Abstract
Desmoplastic melanoma (DM) is a rare variant of melanoma with distinct clinical, histopathologic, and immunohistochemical features. Clinically, DM differs from conventional melanoma by a higher propensity for local recurrence and less frequent metastatic spread to regional lymph nodes. In its pure form, DM has a distinct appearance displaying a low density of fusiform melanocytes in a collagen-rich matrix. While a number of mutations have been identified in primary melanoma, including BRAF, NRAS, GNAQ, GNA11 and KIT, and the occurrence of these mutations has been found to correlate to some extent with the histopathologic features, anatomic site and/or mode of sun exposure, no distinct set of mutations has so far been reported for DM. To study the potential association of neurofibromin (NF1) mutations with DM, we examined 15 desmoplastic and 20 non-desmoplastic melanomas by next-generation sequencing. Mutations of the NF1 gene were found in 14 of 15 (93%) desmoplastic and 4 of 20 (20%) non-desmoplastic melanomas. The high frequency of NF1 mutations in desmoplastic melanomas suggests an important role for NF1 in the biology of this type of melanoma.
Keywords: desmoplastic melanoma, neurofibromin 1, mutation, next-generation sequencing
INTRODUCTION
Desmoplastic melanoma (DM) is a rare variant of primary melanoma with distinct clinical, histopathologic and immunohistochemical features1–4. It tends to affect the head and neck region of elderly individuals1, 3, but may occur anywhere, even in the young5. In its clinical course DM differs from other melanomas by a higher propensity for persistent growth/local recurrence4, 6. DM only rarely metastasizes to regional lymph nodes7. DM has been classified into pure and mixed variants8–11. Among patients with thick tumors, those with pure DM have a more favorable prognosis8–10. Histopathologically, the tumor cells of DM are usually fusiform and amelanotic. In its pure form, the tumor cells are dispersed at a low cell density in a densely collagenous matrix. Immunohistochemically, the tumor cells of DM often fail to react with antibodies to melan-A/MART-1, microphthalmia-associated transcription factor (MITF), tyrosinase, or gp10012, but are usually positive for S100 protein, nerve growth factor receptor (NGFR), and/or SOX1012–15.
Little is known about genetic changes in DM. While a number of mutations have been identified in melanoma, in particular mutations in BRAF (codon V600), NRAS, GNAQ, GNA11, and KIT, they are usually absent in DM. In a recent next-generation sequence analysis of a desmoplastic melanoma with sarcomatoid de-differentiation, we found inactivating mutations of neurofibromin 1 (NF1)16. A subset of desmoplastic melanomas were previously found to be associated with loss of NF1 locus17. Recently, a patient with neurofibromatosis type 1 was found to have developed a DM18. Since NF1 mutations are commonly found in peripheral nerve sheath tumors19, 20, and given the morphologic overlap of such tumors with DM, we reasoned that NF1 mutations may also be associated with DM, and therefore, examined DMs for their mutation status and compared them to a control set of primary and metastatic melanomas without desmoplasia.
MATERIALS AND METHODS
Patients
The study was approved by the institution’s IRB. Fifteen primary desmoplastic and 20 non-desmoplastic melanomas (2 primary, 18 metastatic tumor samples) were randomly selected. Of the 15 DMs, 7 tumors were pure DM characterized by a pauci-cellular fibrosing malignant spindle cell proliferation. Eight tumors were combined or mixed DM with both a classic pauci-cellular as well as a solid spindle cell tumor component (Table 1). One of the tumors was previously published as a case report16. The non-desmoplastic melanomas included 10 superficial spreading melanomas, 5 nodular, 3 acral and 2 lentigo maligna melanomas.
Table 1.
Desmoplastic melanoma – clinical findings, pathologic features, and NF1 mutation status.
| Case # | Age, Sex | Site | MIS | Thickness | Clark Level | DM Type | NF1 Mutation | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Mutation Type | cDNA | Protein | |||||||
| 1 | 82M | Scalp | Yes | 1.9 mm | V | Pure | Nonsense | c.3562C>T | p.Q1188* |
|
| |||||||||
| 2 | 24M | Arm | Yes | 9.5 mm | V | Pure | Nonsense | c.2875C>T | p.Q959* |
|
| |||||||||
| 3 | 85F | Face | Yes | 7.2 mm | V | Pure | Nonsense | c.1555C>T | p.Q519* |
| Splicesite | c.7456-1G>A | p.? (splice) | |||||||
|
| |||||||||
| 4 | 81F | Shoulder | Yes | 1.5 mm | IV | Pure | Missense | c.2530C>T | p.L844F |
| Nonsense | c.2734C>T | p.Q912* | |||||||
|
| |||||||||
| 5 | 77F | Face | Yes | 2.5 mm | IV | Pure | Nonsense | c.1105C>T | p.Q369* |
| Missense | c.2146G>C | p.E716Q | |||||||
|
| |||||||||
| 6 | 66M | Scalp | Yes | 7 mm | V | Pure | Missense | c.6472C>T | p.R2179C |
| Nonsense | c.1713G>A | p.W571* | |||||||
| Missense | c.3496G>A | p.G1166S | |||||||
|
| |||||||||
| 7 | 76M | Scalp | Yes | 1.8 mm | V | Mixed | Missense | c.451A>C | p.N151H |
| Missense | c.6782A>T | p.H2261L | |||||||
|
| |||||||||
| 8 | 85M | Neck | No | 18 mm | V | Mixed | Nonsense | c.3313A>T | p.K1105* |
| Nonsense | c.3826C>T | p.R1276* | |||||||
|
| |||||||||
| 9 | 82M | Scalp | Yes | 16 mm | V | Mixed | Splicesite | c.3198-2A>C | p.? (splice) |
|
| |||||||||
| 10 | 67M | Face | Yes | 3 mm | IV | Mixed | Nonsense | c.532G>T | p.E178* |
| Frameshift | c.1968_1969delCT | p.A656fs | |||||||
|
| |||||||||
| 11 | 58M | Shoulder | No | 11.5 mm | V | Mixed | Splicesite | c.1062+1G>A | p.? (splice) |
|
| |||||||||
| 12 | 66M | Scalp | Yes | 15 mm | V | Mixed | Splicesite | c.4174-2A>T | p.? (splice) |
|
| |||||||||
| 13 | 51M | Chest | Yes | 9.5 mm | V | Mixed | Missense | c.1327T>C | p.F443L |
|
| |||||||||
| 14 | 49F | Nose | Yes | 15 mm | V | Mixed | Splicesite | c.1393-1G>A | p.? (splice) |
| Nonsense | c.2088G>A | p.W696* | |||||||
|
| |||||||||
| 15 | 37F | Trunk | No | 25 mm | V | Pure | - | - | - |
Abbreviations: M = male; F = Female; MIS = melanoma in situ; mm = millimeter; DM = desmoplastic melanoma; NF = neurofibromin
Immunohistochemical Analysis
Five micron thick sections were taken from formalin-fixed, paraffin-embedded tissue, and stained with an automated immunohistochemistry system according to the manufacturer’s guidelines (Ventana BenchMark XT, Tucson, AZ) using a standard avidin-biotin procedure and antibodies for S100 protein and Sox10.
DNA extraction and targeted sequencing
To enrich for the tumor cell population, non-tumor tissue was manually removed from formalin-fixed, paraffin-embedded tissue sections, and areas with at least 50% neoplastic cells were scraped into sterile Eppendorf tubes. DNA was extracted with a QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer’s instructions. The extracted DNA was subjected to deep-coverage targeted sequencing using an assay termed MSK-IMPACT (Memorial Sloan Kettering - Integrated Mutation Profiling of Actionable Cancer Targets).
The MSK-IMPACT assay uses target-specific oligonucleotides (NimbleGen SeqCap) to capture all protein-coding exons and selected introns of 341 cancer-associated genes (oncogenes, tumor suppressor genes, and components of pathways deemed actionable by targeted therapies) and was previously described21. Briefly, 250 ng genomic tumor DNA and a negative control pool of 10 “normal” blood DNAs was fragmented (Covaris E220), and used to prepare barcoded sequencing libraries (New England Biolabs, Kapa Biosystems). The sequencing libraries were pooled at equimolar concentrations (100 ng per library) and used in the capture reaction (NimbleGen SeqCap). To prevent off-target hybridization, we spiked in a pool of blocker oligonucleotides complementary to the full sequences of all barcoded adaptors. The captured libraries were sequenced on an Illumina HiSeq 2500 to generate 75 bp paired-end reads producing an average coverage of >500 per tumor.
Sequence data were de-multiplexed using CASAVA, and aligned to hg19 using BWA22. Local realignment and quality score recalibration were performed using the Genome Analysis Toolkit (GATK) according to GATK best practices23. Sequence data were analyzed to identify single nucleotide variants, small insertions/deletions (indels) using MuTect 24 and GATK, respectively. Single-nucleotide variants and indels were called if the variant allele frequency in the tumor was present in > 5% of sequencing reads, and if the variant allele in the tumor was > 5 times higher than in the negative control. We excluded variant alleles if they were present in more than 2% of reads in the negative control, and if they were previously reported in dpSNP25 or the 1000 genome project26. Single-nucleotide variants and indels were reviewed manually using the Integrative Genomics Viewer27. Oncoprints and MutationMaps were generated in the cBio portal28.
RESULTS
Clinical Characteristics
Fifteen primary cutaneous DM were analyzed. The clinical and pathologic findings are summarized in Table 1. The ages of the patients ranged from 24 – 85 (mean = 65.7 years, median = 67 years). They included 10 men and 5 women. Thirteen tumors occurred in sun-damaged skin of elderly individuals (Fig. 1). Two tumors occurred in skin without evidence of sun-damage in young adults. Ten tumors were located in the head and neck region, three on the upper trunk and two on the upper extremities.
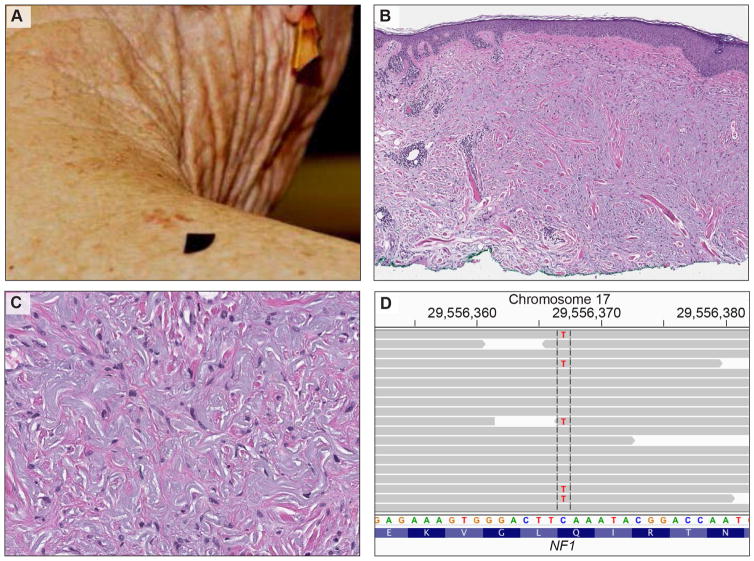
Figure 1.
Clinical, histological, and genetic features of desmoplastic melanoma. A, clinical picture of an 81-year-old woman with an irregular, brown-reddish skin lesion on her right shoulder. B, Histopathology of desmoplastic melanoma: Melanoma in situ (atypical proliferation of solitary units and nests of melanocytes at the dermoepidermal junction) is associated with an infiltrative pauci-cellular fibrosing spindle cell proliferation in the dermis with focal lymphocytic aggregates. C, The dermal spindle cells are dispersed as individual units (not aggregated in sheets or fascicles). They have hyperchromatic nuclei. D, The targeted sequencing data displayed in the Integrative Genomics Viewer (IGV). The gray arrows illustrate the individual sequencing reads aligned to exon 21 of the NF1 gene. The cytosine at base pair position 2734 was replaced by thymine (c.2734C>T) leading to a premature stopcodon (p.Q912*) and to a truncated NF1 protein.
Pathologic and Immunohistochemical Features
Histopathologically, 12 of the 15 DM had associated melanoma in situ (Fig. 1). Tumor thickness ranged from 1.5 – 25 mm (mean = 9.6 mm; median = 9.5 mm). None of the tumors were ulcerated. Four invasive tumors were confined to the dermis. Eleven tumors extended into the subcutis (Clark V).
The DM were strongly positive for S100 protein and SOX10, but negative for melan-A in the invasive tumor component. Melan-A was expressed by intraepidermal and intrafollicular melanoma cells (melanoma in situ).
Molecular Findings
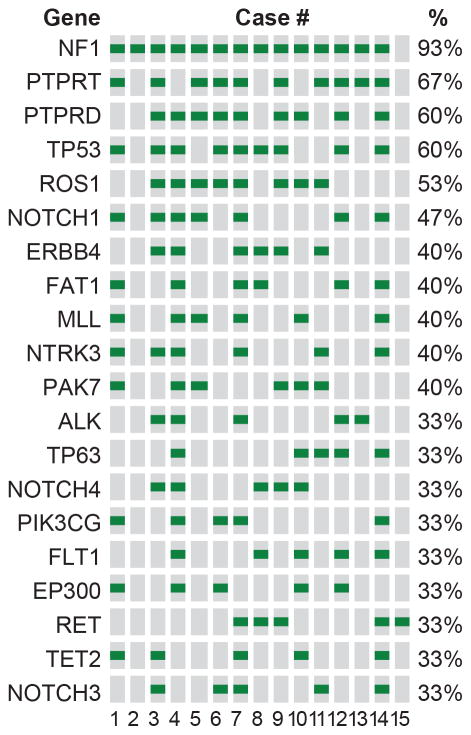
Shared mutations, which were present in at least 30% of tumors, are shown in Figure 2. The gene most commonly mutated in DM was NF1. Mutations of NF1 were detected in 14 of 15 (93%) of primary cutaneous DM. The vast majority of mutations in NF1 were nonsense, frameshift, or splice-site mutations that usually result in truncated non-functional proteins (Fig. 3). Genes mutated in at least 50% of the tumors included protein tyrosine phosphatase, receptor type, T (PTPRT), protein tyrosine phosphatase, receptor type, D (PTPRD), tumor protein p53 (TP53) and Ros protooncogene 1 (ROS1).
Figure 2.
Mutations detected by targeted sequencing and shared by at least 30% of desmoplastic melanomas. The columns denote the samples (according to Table 1), the rows refer to the genes, and the green squares indicate mutations.
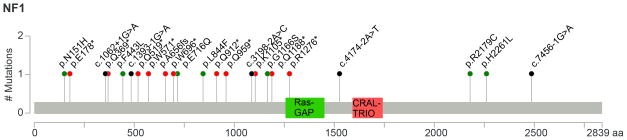
Figure 3.
Mutation pattern in NF1. The y-axis refers to the number of mutations and the x-axis to the amino acids. NF1 contains a Ras-GAP domain (GTPase-activator protein for Ras-like GTPase) between amino acids 1256–1451, and a CRAL-TRIO domain between amino acids 1591–1736. Missense mutations are displayed in green, nonsense mutations in red, and splice-site mutations in black. Missense and nonsense mutations are annotated to the protein level. Splice-site mutations are referenced to the coding DNA sequence.
Mutation in BRAF, GNAQ, GNA11 and KIT were not identified in our cohort of DM. Only one tumor (case #7) had a NRAS mutation. Interestingly, this tumor harbored two missense mutations in NF1, whereas most other DM contained at least one deleterious NF1 mutation (splice-site, non-sense, or frameshift mutations).
NF1 Mutations in Non-desmoplastic Melanoma
To assess the frequency of NF1 mutations in non-desmoplastic cutaneous melanomas, we analyzed 20 tumors (18 metastases from known non-desmoplastic primaries, 2 primary melanomas). The tumors included 10 superficial spreading melanomas, 5 nodular melanomas, 3 acral melanomas and 2 lentigo maligna melanomas. NF1 mutations were detected in 4 tumors (one lentigo maligna melanoma and 3 nodular melanomas) (Table 2). Three of the four non-desmoplastic melanomas with NF1 mutations originated from the skin of the head neck region; one tumor was from the skin of the back. In all non-desmoplastic melanomas of this series, NF1 was associated with another mutation of the RAS/RAF family: two cases had NRAS and another two had BRAF mutations.
Table 2.
NF1 mutations in 4 of 20 non-desmoplastic melanomas.
| Case # | Age, Sex | Site | Thickness | Melanoma Type | NF1 Mutation | |
|---|---|---|---|---|---|---|
|
| ||||||
| cDNA | AAchange | |||||
| 1 | 68 M | Neck | 2.2 mm | Lentigo Maligna | c.C1145T | S382F |
| c.1499_1521delTTCATGCAGATCCAAAGCTCTTG | I500fs | |||||
| c.C1997T | S666F | |||||
| c.C7126T | H2376Y | |||||
| c.C20T | S7F | |||||
|
| ||||||
| 2 | 84M | Ear | 5 mm | Nodular | c.C3097T | Q1033X |
| c.C3652T | Q1218X | |||||
|
| ||||||
| 3 | 60M | Scalp | 6.5 mm | Nodular | c.T3239A | L1080Q |
|
| ||||||
| 4 | 62M | Back | 9.6 mm | Nodular | c.C3028T | Q1010X |
| c.4321delT | L1441X | |||||
Abbreviations: M = male; F = female; mm = millimeter; NF1 = neurofibromin; AA = amino acid
DISCUSSION
We had previously noted inactivating mutations of NF1 in a combined tumor of desmoplastic and dedifferentiated sarcomatoid melanoma16. We now report that NF1 mutations are not unique to that case, but occur at high frequency in DM: NF1 mutations were found in 14 of 15 (93%) of DM.
In our control group of 20 cutaneous melanomas without desmoplasia, NF1 mutations were detected in 4 (20%) cases. Prior studies on the mutation profile of melanomas reported a frequency of NF1 mutations in 16 of 121 (13%) tumors29, and in 13 of 91 (14%)30, respectively. In a recent review the overall frequency of NF1 mutations was estimated at 14% of cutaneous melanomas, with a total of 475 specimens analyzed31. No information was provided about the histopathologic subtype, but it is unlikely that a significant number of DM was included in those studies, because DM is a rare variant of melanoma (approximately 4 % of primary melanomas)3, 32. Thus, compared to those reports and our control group of non-desmoplastic melanomas, NF1 mutations are more commonly found in DM.
NF1 encodes neurofibromin 1, a negative regulator of the Ras/mitogen-activated protein kinase (MAPK) pathway, a key pathway in melanomagenesis31. NF1 is a Ras-specific GTPase-activating protein that accelerates the hydrolysis of the active GTP-bound form of Ras into its inactive GDP-bound form33, but this is not the only mechanism, by which NF1 can downregulate Ras31. Somatic mutations in NF1 have been in a variety of cancers, including melanoma31. Loss of NF1 detected in melanoma was reported in cell lines in the early 1990s 34, 35, and one study reported loss of NF1 locus in a subset of DMs17. Recently, loss of NF1 was shown to prevent BRAF oncogene-induced senescence in melanoma36. In addition, one of the mechanisms of resistance for RAF-inhibitors is thought to occur via loss of NF1 expression37.
NF1 mutations and loss of NF1 protein expression are known to occur in over 90% of neurofibromatosis 1–associated and in over 40% of sporadic malignant peripheral nerve sheath tumors (MPNST)19, 20, 38. MPNST and DM may show considerable morphologic and immunohistochemial overlap. The tumors reported herein are unequivocal melanomas and not MPNSTs, since all but three DM of this series were associated with melanoma in situ, and the invasive component of all tumors was centered in the dermis and showed strong and diffuse labeling for S100 protein and Sox10. Additionally, none of our patients had a history or clinical signs of neurofibromatosis.
A number of additional genes were commonly mutated in the DM of this series (Fig. 2). These are genes known to be mutated in melanoma, but compared to their reported frequency in prior studies29, 30 they seem to be more common in DM. TP53 mutations, for example, were detected in 19% of melanomas by Hodis et al29 and in 12% of tumors by Krauthammer et al30. In our series of DM, TP53 was mutated in 60% of cases (Fig. 2) and interestingly, the majority of TP53 alterations were missense mutations that had clustered in the DNA binding domain.
Other highly mutated genes included the protein tyrosine phosphatase, receptor T (PTPRT, 67%) and D (PTPRD, 60%), which are known as tumor suppressor genes and recognized for their regulatory function on numerous cellular processes including cell growth, cell fate, and cell cycle. Although, the high frequency of mutations in these genes compared to prior sequencing studies of unselected melanoma (Hodis et al29: PTPRT: 26%, PTPRD: 17%; Krauthammer et al30: PTPRT: 0%, PTPRD: 24%) suggests some oncogenic impact in DM, thorough functional studies will be necessary to prove their pathogenetic relevance for several reasons: First, melanoma in general has a high background mutation rate, and it is difficult to distinguish pathogenetic relevant driver-mutations from irrelevant passenger-mutations. Previous studies reported very high point mutation rates in patients with a history of chronic sun exposure, and given the association of DM with chronic UV exposure39, high mutation rates in DM should come as no surprise. Second, whereas most mutations in NF1 were deleterious (nonsense, frameshift, or splice-site mutations), the mutation pattern found in the other genes was mainly composed of missense mutations. Missense mutations usually result in the exchange of one amino acid, and thus may have only minor effects on the protein function. Third, the missense mutations were quite randomly distributed across the genes and no hotspot mutations were found. Consequently, the impact of missense mutations on the protein function is unclear, and functional studies are necessary to analyze their oncogenic consequences.
In summary, having used targeted next-generation sequencing for 341 cancer related genes, we document that inactivating NF1 mutations are more common in DM than in non-desmoplastic melanomas. The finding provides further evidence that DM is, at the molecular level, distinct from conventional melanoma. The fact that NF1 mutations are commonly seen in both pure and mixed DM suggests that they occur early in the evolution of DM and do not merely reflect tumor progression. This is further supported by the fact that NF1 mutations can be found in a lentigo maligna melanoma in situ (KJB, unpublished observation). It is possible that mutations in genes that are not covered by our sequencing assay may also be associated with and play an important role in DM. However, given the high frequency of NF1 mutations in our series of tumors, we believe that NF1 is relevant for the biology of DM and for discussions of targeted treatment of recurrences thereof.
Acknowledgments
The authors wish to thank to Dr. Marghoob for providing us with the clinical picture shown in Fig. 1A. We would also like to thank the following surgeons and dermatologists for their involvement in the care of the patients of this series, including Drs. C. Ariyan, J Chen, DC Coit, J Healey, KS Nehal, E Lee, and S Patel. Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures: This work was supported by grants from the Harry J. Lloyd Trust - Translational Research Grant to K.J.B and T.W., and the Charles H. Revson Senior Fellowship to T.W.
References
- 1.Chen LL, Jaimes N, Barker CA, Busam KJ, Marghoob AA. Desmoplastic melanoma: a review. J Am Acad Dermatol. 2013;68:825–833. doi: 10.1016/j.jaad.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conley J, Lattes R, Orr W. Desmoplastic malignant melanoma (a rare variant of spindle cell melanoma) Cancer. 1971;28:914–936. doi: 10.1002/1097-0142(1971)28:4<914::aid-cncr2820280415>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 3.Busam KJ. Cutaneous desmoplastic melanoma. Adv Anat Pathol. 2005;12:92–102. doi: 10.1097/01.pap.0000155071.86944.a6. [DOI] [PubMed] [Google Scholar]
- 4.Lens MB, Newton-Bishop JA, Boon AP. Desmoplastic malignant melanoma: a systematic review. Br J Dermatol. 2005;152:673–678. doi: 10.1111/j.1365-2133.2005.06462.x. [DOI] [PubMed] [Google Scholar]
- 5.Hadj-Rabia S, Oriot D, Soufir N, et al. Unexpected extradermatological findings in 31 patients with xeroderma pigmentosum type C. Br J Dermatol. 2013;168:1109–1113. doi: 10.1111/bjd.12183. [DOI] [PubMed] [Google Scholar]
- 6.Arora A, Lowe L, Su L, et al. Wide excision without radiation for desmoplastic melanoma. Cancer. 2005;104:1462–1467. doi: 10.1002/cncr.21311. [DOI] [PubMed] [Google Scholar]
- 7.Gyorki DE, Busam K, Panageas K, Brady MS, Coit DG. Sentinel lymph node biopsy for patients with cutaneous desmoplastic melanoma. Ann Surg Oncol. 2003;10:403–407. doi: 10.1245/aso.2003.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Busam KJ, Mujumdar U, Hummer AJ, et al. Cutaneous desmoplastic melanoma: reappraisal of morphologic heterogeneity and prognostic factors. Am J Surg Pathol. 2004;28:1518–1525. doi: 10.1097/01.pas.0000141391.91677.a4. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins WG, Busam KJ, Ben-Porat L, et al. Desmoplastic melanoma: a pathologically and clinically distinct form of cutaneous melanoma. Ann Surg Oncol. 2005;12:207–213. doi: 10.1245/ASO.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 10.George E, McClain SE, Slingluff CL, Polissar NL, Patterson JW. Subclassification of desmoplastic melanoma: pure and mixed variants have significantly different capacities for lymph node metastasis. J Cutan Pathol. 2009;36:425–432. doi: 10.1111/j.1600-0560.2008.01058.x. [DOI] [PubMed] [Google Scholar]
- 11.Pawlik TM, Ross MI, Prieto VG, et al. Assessment of the role of sentinel lymph node biopsy for primary cutaneous desmoplastic melanoma. Cancer. 2006;106:900–906. doi: 10.1002/cncr.21635. [DOI] [PubMed] [Google Scholar]
- 12.Busam KJ, Iversen K, Coplan KC, Jungbluth AA. Analysis of microphthalmia transcription factor expression in normal tissues and tumors, and comparison of its expression with S-100 protein, gp100, and tyrosinase in desmoplastic malignant melanoma. Am J Surg Pathol. 2001;25:197–204. doi: 10.1097/00000478-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Radfar A, Stefanato CM, Ghosn S, Bhawan J. NGFR-positive desmoplastic melanomas with focal or absent S-100 staining: Further evidence supporting the use of both NGFR and S-100 as a primary immunohistochemical panel for the diagnosis of desmoplastic melanomas. Am J Dermatopathol. 2006;28:162–167. doi: 10.1097/01.dad.0000183696.46573.ee. [DOI] [PubMed] [Google Scholar]
- 14.Palla B, Su A, Binder S, Dry S. SOX10 expression distinguishes desmoplastic melanoma from its histologic mimics. Am J Dermatopathol. 2013;35:576–581. doi: 10.1097/DAD.0b013e31827a0b98. [DOI] [PubMed] [Google Scholar]
- 15.Nonaka D, Chiriboga L, Rubin BP. Differential expression of S100 protein subtypes in malignant melanoma, and benign and malignant peripheral nerve sheath tumors. J Cutan Pathol. 2008;35:1014–1019. doi: 10.1111/j.1600-0560.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 16.Kiuru M, McDermott G, Berger M, Halpern AC, Busam KJ. Desmoplastic melanoma with sarcomatoid dedifferentiation. Am J Surg Pathol. 2014;38:864–870. doi: 10.1097/PAS.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutzmer R, Herbst RA, Mommert S, et al. Allelic loss at the neurofibromatosis type 1 (NF1) gene locus is frequent in desmoplastic neurotropic melanoma. Hum Genet. 2000;107:357–361. doi: 10.1007/s004390000374. [DOI] [PubMed] [Google Scholar]
- 18.Rubinstein TJ, Plesec TP, Singh AD. Desmoplastic melanoma of the eyelid and conjunctival melanoma in neurofibromatosis type 1: a clinical pathological correlation. Surv Ophthalmol. 2015;60:72–77. doi: 10.1016/j.survophthal.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Upadhyaya M, Kluwe L, Spurlock G, et al. Germline and somatic NF1 gene mutation spectrum in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs) Hum Mutat. 2008;29:74–82. doi: 10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- 20.Bottillo I, Ahlquist T, Brekke H, et al. Germline and somatic NF1 mutations in sporadic and NF1-associated malignant peripheral nerve sheath tumours. J Pathol. 2009;217:693–701. doi: 10.1002/path.2494. [DOI] [PubMed] [Google Scholar]
- 21.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio Portal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yap YS, McPherson JR, Ong CK, et al. The NF1 gene revisited - from bench to bedside. Oncotarget. 2014;5:5873–5892. doi: 10.18632/oncotarget.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaimes N, Chen L, Dusza SW, et al. Clinical and dermoscopic characteristics of desmoplastic melanomas. JAMA Dermatol. 2013;149:413–421. doi: 10.1001/jamadermatol.2013.2248. [DOI] [PubMed] [Google Scholar]
- 33.Xu GF, O’Connell P, Viskochil D, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 34.Andersen LB, Fountain JW, Gutmann DH, et al. Mutations in the neurofibromatosis 1 gene in sporadic malignant melanoma cell lines. Nat Genet. 1993;3:118–121. doi: 10.1038/ng0293-118. [DOI] [PubMed] [Google Scholar]
- 35.Johnson MR, Look AT, DeClue JE, Valentine MB, Lowy DR. Inactivation of the NF1 gene in human melanoma and neuroblastoma cell lines without impaired regulation of GTP. Ras Proc Natl Acad Sci U S A. 1993;90:5539–5543. doi: 10.1073/pnas.90.12.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maertens O, Johnson B, Hollstein P, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nissan MH, Pratilas CA, Jones AM, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reuss DE, Habel A, Hagenlocher C, et al. Neurofibromin specific antibody differentiates malignant peripheral nerve sheath tumors (MPNST) from other spindle cell neoplasms. Acta Neuropathol. 2014;127:565–572. doi: 10.1007/s00401-014-1246-6. [DOI] [PubMed] [Google Scholar]
- 39.Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]