ABSTRACT
Regulation of central carbon metabolism has long been an important research subject in every organism. While the dynamics of metabolic flows during changes in available carbon sources have been estimated based on changes in metabolism-related gene expression, as well as on changes in the metabolome, the flux change itself has scarcely been measured because of technical difficulty, which has made conclusions elusive in many cases. Here, we used a monitoring system employing Vibrio fischeri luciferase to probe the intracellular metabolic condition in Escherichia coli. Using a batch culture provided with a limited amount of glucose, we performed a time course analysis, where the predominant carbon source shifts from glucose to acetate, and identified a series of sequential peaks in the luciferase activity (peaks 1 to 4). Two major peaks, peaks 1 and 3, were considered to correspond to the glucose and acetate consuming phases, respectively, based on the glucose, acetate, and dissolved oxygen concentrations in the medium. The pattern of these peaks was changed by the addition of a different carbon source or by an increasing concentration of glucose, which was consistent with the present model. Genetically, mutations involved in glycolysis or the tricarboxylic acid (TCA) cycle/gluconeogenesis specifically affected peak 1 or peak 3, respectively, as expected from the corresponding metabolic phase. Intriguingly, mutants for the acetate excretion pathway showed a phenotype of extended peak 2 and delayed transition to the TCA cycle/gluconeogenesis phase, which suggests that peak 2 represents the metabolic transition phase. These results indicate that the bacterial luciferase monitoring system is useful to understand the real-time dynamics of metabolism in living bacterial cells.
IMPORTANCE Intracellular metabolic flows dynamically change during shifts in available carbon sources. However, because of technical difficulty, the flux change has scarcely been measured in living cells. Here, we used a Vibrio fischeri luciferase monitoring system to probe the intracellular metabolic condition in Escherichia coli. Using a limited amount of glucose batch culture, a series of sequential peaks (peaks 1 to 4) in the luciferase activity was observed. Changes in the pattern of these peaks by the addition of extra carbon sources and in mutant strains involved in glycolysis or the TCA cycle/gluconeogenesis gene assigned the metabolic phase corresponding to peak 1 as the glycolysis phase and peak 3 as the TCA cycle/gluconeogenesis phase. Intriguingly, the acetate excretion pathway engaged in peak 2 represents the metabolic transition phase. These results indicate that the bacterial luciferase monitoring system is useful to understand the real-time dynamics of metabolism in living bacterial cells.
INTRODUCTION
Molecular mechanisms of cellular metabolic regulation have been the subject of extensive studies, as understanding these mechanisms is important not only for basic biology but also for practical applications, such as improving the yield of biotechnology products and developing new disease treatments (1, 2). However, while a large amount of information has been accumulated (3, 4), many questions still remain unanswered with respect to the underlying mechanism. Thus far, regulation of metabolic enzyme activity and gene expression has been most intensively studied in Escherichia coli (5). Because the central metabolic pathway is highly conserved among all organisms, information from model organisms, such as E. coli, should be useful for understanding and manipulating metabolic systems in general.
The adaptive response to changing carbon sources was first described by Monod as “diauxie,” which for E. coli means that there are two growth phases when the bacteria are grown in the presence of two sugars (6). Since then, many molecular mechanisms for switching metabolic phases have been described. In the case of diauxie, a time lag between glucose and lactose catabolism was explained by the transcriptional activation of the lactose operon (7). Transcription of the lactose operon is repressed by LacI (a specific, allosteric repressor for lactose) and activated by the cyclic AMP (cAMP) receptor protein (CRP). CRP is a global transcriptional regulator of catabolic enzymes and transporters of nonglucose sugars (8–11), and its genome-wide regulation was identified recently in vivo and in vitro (12, 13). The catabolite repressor activator (Cra) is also important for the transcriptional regulation of genes involved in central carbon metabolism, and it is a key factor for switching between glycolysis and gluconeogenesis (14–16). Other molecular mechanisms for switching between metabolic carbon sources have been described. The phosphotransferase system (PTS), which catalyzes the uptake and concomitant phosphorylation of a number of carbon hydrates, also plays a key role in carbon source repression (reviewed in reference 17). EIIAGlc, encoded by crr, is a PTS protein that is not only important for the PTS but is also critical for regulating the activity, via protein-protein interactions, of the lactose transporter encoded by lacY and adenylate cyclase encoded by cyaA. The expression level of a glucose transporter in bacteria is regulated by mRNA destabilization as well, which is mediated by noncoding RNAs (18). Posttranslational regulation is also important for regulating cellular metabolism. Allosteric control of enzymes in central metabolic pathways, that is, the regulation of enzymatic activity by intracellular intermediate metabolites, has been studied for a long time (reviewed in reference 19). Recently, it has been proposed that many enzymes of the central metabolic pathways are also regulated by posttranslational modifications, such as acetylation and succinylation in E. coli (20–23). In the case of acetylation, it was proposed that acetylphosphate functions as the donor of the acetyl group (24). Physiological roles of acetylation modifications were identified not only in enzymes such as acetyl coenzyme A (acetyl-CoA) synthetase (acs) but also in other proteins, including CheY, a component of the bacterial chemotaxis machinery, and RpoB, a subunit of RNA polymerase (25). The activation of Acs for the utilization of acetate is well defined as the requirement of transcriptional activation of acs by CRP as well as the deacetylation of Acs by CobB (26). Such regulation can change the rate and direction of the metabolic carbon flux, depending on the intracellular and extracellular environments (27–29).
Various systems have been developed to monitor intracellular metabolites or the activity of metabolic pathways. Metabolome analyses, using time of flight mass spectrometry or gas chromatography-mass spectrometry systems, have been used to quantify intermediate metabolites (30, 31). Moreover, carbon metabolic flux has been analyzed using 13C-labeled metabolites (27). In addition to these powerful methods to analyze intermediates from cell extracts, some other methods have enabled the observation of intracellular intermediate metabolites in living cells (32, 33). In a previous study, we observed drastic, physiological phase-dependent changes in light emissions from E. coli cells that expressed Vibrio fischeri luciferase (34). Because the levels of luciferase proteins (LuxA and LuxB) were constant during the growth phase under the examined conditions, changes in the luciferase activity were concluded to have resulted from changes in intracellular redox pools, such as NADH and NADPH, which should have a relationship with the carbon metabolic flux that corresponds to reducing power production (34).
In this study, to evaluate the Vibrio fischeri luciferase monitoring system to observe the carbon metabolic condition in living cells, we cultivated E. coli cells in the presence of limited amounts of glucose and measured the fluctuation of the expressed luciferase activity. It has been well described that E. coli cells first utilize glucose and excrete acetate, and then they take up and utilize acetate subsequent to the depletion of glucose (26, 27). Under glucose-depleted conditions, a series of sequential peaks of luciferase activity appeared from the lag phase to the end of cell growth. Based on the present analyses, we assigned the relevance of each peak to a phase of central carbon metabolism and, thus, confirmed the utility of the luciferase monitoring system.
MATERIALS AND METHODS
Bacterial strains and plasmid.
E. coli BW25113 (W3110 lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) (35) and the Keio collection (a single-gene deletion mutant series derived from strain BW25113) (36) were obtained from the E. coli Stock Center (National Bio-Resource Center, Mishima, Japan). Plasmid pLXUV5, which contains the lacUV5 promoter fragment fused to the V. fischeri luxCDABE genes, was constructed previously to monitor intracellular redox pools (34).
Medium.
Cells were grown in M9 minimal medium (37), supplemented with glucose (0.002%), Casamino Acids (0.2%), and ampicillin (50 μg/ml), called “glucose-limited medium” in this study, at 30°C under constant shaking at 30 rpm using a glass T-shaped connecting tube. For preculture, M9 medium with glucose (0.2%) and Casamino Acids (0.2%) was used, and 1/500 volume of an overnight culture (approximately 20 h) was transferred into fresh medium for the experiments. Depending on the experiment, the concentration of glucose was changed or extra carbon sources were added. Cell growth was monitored by measuring the optical density at 600 nm (OD600).
Measurement of luciferase activity.
Luciferase activity was determined as relative luminescence units (RLUs) by an endpoint assay, based on 10 s of accumulated counting of photons in the raw data measurement mode with a Lumat LB 9507 (Berthold, Bad Wildbad, Germany) luminometer. When cell cultures were used as samples, a 0.1-ml aliquot of the culture was placed in a test tube, and the RLUs were measured immediately. Luciferase activity was calculated as RLUs/OD600 (34).
Measurement of glucose and acetate concentrations in medium.
A 1-ml aliquot of bacterial cultures was collected in a 1.5-ml tube and centrifuged at 20,400 × g for 2 min. The supernatant was transferred to a new 1.5-ml tube for the glucose and acetate measurements. The concentrations of glucose and acetate in the samples were measured by the F-kit for glucose and the F-kit for acetate (Roche, Mannheim, Germany), respectively.
Measurement of the dissolved oxygen concentration in medium.
The dissolved oxygen concentration of the cultures and the cell density in glucose-limited medium were automatically measured for every 1 min by a Fibox 3 oxygen meter (Taitec, Saitama, Japan) at 30°C with aeration and constant stirring at 450 rpm. Seven hundred milliliters of the medium was used, and the culture conditions were controlled by a Bioneer-500L system (B. E. Marubishi, Tokyo, Japan). It was confirmed that the growth curve and the time course of the luciferase activity for the wild-type strain were basically the same as those in small-scale batch cultures in glass T-tubes.
Immunoblot analysis.
Immunoblot analysis was performed as described previously (38) using rabbit anti-LuxB antiserum (34). Two micrograms of total protein was analyzed during the time course.
RESULTS AND DISCUSSION
Time course analysis in glucose-limited medium revealed four peaks of V. fischeri luciferase activity during cell growth.
The monitoring system involving the bacterial luciferase genes was widely used to monitor gene expression in vivo. However, in a previous study, we showed that Vibrio fischeri luciferase, which emits light depending on the level of reduced flavin mononucleotides (FMNH2), can be used to monitor the intracellular redox pool in living bacterial cells (34) (see Fig. S1 in the supplemental material for details). In this study, we examined the utility of Vibrio fischeri luciferase to monitor the carbon metabolic condition in growing bacteria. During glucose catabolism, E. coli activates the glycolytic pathway and excretes acetate, which is known as an overflow metabolism (27). After the exhaustion of utilizable glucose, E. coli switches the metabolic flow to utilize the once excreted acetate as energy and a carbon source (26). During the time course analysis, luciferase activity and cell density were measured at 30-min intervals from 3 to 15 h after inoculation in a minimal medium containing a limited amount (0.002%) of glucose (generically, 0.2% is used for glucose medium in most cases) and 0.2% Casamino Acids. Here, we added Casamino Acids to the medium because glucose and acetate are preferentially used for energy production and not as building blocks, and many mutant strains deficient in central carbon metabolic genes, as shown in a later section, showed poor growth in the absence of Casamino Acids (data not shown), which enabled comparison among mutants (39). Therefore, we used this medium for comparison. The RLUs and the growth curve are shown in Fig. 1A. Cells cultured overnight emitted fewer than 103 RLUs (data not shown), and following 3 h of growth in fresh medium, more than 106 RLUs were emitted. The final cell density reached an OD600 of 0.6 under this glucose-limited medium. The RLUs showed a first peak (peak 1) until 4.5 h, followed by a second tiny peak (peak 2) from 5 to 6.5 h, a third peak from 6.5 to 12 h (peak 3), and a fourth peak from 12 to 14.5 h (peak 4). Therefore, four peaks in total were observed in the wild-type strain during growth under this condition. Peak 1 corresponded to approximately 8 × 106 RLUs until reaching 0.08 OD600 units, peak 2 corresponded to 2 × 106 RLUs from 0.09 to 0.12 OD600 units, peak 3 corresponded to 2 × 106 RLUs from 0.12 to 0.40 OD600 units, and peak 4 corresponded to a constant of around 1 × 104 RLUs from 0.40 to 0.60 OD600 units. After peak 4, the RLUs decreased to less than 1 × 103 when the cell growth finally stopped.
FIG 1.
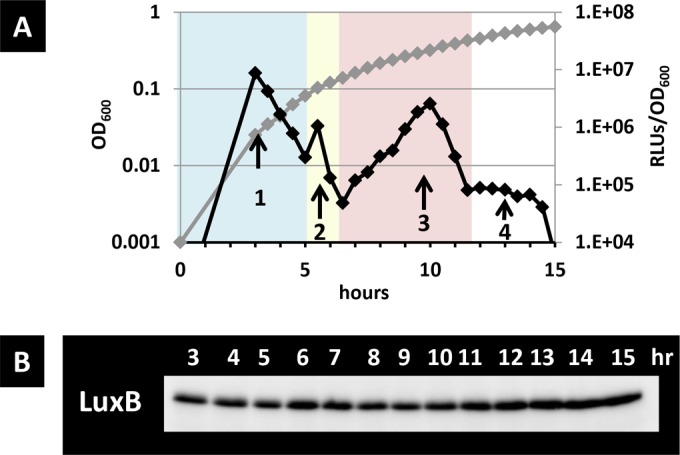
Time course of the luciferase activity and level of LuxB protein in glucose-limited medium. Strain BW25113 (pLXUV5) was grown in M9 glucose-limited medium, and the growth curve and luciferase activity were measured over time (A). The luciferase activity (black rhomboids) and the growth curve (gray rhomboids) were monitored at the indicated times. The metabolic phase is shown by the color of the background. (B) The levels of LuxB are shown.
Luciferase activity can be influenced by enzyme level. To estimate the contribution of the expression level of luciferase, we checked for the accumulation of the LuxB protein by immunoblot analysis at the same condition. The LuxB protein level was constant throughout the experiment (Fig. 1B). These results indicate that the fluctuation of the RLUs corresponded to changes in the carbon metabolism rather than in the luciferase expression level.
The first and third peaks (peaks 1 and 3) corresponded to glycolysis and the tricarboxylic acid (TCA) cycle/gluconeogenesis metabolic phases, respectively.
To correlate the luciferase peaks with the metabolic condition, glucose and acetate concentrations in the medium were measured during the time course analysis. Concentrations of these ingredients were measured at hourly intervals and compared with the luciferase activity (Fig. 2A). After inoculation of cells cultured overnight into fresh medium, the glucose concentration gradually decreased and glucose was depleted after 5 h. Meanwhile, the acetate concentration increased until 5 h and further increased slowly until 8 h, at which point it reached the maximum value. After that, the acetate concentration decreased until 14 h. Comparing the time course of the appearances of RLU peaks shows that the glucose consumption correlated with peak 1 while the acetate consumption correlated with peak 3. These results suggest that each peak corresponded to distinct metabolic phases. Peak 1 corresponds to glycolysis using glucose, and peak 3 corresponds to the TCA cycle and gluconeogenesis using acetate.
FIG 2.
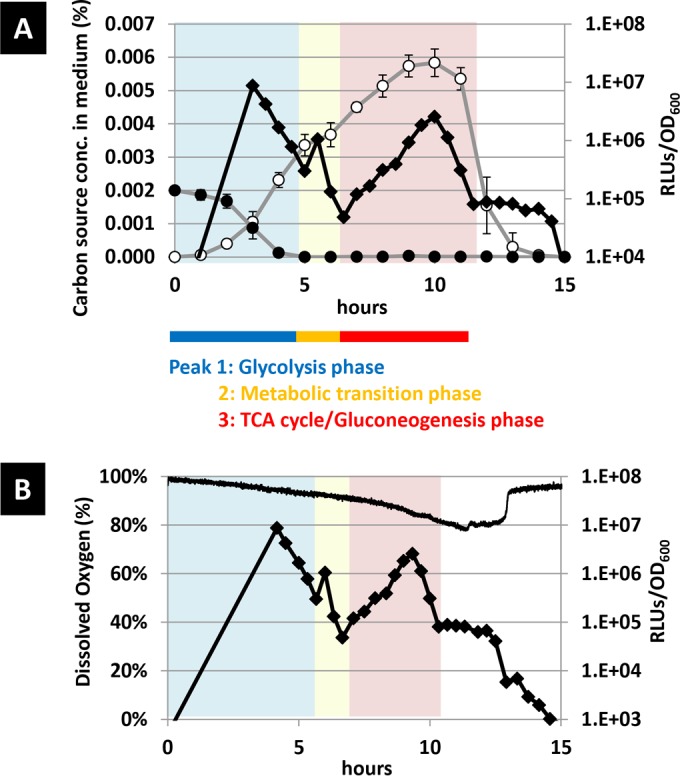
Time course of the luciferase activity and the glucose and acetate concentrations and dissolved oxygen concentration in glucose-limited medium. The glucose and acetate concentrations in medium were measured along with the luciferase activity (A). The dissolved oxygen concentration was directly measured by a nondestructive oxygen concentration meter (B). The luciferase activity (black rhomboids in A and B), and the concentrations of glucose (closed circles in A), acetate (open circles in A), and dissolved oxygen (black line in B) were monitored at the indicated times.
We next measured the dissolved oxygen under the same culture condition. Oxygen is consumed through the respiratory electron transfer chain, and thus the dissolved oxygen concentration in the medium should reflect TCA cycle activity. We measured the dissolved oxygen concentration in the medium every 3 min (Fig. 2B). After inoculation, the dissolved oxygen concentration decreased slowly and reached 90% prior to the occurrence of peak 3 (450 min), and it decreased rapidly to 80% concomitantly with peak 3 (450 to 660 min). This is in good agreement with the features of peak 3 that correspond to the TCA cycle (44, 45). Subsequently, the dissolved oxygen concentration remained constant at 80% for 100 min, indicating that the rate of oxygen dissolution from the air and the rate of consumption by E. coli were in equilibrium here. During peak 4, the RLUs remained constant for a few hours. These results suggest that another metabolic activity follows, presumably related to amino acids catabolism, after the depletion of acetate. At 760 min, the dissolved oxygen concentration suddenly increased from 80% to 95% within 10 min, showing that cell metabolic activity significantly dropped after this point.
Effects of glucose and ammonium concentrations on the luciferase activity profiles.
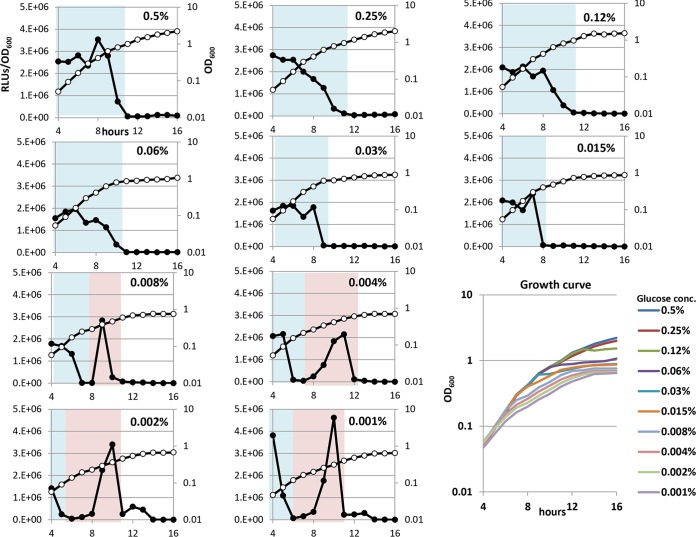
To examine further whether the luciferase peaks represent specific metabolic conditions, M9 medium containing different concentrations of glucose was used to observe the effects. First, luciferase activity and cell density were measured once per hour, from 4 to 16 h, after inoculation in a minimal medium containing glucose concentrations ranging from 0.001% to 0.5%. The final cell density proportionally increased in cases where the initial glucose concentration was less than 0.25% (Fig. 3, lower right), indicating that the final cell density was limited by the input glucose concentration. In contrast, the RLUs exhibited a different pattern, which depended on the initial glucose concentration. The scale of peak 1 increased along with the increasing glucose concentration, while the scale of peak 3 decreased conversely. The RLUs finally declined to less than 103 as cell growth arrested in every case.
FIG 3.
Time course of luciferase activity expressed by the PlacUV5-lux fusion gene under various concentrations of glucose. BW25113 harboring pLXUV5 was grown in M9 medium (31) with glucose and 0.2% Casamino Acids. The glucose concentration ranged from 0.001% to 0.5%. Numbers indicate the concentration of glucose, and luciferase activity (closed circles) and cell density (open circles) were monitored at the indicated times. The metabolic phase is shown by the color of the background. Each cell density was collected in the lower right graph and shown as growth curve.
In contrast, when the RLUs were measured in a minimal medium containing 0.25% glucose and ammonium concentrations ranging from 0.001% to 0.5%, all RLUs showed almost the same pattern, where only the first large peak was always observed (see Fig. S2 in the supplemental material). The final cell density was proportionally increased in cases where the initial ammonium concentration was less than 0.03%, indicating that the cell growth was limited by the input ammonium concentration. Even though the variable concentrations of glucose and ammonium had similar effects on the growth curves, changes of the RLU pattern only depended on the glucose concentration.
Additional carbon sources differently affected the luciferase activity profiles.
Providing the aforementioned glucose-limited medium (0.002% glucose) as the base medium (Fig. 4A), glucose, acetate, succinate, or citrate was added up to a 4-fold molar excess of the initial glucose concentration, and the time course luciferase activity profiles were examined. When glucose was supplemented at 0.01% in total, peak 1 was elongated, peak 2 was unaffected, and peak 3 was shortened (Fig. 4B) as in Fig. 3. By adding acetate, which is a substrate of the TCA cycle and gluconeogenesis, peak 3 increased while peak 1 and peak 2 were unaffected (Fig. 4C). The addition of succinate or citrate, which are intermediates of the TCA cycle, resulted in an enlarged and elongated peak 3, whereas it had little or no effect on peaks 1 and 2 (Fig. 4D and E). The RLUs and the cell optical density at each peak in the presence of extra carbon sources were listed in Table 1. Based on the effects of the additional carbon sources on the intensity of RLUs and cell growth, each peak was again regarded to have resulted from the activity of the corresponding metabolic pathway, with peak 1 resulting from glycolysis and peak 3 resulting from the TCA cycle and gluconeogenesis.
FIG 4.
Effects of additional carbon sources on luciferase activity. BW25113 (pLXUV5) was grown in M9 medium containing glucose (0.002%), which was supplemented with Casamino Acids (0.2%), and luciferase activity is shown (A). In addition to the glucose included in the glucose-limited medium, the carbon sources were added as follows: 0.008% glucose (B), 0.016% acetate (C), 0.016% succinate (D), or 0.016% citrate (E). Luciferase activity (closed circles) and cell density (open circles) were monitored at the indicated times. The metabolic phase is shown by the color of the background. Details are provided in Table 1.
TABLE 1.
Effects of additional carbon sources on bacterial growth and luciferase activity in each peak
| Additional carbon source (%) | Glycolysis phase (peak 1) |
Metabolic transition phase (peak 2) |
TCA cycle/gluconeogenesis phase (peak 3) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Time (h) | Cell growth (OD600) | RLUs | Time (h) | Cell growth (OD600) | RLUs | Time (h) | Cell growth (OD600) | RLUs | |
| Nonea | 4.5 | 0.08 | 4.1.E + 06 | 1.5 | 0.08–0.14 | 2.8.E + 05 | 5.5 | 0.14–0.34 | 4.8.E + 05 |
| Glucose (0.008) | 6.5 | 0.23 | 4.2.E + 06 | 1.5 | 0.23–0.36 | 4.1.E + 05 | |||
| Acetate (0.016) | 4.5 | 0.08 | 5.5.E + 06 | 1.5 | 0.08–0.14 | 2.7.E + 06 | 6 | 0.14–0.46 | 1.4.E + 06 |
| Succinate (0.016) | 4.5 | 0.08 | 5.3.E + 06 | 1.5 | 0.08–0.14 | 7.0.E + 05 | 6 | 0.14–0.46 | 8.0.E + 05 |
| Citrate (0.016) | 4.5 | 0.08 | 4.0.E + 06 | 1.5 | 0.08–0.14 | 5.1.E + 05 | 6 | 0.14–0.44 | 1.0.E + 06 |
An M9 medium containing glucose (0.002%) and Casamino Acids (0.2%) was used as the standard medium.
Effects of mutations of metabolic enzymes and other proteins on the luciferase activity profiles.
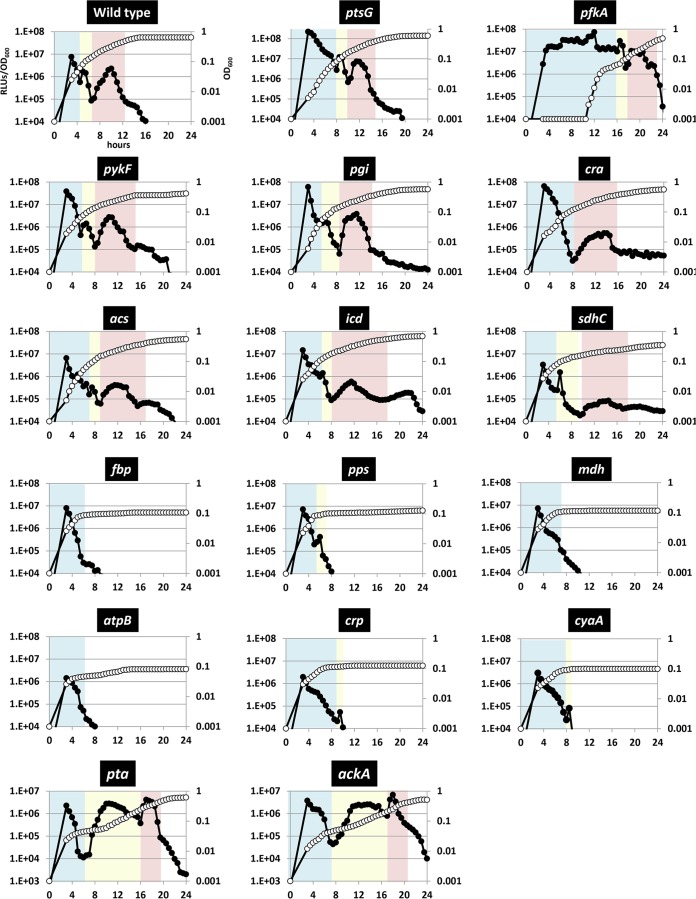
Based on the above results, we concluded that the Vibrio luciferase monitoring system can be used to estimate the real-time carbon metabolic condition in living bacterial cells. To get more insights into the regulation of carbon metabolism, luciferase activity profiling was performed for mutants from the Keio collection (36), each of which was deficient in an enzyme or a regulatory factor involved in the central carbon metabolic pathway. Here, we expected that the growth rate and the intensity of RLUs would be affected when the mutated gene is involved in ongoing metabolic activity. Thus, the influence of each enzyme deficiency should be clarified by comparing the RLU profile of each mutant with that of the wild-type strain (Fig. 5; see also the biological replica in Fig. S3 in the supplemental material). To identify which peak was affected by each mutation, OD600 units were plotted on the x axis for comparison in Fig. S4 in the supplemental material since, interestingly, the appearance of the peaks and the OD600 coincided well in all mutants; peak 1 was observed at an OD600 of ∼0.09, peak 2 was observed at an OD600 of 0.09∼0.12, peak 3 was observed at an OD600 of 0.12∼0.40, and Peak 4 was observed at an OD600 of 0.40∼0.60. In contrast, the intensity of RLUs and the time of appearance of each peak after the inoculation differed depending on the mutated gene (see Table 2 for details). After examining 16 mutant strains, we classified the luciferase profiles as follows.
FIG 5.
Time course of the luciferase-specific activity and cell growth in various enzyme mutants in M9 glucose-limited medium. Cell density (filled symbols) and luciferase-specific activity (open symbols) were monitored periodically for each mutant strain. The name of the mutated gene is shown at the top of each panel. The x axis indicates the time, the left y axis indicates relative light units (luciferase-specific activity), and the right y axis indicates cell density. The metabolic phase is shown by the color of the background. Details are provided in Table 2.
TABLE 2.
Luciferase activity of metabolic mutants
| Gene | Function | Pathway | Glycolysis phase (peak 1) |
Metabolic transition phase (peak 2) |
TCA cycle/gluconeogenesis phase (peak 3) |
|||
|---|---|---|---|---|---|---|---|---|
| RLUs | Time (h) | RLUs | Time (h) | RLUs | Time (h) | |||
| Wild type | 7.6.E+06 | 5 | 1.7.E+06 | 1.5 | 2.3.E+06 | 5 | ||
| ptsG | Glucose permease | Transporter | 2.2.E+08 | 8 | 1.2.E+07 | 2 | 7.4.E+06 | 4 |
| pfkA | 6-Phosphofructokinase I | Glycolysis | 7.3.E+07 | 13 | 3.0.E+07 | 4 | 1.1.E+07 | 5 |
| pykF | Pyruvate kinase I | Glycolysis | 3.9.E+07 | 7 | 1.4.E+06 | 1 | 2.8.E+06 | 6.5 |
| pgi | Phosphoglucose isomerase | Glycolysis/gluconeogenesis | 6.0.E+07 | 7.5 | 1.8.E+06 | 1 | 3.8.E+06 | 5.5 |
| acs | Acetyl-CoA synthetase | Acetate utilization | 6.5.E+06 | 7 | 3.2.E+05 | 1.5 | 4.1.E+05 | 7.5 |
| icd | Isocitrate dehydrogenase | TCA cycle | 1.5.E+07 | 6 | 1.4.E+06 | 1.5 | 5.9.E+05 | 7 |
| sdhC | Succinate dehydrogenase | TCA cycle | 3.3.E+06 | 5.5 | 1.5.E+06 | 2 | 8.5.E+04 | 8.5 |
| cra | Transcription factor | Transcriptional regulation | 6.5.E+07 | 8.5 | NDa | 5.4.E+05 | 7 | |
| fbp | Fructose 1,6-bisphosphatase I | Gluconeogenesis | 8.0.E+06 | 5.5 | NGb | NG | ||
| pps | Phosphoenolpyruvate synthetase | Gluconeogenesis | 7.3.E+06 | 5.5 | 4.3.E+05 | 1 | NG | |
| mdh | Malate dehydrogenase | TCA cycle/gluconeogenesis | 7.3.E+06 | 5.5 | NG | NG | ||
| atpB | ATP synthase | Electron transport chain | 1.4.E+06 | 6 | NG | NG | ||
| crp | Transcription factor | Transcriptional regulation | 2.0.E+06 | 7 | 5.4.E+04 | 1 | NG | |
| cyaA | Adenylate cyclase | Transcriptional regulation | 3.0.E+06 | 7 | 8.1.E+04 | 1 | NG | |
| pta | Phosphate acetyltransferase | Acetate excretion | 2.3.E+06 | 6 | 2.8.E+06 | 10 | 4.2.E+06 | 3.5 |
| ackA | Acetate kinase | Acetate excretion | 3.8.E+06 | 7 | 2.6.E+06 | 9 | 6.9.E+06 | 3 |
ND, not detected.
NG, no growth.
(i) Mutations that affected peak 1.
In the wild-type strain, peak 1 corresponded to approximately 8 × 106 RLUs at an OD600 of approximately 0.09 and continued for 5 h (Fig. 5; Table 2). In comparison, peak 1 for the ptsG mutant lacking the glucose permease exhibited significantly higher RLUs, and the peak continued for 8 h. Mutants deficient in enzymes catalyzing irreversible reactions in the glycolytic pathway, such as 6-phosphofructokinase I (PfkA), pyruvate kinase I (PykF), and phosphoglucose isomerase (Pgi), also exhibited higher RLUs, and peak 1 persisted longer (see peak 1, light blue back color in Fig. 5; see also Fig. S5, left upper panel, in the supplemental material). These results suggested that these enzymes are relevant to the ongoing process during peak 1, and the effects by these mutations are comparable to those of glucose addition (Fig. 3 and 4B). Therefore, peak 1 was again relevant to the glycolytic flux, and here, we designate the metabolic phase corresponding to peak 1 as the glycolysis phase.
(ii) Mutations that showed a lower peak 3.
The numbers of RLUs in the acs (acetyl-CoA synthetase), icd (isocitrate dehydrogenase), and sdhC (succinate dehydrogenase) mutants that were defective in acetate utilization and TCA cycle were lower than those of the wild-type strain at peak 3 (see peak 3, light red background color in Fig. 5; see also Fig. S5, right upper panel, in the supplemental material). The mutants of icd and sdhC could use the glyoxylate pathway instead of the TCA cycle, and this resulted in depression of NADPH and ATP production (40). Thus, these mutants were considered to show lower luciferase activity and slow growth at peak 3. In addition, the cra mutant affected both peak 1 and peak 3 because cra encodes a dual transcriptional regulator that plays repressor for glycolysis and activator for the gluconeogenesis pathway.
(iii) Mutations that showed arrested growth after peak 1.
The growth of mutants deficient in fbp (fructose 1,6-bisphosphatase I), pps (phosphoenolpyruvate synthetase), mdh (malate dehydrogenase), atpB (a subunit of ATP synthase), crp (cAMP receptor protein), and cyaA (adenylate cyclase) was arrested after OD600 reached 0.1 (Fig. 5; see also Fig. S5, left lower panel, in the supplemental material). These genes encode components required for gluconeogenesis, ATP synthase, and a transcription factor that regulates carbon source-related metabolic genes in the absence of glucose. The growth arrest of these mutants again strongly supported the proposal that peak 1 corresponds to the phase of glycolysis and that peak 3 corresponds to the phase of the TCA cycle and gluconeogenesis. Note that mutants deficient in the TCA cycle showed slow growth and that mutants deficient in gluconeogenesis and ATP synthase could not even move to the TCA cycle and gluconeogenesis phase.
Acetate excretion pathway is required to switch the metabolism from glycolysis phase to TCA cycle/gluconeogenesis phase.
Subsequent to the depletion of available glucose, E. coli cells switch the major fueling pathway from glycolysis to the TCA cycle. Until now, it has been considered that the main mechanism for this switch is based on transcriptional regulation mediated by cAMP-CRP, which is activated by glucose depletion (8, 9, 13). As described above, a series of peaks in the luciferase activity were identified during the time course analysis in the glucose-limited medium. Here, with respect to peak 2 appearing 1.5 h after peak 1 and positioned around the metabolic transition, we unexpectedly found that the appearance of peak 2 was greatly affected in mutants deficient in either pta or ackA, which encode phosphate acetyltransferase and acetate kinase, respectively (see peak 2, light yellow background color in Fig. 5; see also Fig. S5, right lower panel, in the supplemental material). These enzymes constitute the acetate excretion pathway that converts acetyl-CoA to acetate. In these mutants, peak 2 and the diauxic growth arrest lasted for about 10 h, which appears to be a very long lag phase, without affecting peaks 1 and 3.
It has been considered that the acetate excretion pathway functions as a carbon overflow drain during glucose catabolism (29, 41). However, as we found in this study, this pathway is likely essential for the smooth metabolic phase transition from the glucose-fueled metabolic phase into the acetate-fueled TCA cycle and gluconeogenesis phase. These results suggest that some important process is occurring during the transition from glycolytic to TCA cycle metabolism, which employs the acetate excretion pathway. The addition of 0.03 to 1 mM cAMP into the medium did not affect the long growth arrest in the pta mutant (see Fig. S6 in the supplemental material), suggesting that the role of the acetate pathway is independent of transcriptional regulation mediated by cAMP-CRP. While the importance of this acetate excretion pathway, such as for biomass production and controlling cell density, has been repeatedly described in previous studies (42, 43), its basis at the molecular level has been elusive thus far. In this study, we succeeded in identifying the regulatory significance, and thus future elucidation of the underlying mechanism may open a way for various applications.
Conclusions.
In this study, we analyzed the metabolic flux through the central carbon metabolic pathway in living E. coli cells using the V. fischeri luciferase monitoring system. Luciferase activity peaks 1 and 3 corresponded to the consumption of glucose and acetate, respectively, in the medium. In addition, we found a novel mechanism involved in the acetate excretion pathway, which is essential to shift the metabolic phase from glycolysis to the TCA cycle and gluconeogenesis. Identifying the underlying molecular mechanism of this switch will require further study. The luciferase activity data set for metabolic gene mutants grown in the presence of different carbon sources was useful to understand which gene functions are relevant to each metabolic phase. The luciferase monitoring system can be used to conduct time-lapse monitoring of metabolic activity, even in a single cell (unpublished data), and it may provide useful information for understanding single-cell metabolic behavior in any cell population. Taken together, we propose that bacterial luciferase can be used as a gauge to monitor the real-time dynamics of carbon metabolism in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We thank Akira Ishihama for critically reading the manuscript. We also thank Yoshinori Ohsumi and laboratory members (Tokyo Institute of Technology) for technical support during the monitoring of dissolved oxygen concentrations as well as the Biomaterial Analysis Center, Technical Department of Tokyo Institute of Technology, for technical support and the National BioResouce Project, NIG, Japan, for providing the E. coli Keio collection.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01400-16.
REFERENCES
- 1.National Research Council Committee on Biobased Industrial Products. 2000. Biobased industrial products: priorities for research and commercialization. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 2.Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH. 2016. Genereviews. University of Washington, Seattle, WA. [Google Scholar]
- 3.Berg JM, Tymoczko JL, Stryer L. 2010. Biochemistry, 7th ed W. H. Freeman, New York, NY. [Google Scholar]
- 4.Brady ST, Sigel GJ, Albers RW, Price DL. 2011. Basic neurochemistry: principles of molecular, cellular and medical meurobiology. 8th ed Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- 5.Sauer U. 14 August 2007, posting date Introduction and perspectives. EcoSal Plus 2007. doi: 10.1128/ecosal.3.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Monod J. 1942. Recherches sur la croissance des cultures bacteriennes. Universite de Paris Press, Paris, France. [Google Scholar]
- 7.Dickson RC, Abelson J, Barnes WM, Reznikoff WS. 1975. Genetic regulation: the Lac control region. Science 187:27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- 8.Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol 11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Fic E, Bonarek P, Grecki A, Kedracka-Krok S, Mikolajczak J, Polit A, Tworzydlo M, Dziedzicka-Wasylewska M, Wasylewski Z. 2009. cAMP receptor protein from Escherichia coli as a model of signal transduction in proteins—a review. J Mol Microbiol Biotechnol 17:1–11. doi: 10.1159/000178014. [DOI] [PubMed] [Google Scholar]
- 10.Pyles EA, Lee JC. 1996. Mode of selectivity in cyclic AMP receptor protein-dependent promoters in Escherichia coli. Biochemistry 35:1162–1172. doi: 10.1021/bi952187q. [DOI] [PubMed] [Google Scholar]
- 11.Zheng D, Constantinidou C, Hobman JL, Minchin SD. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res 32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci U S A 102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada T, Fujita N, Maeda M, Ishihama A. 2005. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells 10:907–918. doi: 10.1111/j.1365-2443.2005.00888.x. [DOI] [PubMed] [Google Scholar]
- 14.Geerse RH, van der Pluijm J, Postma PW. 1989. The repressor of the PEP-fructose phosphotransferase system is required for the transcription of the pps gene of Escherichia coli. Mol Gen Genet 218:348–352. doi: 10.1007/BF00331288. [DOI] [PubMed] [Google Scholar]
- 15.Shimada T, Fujita N, Yamamoto K, Ishihama A. 2011. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6:e20081. doi: 10.1371/journal.pone.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada T, Yamamoto K, Ishihama A. 2011. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J Bacteriol 193:649–659. doi: 10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita T, Maki K, Aiba H. 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanwal BD. 1970. Allosteric controls of amphilbolic pathways in bacteria. Bacteriol Rev 34:20–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Y, Wan X, Chen Y, Cha YH, Lin H, Zhao Y, Tan M. 2013. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics 12:3509–3520. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan KL, Xiong Y. 2011. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci 36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosono S, Tamura M, Suzuki S, Kawamura Y, Yoshida A, Nishiyama M, Yoshida M. 2015. Changes in the acetylome and succinylome of Bacillus sublilis in response to carbon source. PLoS One 10:e0131169. doi: 10.1371/journal.pone.0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. 2008. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol 18:1529–1536. [PubMed] [Google Scholar]
- 24.Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, Nyström T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Bernal V, Castaño-Cerezo S, Gallego-Jara J, Écija-Conesa A, de Diego T, Iborra JL, Canovas M. 2014. Regulation of bacterial physiology by lysine acetylation of proteins. N Biotechnol 31:586–595. doi: 10.1016/j.nbt.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Castaño-Cerezo SV, Bernal V, Blanco-Catalá J, Iborra JL, Cánovas M. 2011. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol Microbiol 82:1110–1128. doi: 10.1111/j.1365-2958.2011.07873.x. [DOI] [PubMed] [Google Scholar]
- 27.Haverkorn van Rijsewijk BR, Nanchen A, Nallet S, Kleijn RJ, Sauer U. 2011. Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Mol Syst Biol 7:477. doi: 10.1038/msb.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pisithkul T, Patel NM, Amador-Noguez D. 2015. Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr Opin Microbiol 24:29–37. doi: 10.1016/j.mib.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Wegner A, Meiser J, Weindl D, Hiller K. 2015. How metabolites modulate metabolic flux. Curr Opin Biotechnol 34:16–22. doi: 10.1016/j.copbio.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, Tomita M, Soga T. 2008. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol Biosyst 4:135–147. doi: 10.1039/B714176A. [DOI] [PubMed] [Google Scholar]
- 31.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. 2003. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res 2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 32.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. 2009. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A 106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lüddecke J, Forchhammer K. 2013. From PII signaling to metabolite sensing: a novel 2-oxoglutarate sensor that details PII-NAGK complex formation. PLoS One 8:e83181. doi: 10.1371/journal.pone.0083181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koga K, Harada T, Shimizu H, Tanaka K. 2005. Bacterial luciferase activity and the intracellular redox pool in Escherichia coli. Mol Genet Genomics 274:180–188. doi: 10.1007/s00438-005-0008-5. [DOI] [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 38.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, σ38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A 90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ip K, Donoghue N, Kim MK, Lun DS. 2014. Constraint-based modeling of heterologous pathways: application and experimental demonstration for overproduction of fatty acids in Escherichia coli. Biotechnol Bioeng 111:2056–2066. doi: 10.1002/bit.25261. [DOI] [PubMed] [Google Scholar]
- 40.Kabir MM, Shimizu K. 2004. Metabolic regulation analysis of icd-gene knockout Escherichia coli based on 2D electrophoresis with MALDI-TOF mass spectrometry and enzyme activity measurements. Appl Microbiol Biotechnol 65:84–96. doi: 10.1007/s00253-004-1627-1. [DOI] [PubMed] [Google Scholar]
- 41.Haeusser DP, Levin PA. 2008. The great divide: coordinating cell cycle events during bacterial growth and division. Curr Opin Microbiol 11:94–99. doi: 10.1016/j.mib.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holms WH. 1986. The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass, and excretion of acetate. Curr Top Cell Regul 28:69–105. doi: 10.1016/B978-0-12-152828-7.50004-4. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyström T, Larsson C, Gustafsson L. 1996. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J 15:3219–3228. [PMC free article] [PubMed] [Google Scholar]
- 45.Portnoy VA, Scott DA, Lewis NE, Tarasova Y, Osterman AL, Palsson BØ. 2010. Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655. Appl Environ Microbiol 76:6529–6540. doi: 10.1128/AEM.01178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.