lifecycle stage | Reference |
|---|---|---|---|
| Measles virus | Tubulin | Replication | [76, 95] |
| Microfilaments | Budding | ||
| Sendai virus | Actin | Transcription | [71, 76] |
| Tubulin | Transcription | ||
| Microfilaments | Assembly | ||
| Respiratory syncytial virus | Actin | Transcription | [33, 82, 84, 96, 97] |
| Tubulin | Transcription | ||
| Microfilaments | Entry, Assembly | ||
| Human Parainfluenza virus type 3, 5 | Actin | Transcription | [98, 99] |
| Microfilaments | Assembly |
Paramyxovirus M protein and cytoskeleton
The interaction of Newcastle disease virus (NDV) and SeV M proteins with actin filaments is important during virion maturation [70]. Expression of SeV M protein is essential for the actin restructuring that is observed during infection and virion release. MeV M-RNP complexes use the F-actin network to move to the plasma membrane; however, actin dynamics are required for viral budding [69].
Cytoskeleton in RSV infection
RSV is known to interact with the cytoskeleton in several ways and at various stages throughout its life cycle. In addition to transcriptional regulation, the cytoskeleton is also likely to be involved in the directional movement of RSV components. Live video microscopy showing filamentous virus budding from the same region of cell membrane at a rate of several virions per minute suggests directed transport [80]. The picture that has emerged so far seems to be that of the three cytoskeleton components, actin is the most important in RSV pathogenesis. However, the microtubule inhibitor nocodazole has been shown to reduce RSV titre by almost as much as cytochalasin D (9 fold reduction compared to 10 fold reduction) [81]. In the following sections, we present an overview of the known interactions, focussing on M and F proteins.
Transcription
Actin is present as an internal component in purified RSV virions [34] and antibodies against actin inhibit RSV transcription [33]. In a study using microarrays to analyse host gene expression in RSV infected cells, it was found that five genes associated with the cytoskeleton were upregulated shortly after infection [82]. There is evidence that RSV is indirectly involved with the cytoskeleton through interaction with actin associated proteins. Profilin, an actin-modulatory protein, has a significant role in RSV transcription [15, 32]. Although the RSV polymerase complex is able to mediate transcription in the absence of any cellular factors, actin is required for optimal activity. Current understanding of paramyxovirus transcription suggests that monomeric actin is required for transcriptase activity; the presence of actin in the virions likely reflects the involvement of actin in RSV transcription [75].
Assembly
The cytoskeleton has been suggested to play a role in transport of RNPs to RSV budding sites; RNPs display a myosin motor driven directional movement on the actin cytoskeleton [77]. Using live cell tracking of RNPs in cells infected with a recombinant RSV that expressed GFP, Santangelo and Bao [77] showed that RNPs move from the vicinity of filamentous virus at the apical surface and formed circular, inclusion-like structures in the cytoplasm on treatment with cytochalasin D. Importantly, myosin colocalised strongly with the virus filaments but not with cytoplasmic inclusion-like structures; suggesting myosin’s role in movement of RNPs to the budding site. Given the key role of M and actin in RSV assembly, it is important to understand the interactions between M and actin which underlie M’s role as facilitator and bringing together the RNP and envelope glycoproteins for virus assembly at the plasma membrane.
Budding
That microfilaments are required for optimal RSV budding and release is shown by the fact that virus release is inhibited by the microfilament destabilizing drug cytochalasin D, as mentioned previously [34, 75, 83].
In view of the connection between lipid rafts and the actin network [84], and that RSV buds from lipid raft domains of the plasma membrane [37, 85], myosin-driven transport likely also plays a role in RSV budding and release [77]. Increased F-actin is also observed at the lipid raft domain budding sites of human metapneumovirus [86], suggesting assembly/budding mechanism similar to RSV. Actin-modulatory proteins profilin and RhoGTPase are associated with RSV budding. As an actin-modulatory protein, profilin plays a role in promoting the formation and stabilization of stress fibres. In RSV infected cells, it has a role in virion morphogenesis and cell fusion in addition to enhancing the transcriptase activity [71]. RhoA GTPase has a variety of functions in the cell, including organisation of the actin cytoskeleton; blocking its action has a deleterious effect on RSV budding [87, 88] and RSV infection results in activation of RhoA, leading to rearrangement of the actin cytoskeleton. Interestingly, inhibition of RhoA leads to virus filaments adopting a more spherical particle morphology rather than a blunted one [89]. Phosphoinositide-3-kinase (PI3K) signalling, which has an important role in activation of F-actin associated signalling pathways that lead to structural changes in the F-actin network during infection, has a role in the organisation of RSV proteins at assembly sites [83]. In addition, PI3K activity may be involved in the formation of pneumovirus filaments that enable cell-to-cell viral spread [71, 89].
Although cytoskeletal components are clearly associated with RSV assembly and budding, studies undertaken on the effects of actin or tubulin disruption have suggested that RSV may assemble into filaments using a mechanism that does not depend on the host cytoskeleton [89].
RSV F protein
The F protein is essential for assembly and budding of RSV. The F-protein cytoplasmic tail (FCT) has been shown to be required for efficient replication as studies on viruses lacking this domain have effectively demonstrated a 100- to 1000-fold decrease in viral titres. RSV infection induces the formation of filaments, which have been shown to contain the viral proteins F, M, N, and P [89]. The formation of RSV filaments appears to depend on the FCT, especially a specific phenyalanine (Phe) residue at position 22 [13, 89, 90]. Interestingly, a mutation in Phe22 leads to the inability of RSV F to recruit viral proteins into VLPs and, thus, form filaments [89]. Therefore, the F protein plays a role in coordinating the assembly of the virus filaments at the cell surface [13, 89, 90].
The F protein is key to the formation of syncytia that are characteristic of RSV infection. F interacts directly with RhoA in vitro and inhibition of this interaction in infected cells results in abrogation of syncytia formation [39]; clearly, RhoA-F interaction drives cell to cell spread of RSV.
RSV M and F protein
The minimum RSV assembly complex contains F, M and the RNP core [91] and is formed in cytoplasmic inclusions [90]. Although cytoplasmic inclusions are characteristic of RSV infected cells, their role in infection is not completely understood [27, 83, 89]. It has been suggested that inclusions are sites of replication and/or transcription, sites for dead-end product accumulation, or sites for morphogenic intermediates [27]. RSV M protein associates with cytoplasmic inclusions late in infection through its interaction with the M2-1 protein and has been suggested to play a role in the switch between transcription and assembly [92, 93]. One study has effectively shown that both M and F protein association with cytoplasmic inclusions is critical in the assembly process, a step preceding filamentous virion formation, supporting the idea that viral filaments are formed or initiated from inclusions acting as scaffolds [90]. These associations with inclusions have been shown to be related to FCT-dependent changes, where increased amounts of F protein accumulated at the inclusions in the absence of FCT. In addition, the proportion of M associated with inclusions has been shown to be low when there is an abundance of filamentous (as opposed to spherical) virus. In turn, the M is increased in the cytoplasm in studies that involved the deletion of FCT, suggesting that the cytoplasmic M requires association with inclusions in order for incorporation into filaments [90].
F protein targets virus components to the lipid rafts, sites of virus budding [39, 89]. F and G glycoproteins as well as M are sorted into lipid rafts in RSV infected cells [38], with M forming structures suggesting an interaction with the cytoskeleton [89]. Inhibition of the Golgi secreting pathway prevents M being sorted into lipid rafts. M and F are co-localised to lipid rafts in cells co-expressing the two proteins and in RSV infected cells.
RSV M protein and actin
The M protein has been suggested to interact with actin for assembly and budding of RSV virions [94]. Both the M protein and the actin cytoskeleton have previously been described to individually play a role in the assembly of RSV. As mentioned, the M protein interacts with the RNP complex to inhibit the transcriptase activity probably in preparation for packaging [92]. As M also interacts with the host cytoskeleton, studies have suggested that there is an important role for this cellular component in RNP transport to the plasma membrane [77]. Current knowledge suggests that the M-containing complex is anchored onto the microfilament network that the nucleocapsid takes advantage of to reach RSV budding sites [89, 90]. The formation of filamentous virus is probably mediated through the interaction between RNPs, envelope glycoprotein complex and the plasma membrane by association of M with FCT [6, 37].
Conclusions: proposed model for the role of actin in RSV assembly and budding
Based on current literature on RSV as discussed above, it appears that the RNPs of RSV use microfilaments as a means of transport to reach assembly sites as shown for the closely related paramyxoviruses (Fig. 3); this movement may be mediated by M’s interaction with microfilaments, ensuring that only mature RNPs move to the plasma membrane. F protein probably also associates with the complex at this time mediated by M-FCT interaction to target the complex to the lipid rafts. At the cell surface assembly site, M-RNPs (+/− F) may associate with envelope glycoproteins via M’s self-oligomerising activity that allows it to bind to the M-F/G complexes (formed through M’s interactions with cytoplasmic tail of G, and F) forming the budding particle. FCT functions to engage RhoA, leading to extension of the virus filaments, followed by an as yet unknown scission mechanism that releases the RSV particles.
Fig. 3.
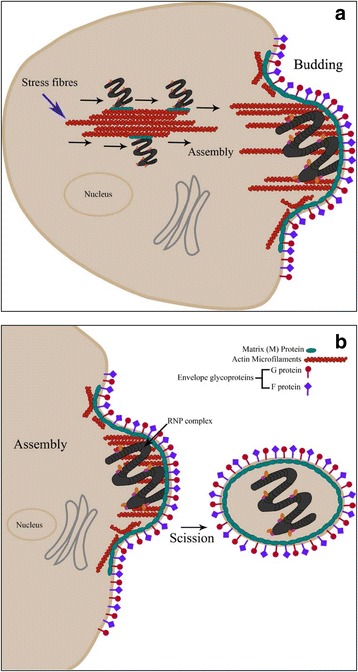
Model for RSV use of microfilaments in viral assembly. a The M-containing RNPs of RSV utilise microfilaments (stress fibres) as a means of transport to reach assembly sites; this movement is facilitated by the M protein interaction with microfilaments. At the assembly site, the RNPs associate with bundles of actin filaments and the envelope glycoproteins to complete viral assembly and initiate budding. b The growth of the viral filament is supported by microfilaments and actin-modulatory proteins; mature virions are released through as yet unknown mechanism of membrane scission
Acknowledgements
The authors thank Ms Melina Samimi for her expert assistance in in the production of the figures in this review.
Funding
Not applicable.
Availability of data and material
Not applicable.
Authors’ contributions
SS and RG wrote the majority of the manuscript, JG wrote the section on exploitation of the cytoskeleton by different virus families, SS generated the figures. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ARE
Apical recycling endosome
- ATP
Adenosine triphosphate
- CEV
Cell-associated enveloped virions
- ESCRT
Endosomal sorting complexes required for transport
- F-actin
Filamentous actin
- FCT
F-protein cytoplasmic tail
- G-actin
Globular actin
- GAGs
Glycosaminoglycans
- GFP
Green fluorescent protein
- hPIV3
Parainfluenza virus 3
- HSV
Herpes simplex virus
- IEV
Intracellular enveloped virions
- IMV
Intracellular mature virus
- LRTI
Lower respiratory tract infection
- MeV
Measles virus
- MTOC
Microtubule organizing centre
- MVb
Myosin Vb
- NDV
Newcastle disease virus
- PI3K
Phosphoinosulfide-3-kinase
- Rab11A
Ras-related protein
- Rab11A-FIP2
Rab11 family-interacting protein 2
- RNPs
Ribonucleoprotein complexes
- RSV
Respiratory syncytial virus
- SeV
Sendai virus
- VLPs
Virus-like particles
References
- 1.Malloy AM, Falsey AR, Ruckwardt TJ. Consequences of immature and senescent immune responses for infection with respiratory syncytial virus. Curr Top Microbiol Immunol. 2013;372:211–231. doi: 10.1007/978-3-642-38919-1_11. [DOI] [PubMed] [Google Scholar]
- 2.Turner T, Kopp B, Paul G, Hayes D, Jr, Thompson R, Landgrave L. Respiratory syncytial virus: current and emerging treatment options. Clinicoecon Outcomes Res. 2014;6:217. doi: 10.2147/CEOR.S60710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins PL, Graham BS. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol. 2008;82:2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, et al. Taxonomy of the order Mononegavirales: update 2016. Arch Virol. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins PL, Crowe JEJ: Respiratory syncytial virus and metapneumovirus. 2007:1601–1646.
- 6.Fontana JM, Bankamp B, Rota PA. Inhibition of interferon induction and signaling by paramyxoviruses. Immunol Rev. 2008;225:46–67. doi: 10.1111/j.1600-065X.2008.00669.x. [DOI] [PubMed] [Google Scholar]
- 7.Wu W, Tran KC, Teng MN, Heesom KJ, Matthews DA, Barr JN, Hiscox JA. The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J Virol. 2012;86:7777–7789. doi: 10.1128/JVI.00460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyapalle S, Wong T, Garay J, Teng M, San Juan-Vergara H, Mohapatra S, Mohapatra S. Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection. PLoS One. 2012;7:e29386. doi: 10.1371/journal.pone.0029386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teng MN, Whitehead SS, Bermingham A, St Claire M, Elkins WR, Murphy BR, Collins PL. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J Virol. 2000;74:9317–9321. doi: 10.1128/JVI.74.19.9317-9321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling Z, Tran KC, Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol. 2009;83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng MN, Collins PL. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J Virol. 1999;73:466–473. doi: 10.1128/jvi.73.1.466-473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Najjar F, Schmitt AP, Dutch RE. Paramyxovirus glycoprotein incorporation, assembly and budding: A three way dance for infectious particle production. Viruses. 2014;6:3019–3054. doi: 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anomasiri WT, Tovell DR, Tyrrell DL. Paramyxovirus membrane protein enhances antibody production to new antigenic determinants in the actin molecule: a model for virus-induced autoimmunity. J Virol. 1990;64:3179–3184. doi: 10.1128/jvi.64.7.3179-3184.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke E, Mahoney NM, Almo SC, Barik S. Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J Virol. 2000;74:669–675. doi: 10.1128/JVI.74.2.669-675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyer SA, Baker SC, Lessard JL. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc Natl Acad Sci U S A. 1986;83:5405–5409. doi: 10.1073/pnas.83.15.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutch RE. Entry and fusion of emerging paramyxoviruses. PLoS Pathog. 2010;6:e1000881. doi: 10.1371/journal.ppat.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakker SE, Duquerroy S, Galloux M, Loney C, Conner E, Eleouet JF, Rey FA, Bhella D. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J Gen Virol. 2013;94:1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng MN, Whitehead SS, Collins PL. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology. 2001;289:283–296. doi: 10.1006/viro.2001.1138. [DOI] [PubMed] [Google Scholar]
- 20.Hallak LK, Kwilas SA, Peeples ME. Interaction between respiratory syncytial virus and glycosaminoglycans, including heparan sulfate. Methods Mol Biol (Clifton, NJ) 2007;379:15–34. doi: 10.1007/978-1-59745-393-6_2. [DOI] [PubMed] [Google Scholar]
- 21.Hallak LK, Spillmann D, Collins PL, Peeples ME. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol. 2000;74:10508–10513. doi: 10.1128/JVI.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17:1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 23.Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng JS, Krust B. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res. 2000;261:312–328. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- 24.Gutiérrez-Ortega A, Sánchez-Hernández C, Gómez-García B. Respiratory syncytial virus glycoproteins uptake occurs through clathrin-mediated endocytosis in a human epithelial cell line. Virol J. 2008;5:127. doi: 10.1186/1743-422X-5-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolokoltsov AA, Deniger D, Fleming EH, Roberts NJ, Karpilow JM, Davey RA. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J Virol. 2007;81:7786–7800. doi: 10.1128/JVI.02780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22 K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 27.Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE, Santangelo PJ. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol. 2012;86:8245–8258. doi: 10.1128/JVI.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins PL, Dickens LE, Buckler-White A, Olmsted RA, Spriggs MK, Camargo E, Coelingh KV. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci U S A. 1986;83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo L, Fearns R, Collins PL. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fearns R, Collins PL. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73:388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bermingham A, Collins PL. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc Natl Acad Sci U S A. 1999;96:11259–11264. doi: 10.1073/pnas.96.20.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitko V, Oldenburg A, Garmon NE, Barik S. Profilin is required for viral morphogenesis, syncytium formation, and cell-specific stress fiber induction by respiratory syncytial virus. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252:137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- 34.Ulloa L, Serra R, Asenjo A, Villanueva N. Interactions between cellular actin and human respiratory syncytial virus (HRSV) Virus Res. 1998;53:13–25. doi: 10.1016/S0168-1702(97)00121-4. [DOI] [PubMed] [Google Scholar]
- 35.Roberts SR, Compans RW, Wertz GW. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J Virol. 1995;69:2667–2673. doi: 10.1128/jvi.69.4.2667-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76:5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marty A, Meanger J, Mills J, Shields B, Ghildyal R. Association of matrix protein of respiratory syncytial virus with the host cell membrane of infected cells. Arch Virol. 2004;149:199–210. doi: 10.1007/s00705-003-0183-9. [DOI] [PubMed] [Google Scholar]
- 38.McCurdy LH, Graham BS. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J Virol. 2003;77:1747–1756. doi: 10.1128/JVI.77.3.1747-1756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utley TJ, Ducharme N, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci U S A. 2008;105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaikh FY, Utley TJ, Craven RE, Rogers MC, Lapierre LA, Goldenring JR, et al. Respiratory syncytial virus assembles into structured filamentous virion particles independently of host cytoskeleton and related proteins. PLoS ONE. 2012;7(7):e40826. [DOI] [PMC free article] [PubMed]
- 41.Ward BM. The taking of the cytoskeleton one two three: how viruses utilize the cytoskeleton during egress. Virology. 2011;411:244–250. doi: 10.1016/j.virol.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/S0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 43.Sodeik B. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 2000;8:465–472. doi: 10.1016/S0966-842X(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 44.Cooper GM, Hausman RE. The Cell: A Molecular Approach. 2009. [Google Scholar]
- 45.Wear MA, Schafer DA, Cooper JA. Actin dynamics: Assembly and disassembly of actin networks. Curr Biol. 2000;10:891–895. doi: 10.1016/S0960-9822(00)00845-9. [DOI] [PubMed] [Google Scholar]
- 46.Döhner K, Sodeik B. The role of the cytoskeleton during viral infection. In membrane trafficking in viral replication. Edited by Marsh M. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005:67–108. [DOI] [PubMed]
- 47.Cramer LP. Organization and polarity of actin filament networks in cells: implications for the mechanism of myosin-based cell motility. Biochem Soc Symp. 1999;65:173–205. [PubMed] [Google Scholar]
- 48.Carlier M-F, Wiesner S, Le Clainche C, Pantaloni D. Actin-based motility as a self-organized system: mechanism and reconstitution in vitro. C R Biol. 2003;326:161–170. doi: 10.1016/S1631-0691(03)00067-2. [DOI] [PubMed] [Google Scholar]
- 49.Marsh M, Bron R. SFV infection in CHO cells: cell-type specific restrictions to productive virus entry at the cell surface. J Cell Sci. 1997;11(Pt 1):95–103. doi: 10.1242/jcs.110.1.95. [DOI] [PubMed] [Google Scholar]
- 50.Digard P, Elton D, Bishop K, Medcalf E, Weeds A, Pope B. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J Virol. 1999;73:2222–2231. doi: 10.1128/jvi.73.3.2222-2231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldstein LSB, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 52.Döhner K, Nagel C-H, Sodeik B, Luby-Phelps K, Sodeik B, Ploubidou A, Way M, Smith GA, Enquist LW, Döhner K, et al. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 2005;13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 53.Greber UF, Way M. A superhighway to virus infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Chen PH, Ornelles DA, Shenk T. The adenovirus L3 23-kilodalton proteinase cleaves the amino-terminal head domain from cytokeratin 18 and disrupts the cytokeratin network of HeLa cells. J Virol. 1993;67:3507–3514. doi: 10.1128/jvi.67.6.3507-3514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferreira LR, Moussatché N, Moura Neto V. Rearrangement of intermediate filament network of BHK-21 cells infected with vaccinia virus. Arch Virol. 1994;138:273–285. doi: 10.1007/BF01379131. [DOI] [PubMed] [Google Scholar]
- 56.Smith GL, Vanderplasschen A, Law M. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol. 2002;83:2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- 57.Sanderson CM, Hollinshead M, Smith GL. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J Gen Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 58.Geada MM, Galindo I, Lorenzo MM, Perdiguero B, Blasco R. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J Gen Virol. 2001;82:2747–2760. doi: 10.1099/0022-1317-82-11-2747. [DOI] [PubMed] [Google Scholar]
- 59.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 60.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging Poliovirus Entry in Live Cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaughan JC, Brandenburg B, Hogle JM, Zhuang X. Rapid actin-dependent viral motility in live cells. Biophys J. 2009;97:1647–1656. doi: 10.1016/j.bpj.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyoshi J, Takai Y. Nectin and nectin-like molecules: biology and pathology. Am J Nephrol. 2007;27:590–604. doi: 10.1159/000108103. [DOI] [PubMed] [Google Scholar]
- 63.De Regge N, Nauwynck HJ, Geenen K, Krummenacher C, Cohen GH, Eisenberg RJ, Mettenleiter TC, Favoreel HW. α-Herpesvirus glycoprotein D interaction with sensory neurons triggers formation of varicosities that serve as virus exit sites. J Cell Biol. 2006;174:267–275. doi: 10.1083/jcb.200510156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dohner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feierbach B, Piccinotti S, Bisher M, Denk W, Enquist LW. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2006;2:e85. doi: 10.1371/journal.ppat.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forest T, Barnard S, Baines JD. Active intranuclear movement of herpesvirus capsids. Nat Cell Biol. 2005;7:429–431. doi: 10.1038/ncb1243. [DOI] [PubMed] [Google Scholar]
- 67.Bjerke SL, Roller RJ. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology. 2006;347:261–276. doi: 10.1016/j.virol.2005.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reynolds AE, Liang L, Baines JD. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J Virol. 2004;78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dietzel E, Kolesnikova L, Maisner A. Actin filaments disruption and stabilization affect measles virus maturation by different mechanisms. Virol J. 2013;10:249. doi: 10.1186/1743-422X-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miazza V, Mottet-Osman G, Startchick S, Chaponnier C, Roux L. Sendai virus induced cytoplasmic actin remodeling correlates with efficient virus particle production. Virology. 2011;410:7–16. doi: 10.1016/j.virol.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 71.Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 73.Sit S-T, Manser E, Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR, Alexandrova AY, Arnold K, Schaub S, et al. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 74.Chambers R, Takimoto T. Trafficking of Sendai Virus Nucleocapsids Is Mediated by Intracellular Vesicles. PLoS One. 2010;5:e10994. doi: 10.1371/journal.pone.0010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106:133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 76.Mitra R, Baviskar P, Duncan-Decocq RR, Patel D, Oomens AGP. The human respiratory syncytial virus matrix protein is required for maturation of viral filaments. J Virol. 2012;86:4432–4443. doi: 10.1128/JVI.06744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santangelo PJ, Bao G. Dynamics of filamentous viral RNPs prior to egress. Nucleic Acids Res. 2007;35:3602–3611. doi: 10.1093/nar/gkm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: Building particles that transmit infections. Int J Biochem Cell Biol. 2010;42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. Role of matrix and fusion proteins in budding of Sendai virus. J Virol. 2001;75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bächi T. Direct observation of the budding and fusion of an enveloped virus by video microscopy of viable cells. J Cell Biol. 1988;107:1689–1695. doi: 10.1083/jcb.107.5.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kallewaard NL, Bowen AL, Crowe JE. Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology. 2005;331:73–81. doi: 10.1016/j.virol.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Martínez I, Lombardía L, García-Barreno B, Domínguez O, Melero JA. Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells. J Gen Virol. 2007;88:570–581. doi: 10.1099/vir.0.82187-0. [DOI] [PubMed] [Google Scholar]
- 83.Jeffree CE, Brown G, Aitken J, Su-Yin DY, Tan BH, Sugrue RJ. Ultrastructural analysis of the interaction between F-actin and respiratory syncytial virus during virus assembly. Virology. 2007;369:309–323. doi: 10.1016/j.virol.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 84.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henderson G, Murray J, Yeo RP. Sorting of the respiratory syncytial virus matrix protein into detergent-resistant structures is dependent on cell-surface expression of the glycoproteins. Virology. 2002;300:244–254. doi: 10.1006/viro.2002.1540. [DOI] [PubMed] [Google Scholar]
- 86.Jumat MR, Nguyen Huong T, Wong P, Loo LH, Tan BH, Fenwick F, Toms GL, Sugrue RJ. Imaging analysis of human metapneumovirus-infected cells provides evidence for the involvement of F-actin and the raft-lipid microdomains in virus morphogenesis. Virol J. 2014;11:198. doi: 10.1186/s12985-014-0198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gower TL, Pastey MK, Peeples ME, Collins PL, McCurdy LH, Hart TK, Guth A, Johnson TR, Graham BS. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J Virol. 2005;79:5326–5336. doi: 10.1128/JVI.79.9.5326-5336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gower TL, Peeples ME, Collins PL, Graham BS. RhoA is activated during respiratory syncytial virus infection. Virology. 2001;283:188–196. doi: 10.1006/viro.2001.0891. [DOI] [PubMed] [Google Scholar]
- 89.Shaikh FY, Cox RG, Lifland AW, Hotard AL, Williams JV, Moore ML, Santangelo PJ, Crowe JE. A critical phenylalanine residue in the respiratory syncytial virus fusion protein cytoplasmic tail mediates assembly of internal viral proteins into viral filaments and particles. MBio. 2012;3:1–10. doi: 10.1128/mBio.00270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baviskar PS, Hotard AL, Moore ML, Oomens AGP. The respiratory syncytial virus fusion protein targets to the perimeter of inclusion bodies and facilitates filament formation by a cytoplasmic tail -dependent mechanism. J Virol. 2013;87:10730–10741. doi: 10.1128/JVI.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teng MN, Collins PL. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ghildyal R, Mills J, Murray M, Vardaxis N, Meanger J. Respiratory syncytial virus matrix protein associates with nucleocapsids in infected cells. J Gen Virol. 2002;83:753–757. doi: 10.1099/0022-1317-83-4-753. [DOI] [PubMed] [Google Scholar]
- 93.Li D, Jans DA, Bardin PG, Meanger J, Mills J, Ghildyal R. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2-1 protein. J Virol. 2008;82:8863–8870. doi: 10.1128/JVI.00343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oliveira AP, Simabuco FM, Tamura RE, Guerrero MC, Ribeiro PGG, Libermann TA, Zerbini LF, Ventura AM. Human respiratory syncytial virus N, P and M protein interactions in HEK-293 T cells. Virus Res. 2013;177:108–112. doi: 10.1016/j.virusres.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 95.Stallcup KC, Raine CS, Fields BN. Cytochalasin B inhibits the maturation of measles virus. Virology. 1983;124:59–74. doi: 10.1016/0042-6822(83)90290-8. [DOI] [PubMed] [Google Scholar]
- 96.Harpen M, Barik T, Musiyenko A, Barik S. Mutational analysis reveals a noncontractile but interactive role of actin and profilin in viral RNA-dependent RNA synthesis. J Virol. 2009;83:10869–76. doi: 10.1128/JVI.01271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kipper S, Hamad S, Caly L, Avrahami D, Bacharach E, Jans Da, Gerber D, Bajorek M. New host factors important for Respiratory Syncytial Virus (RSV) replication revealed by a novel microfluidics screen for interactors of matrix (M) protein. Mol. Cell Proteomics. 2015;14:532–43. doi: 10.1074/mcp.M114.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pastey MK, Gower TL, Spearman PW, Crowe JE, Graham BS. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat Med. 2000;6:35–40. doi: 10.1038/71503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wurth MA, Schowalter RM, Smith EC, Moncman CL, Dutch RE, McCann RO. The actin cytoskeleton inhibits pore expansion during PIV5 fusion protein-promoted cell-cell fusion. Virology. 2010;404:117–26. doi: 10.1016/j.virol.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


