ABSTRACT
Genome sequencing projects in the last decade revealed numerous cryptic biosynthetic pathways for unknown secondary metabolites in microbes, revitalizing drug discovery from microbial metabolites by approaches called genome mining. In this work, we developed a heterologous expression and functional screening approach for genome mining from genomic bacterial artificial chromosome (BAC) libraries in Streptomyces spp. We demonstrate mining from a strain of Streptomyces rochei, which is known to produce streptothricins and borrelidin, by expressing its BAC library in the surrogate host Streptomyces lividans SBT5, and screening for antimicrobial activity. In addition to the successful capture of the streptothricin and borrelidin biosynthetic gene clusters, we discovered two novel linear lipopeptides and their corresponding biosynthetic gene cluster, as well as a novel cryptic gene cluster for an unknown antibiotic from S. rochei. This high-throughput functional genome mining approach can be easily applied to other streptomycetes, and it is very suitable for the large-scale screening of genomic BAC libraries for bioactive natural products and the corresponding biosynthetic pathways.
IMPORTANCE Microbial genomes encode numerous cryptic biosynthetic gene clusters for unknown small metabolites with potential biological activities. Several genome mining approaches have been developed to activate and bring these cryptic metabolites to biological tests for future drug discovery. Previous sequence-guided procedures relied on bioinformatic analysis to predict potentially interesting biosynthetic gene clusters. In this study, we describe an efficient approach based on heterologous expression and functional screening of a whole-genome library for the mining of bioactive metabolites from Streptomyces. The usefulness of this function-driven approach was demonstrated by the capture of four large biosynthetic gene clusters for metabolites of various chemical types, including streptothricins, borrelidin, two novel lipopeptides, and one unknown antibiotic from Streptomyces rochei Sal35. The transfer, expression, and screening of the library were all performed in a high-throughput way, so that this approach is scalable and adaptable to industrial automation for next-generation antibiotic discovery.
INTRODUCTION
Microbial natural products have been a main source of drugs during the last century due to their diverse structures and bioactivities (1). The natural product drug pipeline seems to have stalled in the last 3 decades, even though recent Streptomyces genome sequencing projects have uncovered multiple biosynthetic gene clusters (BGCs) for undiscovered natural products (secondary metabolites), far exceeding the number of known metabolites (2–4). However, those BGCs are usually cryptic under laboratory conditions, producing very little or no corresponding secondary metabolites, which are needed for biological testing (5, 6).
Different approaches have been developed to activate the cryptic BGCs for coelichelin, clostrubin, stambomycin, and malleilactone in the original strains, using medium optimization, regulator engineering, and feeding of chemical elicitors, respectively (6–9). Alternatively, entire biosynthetic gene clusters were cloned and transferred to surrogate expression hosts for the production of natural products. BGCs of interest are often cloned using cosmid or bacterial artificial chromosome (BAC) libraries. For instance, the arimetamycin BGC (40 kb) was identified from cosmid libraries of soil metagenomes (10). Bacterial BGCs are usually 10 to >100 kb in length. Cosmids can take DNA inserts up to 45 kb. BACs can take inserts up to 490 kb (11). Therefore, BAC libraries were used to capture large BGCs: for example, the giant daptomycin BGC (128 kb) from Streptomyces roseosporus NRRL11379 (12). BGCs of interest can also be cloned by other methods, such as restriction digestion and ligation to a BAC vector, pSBAC (meridamycin [95 kb]) (13), transformation-associated recombination in yeast (taromycin [67 kb]) (14), or linear-plus-linear homologous recombination (15). Since Escherichia coli cloning host is not suitable to express high-GC-percentage Streptomyces DNA (16, 17), the cloned large BGCs should be introduced into the Streptomyces host for production of natural products. Streptomyces coelicolor M1152/M1154 (18), Streptomyces avermitilis SUKA17/22 (19), and Streptomyces lividans SBT5 (20) are good expression hosts that lack competing BGCs, which greatly facilitates the purification of the bioactive compounds expressed from cloned BGCs by virtue of the clean background. In addition, large genomic deletion (1.4 Mb) in S. avermitilis SUKA17/22, point mutations of RNA polymerase β subunit (C1298T) and ribosomal protein S12 (A262G) in S. coelicolor M1152/M1154, mutation of ribosomal protein S12 (K88E), and introduction of two heterologous global activators (afsRScla) in S. lividans SBT5 greatly improved the expression of exogenous BGCs (18–20).
The sequence-led cloning approaches described above depend on bioinformatics analysis to predict potentially interesting BGCs. It would be preferable and potentially less expensive to express multiple large fragments of Streptomyces DNA in expression hosts and start with the screening for biological activity. This could reveal novel types of compounds that cannot be predicted by sequence analysis (16, 21). To achieve effective functional screening, the arrayed BAC genomic library in E. coli must be transferred into a suitable Streptomyces expression host in a high-throughput way. In a pioneer metagenomic study using large DNA fragments for bioprospecting, Martinez et al. introduced a high-throughput method for library transfer. Pooled DNA from the BAC library was electroporated into E. coli ET12567/pUB307 to make an intermediate library, which was used as a conjugation donor or donors to transfer the BAC DNA efficiently into S. lividans by high-throughput biparental mating (22). ET12567/pUB307 was used as an intermediate host to provide the conjugation helper plasmid and nonmethylated BAC DNA that is less restricted by many Streptomyces strains (23). However, electroporation of pooled DNA favors smaller BACs (24), which are less likely to contain complete BGCs DNA, and the dam mutation in E. coli ET12567 increases mutation frequency, making it undesirable for the maintenance especially of large gene clusters (25).
Here, we describe a high-throughput genomic library expression analysis system (LEXAS) for efficient, function-driven discovery of cryptic and new antibiotics from Streptomyces spp., which are known producers of varied antibiotics. Each BAC clone was transferred individually into an engineered antibiotic overproduction host, avoiding preference for smaller BACs. The LEXAS captured two known antibiotics, identified two novel lipopeptides and their BGCs which were not produced/expressed in the native Streptomyces rochei strain, and revealed a cryptic BGC for an unknown antibiotic.
METHODS AND MATERIALS
Bacterial strains, plasmids, and culture conditions.
Streptomyces rochei Sal35 (China Center for Type Culture Collection no. AA97007) was isolated from forest soil of Shengnongjia (Eastern Hubei Province). S. lividans SBT5 [Δact ΔredKL ΔcdaPS3-SLI3600::afsRScla rpsL(K88E)] (20) was used as the expression host for the genomic library in the LEXAS procedure. S. lividans SBT5 was derived from S. lividans TK24 [rpsL(K88E)], and three endogenous antibiotic pathways were disrupted by deleting the entire act gene cluster (removal of 22-kb StuI fragments), deleting the two red genes redKL (removal of 5.8-kb AscI fragments), and replacing the two cda genes cdaPS3-SLI3600 (removal of a 7.1-kb PstI-EcoRI fragment) with the two global activator genes afsRScla by homologous recombination (20). S. lividans SBT18 was derived from S. lividans SBT5 by inserting an extra copy of afsRScla in the chromosome at the ΦBT1 attachment site using pAfsRScla (26). S. lividans SBT18, S. coelicolor M1152 [Δact Δred Δcpk Δcda rpoB(C1298T)], and S. coelicolor M1154 [Δact Δred Δcpk Δcda rpoB (C1298T) rpsL(A262G)] (18) were used as heterologous expression hosts to optimize metabolite production. E. coli DH10B was used as the host for BAC library construction. E. coli ET12567 bearing the RK2-derived helper plasmid pUB307 (23) was used to facilitate the intergeneric triparental conjugation from E. coli DH10B/BACs to Streptomyces. Staphylococcus aureus, Bacillus mycoides, Mycobacterium smegmatis mc2155, and Saccharomyces sake were used as indicators for the bioassay experiments.
pHL921 (see Fig. S1 in the supplemental material), bearing the origin of transfer from RK2 (oriTRK2) and int and attP from the Streptomyces phage ΦC31, was used for the construction of a genomic BAC library so that the resultant BAC clones can be mobilized and integrated into the Streptomyces chromosome at the site of attachment, attB. 2B8 and 8E6 were BAC clones bearing the streptothricin gene cluster. 8H1 was the BAC clone bearing the borrelidin gene cluster. 2A6, 3E5, 5A8, 8C10, and 8D1 were BAC clones bearing the cda2 gene cluster. 2F3, 6E1, 6F11, and 8A11 were BAC clones bearing the gene cluster for the unknown antibiotic. All strains, plasmids, and BACs used in this study are listed in Table 1.
TABLE 1.
Strains, plasmids, and BACs used in this study
| Strain, plasmid, or BAC | Description | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH10B | General cloning and plasmid maintenance | Invitrogen |
| ET12567 | dam dcm hsdM hsdS hsdR cat tet | 50 |
| Streptomyces rochei Sal35 | Wild-type strain for genomic BAC library construction | CCTCC |
| Streptomyces lividans | ||
| SBT5 | Δact ΔredKL ΔcdaPS3::afsRScla rpsL(K88E) | 20 |
| SBT18 | Δact ΔredKL ΔcdaPS3::afsRScla afsRScla rpsL(K88E) | 26 |
| Streptomyces coelicolor | ||
| M1152 | M145 Δact Δred Δcpk Δcda rpoB(C1298T) | 18 |
| M1154 | M145 Δact Δred Δcpk Δcda rpoB(C1298T) rpsL(A262G) | 18 |
| YF11 | M145 Δact ΔredL | 51 |
| Streptomyces avermitilis NRRL8165 | Wild-type strain | NRRL |
| Staphylococcus aureus CICC 10201 | Sensitive strain for bioassay in LEXAS screening | CICC |
| Bacillus mycoides | Sensitive strain for bioassay in LEXAS screening | CCTCC |
| Mycobacterium smegmatis mc2155 | Sensitive strain for bioassay in LEXAS screening | 52 |
| Saccharomyces sake | Sensitive strain for bioassay in LEXAS screening | Delin You, SJTUa |
| Plasmids | ||
| pHL921 | oriTRK2 int-attPΦC31 aac(3)-IV aadA parA parB parC; E. coli-Streptomyces shuttle vector for BAC library construction | This work |
| pUB307 | neo; RK2-derived self-mobilizable plasmid | 23 |
| BACs | ||
| 8H1 | Borrelidin-producing BAC from contig 1 | This work |
| 2B8 | Streptothricin-producing BAC from contig 2 | This work |
| 8E6 | Streptothricin-producing BAC from contig 2 | This work |
| 2A6 | Linear lipopeptide-producing BAC from contig 3 | This work |
| 3E5 | Linear lipopeptide-producing BAC from contig 3 | This work |
| 5A8 | Linear lipopeptide-producing BAC from contig 3 | This work |
| 8C10 | Linear lipopeptide-producing BAC from contig 3 | This work |
| 8D1 | Linear lipopeptide-producing BAC from contig 3 | This work |
| 2F3 | BAC from contig 4 producing unidentified metabolites | This work |
| 6E1 | BAC from contig 4 producing unidentified metabolites | This work |
| 6F11 | BAC from contig 4 producing unidentified metabolites | This work |
| 8A11 | BAC from contig 4 producing unidentified metabolites | This work |
SJTU, Shanghai Jiao Tong University.
E. coli strains were grown in Luria-Bertani (LB) broth at 37°C. Streptomyces strains were grown on soya flour medium (SFM) agar (27) at 30°C for sporulation. Streptomyces spores were suspended in 2× YT (yeast extract-tryptone) broth and heat shocked for 10 min at 50°C for conjugation (27). The E. coli-Streptomyces conjugation mixture was plated on SFM agar supplemented with MgSO4 to 20 mM. SFM (27), GYM (28), YBP (29), and R3 (28) media were used for the fermentation of Streptomyces. S. aureus and B. mycoides were grown in LB at 37°C, M. smegmatis mc2155 was grown in LB with 1% glycerol and 0.1% Tween 80 at 37°C, and S. sake was grown in potato dextrose broth (PDB) at 30°C. LB soft agar, LB (with 1% glycerol) soft agar, and PDB soft agar (0.5% agar for each) were used for the overlaying of indicators onto the microfermentation plates.
Metabolic analysis of S. rochei Sal35 and BAC-containing Streptomyces strains.
S. rochei Sal35 and BAC-containing Streptomyces strains were fermented on agar-solidified medium (35 ml medium per 9-cm petri dish) for 6 days at 30°C. S. rochei Sal35 was fermented on SFM, GYM, YBP, and R3 media. S. coelicolor M1154/8E6 was fermented on R3 medium. S. coelicolor M1152/8H1 was fermented on GYM medium. S. coelicolor M1152/8D1, S. coelicolor M1154/8D1, and S. lividans SBT18/8D1 were fermented on R3 medium. Solid ferment from each dish was sliced individually and extracted with 35 ml methanol-ethylacetate-acetic acid in an 80:20:5 mixture for three times. The extract was concentrated under reduced rotary evaporation (Buchi R210 rotary evaporator) at 40°C and dissolved in 1 ml methanol (MeOH). The crude extract (20 μl) was analyzed by high-performance liquid chromatography coupled with a diode array detector and followed by mass spectrometry (HPLC-DAD-MS). Borrelidin and lipopeptides were analyzed on a reverse-phase C18 column (Agilent Zorbax ODS C18, 5 μm, 4.6 by 250 mm). The mobile phases used were H2O (0.1% formic acid [A]) and acetonitrile (ACN [B]). The elution gradient was 0 to 5 min with 95% A–5% B, 20 min with 60% A–40% B, 30 to 35 min with 100% B, and 36 to 45 min with 95% A–5% B at a flow rate of 0.6 ml/min. Under this condition, borrelidin was eluted at 33.6 min. Lipopeptides 8D1-1 and 8D1-2 were eluted at 27.8 and 28.2 min, respectively. Streptothricins were analyzed on a reverse-phase C18 column (Agilent Zorbax ODS C18, 3.5 μm, 2.1 by 150 mm) eluted with H2O (0.05% formic acid and 0.05% n-heptafluorobutyric acid [A]) and ACN (B) under the following gradient: 0 min with 90% A–10% B, 3 min with 75% A–25% B, 12 min with 70% A–30% B, 16 to 18 min with 100% B, and 18.1 to 25 min with 90% A–10% B at a flow rate of 0.3 ml/min (30).
Construction of the genomic BAC library of S. rochei Sal35.
The genomic BAC library of S. rochei Sal35 was constructed according to the standard protocol (31). Genomic DNA of S. rochei Sal35 was prepared in agarose plugs and then partially digested with Sau3AI in situ and separated by pulsed-field gel electrophoresis (PFGE). High-molecular-weight DNA fragments were recovered from the agarose gel by electroelution using a Bio-Rad model 422 Electro-Eluter at 10 mA/tube for 2 h at 4°C in 1× TAE buffer (0.04 M Tris-acetate, 1 mM EDTA). pHL921 (see Fig. S1 in the supplemental material) was digested with BamHI, and the 13.7-kb fragment was gel purified, dephosphorylated, and ligated with the partially digested genomic DNA. The ligation mixture was desalted and concentrated on a Millipore 0.025-μm-pore filter with 10% polyethylene glycol (PEG) 8000 for 2 h at 4°C and electroporated into E. coli DH10B competent cells. The electroporation mixture was incubated for 1 h in LB at 37°C and then spread onto LB agar plates containing 50 μg/ml apramycin for selection of BAC constructs.
LEXAS screening of the S. rochei Sal35 BAC genomic library.
The mobilization of the BAC clones from E. coli host to Streptomyces was accomplished using a high-throughput triparental conjugation approach, including E. coli ET12567/pUB307 (helper), DH10B/BAC strains (donors), and S. lividans SBT5 (recipient). The BAC library (conjugation donor) was inoculated into 96-well plates with LB (130 μl per well) and cultured for 4 to 6 h to an optical density at 600 nm (OD600) of 0.4 to 0.6. The helper ET12567/pUB307 was also cultured in LB (100 ml) with 50 μg/ml kanamycin to an OD600 of 0.4 to 0.6, collected by centrifugation, washed three times with fresh LB, and resuspended in 15 ml LB. Then, 20 μl of concentrated ET12567/pUB307 was pipetted into each DH10B/BAC well, resulting in a helper/donor ratio of approximately 1:1, and mixed thoroughly by shaking on a rotary shaker (Lab-Therm LT-X; Kuhner, Basel, Switzerland) at 200 rpm for 5 min. In the meantime, fresh S. lividans SBT5 spores were collected from two petri dishes of fully sporulated culture (on SFM, 30°C, 5 to 6 days) and suspended in 4 ml 2× YT medium to yield a spore suspension of ca. 109 CFU/ml. The spore suspension was incubated in a water bath for 10 min at 50°C, cooled down to room temperature, and spread on MgSO4-supplemented SFM plates (9-cm petri dishes) at 200 μl/dish. The spore-coated SFM plates were blow-dried in a laminar hood for 30 min. The E. coli donor-helper mixtures were then transferred by a 48-pin replicator onto the spore-coated plates. S. lividans SBT5 spores collected from two SFM agar dishes were sufficient for the triparental conjugation of a genomic library with 960 clones stored in 96-well plates. After the E. coli cell mixtures were transferred to the spore-coated plates, the RK2-derived helper plasmid pUB307 in ET12567 would transfer itself into the DH10B/BAC cells to induce the mobilization of the oriTRK2-containing BACs for conjugative transfer to S. lividans SBT5. After 12 to 16 h of incubation at 30°C, the conjugation plates were flooded with apramycin and spectinomycin to a final concentration of 50 μg/ml for selection of exconjugants containing intact BAC clones and trimethoprim (50 μg/ml) to remove the E. coli donor and helper strains and then incubated at 30°C for another 4 to 6 days. Exconjugants, when sporulated well, were transferred with a 48-pin replicator onto SFM plates containing apramycin (50 μg/ml) and nalidixic acid (25 μg/ml) to remove possible residual E. coli donor and helper strains and S. lividans SBT5.
The resultant library of S. lividans SBT5 exconjugants was then replicated to the selected productive media SFM, GYM, R3, and YBP for a 6-day microfermentation at 30°C. Indicator strains premixed with soft agar were overlaid onto the fermentation plates and incubated for 24 to 48 h for the observation of inhibition zones.
Identification of streptothricins and borrelidin.
Specific PCR primers designed for amplification of DNA fragments from the stt and bor pathway and LC-MS spectrometry were used to identify streptothricins and borrelidin.
DNA sequence.
The BAC DNA sequences were determined by using the Roche/454 GS (FLX Titanium) sequencing platform, and the assembled DNA sequences were submitted to antiSMASH (http://antismash.secondarymetabolites.org/) for secondary metabolite BGC analysis (32). The 16S rRNA gene was obtained by sequencing the PCR product amplified from S. rochei Sal35 genomic DNA using primers Fwd1 (AGAGTTTGATCATGGCTCAG) and Rev1 (GGTTACCTTGTTACGACTT).
Isolation and purification of linear lipopeptides 8D1-1 and 8D1-2.
Fresh spores of S. coelicolor M1152/8D1 were spread uniformly onto 30 liters of solidified R3 medium and cultured for 6 days at 30°C. The fermented culture was extracted three times with equal volumes of methanol-ethylacetate-acetic acid in an 80:20:5 mixture. The organic extract was concentrated under reduced pressure at 40°C in a rotary evaporator (Heidolph Laborota 20 control safety), dissolved in 3 liters of sterile water, and flushed through a CHP20P column eluted with an H2O-MeOH gradient. The fractions containing 8D1-1 and 8D1-2 were collected, loaded onto a C18 reverse-phase column, and eluted with the H2O-MeOH mixture from 50% to 100% MeOH. The 60% and 70% MeOH eluents containing 8D1-1 and 8D1-2 were collected, injected into a semipreparative C18 reverse-phase column (Agilent Zorbax ODS C18, 5 μm, 9.4 by 250 mm), and eluted with H2O (0.1% formic acid [A]) and ACN (B). A 15-min isocratic elution with 48% B at a flow rate of 2 ml/min was used. 8D1-1 and 8D1-2 were eluted at 5.7 and 8.9 min, respectively. Finally, 20 mg of 8D1-1 and 15 mg of 8D1-2 were prepared. Chemical structures of 8D1-1 and 8D1-2 were determined by high-resolution mass spectrometry, tandem MS (MS/MS), and one-dimensional (1D) and 2D nuclear magnetic resonance (NMR) spectrometry.
Marfey assay for chiral analysis of amino acids.
For determination of the configuration of 4-hydroxyphenylglycine 6 (Hpg6) and 3-OH-Asn9, a sample of ca. 0.1 mg 8D1-1 was hydrolyzed in 2 ml 6 N HCl in a sealed tube at 120°C for 24 h. The hydrolysate was dried under reduced pressure at 50°C in a Buchi R210 rotary evaporator, dissolved in 20 μl double-distilled water (ddH2O), and transferred to a 1.5-ml Eppendorf tube, and 40 μl of 1% acetone solution of N-(5-fluoro-2, 4-dinitrophenyl)-d-alaninamide (FDAA [Marfey's reagent]) and 8 μl of 1 M NaHCO3 solution were added to the hydrolysate for derivatization. The reaction mixture was heated at 40°C with frequent shaking for 1 h over a hot plate and then cooled to room temperature. The reaction was quenched by adding 4 μl of 2 N HCl and diluted with 200 μl MeOH for LC-MS analysis (33). To determine the configuration of Trp3, a sample of 0.1 mg 8D1-1 was hydrolyzed in a 2-ml solution of trifluoroacetic acid-HCl at 1:2 (5% thioglycolic acid) in a sealed tube at 166°C for 0.5 h (34). The hydrolysate was then dried under reduced pressure at 60°C and treated by the same procedure as above. Standards (l/d-Hpg, 3-OH-l-Asp, and l/d-Trp) were derivatized using the same procedure.
LC-MS analysis for FDAA-derivatized amino acids.
FDAA-derivatized amino acids (2 μl) were injected to Agilent G6530 high-resolution electrospray ionization-quadrupole time of flight (HRESI-QTOF) mass spectrometer equipped with an Agilent 1260 HPLC system on a C18 reverse-phase column (Agilent Zorbax ODS C18, 3.5 μm, 2.1 by 150 mm). An elution gradient of 0 to 20 min with 10 to 80% B, 20 to 25 min with 80 to 100% B, and 26 to 35 min with 10% B was applied, with H2O (0.1% formic acid [A]) and ACN (B) as the mobile phases at a flow rate of 0.2 ml/min. The mass spectrometer was operated in positive mode, recording a mass range of m/z 50 to 1,700. Molecular masses of 457.1 ± 0.2 Da, 420.1 ± 0.2 Da, and 402.1 ± 0.2 Da corresponding to the FDAA-Trp, FDAA-Hpg, and FDAA-3-OH-Asp residues, respectively, were extracted for data analysis.
Accession number(s).
The DNA sequences obtained in this research have been deposited in the NCBI database under GenBank accession no. KX440952 (16S rRNA gene), KP823602 (pHL921), KT362046 (borrelidin BGC), KT362049 (streptothricin BGC), KT362047 (linear lipopeptide BGC), and KX346560 (unknown antibiotic).
RESULTS
LEXAS procedure to discover antibiotics and biosynthetic gene clusters in Streptomyces.
LEXAS is designed to discover antibiotics and BGCs in Streptomyces. The procedure (Fig. 1) involves an engineered antibiotic-overproducing S. lividans strain, SBT5, that allows high-frequency conjugative DNA transfer from E. coli (cloning host of the BAC library) and stimulates production of antibiotics encoded by heterologous BGCs from genomic libraries of Streptomyces strains. We developed a high-throughput library conjugation procedure by modifying the standard triparental conjugation protocol (27) to guarantee that each primary BAC clone in E. coli results in a single exconjugant Streptomyces culture. As a result, multiple cryptic BGCs from a natural strain are separated and distributed into an array of discrete exconjugants suitable for optimized heterologous expression. This enables bioassay-guided high-throughput screening of the library specifically for BGCs producing antibiotics. Because each BAC clone is transferred to Streptomyces individually in an orderly manner, it is simple to trace active clones back to the original E. coli transformants in 96-well plates, allowing facile DNA sequencing and making genetic modifications.
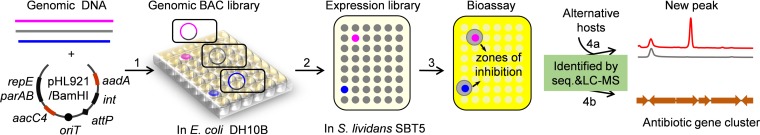
FIG 1.
Schematic representation of the LEXAS library expression and analysis procedure. (Step 1) Streptomyces DNA fragments of ca. 100 kb are cloned into the mobilizable BAC vector pHL921. (Step 2) Using a triparental mating, the pHL921-derived BAC clones are transferred well to spot from E. coli DH10B to the expression host S. lividans SBT5. DNA and clones containing antibiotic biosynthetic gene clusters are highlighted in purple or blue. (Step 3) Replicas of the SBT5 clones are screened for antimicrobial activity using multiple indicator strains. (Steps 4a and b) HPLC-MS analysis of metabolites and DNA sequence analyses of active clones.
The LEXAS procedure comprises four steps. Step 1 is to construct a BAC genomic library in E. coli DH10B using high-molecular-weight DNA fragments from the target Streptomyces strain. E. coli DH10B maintains the cloned DNA stably, but it does not support expression of the Streptomyces genes. The E. coli-Streptomyces shuttle vector pHL921 was specifically designed to carry the entire long insert DNA into Streptomyces for heterologous expression (Fig. 1; see Fig. S1 in the supplemental material). In pHL921, two antibiotic resistance genes, aacC4 and aadA, were placed on both sides of oriTRK2 to provide dual selection to avoid previously reported abortive conjugation (22).
In step 2, the BAC clones are mobilized to Streptomyces in a high-throughput way. Three Streptomyces species were tested as recipients using DH10B/pHL921 cultured in 96-well microtiter plates. S. lividans SBT5 produced exconjugants corresponding to all wells, while S. coelicolor and S. avermitilis produced none. S. lividans SBT5 was therefore selected as the primary host for high-throughput expression of BAC libraries. Because of engineered gene deletions, S. lividans SBT5 has lost the ability to produce the endogenous actinorhodin, undecylprodigiosin, and calcium-dependent antibiotics, but it has three global activators that favor the production of antibiotics from cloned exogenous BGCs (20).
Step 3 is a bioassay for antimicrobial activity. The resulting exconjugants, each containing a single BAC clone inserted site specifically into the S. lividans SBT5 chromosome, are replica plated to different agar media for microfermentation. Overlaying with antibiotic-sensitive indicator strains reveals growth inhibition zones around exconjugants that produce an antibiotic encoded by the BAC clones.
Step 4 is divided into two parts for identification of secondary metabolites (4a) and corresponding BGCs (4b). Step 4a involves testing of active clones in alternative expression hosts such as S. coelicolor M1152, S. coelicolor M1154, and S. lividans SBT18, to achieve optimal antibiotic production for structure elucidation and biological testing. Conjugative transfer of specific pHL921-derived BAC clones into strains other than S. lividans SBT5 is possible using more cells and one petri dish for each standard triparental mating. In step 4b, related, overlapping clones are identified using restriction enzyme digestion, PCR, and sequencing.
Production of streptothricins and borrelidin by S. rochei Sal35 and the LEXAS clones.
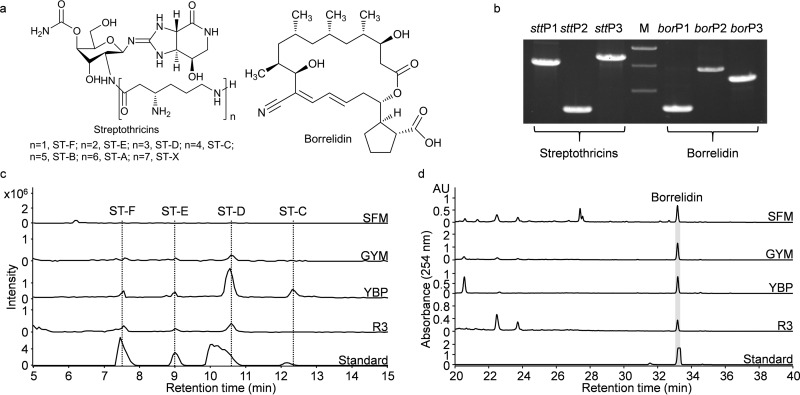
As a test for the validation of the LEXAS procedure, we chose S. rochei Sal35, recently isolated from Shennongjia (Eastern Hubei) forest soil. Other S. rochei strains have previously been reported to hold BGCs for the production of streptothricin (30), borrelidin (35), lankacidin (36), and lankamycin (37). PCR primers were designed to detect the homologous BGCs in S. rochei Sal35. Streptothricin primers (stt) targeting the two stand-alone adenylation domain-coding genes orf5 and orf19 and the resistance gene sttA resulted in specific PCR products of 1.5, 0.7, and 1.7 kb, as expected from the documented stt gene cluster (30). Borrelidin primers (bor) targeting the precursor trans-cyclopentane dicarboxylic acid synthesis gene borE, polyketide synthase (PKS) gene borA2, and postmodification of nitrile group introduction gene borJ resulted in the expected PCR products of 0.7, 1.5, and 1.2 kb (35) (Table 2 and Fig. 2b). The sequences of all the PCR products confirmed the presence of the expected stt and bor genes in S. rochei Sal35. Lankacidin primers (lkc) targeting the PKS genes lkcC and lkcF and amine oxidase gene lkcE and lankamycin primers (lkm) targeting the two P450 hydroxylase genes lkmF and lkmK and the β-glucosidase gene lkmH were also designed to probe the lkc and lkm pathways in S. rochei Sal35 (Table 2). However, the lankacidin (lkc) and lankamycin (lkm) primers did not give specific PCR products, suggesting the absence of lkc and lkm pathways in S. rochei Sal35. The lkc and lkm pathways were reported to be located on a 210-kb linear plasmid pSAL2-L in S. rochei 7434AN4 (36, 37), while no plasmid band was detected by PFGE in S. rochei Sal35, further supporting the absence of lkc and lkm pathways. In addition to the PCR amplifications, the crude extracts of S. rochei Sal35 ferments were also analyzed by HPLC-DAD-MS. In accordance with the PCR results, streptothricins (ST-B, ST-C, ST-D, ST-E, and ST-F) and borrelidin, but not lankacidin or lankamycin, were detected in the methanol extracts, which indicated that S. rochei Sal35 could only produce streptothricins and borrelidin (Fig. 2c and d; see Fig. S2 in the supplemental material).
TABLE 2.
PCR primers designed to verify the stt, bor, lkc, and lkm pathways
| Primer paira | Primer sequence (5′→3′) | Product size (bp) |
|---|---|---|
| borP1-F | AACTGGCAGGACTACAACACC | 743 |
| borP1-R | GGGAGACGGCCCTTCTC | |
| borP2-F | TCACCTGGTAACAGCCACCA | 1,480 |
| borP2-R | CATCAGCCCTTTCACTCTTCG | |
| borP3-F | TCGTGTCGTGCCGTCAT | 1,201 |
| borP3-R | GCTGAAGGAGGAAGCGTGA | |
| lkcP1-F | TCCGTGTCTTCGTCCCTGA | 1,429 |
| lkcP1-R | CCGATGTCCCGCTCCTT | |
| lkcP2-F | GTCGCCCCATGCCACCACCAG | 1,835 |
| lkcP2-R | CGCCCACCAAGCCCGTCGTCT | |
| lkcP3-F | GTGGGGCTGAGGAATCTGC | 1,271 |
| lkcP3-R | GCCTGCGACACGCTGCT | |
| lkmP1-F | CGGGAAGCAGACGAGCC | 1,612 |
| lkmP1-R | CGACCAAGGGCAATGTGAC | |
| lkmP2-F | GAATGACGACCTGAGGAAATGC | 675 |
| lkmP2-R | CCGCCAGAAGCCGAGAAC | |
| lkmP3-F | CTGTGGGCGATGGTGAGC | 1,807 |
| lkmP3-R | CGATGTCGGCGATGCG | |
| sttP1-F | CGCCAATATTCCAACAGAATC | 1,536 |
| sttP1-R | CCACGGTGCCCAGGTAG | |
| sttP2-F | GCATGCTCATGTAGAGCGC | 727 |
| sttP2-R | GAACAGTCGTCGAAATGGG | |
| sttP3-F | ACCGATGCGTCGAAGAGC | 1,710 |
| sttP3-R | CCTGCCATGACCCAGCC |
Primer orientation: F, forward; R, reverse.
FIG 2.
Identification of streptothricins and borrelidin from S. rochei Sal35. (a) Chemical structures of streptothricins and borrelidin produced by S. rochei Sal35. (b) PCR amplification of specific DNA fragments in the stt and bor pathways from S. rochei Sal35 genomic DNA. The DNA marker was the 1-kb DNA ladder from Dongsheng Biotech, and the indicative sizes of the bands are 2, 1.5, and 1 kb in panel b. Sequences of PCR primers are listed in Table 2. (c) Extracted ion chromatogram spectra of streptothricins produced by S. rochei Sal35 fermented on SFM, GYM, YBP, and R3 media. ST-F (m/z, 503.4), ST-E (m/z, 631.6), ST-D (m/z, 759.7), and ST-C (m/z, 887.8) were detected and compared to the streptothricin standards; the mass spectra of each compound are shown in Fig. S2 in the supplemental material. (d) HPLC profiles of borrelidin production in S. rochei Sal35 fermented on SFM, GYM, YBP, and R3 media. AU, absorbance units. Note the different scale on the y axis.
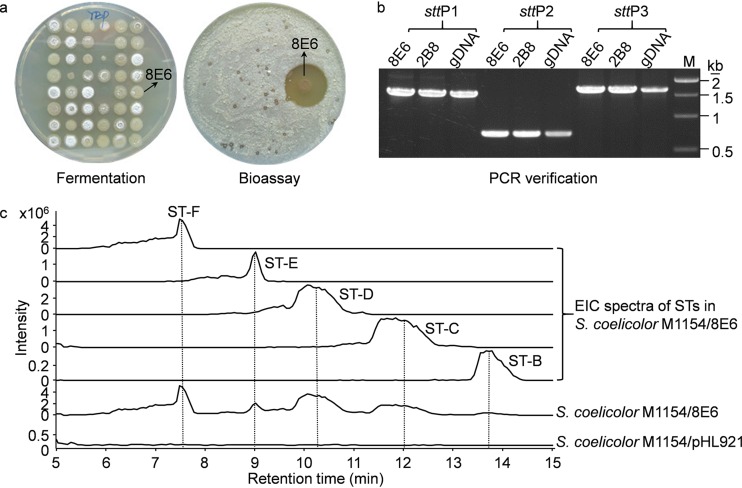
To test the LEXAS, a BAC genomic library of S. rochei Sal35 was constructed with 768 independent E. coli clones with an average insert size of ca.100 kb, providing ca. 8 genome equivalents of cloned DNA (see Fig. S3 in the supplemental material). The high-throughput library conjugation gave rise to S. lividans exconjugants for 93% of all BAC clones (Fig. 3a). After microfermentation and antimicrobial screening, 12 (1.7%) S. lividans SBT5 exconjugants from the BAC library exhibited antimicrobial activity (Fig. 3a and Table 3; see Fig. S4 in the supplemental material). All of the 12 BAC clones were extracted for restriction enzyme analysis and grouped into 4 contigs according to the restriction patterns (Table 3 and Fig. 4a and b; see Fig. S5 in the supplemental material). PCR detection with specific stt and bor primers revealed the presence of the bor pathway in contig 1 (8H1) and the stt pathway in contig 2 (2B8 and 8E6) (Fig. 3b; see Fig. S6a in the supplemental material). Metabolic profiling of the Streptomyces exconjugants of 8E6 and 8H1 also revealed the accumulation of streptothricins and borrelidin, respectively (Fig. 3c; see Fig. S6b). In addition, compared to the original strain, S. rochei Sal35, S. coelicolor M1154/8E6 produced more streptothricin components (Fig. 3c; see Fig. S2a in the supplemental material). This demonstrated that LEXAS could be used to capture and express large BGCs.
FIG 3.
LEXAS screening of S. rochei Sal35 BAC genomic library revealed the heterologous expression of the background stt pathway. (a) Selected microferments and bioassay plates from the LEXAS screening procedure. The positive 8E6 BAC exconjugant exhibited strong antibacterial activity against M. smegmatis mc2155. (b) PCR verification of the stt pathway in 2B8 and 8E6. Genomic DNA (gDNA) was set as the control. (c) Comparative metabolic profiling of S. coelicolor M1154/8E6 and S. coelicolor M1154/pHL921 propagated in R3 medium. EIC, extracted ion chromatography. Note the different scale on the y axis.
TABLE 3.
Positive BAC clone hits from S. rochei Sal35 by LEXAS
| Contig no. | BAC clone(s) | Antimicrobial spectruma | Metabolite(s) |
|---|---|---|---|
| 1 | 8H1 | S. aureus | Borrelidin |
| 2 | 2B8, 8E6 | S. aureus, B. mycoides, M. smegmatis mc2155, S. sake | Streptothricins |
| 3 | 2A6, 3E5, 5A8, 8C10, 8D1 | S. aureus | Linear lipopeptides |
| 4 | 2F3, 6E1, 6F11, 8A11 | S. aureus, B. mycoides | Unknownb |
Inhibition zone formation observed in the bioactivity screening to indicator strains.
The metabolite encoded by contig 4 was not detected in this work.
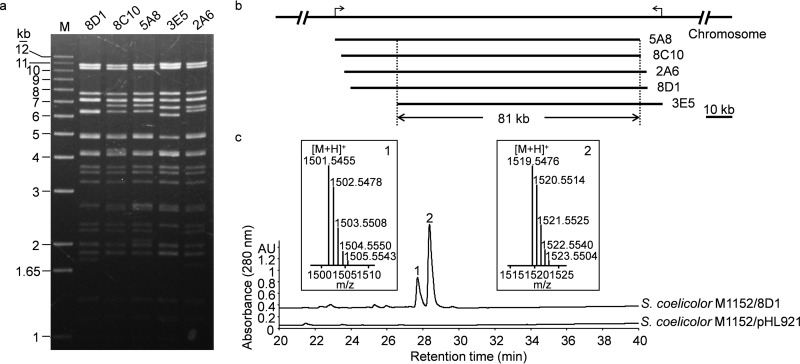
FIG 4.
Discovery of a cryptic BGC from S. rochei Sal35 producing two lipopeptide compounds. (a) PvuII digestion of BAC clones from contig 3 showing multiple fragments of identical sizes. (b) Organization of the BAC clones in the chromosome. The two black arrows on the S. rochei Sal35 chromosomal DNA indicate the boundary of contig 3. The overlapping region of 81 kb contains the cryptic BGC. (c) Comparative metabolic profiling of S. coelicolor M1152/8D1 and S. coelicolor M1152/pHL921 by HPLC. Two differential peaks were eluted at 28 min, and the corresponding MS spectra of 8D1-1 (m/z, 1,501.5455) and 8D1-2 (m/z, 1,519.5476) are shown as inserts. AU, absorbance units.
Furthermore, BAC clones of contigs 3 and 4 gave no band in PCR amplification when stt or bor primers were used. When isolated BAC DNA of these contigs was transferred to E. coli and conjugated to S. lividans SBT5, all resulting exconjugants displayed antibacterial activity. These findings suggested that contigs 3 and 4 might carry cryptic BGCs for previously unreported metabolites in S. rochei strains.
Identification of two linear lipopeptides from BAC clone of contig 3.
One of the active BAC clones, 8D1 of contig 3, was introduced into S. lividans SBT18, S. coelicolor M1152, and S. coelicolor M1154. The exconjugants were fermented on R3 medium, extracted, and analyzed by HPLC and LC-MS to detect new compounds. Compared with the vector control, two differential HPLC peaks were observed in S. coelicolor M1154/8D1 and S. coelicolor M1152/8D1 (Fig. 4c). The molecular formula was determined using HRESI-MS to be C66H80N14O27 for peak 1 compound (named 8D1-1; m/z, 1,501.5455 [M+H]+) and C66H82N14O28 for peak 2 compound (named 8D1-2; m/z, 1,519.5476 [M+H]+) (Fig. 4c), None of these was represented in the SciFinder or PubChem databases (38). 8D1-1 and 8D1-2 were also detected in the extract of S. lividans SBT18/8D1 by HPLC (see Fig. S7 in the supplemental material). Furthermore, calcium-dependent antibiotic (CDA) components CDA3a, CDA3b, CDA4a, and CDA4b were detected in S. lividans SBT18/8D1 by HRESI-MS (see Fig. S7), suggesting that BAC clone 8D1 complemented the CDA production in S. lividans SBT18, whose endogenous CDA biosynthetic pathway has been disrupted (see Fig. S8 in the supplemental material). In addition, HPLC and MS analyses of S. rochei Sal35 extracts, fermented in four different production media, indicated that the native strain did not produce these compounds (see Fig. S9 in the supplemental material).
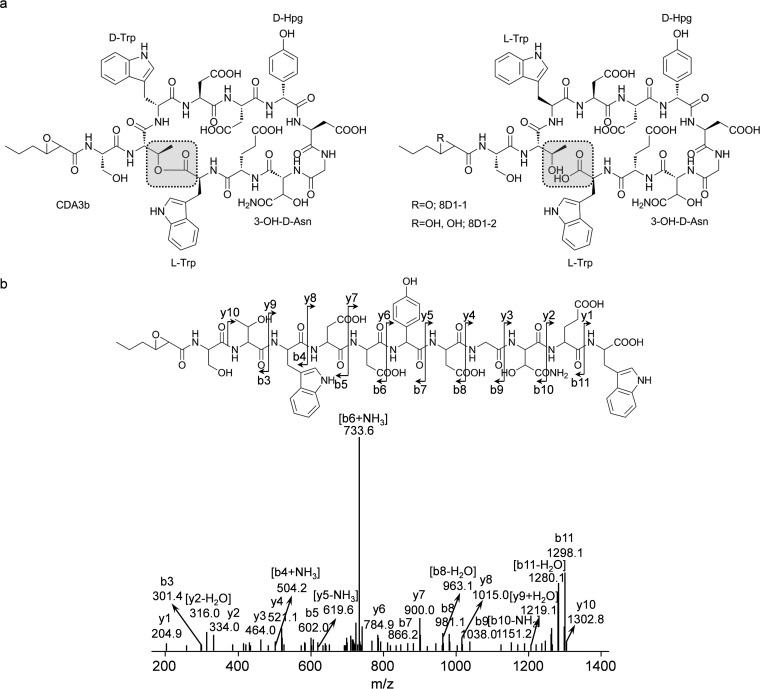
To elucidate the chemical structure of these new compounds, 20 mg of 8D1-1 and 15 mg of 8D1-2 pure compounds were prepared by macroporous resin column and reverse-phase column chromatography from S. coelicolor M1152/8D1 extract. The structure of 8D1-1 was determined by MS/MS, 1H-NMR, 13C-NMR, and 2D NMR as a linear lipopeptide with an undecapeptide, Ser1-Thr2-Trp3-Asp4-Asp5-Hpg6-Asp7-Gly8-Han9-Glu10-Trp11, linked at the NH2 terminus by a 3-propyloxirane-2-carboxylic acid (Poc) with an amide bond, where “Hpg” is 4-hydroxyphenylglycine and “Han” is 3-hydroxyasparagine (3-OH-Asn) (Fig. 5a and b; see Table S1 in the supplemental material). The structure of 8D1-2 was determined as an epoxy-ring-opened derivative of 8D1-1: i.e., a 2,3-dihydroxyhexanoic acid (Dhh) unit linked with the same undecapeptide (Fig. 5a; see Table S2 in the supplemental material). The detailed NMR data of 8D1-1 and 8D1-2 are summarized in Tables S1 and S2.
FIG 5.
Structure determination of the linear lipopeptides 8D1-1 and 8D1-2. (a) Chemical structures of CDA3b from S. coelicolor A3(2) and 8D1-1 and 8D1-2 from S. rochei Sal35. CDA3b is cyclic, and 8D1-1 and 8D1-2 are linear without an ester bond (gray highlights). 8D1-1 and CDA3b contain an epoxide group, which is hydrolyzed in 8D1-2. (b) MS/MS analysis of 8D1-1 showing the “b/y” fragments.
The primary structure of 8D1-1 is highly similar to that of the cyclic depsipeptide calcium-dependent antibiotic CDA3b produced by S. coelicolor and S. lividans, in which the Trp3, Hpg6, and 3-OH-Asn residues are epimerized to a d-configuration during their incorporation into the final product by the CDA nonribosomal peptide synthetase (NRPS) assembly line (39). To determine the configuration of these residues in the linear lipopeptides, 8D1-1 was hydrolyzed, derivatized with FDAA, and analyzed by LC-MS in parallel with standard stereoisomeric monomers. Trp3, Hpg6, and 3-OH-Asn of 8D1-1 were determined to be in the l-, d-, and d-configurations, respectively (Fig. 6; see Fig. S10 in the supplemental material). In summary, 8D1-1 was a linear and stereoisomeric lipopeptide analog of CDA3b.
FIG 6.
Chirality of Trp3 in 8D1-1. (a) Marfey's reagent FDAA was used to derivatize d- and l-tryptophan and the 8D1-1 acid hydrolysate. Both Trp3 and Trp11 residues in 8D1 were mainly in the l-configuration, while Trp3 and Trp11 in CDA3b should be in the d-configuration and l-configuration, respectively. In addition, a 1:1 ratio of l-Trp to d-Trp in CDA3b should be expected (39). Small amounts of d-tryptophan were also detected in 8D1-1, possibly because of partial activity of E1 domain. (b) Mass spectrum of FDAA-l/d-Trp.
Bioinformatic analysis of the cda2 gene cluster from S. rochei Sal35 for the biosynthesis of 8D1-1/2.
All five BAC clones of contig 3 produced 8D1-1 or -2 in S. coelicolor M1152. Restriction digestion indicated an overlapping region of ca. 81 kb (Fig. 4a and b). DNA sequence analysis revealed that the 81-kb overlapping region contained a cluster of genes similar to the CDA biosynthetic gene cluster from S. coelicolor A3(2), and the comparison of the two BGCs is summarized in Fig. S11 in the supplemental material. Therefore, the name cda2 was given to this gene cluster originating from S. rochei Sal35. cda2 contained all biosynthetic and regulatory genes of the cda pathway that had been reported to be important for the production of CDA3b in S. coelicolor A3(2) (40), including three NRPS genes (cda2PS1, cda2PS2, and cda2PS3), homologous genes for the biosynthesis of trans-2,3-epoxyhexanoyl-coenzyme A (CoA), and the nonproteinogenic amino acid 4-OH-Hpg, the modification of Asn9, and transcriptional regulation (see Fig. S11). In addition, the NRPS domains in Cda2PS1, Cda2PS2, and Cda2PS3 were organized the same as the counterparts in the S. coelicolor pathway. Specifically, three epimerization domains of the CDA peptide synthases in S. coelicolor A3(2) were also present in Cda2PS1 and Cda2PS2, according to the prediction by the NRPS analysis (41). The three epimerization domains of CDA NRPSs (two in CdaPS1 and one in CdaPS2) were responsible for the l- to d-configuration conversions of the Trp3, Hpg6, and 3-OH-Asn residues in CDAs (40). However, the preservation of l-Trp3 in 8D1-1/2 suggested that the corresponding epimerization domain in CdaPS1 did not function during the assembly of these linear lipopeptides. Multiple-sequence alignment of the epimerization domains indicated those functionally important residues documented previously were conserved in the E1 epimerization domain of Cda2PS1, while one residue, Val159, was mutated to Met159 (42) (see Fig. S12 in the supplemental material). This V159M point mutation, probably along with other point mutations, might be responsible for the loss of function of the Cda2PS1 E1 domain leading to the preservation of l-Trp3 during assembly of the lipopeptide intermediates.
Bioinformatic analysis of contig 4.
In S. lividans SBT5, the four BAC clones of contig 4 (2F3, 6E1, 6F11, and 8A11) inhibited S. aureus and B. mycoides. The alternative host, S. lividans SBT18/8A11, produced the largest inhibition zones and was used to accumulate corresponding metabolites. Unfortunately, no differential HPLC or MS signals were observed by comparative metabolic profiling to S. lividans SBT18/pHL921. We also tried to purify the bioactive metabolites by tracing bioactivity to Bacillus cereus, but we failed to obtain a pure compound because the activity was lost during vacuum drying or lyophilization, which indicates that the compound might have been volatile.
The four BAC clones of contig 4 shared a 68-kb overlapping DNA region (see Fig. S5b in the supplemental material), in which a hybrid arylpolyene-lanthipeptide gene cluster (3) was predicted by antiSMASH (32). The predicted gene cluster covered a ca. 25-kb DNA region, including homologous genes for a long-chain acyl-CoA synthetase (CoA ligase), two β-ketoacyl-acyl carrier protein (ACP) synthases, one β-ketoacyl-ACP reductase, one enoyl-ACP reductase, a stand-alone ACP, and a multidomain protein containing a middle Ser/Thr protein kinase domain and a C-terminal lanthionine synthetase C (LanC)-like domain. The predicted functions of the hypothetical proteins are listed in Table S3 in the supplemental material.
DISCUSSION
The millions of cryptic BGCs in microbial genomes are a potential treasure trove for future drug development (3–5, 43, 44). However, most BGCs in natural strains are silent under laboratory conditions. The coexistence of multiple secondary metabolite BGCs in one microbial strain imposes an additional tier of complications to antibiotic discovery. The LEXAS procedure described here is essentially a heterologous expression approach for activating and mining of cryptic metabolites from Streptomyces, the most productive genus of bacteria. The library conjugation procedure of LEXAS allowed partitioning of multiple cryptic BGCs of natural Streptomyces strains into arrays of single Streptomyces exconjugants for heterologous expression, thereby eliminating potential interpathway competition for common biosynthetic substrates, cross talk or cross regulation, and interference in bioassays. Furthermore, the utilization of the engineered antibiotic overproduction host S. lividans SBT5 allowed efficient activation of production of otherwise cryptic compounds, such as the linear lipopeptides 8D1-1 and -2 in this study. One important feature of LEXAS is the high-throughput bulk transfer of the genomic BAC library in 96-well plates to S. lividans SBT5 to generate a “well-to-spot” copy of the exconjugant library. Therefore, the subsequent bioactive hits could be easily traced back to the E. coli clones in the 96-well plates. Consequently, the positive clones could be easily sequenced, genetically modified, or transferred to surrogate hosts for optimized production of secondary metabolites.
LEXAS is based on the BAC library, which can bear exogenous DNA fragments of 100 kb and more (11); therefore, it is suitable to mine secondary metabolites encoded by large BGCs such as the giant PKS and NRPS gene clusters, as demonstrated here by the large gene clusters of streptothricins (∼40 kb), borrelidin (∼70 kb), and linear lipopeptides 8D1-1 and -2 (∼80 kb). In addition, LEXAS was very efficient because conjugative transfer, microfermentation, and the bioassay were all performed in a high-throughput way using 48 different BAC clones representing almost half of a Streptomyces genome per standard petri dish. The 1.7% hit rate in BAC clones and 3 to 4 bioactive contigs from the BAC genomic library of one Streptomyces strain (Sal35) are good signs, given that tens of thousands of strains have been collected by pharmaceutical companies and others. Most importantly, LEXAS is a bioactivity-guided functional mining system, which is independent of sequence analysis. Therefore, it is promising to use LEXAS for discovery of novel antibiotics synthesized by entirely novel enzymes that are not readily predicted bioinformatically.
LEXAS will, of course, rediscover many known antibiotics like streptothricins and borrelidin here. In this work, PCR, DNA sequencing, and LC-MS analysis are used to recognize known BGCs and compounds. However, Baltz developed an E. coli K-12 indicator strain that is resistant to most commonly rediscovered antibiotics, such as streptothricin, streptomycin, tetracycline, chloramphenicol, ampicillin, bleomycin, rifampin, and aminoglycosides, by inserting their resistance genes into the chromosome (45). This strain could also be introduced to the LEXAS procedure to exclude known antibiotics.
Three routes have been reported to generate linear peptide carboxylates instead of cyclic peptide lactones/lactams in microbes: (i) a C-terminal thioesterase (TE) domain of ACV synthetase catalyzed hydrolysis of the enzyme-S-peptide intermediate in the penicillin biosynthetic pathway (46), (ii) a serine protease catalyzed hydrolysis of the ester bond of the cyclic depsipeptide daptomycin in actinomycetes (47), and (iii) mutations in epimerization domain disabled the formation of d-configured amino acid residues (42), which might influence proper folding of the acyl-peptide intermediate required for the intramolecular cyclization reaction to proceed (48). In this study, Cda2PS3 of the 8D1-1/2 biosynthetic pathway has a C-terminal TE domain and the same NRPS domain architecture as CdaPS3: i.e., C-A-T-C-A-T-TE. The restored production of cyclized CDA3a/b and CDA4a/b in S. lividans SBT18/8D1 suggested that the TE domain of Cda2PS3 possessed cyclization activity like CdaPS3-TE (49). Based on the genetic complementation data and the fact that the linear lipopeptides 8D1-1 and -2 contain an l-Trp3 in place of d-Trp3, here we propose that the third route is responsible for the generation of the linear products in the cda2 pathway: the Cda2 NRPS machinery generates l-Trp3-lipopeptide-enzyme intermediates, which cannot be cyclized by Cda2PS3-TE, and consequently, hydrolysis of the intermediates releases the linear products. Because both S. lividans and S. coelicolor are native producers of CDAs and no linear derivative has been reported in either strain, it is unlikely that 8D1-1 or 8D1-2 is generated from hydrolysis by an endogenous serine protease as in the case of daptomycin hydrolysis (47).
In conclusion, the LEXAS procedure established in this study is ready for functional genome mining, its usefulness was demonstrated in the discovery of metabolites encoded by very large BGCs, including an aminoglycoside (streptothricins), a polyketide (borrelidin), two linear lipopeptides (8D1-1 and -2), and one unknown antibiotic, of which the last two were reported for the first time in strains of S. rochei.
Supplementary Material
ACKNOWLEDGMENTS
We thank Keith Chater, David Hopwood, and Tobias Kieser for critical reading and correction of the manuscript, Mervyn Bibb, Gerard Wright, and Julian Davies for helpful discussion at the 2012 John Innes-Rudjer Bošković Summer School in Dubrovnik, and Mervyn Bibb, Songwang Hou, Colin Smith, and Tianshen Tao for gifts of plasmids and bacterial strains.
This work was supported by the Ministry of Science and Technology (973: 2009CB118905 and 863: 2010AA10A201), the National Science Foundation of China (31370134), and the Ministry of Education (NCET-08-0778-533-09001) of China, and the Science and Technology Commission of Shanghai Municipality (15JC1400401).
Funding Statement
This work, including the efforts of Meifeng TAO, was funded by the Natural Science Foundation of the People’s Republic of China (31370134), the Ministry of Education of the People’s Republic of China (MOE) (NCET-08-0778-533-09001), the Science and Technology Commission of Shanghai Municipality (STCSM) (15JC1400401), and the Ministry of Science and Technology of the People’s Republic of China (MOST) (2009CB118905 and 2010AA10A201). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01383-16.
REFERENCES
- 1.Newman DJ, Cragg GM. 2012. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 3.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JW, Vederas JC. 2009. Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 5.Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seyedsayamdost MR. 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A 111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lautru S, Deeth RJ, Bailey LM, Challis GL. 2005. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol 1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 8.Pidot S, Ishida K, Cyrulies M, Hertweck C. 2014. Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew Chem Int Ed Engl 53:7856–7859. doi: 10.1002/anie.201402632. [DOI] [PubMed] [Google Scholar]
- 9.Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. 2011. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci U S A 108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang HS, Brady SF. 2013. Arimetamycin A: improving clinically relevant families of natural products through sequence-guided screening of soil metagenomes. Angew Chem Int Ed Engl 52:11063–11067. doi: 10.1002/anie.201305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmer R, Verrinder Gibbins AM. 1997. Construction and characterization of a large-fragment chicken bacterial artificial chromosome library. Genomics 42:217–226. doi: 10.1006/geno.1997.4738. [DOI] [PubMed] [Google Scholar]
- 12.Miao V, Coeffet-Legal MF, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, Silva CJ, Wrigley SK, Baltz RH. 2005. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Jiang H, Haltli B, Kulowski K, Muszynska E, Feng X, Summers M, Young M, Graziani E, Koehn F, Carter GT, He M. 2009. Rapid cloning and heterologous expression of the meridamycin biosynthetic gene cluster using a versatile Escherichia coli-Streptomyces artificial chromosome vector, pSBAC. J Nat Prod 72:389–395. doi: 10.1021/np8006149. [DOI] [PubMed] [Google Scholar]
- 14.Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. 2014. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A 111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y. 2012. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 16.McMahon MD, Guan C, Handelsman J, Thomas MG. 2012. Metagenomic analysis of Streptomyces lividans reveals host-dependent functional expression. Appl Environ Microbiol 78:3622–3629. doi: 10.1128/AEM.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabor EM, Alkema WB, Janssen DB. 2004. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 6:879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Escribano JP, Bibb MJ. 2011. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-ya K, Cane DE, Ikeda H. 2013. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol 2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai T, Yu Y, Xu Z, Tao M. 2014. Construction of Streptomyces lividans SBT5 as an efficient heterologous expression host. J Huazhong Agric Univ 33:1–6. [Google Scholar]
- 21.Handelsman J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez A, Kolvek SJ, Yip CL, Hopke J, Brown KA, MacNeil IA, Osburne MS. 2004. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70:2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flett F, Mersinias V, Smith CP. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 24.Sheng Y, Mancino V, Birren B. 1995. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res 23:1990–1996. doi: 10.1093/nar/23.11.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barras F, Marinus MG. 1989. The great GATC: DNA methylation in E. coli. Trends Genet 5:139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Z, Shi T, Xu M, Brock NL, Zhao YL, Wang Y, Deng Z, Pang X, Tao M. 2016. Hybrubins: bipyrrole tetramic acids obtained by crosstalk between a truncated undecylprodigiosin pathway and heterologous tetramic acid biosynthetic genes. Org Lett 18:572–575. doi: 10.1021/acs.orglett.5b03609. [DOI] [PubMed] [Google Scholar]
- 27.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 28.Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol 178:7276–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou X, Zhang B, Zhang L, Zhao G, Ding X. 2009. Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl Environ Microbiol 75:2158–2165. doi: 10.1128/AEM.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maruyama C, Toyoda J, Kato Y, Izumikawa M, Takagi M, Shin-ya K, Katano H, Utagawa T, Hamano Y. 2012. A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat Chem Biol 8:791–797. doi: 10.1038/nchembio.1040. [DOI] [PubMed] [Google Scholar]
- 31.Luo M, Wing RA. 2003. An improved method for plant BAC library construction. Methods Mol Biol 236:3–20. [DOI] [PubMed] [Google Scholar]
- 32.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. 2013. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhushan R, Bruckner H. 2004. Marfey's reagent for chiral amino acid analysis: a review. Amino Acids 27:231–247. doi: 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- 34.Yokote Y, Arai KM, Akahane K. 1986. Recovery of tryptophan from 25-minute acid hydrolysates of protein. Anal Biochem 152:245–249. doi: 10.1016/0003-2697(86)90405-7. [DOI] [PubMed] [Google Scholar]
- 35.Olano C, Wilkinson B, Sanchez C, Moss SJ, Sheridan R, Math V, Weston AJ, Brana AF, Martin CJ, Oliynyk M, Mendez C, Leadlay PF, Salas JA. 2004. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: cluster analysis and assignment of functions. Chem Biol 11:87–97. [DOI] [PubMed] [Google Scholar]
- 36.Tatsuno S, Arakawa K, Kinashi H. 2007. Analysis of modular-iterative mixed biosynthesis of lankacidin by heterologous expression and gene fusion. J Antibiot (Tokyo) 60:700–708. doi: 10.1038/ja.2007.90. [DOI] [PubMed] [Google Scholar]
- 37.Arakawa K, Kodama K, Tatsuno S, Ide S, Kinashi H. 2006. Analysis of the loading and hydroxylation steps in lankamycin biosynthesis in Streptomyces rochei. Antimicrob Agents Chemother 50:1946–1952. doi: 10.1128/AAC.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. 2016. PubChem Substance and Compound databases. Nucleic Acids Res 44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kempter C, Kaiser D, Haag S, Nicholson G, Gnau V, Walk T, Gierling KH, Decker H, Zähner H, Jung G. 1997. CDA: calcium-dependent peptide antibiotics from Streptomyces coelicolor A3(2) containing unusual residues. Angew Chem Int Ed Engl 36:498–501. doi: 10.1002/anie.199704981. [DOI] [Google Scholar]
- 40.Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BA, Hayes MA, Smith CP, Micklefield J. 2002. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9:1175–1187. doi: 10.1016/S1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- 41.Bachmann BO, Ravel J. 2009. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol 458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 42.Stachelhaus T, Walsh CT. 2000. Mutational analysis of the epimerization domain in the initiation module PheATE of gramicidin S synthetase. Biochemistry 39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 43.Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. 2014. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachmann BO, Van Lanen SG, Baltz RH. 2014. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41:175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baltz RH. 2007. Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131. [Google Scholar]
- 46.Byford MF, Baldwin JE, Shiau CY, Schofield CJ. 1997. The mechanism of ACV synthetase. Chem Rev 97:2631–2650. doi: 10.1021/cr960018l. [DOI] [PubMed] [Google Scholar]
- 47.D'Costa VM, Mukhtar TA, Patel T, Koteva K, Waglechner N, Hughes DW, Wright GD, De Pascale G. 2012. Inactivation of the lipopeptide antibiotic daptomycin by hydrolytic mechanisms. Antimicrob Agents Chemother 56:757–764. doi: 10.1128/AAC.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruner SD, Weber T, Kohli RM, Schwarzer D, Marahiel MA, Walsh CT, Stubbs MT. 2002. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure 10:301–310. doi: 10.1016/S0969-2126(02)00716-5. [DOI] [PubMed] [Google Scholar]
- 49.Grunewald J, Sieber SA, Marahiel MA. 2004. Chemo- and regioselective peptide cyclization triggered by the N-terminal fatty acid chain length: the recombinant cyclase of the calcium-dependent antibiotic from Streptomyces coelicolor. Biochemistry 43:2915–2925. doi: 10.1021/bi036140d. [DOI] [PubMed] [Google Scholar]
- 50.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. doi: 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H, Wang Y, Yu Y, Bai T, Chen L, Liu P, Guo H, Zhu C, Tao M, Deng Z. 2012. A non-restricting and non-methylating Escherichia coli strain for DNA cloning and high-throughput conjugation to Streptomyces coelicolor. Curr Microbiol 64:185–190. doi: 10.1007/s00284-011-0048-5. [DOI] [PubMed] [Google Scholar]
- 52.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.