Abstract
DNA double strand breaks (DSB)s often require end processing prior to joining during their repair by non-homologous end joining (NHEJ). Although the yeast proteins, Pol4, a Pol X family DNA polymerase, and Rad27, a nuclease, participate in the end processing reactions of NHEJ, the mechanisms underlying the recruitment of these factors to DSBs are not known. Here we demonstrate that Nej1, a NHEJ factor that interacts with and modulates the activity of the NHEJ DNA ligase complex (Dnl4/Lif1), physically and functionally interacts with both Pol4 and Rad27. Notably, Nej1 and Dnl4/Lif1, which also interacts with both Pol4 and Rad27, independently recruit the end processing factors to in vivo DSBs via mechanisms that are additive rather than redundant. As was observed with Dnl4/Lif1, the activities of both Pol4 and Rad27 were enhanced by the interaction with Nej1. Furthermore, Nej1 increased the joining of incompatible DNA ends in reconstituted reactions containing Pol4, Rad27 and Dnl4/Lif1, indicating that the stimulatory activities of Nej1 and Dnl4/Lif1 are also additive. Together our results reveal novel roles for Nej1 in the recruitment of Pol4 and Rad27 to in vivo DSBs and the coordination of the end processing and ligation reactions of NHEJ.
Keywords: DNA damage, DNA double-strand break, DNA nuclease, DNA ligase, DNA polymerase, genomic instability, protein-protein interaction
1. Introduction
DNA double strand breaks (DSB)s are cytotoxic and mutagenic lesions that must be repaired to maintain genomic integrity. Although multiple pathways have evolved to repair DSBs, these can be divided into two groups depending upon whether extensive DNA sequence homology is used to guide the repair. In mammalian cells, the majority of DSBs are repaired by non-homologous end joining (NHEJ) whereas this type of repair makes relatively minor contribution to DSB repair in the yeast, Saccharomyces cerevisiae (1–3).
The major NHEJ pathways in mammals and yeast share several functionally homologous components, including Ku70-Ku80 (yKu70-yKu80), DNA ligase IV-XRCC4 (Dnl4/Lif1) and XLF (Nej1) (1–3). DSB repair by NHEJ is initiated by the binding of the Ku heterodimer to DNA ends followed by protein-mediated DNA end-bridging. In mammals, this is carried out by the catalytic subunit of the DNA-dependent protein kinase (DNA PKcs), which binds to Ku-bound DNA ends and then juxtaposes the ends via an interaction between DNA PKcs molecules bound to two different DNA ends (4). End-bridging in yeast, which lacks a homolog of DNA PKcs, is carried out by the Mre11/Rad50/Xrs2 complex (5). DNA end joining is carried out by DNA ligase IV/XRCC4 (Dnl4/Lif1) together with XLF (Nej1) (6–20). While the binding of Ku to a DNA end serves as the signal for the recruitment of the other NHEJ factors (21–26), studies in yeast have shown that both Dnl4/Lif1 and Nej1 contribute to the stable association of yKu70-yKu80 with DSBs in vivo (15,26).
Most DSB ends generated by ionizing radiation and/or oxygen free radicals have termini that cannot be directly ligated. The processing of juxtaposed DNA ends by nucleases and Pol X family DNA polymerases to generate ligatable termini results in small deletions and insertions at the repair site (1–3). Pol X family members, Pol mu, Pol lambda and terminal transferase in mammals and Pol4 in yeast, have been implicated in the addition of nucleotides to DSB termini (1–3,27). The nuclease Artemis, which binds to DNA PKcs, plays a role in the processing of a subset of DSBs in mammalian cells (28). Genetic studies in yeast have implicated a DNA replication protein, Rad27, a homolog of mammalian flap endonuclease 1 (29), in the processing of a subset of DSBs with incompatible DNA ends (30). A later study using a different assay failed to reproduce these findings, suggesting that Rad27 either is not involved in end processing or is functionally redundant with other nucleases in the end processing step of this repair pathway (31). In support of the idea that Rad27 participates in the end processing reactions, we previously characterized physical and functional interactions among Pol4, Rad27 and Dnl4/Lif1 that coordinate end processing and ligation (32,33). Here we demonstrate that Nej1 also interacts with Pol4 and Rad27, modulating their DNA synthesis and nuclease activities, respectively. In addition, we show that interactions with both Nej1 and Dnl4/Lif1 contribute to the recruitment of Pol4 and Rad27 to in vivo DSBs.
2. Materials and methods
2.1. Yeast Strains
The yeast strains SLY1A, SLY1A lif1Δ and SLY1A nej1Δ have been described previously (15,34). To generate a strain lacking both Lif1 and Nej1, the plasmid pRS306 was used to generate URA3 marked deletions of LIF1 in the SLY1A nej1Δ strain.
A sequence encoding a 9x Myc tag was added to the C-termini of both Pol4 and Rad27 proteins by the polymerase chain reaction using primers flanking the 3′ end of either the POL4 or RAD27 genes and plasmid pYM20-9Myc-hph as the template. The PCR fragments were used to replace the wild type POL4 and RAD27 genes of SLY1A with versions encoding Pol4 and Rad27 proteins with a C-terminal c-Myc tag.
2.2. Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described (26,35). Briefly, 2% galatose was added to mid-log phase cultures grown in pre-induction medium (3% Glycerol/2% Lactic acid/ 0.05% Glucose) at 30°C to induce HO endonuclease expression. After in vivo cross-linking with 1% formaldehyde (v/v), cells were lysed and genomic DNA was sonicated to yield fragments with an average size of 500–700 base pair. For immunoprecipitation, the sonicated extracts were incubated with anti-Myc antibody (Millipore) at 4°C overnight, followed by 1 h incubation with Protein A/G beads (Santa Cruz). After cross-link reversal, genomic DNA was purified and amplified by real-time quantitative PCR (StepOne, Applied Biosystems) using a primer set (5′ CCCTGGTTTTGGTTTTGTAG 3′ and 5′CACATCTTCCCAATATCCGTCACC 3′) that annealed at a site adjacent to the HO-induced DSB and primers specific for the PRE1 gene situated on chromosome V (5′ CCCACAAGTCCTCTGATTTACATTCG 3′, and 5′ GGAATTCACCGCATGGTTTTCATAAGAG 3′). The recruitment of protein to the HO break is expressed as relative immunoprecipitation, which represents the ratio of the specific signal at the HO break to the nonspecific signal at the PRE1 locus, normalized to the value obtained from an uninduced sample.
2.3. Plasmids
Plasmids pET-28b-Pol4, pET-28b-Pol4ΔBRCT, pGST-Pol4, pET-Rad27 and pGST-Rad27 have been described previously (32,33). DNA sequences encoding full length Nej1 and derivatives lacking either the N-terminal 129 amino acids (ΔN-Nej1) or the C-terminal 120 amino acids (Nej1-ΔC) were amplified by PCR from pYes2.1-Nej1 (15) and subcloned into the E. coli expression plasmid pET28a(+) to generate pET28a-Nej1, pET28a-ΔN-Nej1, pET28a-Nej1-ΔC. The sequences of the plasmid inserts were verified.
2.4. Protein purification
Calmodulin binding peptide (CBP)-tagged Nej1, his-tagged Dnl4/Lif1 and his-tagged human XLF were purified as described previously (5,15,36). To purify his-tagged versions of Pol4, Rad27 and Nej1, E. coli Rosetta cells (Novagen) harboring expression plasmids were grown at 37°C in 2xYT medium (16 g tryptone, 10 g yeast extract and 5 g NaCl per liter) containing 100 μg/ml kanamycin and 34 μg/ml chloramphenicol. At an optical density of 0.5 at 600 nm, isopropyl β-D-1-thiogalactopyranoside was added to a final concentration of 0.2 mM and growth was continued overnight at 16°C. Cells were harvested, resuspended in lysis buffer (50 mM Tris-HCl pH7.5, 50 mM NaCl, 1 mM EDTA, 5% glycerol, 0.2% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine-HCl, 2 μg/ml aprotinin) and lysed by sonication. After centrifugation, his-tagged proteins were purified from the cleared lysate by phosphocellulose (P11, Whatman) and nickel-nitrilotriacetic acid-agarose (Qiagen). His-tagged versions of Pol4 and Rad27 were further purified by SP Sepharose FF (GE Healthcare) column chromatography whereas his-tagged versions of full length Nej1, and N- and C-terminal fragments of Nej1 were further purified by Mono Q (GE Healthcare) column chromatography. Dnl4/Lif1 and yeast Ku were purified from yeast cells as described previously (5).
To purify glutathione-S-transferase (GST)-tagged proteins, E. coli DH5 cells harboring either pGST-Pol4 or pGST-Rad27 were grown in 2xYT medium containing 100 μg/ml ampicillin and harvested as described above. Cell pellets were resuspended in GST binding buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) containing protease inhibitors and 0.2% Triton X100 and then sonicated. After clarification, the lysate was incubated with Glutathione Sepharose beads that had been pre-equilibrated with GST binding buffer at 4°C for 2 hours. The beads were washed with GST binding buffer prior followed by elution of bound proteins with 10 mM glutathione in GST binding buffer. Protein concentrations were measured by the Bradford assay (37) using bovine serum albumin as the standard. Purified proteins were dispensed in small aliquots, flash-frozen in liquid nitrogen and stored at −80°C.
2.5. Pull down assays
Purified his-tagged Pol4, Pol4ΔBRCT or Rad27 (1 μg) were immobilized on nickel beads and then incubated with extracts (200 μg) from yeast cells expressing CBP-tagged Nej1 (15) in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5% Glycerol, 0.2% NP-40 containing a mixture of protease inhibitors. After washing with the same buffer containing 30 mM imidozale, proteins bound to the beads were eluted with SDS sample buffer. CBP-Nej1 was detected by immunoblotting with CBP antibody (Millipore). To determine the region of Nej1 that interacts with Pol4 and Rad27, nickel beads in the absence or presence of his-tagged full-length Nej1, Nej1 C-terminal domain (ΔN-Nej1), or Nej1 N-terminal domain (Nej1-ΔC) were incubated with 1 μg of purified GST-tagged Pol4 or GST-tagged Rad27 as described above. GST fusion proteins bound to the beads was detected by immunoblotting with GST antibody (Cell Signaling).
2.6. Gap-filling DNA synthesis assays
DNA polymerase assays with Pol4 were carried out as described previously with DNA substrates that contained either a 1-nucleotide gap within a DNA duplex or a 3-nucleotide gap generated by the annealing of the partially complementary single strand ends of two DNA duplexes (32). In each case, the oligonucleotide providing the 3′ HO primer at the gap was 5′ end-labeled. The labeled DNA substrates (0.1 pmol or 1 pmol) were incubated for 1 h at 25°C with Pol4 (0.4 pmol) in the absence or presence of Nej1 in reactions (10 μl) containing 35 mM Tris-HCl, pH7.5, 10 mM MgCl2, 0.05 mM of each of the four dNTPs, and 1 mM ATP. Reactions were stopped by addition of 20 μl gel loading buffer (95% (v/v) formamide, 0.5% SDS, 20 mM EDTA, 0.5% (w/v) bromphenol blue, and 0.5% (w/v) xylene cyanol). After separation by denaturing gel electrophoresis, labeled DNA molecules were detected and quantitated by phosphorimager analysis.
2.7. DNA flap endonuclease assays
The ability of Rad27 to remove 3-nucleotide 5′ single strand flaps was measured as described previously using DNA substrates with either the 5′ flap within a duplex or the 5′ flap generated by the annealing of the partially complementary single strand ends of two DNA duplexes (32). In each case, the oligonucleotide providing the 5′ flap was 5′ end-labeled. The labeled DNA substrates (0.1 pmol or 1 pmol) were incubated for 1 h at 25°C with Rad27 (0.4 pmol) in the absence or presence of Nej1 in reactions (10 μl) containing 35 mM Tris-HCl, pH7.5, 10 mM MgCl2 and 1 mM ATP. Reactions were stopped and labeled DNA fragments were separated and detected as described above.
2.8. Ligation of DNA substrates with incompatible DNA ends
To measure the joining of DNA molecules that require both gap-filling DNA synthesis and flap removal, we constructed DNA substrates with partially complementary ends that when annealed generate a 3-nucleotide 5′ single strand flap and a 3-nucleotide gap. Equal amounts (0.1 pmol) of the labeled and unlabeled duplexes were incubated with 0.4 pmol Rad27, 0.4 pmol Pol4, 0.4 pmol Dnl4/Lif1 and increasing amount of Nej1 in a reaction mixture (10 μl) containing 35 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.05 mM of each of the four dNTPs, and 1 mM ATP for 1 h at 25°C. Reactions were stopped and labeled DNA fragments were separated and detected as described above.
3. Results
3.1 Both Lif1 and Nej1 are required for the efficient recruitment of end processing factors, Pol4 and Rad27, to in vivo DSBs
Although previous genetic studies have implicated the Pol X family DNA polymerase, Pol4, and the DNA 5′ flap endonuclease, Rad27, in the repair of subset of DSBs by NHEJ (27,30,32,33), the localization of these factors to in vivo DSBs has not been demonstrated. Using ChIP, we examined the steady state levels of Pol4 and Rad27 at a site-specific DSB generated within the MAT locus by expression of HO endonuclease in a donorless yeast strain. In accord with previous studies (15,26), more than 90% of the HO endonuclease sites at the MAT locus were cleaved 30 min after induction with galactose in the nej1Δ and lif1Δ strains (Fig. S1). The kinetics of cleavage of the HO endonuclease site in the lif1Δnej1Δ strain was indistinguishable from the single mutant strains and from the wild type SLY1A strain (Fig. S1). Similar to Mre11/Rad50/Xrs2 and Lif1 (15,26), Pol4 (Fig. 1A) and Rad27 (Fig. 1B) were rapidly recruited to the MAT DSB and continued to accumulate for 2 h after induction of HO endonuclease. Since both Pol4 and Rad27 physically and functionally interact with Dnl4/Lif1 in vitro (32,33), we examined the role of this complex and its associated protein Nej1 in the recruitment of Pol4 and Rad27 to in vivo DSBs. In a lif1Δ strain that lacks a functional Dnl4/Lif1 complex (15), the rate of recruitment of both Pol4 (Fig. 1A) and Rad27 (Fig. 1B) was reduced. The absence of Nej1 had a similar effect on the recruitment of both Pol4 (Fig. 1A) and Rad27 (Fig. 1B) to that observed in the strain lacking a functional Dnl4/Lif1 complex whereas no significant recruitment of either Pol4 (Fig. 1A) or Rad27 (Fig. 1B) was observed in the lif1Δ nej1Δ strain. Thus, we conclude that that Nej1 and Dnl4/Lif1 independently contribute to the localization of both Pol4 and Rad27 at an in vivo DSB.
Fig. 1. Recruitment of Pol4 and Rad27 to an in vivo DSB: Effect of genetic inactivation of NEJ1 and LIF1.
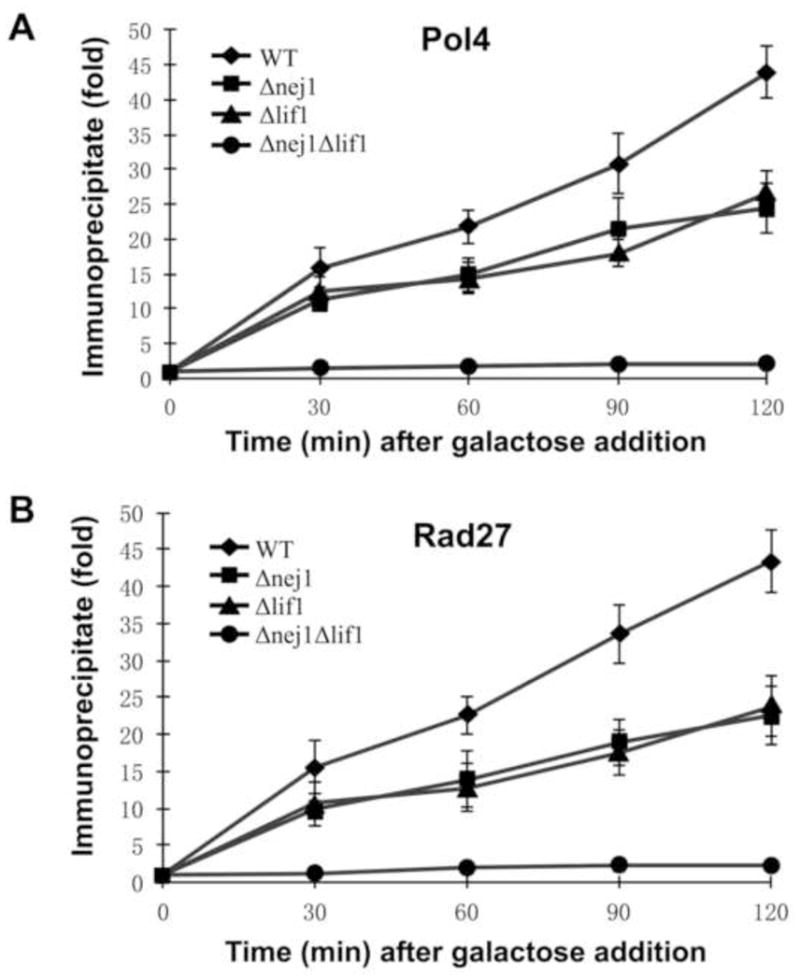
Kinetics of recruitment of; (A), Pol4; (B), Rad27 to the MAT DSB in the wild type SLY1A strain (diamond), SLY1A nej1Δ (square), SLY1A lif1Δ (triangle) and SLY1A nej1Δlif1Δ (circle) mutant strains was measured by chromatin immunoprecipitation as described in Experimental Procedures. Data represent the mean± S.D. of three or more independent experiments.
3.2. Nej1 interacts with Pol4 and Rad27
The Dnl4/Lif1-mediated recruitment of Pol4 and Rad27 to an in vivo DSB is consistent with previous biochemical studies demonstrating that Dnl4/Lif1 interacts with both Pol4 and Rad27 (32,33). This prompted us to ask whether Nej1 also interacts with these proteins. In initial studies, we purified his-tagged versions of full length Pol4 and Rad27 (Fig. S2, lanes 2 and 6, respectively) and found that CBP-tagged Nej1 in yeast cell extracts was specifically retained by nickel beads liganded by either Pol4 (Fig. 2A, lane 6) or Rad27 (Fig. 2A, lane 5). The non-catalytic N-terminal region of Pol4 contains a breast cancer susceptibility protein 1 C-terminal (BRCT) domain that interacts with Dnl4/Lif1 (32). To address the possible role of the Pol4 BRCT domain in the interaction with Nej1, we purified a his-tagged version of Pol4 lacking the BRCT domain (Fig. S2, lane 4) and found that, unlike full-length Pol4, this truncated protein was unable to direct the specific binding of CBP-Nej1 in yeast extracts to nickel beads (Fig. 2B, compare lanes 5 and 6), indicating that the Pol4 BRCT domain is required for the interaction with Nej1.
Fig. 2. Nej1 physically interacts with both Pol4 and RAD27; interaction of Nej1 with Pol4 is dependent upon the Pol4 BRCT domain.
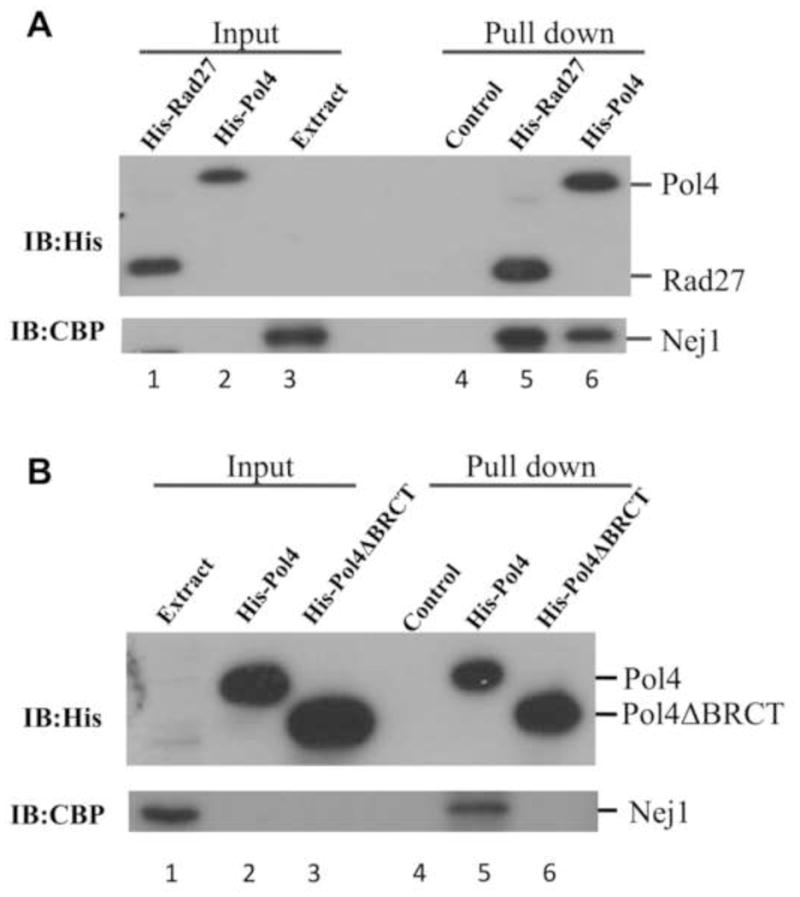
(A) Extracts (200 μg) from a yeast strain expressing CBP-Nej1 were incubated with; lane 4, nickel beads (Control); lane 5, nickel beads liganded by his-tagged Rad27 (1 μg, His-Rad27); lane 6, his-tagged Pol4 (1 μg, His-Pol4). Lane 1, 50 ng Rad27; lane 2, 50 ng Pol4 and lane 3, 10 μg yeast extract. (B) Extracts (200 μg) from yeast strain expressing CBP-Nej1 were incubated with; lane 4, nickel beads (Control); lane 5, nickel beads liganded by His-tagged Pol4 (1 μg, His-Pol4); lane 6, His-tagged Pol4 lacking the C-terminal BRCT domain (1 μg, His-Pol4ΔBRCT). Lane 1, 10 μg yeast extract Rad27; lane 2, 50 ng Pol4 and lane 3, 50 ng Pol4ΔBRCT. After washing, proteins were eluted with SDS sample buffer. Rad27, Pol4 and Nej1 were detected in the eluates by immunoblotting with anti-His (upper panel) and anti-CBP antibody (lower panel).
To determine if Nej1 interacts directly with both Pol4 and Rad27, we purified his-tagged versions of full-length Nej1 (Fig. S3, lane 2), an N-terminal fragment (residues 1–223) that interacts with yKu70–yKu80 (15) (Fig. S3, lane 4), and a C-terminal fragment (residues 129–343) that interacts with Lif1 (38,39) (Fig. S3, lane 6), and examined their interactions with GST-tagged versions of both Pol4 and Rad27. GST-Pol4 was specifically retained by nickel beads liganded by either full-length Nej1 or its C-terminal fragment (Fig. 3A). Similarly, GST-Rad27 was specifically retained by nickel beads liganded by either full-length Nej1 or its C-terminal fragment (Fig. 4). Thus, both Pol4 and Rad27 bind to the same C-terminal region of Nej1 as Lif1 (38,39). To confirm that the N-terminal BRCT domain of Pol4 interacts with Nej1, we purified the Pol4 BRCT domain as a GST fusion protein (GST-Pol4BRCT) and examined its binding to nickel beads in the presence or absence of his-tagged Nej1. As expected, GST-Pol4BRCT but not GST was specifically retained by nickel beads liganded by full-length Nej1 (Fig. 3B, compare lanes 3 and 7). In addition, this interaction was not reduced by ethidium bromide (Fig. 3B, lane 8), indicating that it is unlikely to be mediated by nucleic acid. Since the interaction between Pol4 and Nej1 was detected with proteins purified from E. coli, we conclude that it is not dependent upon post-translational modifications, such as phosphorylation.
Fig. 3. The C-terminal region of Nej1 mediates the interaction with Pol4.
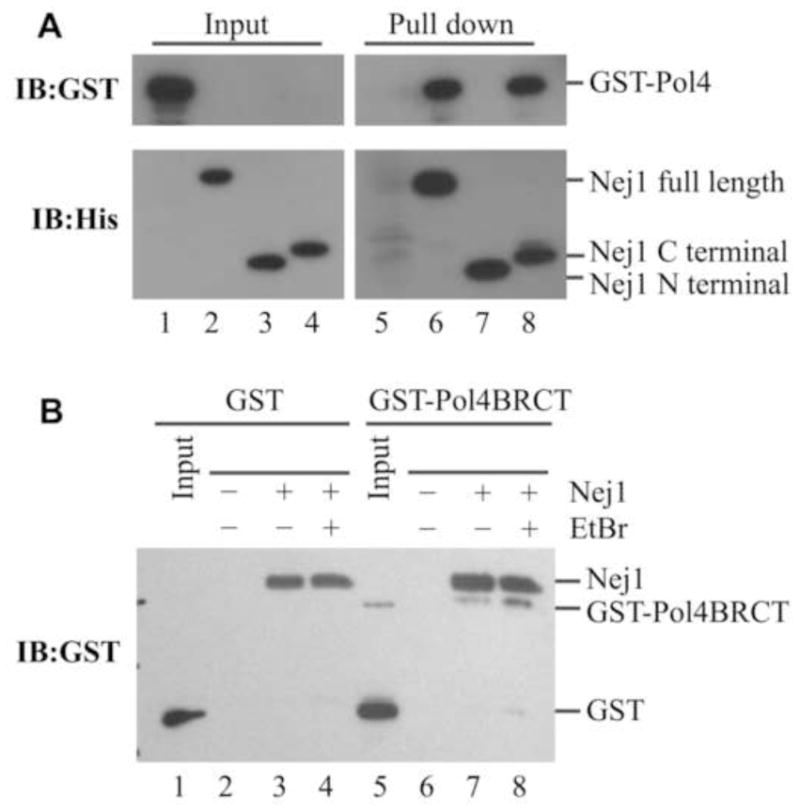
(A) Purified proteins (50 ng of each); GST-Pol4 (lane 1, 5% of input); full-length Nej1 (lane 2); Nej1 N-terminal fragment (lane 3); Nej1 C-terminal fragment (lane 4). Purified GST-Pol4 (1 μg) was incubated with nickel beads liganded by; lane 5, no protein; lane 6, full-length Nej1 (Nej1 full length); lane 7, N-terminal fragment of Nej1 (Nej1 N terminal); lane 8, C-terminal fragment of Nej1( Nej1 C terminal) as described in Experimental Procedures. (B) Purified GST protein (lane 1, 4% of input) was incubated with nickel beads liganded by; no protein (lane 2); his-tagged Nej1 (lane 3); his-tagged Nej1 in the presence of 50 μg/ml ethidium bromide (lane 4). Purified GST-Pol4BRCT protein (lane 5, 4% of input) was incubated with nickel beads liganded by; no protein (lane 6); his-tagged Nej1 (lane 7); his-tagged Nej1 in the presence of 50 μg/ml ethidium bromide (lane 8). GST tagged proteins were detected by immunoblotting with GST antibodies. Under these conditions, Nej1 cross-reacted with the GST antibody.
Fig. 4. The C-terminal region of Nej1 mediates the interaction with Rad27.
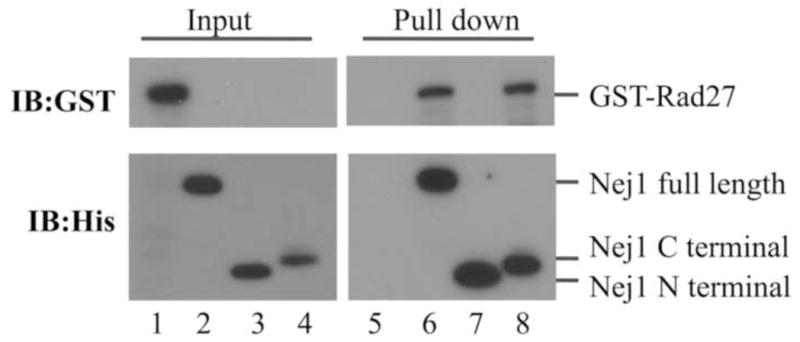
Purified proteins (50 ng of each); GST-Rad27 (lane 1, 5% of input); full-length Nej1 (lane 2); Nej1 N-terminal fragment (lane 3); Nej1 C-terminal fragment (lane 4). Purified GST-Pol4 (1 μg) was incubated with nickel beads liganded by; lane 5, no protein; lane 6, full-length Nej1 (Nej1 full length); lane 7, N-terminal fragment of Nej1 (Nej1 N terminal); lane 8, C-terminal fragment of Nej1 (Nej1 C terminal). GST- and his-tagged fusion proteins were detected by immunoblotting with GST (upper panel) and His (lower panel) antibodies.
3.3. Nej1 stimulates DNA synthesis by Pol4
To investigate the effect of Nej1 on Pol4 activity, we constructed two DNA duplexes with partially overlapping single-strand ends that when annealed generate a nick and a 3-nucleotide gap to mimic gap structures at aligned DNA ends (Fig. 5A). The addition of Nej1 stimulated gap-filling synthesis in a concentration-dependent manner, resulting in an ~5-fold increase in synthesis at a 2:1 ratio of Nej1 to Pol4. To address the role of the interaction between Nej1 and Pol4 in this process, we examined the effects of an N-terminal fragment of Nej1, which binds to yKu70–yKu80 (15), and a C-terminal fragment, which binds to both Pol4 and Lif1 and also has DNA binding activity (38,39), on DNA synthesis. While the Nej1 C-terminal fragment enhanced gap-filling synthesis by Pol4 to an even greater extent than full-length Nej1, the Nej1 N-terminal fragment did not stimulate gap-filling synthesis (Fig. 5A). As it is possible that Nej1 stimulates DNA synthesis by enhancing the annealing and/or end-bridging of the DNA duplexes, we carried out similar experiments with a single DNA duplex containing a 1-nucleotide gap (Fig. 5B). With this substrate, full length Nej1 and the C-terminal fragment promoted strand-displacement DNA synthesis by Pol4 whereas the N-terminal fragment did not (Fig. 5B) although the extent of the stimulation (about 4-fold for the C-terminal fragment of Nej1 and about 3-fold for full length Nej1 at a 2:1 ratio to Pol4) was less than that observed on the DNA substrate that required annealing to generate the gap. As expected, purified Nej1 had no detectable DNA polymerase activity (Fig. S4A). To demonstrate the specificity of the effect of Nej1 on Pol4 activity, we carried out similar reactions with human XLF, which is likely to be structurally and functionally homologous to Nej1 but shares very little similarity at the amino acid sequence level (40). In contrast to Nej1, XLF did not enhance gap-filling synthesis on either the DNA substrate with a nick and a 3-nucleotide gap (Fig. 6A) or the DNA substrate with a 1-nucleotide gap (data not shown). Together, these results demonstrate that the C-terminal domain of Nej1 interacts with and specifically stimulates the DNA synthesis activity of Pol4. In previous studies, yeast Ku inhibited ligation by Dnl4/Lif1 (5), prompting us to examine the effect of Ku on gap-filling DNA synthesis. At a ratio of one Ku per DNA end, there was no detectable effect on DNA synthesis by Pol4 either in the absence (data not shown) or presence of Nej1 (Fig. 6B).
Fig. 5. Nej1 stimulates gap-filling DNA synthesis by Pol4.
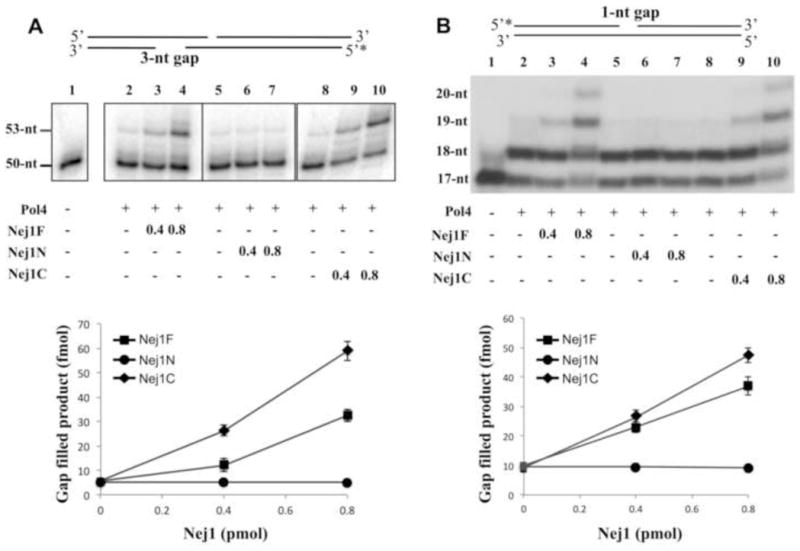
(A) One of two linear DNA duplexes with partially complementary ends that anneal to form a 3-nucleotide gap was end-labeled as indicated (0.1 pmol of each). The DNA substrate (0.1 pmol, lane 1) was incubated for 1 h at 25oC with Pol 4 (0.4 pmol, lanes 2–10) in the absence (lanes 2, 5 and 8) and presence of; lane 3, 0.4 pmol full length Nej1 (Nej1F); lane 4, 0.8 pmol full length Nej1; lane 6, 0.4 pmol Nej1 N-terminal fragment (Nej1N); lane 7, 0.8 pmol Nej1 N-terminal fragment; lane 9, 0.4 pmol Nej1 C-terminal fragment (Nej1C); lane 10, 0.8 pmol Nej1 C-terminal fragment. (B) Linear DNA duplex with an internal 1-nucleotide gap was end-labeled as indicated. The DNA substrate (1 pmol, lane1) was incubated for 1 h at 25°C with Pol4 (0.4 pmol, lanes 2–10) in the absence (lanes 2, 5 and 8) and presence of; lane 3, 0.4 pmol full length Nej1 (Nej1F); lane 4, 0.8 pmol full length Nej1; lane 6, 0.4 pmol Nej1 N-terminal fragment (Nej1N); lane 7, 0.8 pmol Nej1 N-terminal fragment; lane 9, 0.4 pmol Nej1 C-terminal fragment (Nej1C); lane 10, 0.8 pmol Nej1 C-terminal fragment. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The boxes in Panel A indicate the grouping of images from different parts of the same gel. The positions of the substrate and extended products are indicated on the left. The results of three independent experiments are shown graphically with error bars indicating the standard deviation from the mean.
Fig. 6. Effects of human XLF and yeast Ku on gap-filling DNA synthesis by Pol4.
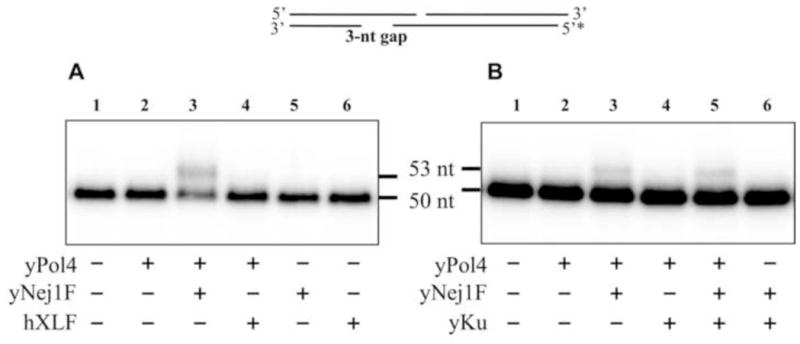
One of two linear DNA duplexes with partially complementary ends that anneal to form a 3-nucleotide gap was end-labeled as indicated (0.1 pmol of each). (A) The DNA substrate (0.1 pmol, lane 1) was incubated for 1 h at 25°C with Pol 4 (0.3 pmol), full length yeast Nej1 (yNej1F, 1.2 pmol) and human XLF (hXLF, 1.2 pmol) as indicated. (B) Similar reactions were carried out in the absence or presence of yKu70–yKu80 (0.4 pmol) as indicated. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The positions of the substrate and extended products are indicated on the left.
3.4. Nej1 stimulates flap cleavage by Rad27
Rad27 is a DNA structure-specific endonuclease that cleaves 5′ single-strand flaps. We examined the effect of Nej1 on the cleavage of 3-nucleotide 5′ flaps either formed by the annealing of two DNA duplexes with partially complementary single-strand ends (Fig. 7A) or within a single DNA duplex (Fig. 7B). As expected, purified Nej1 had no detectable nuclease activity with a flap substrate (Fig. S4B). Nej1 stimulated the activity of Rad27 on both types of DNA substrate with the stimulatory activity residing within the C-terminal fragment that binds to Rad27 (Fig. 7). Human XLF did not increase Rad27 activity (Fig. 8A), indicating that the stimulation by Nej1 is specific. In contrast to Pol4, yeast Ku inhibited Rad27 activity on the substrate formed by the annealing of two DNA duplexes with partially complementary single-strand ends either in the presence or absence of Nej1 (Fig. 8B).
Fig. 7. Nej1 stimulates Rad27 nuclease activity.
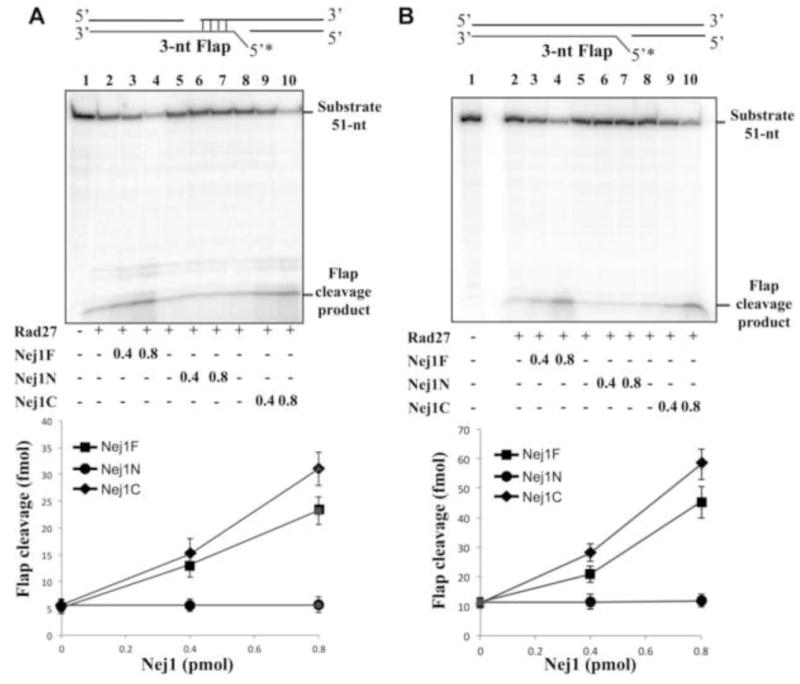
(A) One of two linear DNA duplexes with partially complementary ends that anneal to form a 5′ 3-nucleotide flap was end-labeled as indicated (0.1 pmol of each). The DNA substrate (0.1 pmol, lane 1) was incubated for 1 h at 25°C with Rad27 (0.4 pmol, lanes 2–10) in the absence (lanes 2, 5 and 8) and presence of; lane 3, 0.4 pmol full length Nej1 (Nej1F); lane 4, 0.8 pmol full length Nej1; lane 6, 0.4 pmol Nej1 N-terminal fragment (Nej1N); lane 7, 0.8 pmol Nej1 N-terminal fragment; lane 9, 0.4 pmol Nej1 C-terminal fragment (Nej1C); lane 10, 0.8 pmol Nej1 C-terminal fragment. (B) Linear DNA duplex with an internal 5′ 3-nucleotide flap was end-labeled as indicated (1 pmol). The DNA substrate (1 pmol, lane 1) was incubated for 1 h at 25°C with Rad27 (0.4 pmol, lanes 2–10) in the absence (lanes 2, 5 and 8) and presence of; lane 3, 0.4 pmol full length Nej1 (Nej1F); lane 4, 0.8 pmol full length Nej1; lane 6, 0.4 pmol Nej1 N-terminal fragment (Nej1N); lane 7, 0.8 pmol Nej1 N-terminal fragment; lane 9, 0.4 pmol Nej1 C-terminal fragment (Nej1C); lane 10, 0.8 pmol Nej1 C-terminal fragment. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The positions of the substrate and products of nucleolytic digestion are indicated on the right. The results of three independent experiments are shown graphically with the error bars indicating the standard deviation from the mean.
Fig. 8. Effects of human XLF and yeast Ku on Rad27 nuclease activity.
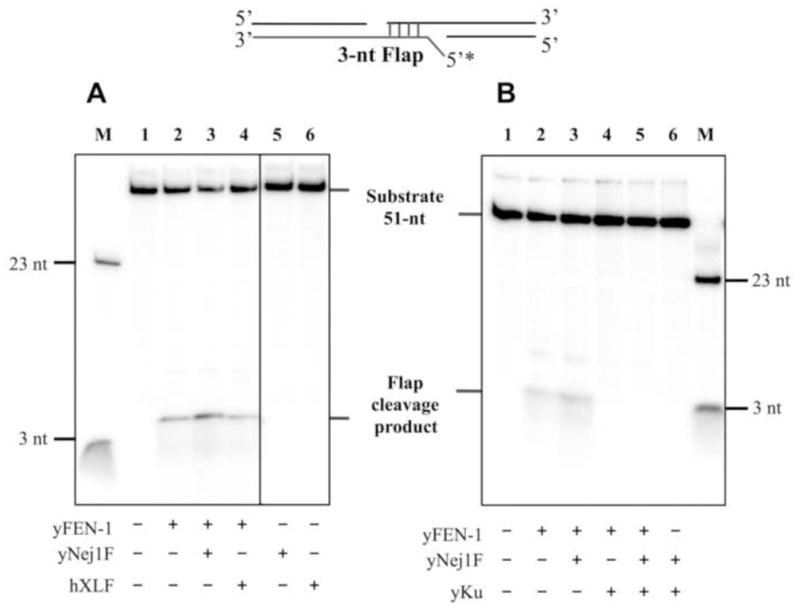
(A) One of two linear DNA duplexes with partially complementary ends that anneal to form a 5′ 3-nucleotide flap was end-labeled as indicated (0.1 pmol of each). The DNA substrate (0.1 pmol, lane 1) was incubated for 1 h at 25°C with Rad27 (1.2 pmol), full length yeast Nej1 (yNej1F, 2.4 pmol) and human XLF (hXLF, 2.4 pmol) as indicated. (B) Similar reactions were carried out in the absence or presence of yKu70–yKu80 (0.4 pmol) as indicated. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The positions of the substrate, flap cleavage products and molecular mass markers of 3 and 23 nucleotides are indicated. The boxes in panel A indicate the grouping of images from different parts of the same gel.
3.5. Effect of Nej1 on the ligation of DNA substrates that require both flap removal and gap-filling synthesis
Previously, we described a series of pairwise interactions among Pol4, Rad27 and Dnl4-Lif1 and demonstrated that these factors coordinately processed and joined DNA molecules with incompatible 5′ ends (32,33). No ligation product was detected in reactions that lacked either Pol4 or Rad27 (33). As Nej1 also interacts with Pol4 and Rad27 in addition to the Lif1 subunit of Dnl4/Lif1 (12,14,19,20,38,39), we examined the effect of Nej1 on reconstituted reactions with these factors and a DNA substrate with incompatible 5′ ends (Fig. 9). When added in an equal amount to the other factors, Nej1 enhanced 5′ flap removal (Fig. 9A), gap-filling DNA synthesis and ligation (Fig. 9B), indicating that the stimulatory effects of Dnl4/Lif1 and Nej1 on the activities of Pol4 and RAD27 are additive and that Pol4, Rad27, Nej1 and Dnl4/Lif1 act together to process and join incompatible DNA ends.
Fig. 9. Effect of Nej1 on the processing and ligation of NHEJ intermediate with a mismatched 5′ end in reconstituted reactions with Pol4, Rad27 and Dnl4/Lif1.
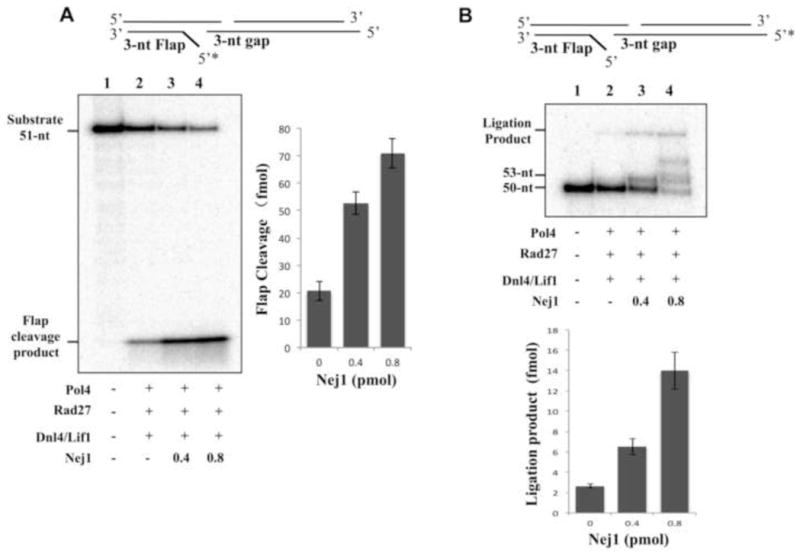
(A) One of two linear DNA duplexes with partially complementary ends that anneal to generate a 3-nucleotide gap and 3-nucleotide 5′ flap was end-labeled as indicated (0.1 pmol of each) to monitor removal of the mismatched 5′ flap. The labeled DNA substrate (0.1 pmol, lane 1) was incubated at 25°C for 1 hour with 0.4 pmol Rad27, 0.4 pmol Pol4 and 0.4 pmol Dnl4/Lif1 in the absence (lane 2) and presence of Nej1 (lane 3, 0.4 pmol; lane 4, 0.8 pmol) as described in Experimental Procedures. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The positions of the 51-nt substrate and 3-nt cleavage product are indicated on the right. The results of three independent experiments are shown graphically with the error bars indicating the standard deviation from the mean. (B) To monitor gap-filling DNA synthesis and ligation, the same DNA substrate (0.1 pmol, lane 1) was end-labeled as indicated and then incubated at 25°C for 1 hour with 0.4 pmol Rad27, 0.4 pmol Pol4 and 0.4 pmol Dnl4/Lif1 in the absence (lane 2) and presence of Nej1 (lane 3, 0.4 pmol; lane 4, 0.8 pmol) as described in Experimental Procedures. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The positions of the 50-nt substrate, 53-nt intermediate generated by gap-filling DNA synthesis and the 89-nt ligation product are indicated on the right. The results of three independent experiments are shown graphically with the error bars indicating the standard deviation from the mean.
4. Discussion
The majority of DSBs repaired by NHEJ require processing prior to ligation. Although a large number of enzymes including polynucleotide kinase phosphatase (41) and several nucleases (28,30) and DNA polymerases (27,42–46) have been implicated in these processing reactions, relatively little is known about the mechanism underlying the localization and coordination of end processing factors at DSBs. In yeast, Pol4, a Pol X family member, and Rad27, a DNA structure-specific endonuclease involved in Okazaki fragment processing, have been implicated in the repair of a subset of DSBs by NHEJ (27,30). Previous studies have shown that the recruitment of core NHEJ factors, Mre11/Rad50/Xrs2, Dnl4/Lif1 and Nej1, to DSBs is dependent upon yKu70–yKu80 and that pairwise interactions among the core NHEJ factors contribute to their assembly at the DSB (5,15,22,24,26,38). Here we have identified novel physical and functional interactions between the Dnl4/Lif1-asociated factor Nej1 and both Rad27 and Pol4 and shown that both Nej1 and Dnl4/Lif1 contribute to the recruitment of these end processing factors to in vivo DSBs.
Although the ends generated by HO endonuclease cleavage at the MAT locus can be rejoined without processing, both Pol4 and Rad27 were rapidly recruited to the site-specific DSB. This is consistent with studies showing that the human Pol X family member, Pol mu is recruited to DSBs generated by ionizing radiation (47). Although most DSBs generated by HO endonuclease cleavage are accurately repaired, resulting in further cycles of cleavage and rejoining, relatively rare inaccurate joints that prevent subsequent cleavage by HO endonuclease are generated as a result of end-processing by factors such as Rad27 and Pol4 (48). Notably, inactivation of Pol4 in a rad52 strain results in a marked reduction in CA insertions at the HO site, one of the major types of DNA sequence change observed at misrepaired HO endonuclease sites (27,48) whereas Rad27 is involved in the repair of NHEJ events that involve removal of 5′ flap intermediates (30). While the loss of either Nej1 or the Dnl4/Lif1 complex partially reduced recruitment of Pol4 and Rad27, loss of both Nej1 and the Dnl4/Lif1 complex essentially abolished their recruitment. These results are consistent with a model in which Nej1 and Dnl4/Lif1 independently recruit Pol4 and Rad27 to DSBs via protein-protein interactions (32,33) and provide a molecular explanation as to why pol4 mutant alleles, which encode truncated polypeptides lacking the BRCT domain that mediates the interaction with both Nej1 and Dnl4/Lif1, have the same phenotype as pol4 null mutants (27). It should, however, be noted that, since both Nej1 and Dnl4/Lif1 appear to stabilize the binding of yKu70–yKu80 at DSBs (15,26), the effect of these factors on the recruitment of Pol4 and Rad27 could also be due to the reduced stability of NHEJ protein complexes assembled at DSBs.
XLF and XRCC4 form structurally similar homodimers with N-terminal globular head domains and C-terminal α-helical tails (49–51). It is assumed that the structures of the functionally homologous yeast proteins, Nej1 and Lif1, will be similar. The amino terminal 244 residues of Nej1, which contain the globular head domain and first part of the coiled-coil region, form a stable structure that is resistant to proteolysis (39). This fragment, which homodimerizes via the globular head domains (39), interacts with yKu70–yKu80 and binds to DNA ends in a yKu70–yKu80 dependent manner (15). Although apparently unstructured, the C-terminal residues of Nej1 (aa 270–342) are required for DNA and Lif1 binding in vitro and for both nuclear localization and NHEJ activity in vivo (39,52). Notably, Nej1 enhances inter- but not intra-molecular ligation by Dnl4/Lif1, suggesting that Nej1 has end-bridging activity (15). Here we have found that both Pol4 and Rad27 also interact with and are stimulated by the C-terminal region of Nej1. Unlike ligation (15), the enhancement of gap-filling synthesis and flap cleavage by Nej1 was also observed with DNA substrates that did not require end-bridging to generate gaps and flaps, indicating that the increased activity is at least partly due to the direct interaction of Nej1 with Pol4 and Rad27.
Previously, we have shown that both Pol4 and Rad27 interact with the Dnl4 but not the Lif1 subunit of the Dnl4/Lif1 complex (32,33). As with Nej1, the stimulation of Pol4 activity by Dnl4/Lif1 was independent of end-bridging (32). In accord with the pairwise interactions among Dnl4/Lif1, Pol4 and Rad27, these factors act together to join DNA molecules that require flap removal and gap filling synthesis prior to ligation (32,33). Although it is not known whether the interactions of Rad27 and Pol4 with either Nej1 or Dnl4/Lif1 are mutually exclusive, the addition of Nej1 to these reactions enhanced flap cleavage, gap-filling synthesis and ligation, indicating that the stimulatory effects of Nej1 and Dnl4/Lif1 are additive. While yKu70–yKu80 had no effect on Pol4 activity at juxtaposed DNA ends, it inhibited Rad27 flap cleavage activity. It has been shown previously that yKu70–yKu80 also inhibits ligation by Dnl4/Lif1 (5). However, in the presence of Mre11/Rad50/Xrs2, yKu70–yKu80 stimulatesd intermolecular ligation (5). Further studies are needed to determine the stoichiometry and architecture of the complexes formed by Dnl4/Lif1, Nej1, Pol4 and Rad27 on DNA ends in the absence and presence of yKu70–yKu80 and the effect of Mre11/Rad50/Xrs2 on complex formation on DNA ends and end-processing. Interestingly, homodimers of the functionally homologous human proteins, XLF and XRCC4, form filaments that bind to and bridge DNA ends (53–55). It should be noted that, while XRCC4 homodimers are likely to be present in cells in addition to DNA ligase IV/XRCC4 complexes (56), the functional relevance of XRCC4 homodimers and XLF/XRCC4 filaments has not yet been demonstrated. Nonetheless it is possible that these filaments (XLF/XRCC4 in humans and Nej1/Lif1 in yeast) maintain the alignment of the ends of the DNA duplexes after initial end bridging by DNA PKcs in humans and Mre11/Rad50/Xrs2 in yeast, providing the framework for the end-processing and ligation reactions. In this scenario, Dnl4/Lif1 complexes are likely to be located next to the aligned DNA ends, combining with adjacent Nej1 molecules in the filament to provide binding site for sites for Pol4 and Rad27. Presumably these filaments provide additional binding sites for the other DNA polymerases and nucleases that have also been implicated in the end-processing reactions of NHEJ.
Supplementary Material
Yeast Nej1 and Dnl4/Lif1 independently recruit Pol4 and Rad27 to in vivo DNA double strand breaks
Nej1 interacts with and stimulates gap-filling DNA synthesis by Pol4
Nej1 interacts with and stimulates 5′ flap cleavage by Rad27
Nej1 stimulates the processing and joining of incompatible DNA ends by Pol4, Rad27 and Dnl4/Lif1
Acknowledgments
We thank S. Fang for purification of XLF. This work was supported by grant 13639 from the Canadian Institutes of Health Research (SPLM) and grants R01 GM47251 (AET) and P01 CA92584 (AET and SPLM) from the National Institutes of Health.
Abbreviations
- BRCT
breast cancer susceptibility protein 1 C-terminal domain
- CBP
calmodulin binding peptide
- ChIP
chromatin immunoprecipitation
- DNA PKcs
DNA-dependent protein kinase
- DSB
DNA double strand break
- GST
glutathione-S-transferase
- NHEJ
non-homologous end joining
- PCR
polymerase chain reaction
Footnotes
All the authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Wilson TE. SnapShot: Nonhomologous DNA end joining (NHEJ) Cell. 142:496–496. e491. doi: 10.1016/j.cell.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. Embo J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 6.Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- 7.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 9.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci U S A. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrmann G, Lindahl T, Schar P. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. Embo J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Tomkinson AE. Yeast nej1 is a key participant in the initial end binding and final ligation steps of nonhomologous end joining. J Biol Chem. 286:4931–4940. doi: 10.1074/jbc.M110.195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 17.Grawunder U, Zimmer D, Kulesza P, Lieber MR. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J Biol Chem. 1998;273:24708–24714. doi: 10.1074/jbc.273.38.24708. [DOI] [PubMed] [Google Scholar]
- 18.Wei YF, Robins P, Carter K, Caldecott KW, Papin DJC, Yu G-L, Wang R-P, Shell BK, Nash RA, Schar P, Barnes DE, Haseltine WA, Lindahl T. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA igase III, an enzyme active in DNA repair and genetic recombination. Mol Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kegel A, Sjostrand JO, Astrom SU. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol. 2001;11:1611–1617. doi: 10.1016/s0960-9822(01)00488-2. [DOI] [PubMed] [Google Scholar]
- 20.Ooi SL, Shoemaker DD, Boeke JD. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science. 2001;294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- 21.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmbos PL, Daley JM, Wilson TE. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol. 2005;25:10782–10790. doi: 10.1128/MCB.25.24.10782-10790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics. 2008;180:1809–1819. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Topper LM, Wilson TE. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008;178:1237–1249. doi: 10.1534/genetics.107.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 27.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 28.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 29.Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci U S A. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daley JM, Wilson TE. Evidence that base stacking potential in annealed 3′ overhangs determines polymerase utilization in yeast nonhomologous end joining. DNA Repair (Amst) 2008;7:67–76. doi: 10.1016/j.dnarep.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng HM, Tomkinson AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J Biol Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- 33.Tseng HM, Tomkinson AE. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J Biol Chem. 2004;279:47580–47588. doi: 10.1074/jbc.M404492200. [DOI] [PubMed] [Google Scholar]
- 34.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 35.Trujillo KM, Osley MA. A Role for H2B Ubiquitylation in DNA Replication. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Deshpande RA, Wilson TE. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 2007;6:1507–1516. doi: 10.1016/j.dnarep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulek M, Yarrington R, McGibbon G, Boeke JD, Junop M. A critical role for the C-terminus of Nej1 protein in Lif1p association, DNA binding and non-homologous end-joining. DNA Repair (Amst) 2007;6:1805–1818. doi: 10.1016/j.dnarep.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. Cernunnos interacts with the XRCC4-DNAligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 41.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. Embo J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 45.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capp JP, Boudsocq F, Besnard AG, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. Involvement of DNA polymerase mu in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 2007;35:3551–3560. doi: 10.1093/nar/gkm243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 50.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. Embo J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. Embo J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahaney BL, Lees-Miller SP, Cobb JA. The C-terminus of Nej1 is critical for nuclear localization and non-homologous end-joining. DNA Repair (Amst) 2014;14:9–16. doi: 10.1016/j.dnarep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012;40:1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF regulates filament architecture of the XRCC4.ligase IV complex. Structure. 2010;18:1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mani RS, Yu Y, Fang S, Lu M, Fanta M, Zolner AE, Tahbaz N, Ramsden DA, Litchfield DW, Lees-Miller SP, Weinfeld M. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J Biol Chem. 2010;285:37619–37629. doi: 10.1074/jbc.M109.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


