Abstract
Oral cancer, primarily oral squamous cell carcinoma (OSCC), continues to be a major global health problem with high incidence and low survival rates. While the major risk factors for this malignancy, mostly lifestyle related, have been identified, around 15% of oral cancer cases remain unexplained. In light of evidence implicating bacteria in the aetiology of some cancer types, several epidemiological studies have been conducted in the last decade, employing methodologies ranging from traditional culture techniques to 16S rRNA metagenomics, to assess the possible role of bacteria in OSCC. While these studies have demonstrated differences in microbial composition between cancerous and healthy tissues, they have failed to agree on specific bacteria or patterns of oral microbial dysbiosis to implicate in OSCC. On the contrary, some oral taxa, particularly Porphyromonas gingivalis and Fusobacterium nucleatum, show strong oral carcinogenic potential in vitro and in animal studies. Bacteria are thought to contribute to oral carcinogenesis via inhibition of apoptosis, activation of cell proliferation, promotion of cellular invasion, induction of chronic inflammation, and production of carcinogens. This narrative review provides a critical analysis of and an update on the association between bacteria and oral carcinogenesis and the possible mechanisms underlying it.
Keywords: bacteria, carcinoma, dysbiosis, inflammation, microbiome, mouth, squamous cell
Oral cancer encompasses malignant neoplasms, predominantly oral squamous cell carcinoma (OSCC), evolving from the lining mucosae of the lips and the mouth (oral cavity) including the anterior two-thirds of the tongue as defined by the International Classification of Disease (1). Along with oropharyngeal cancer, it constitutes an important global public health problem, ranking the eighth most common cancer in the world. In 2012, an estimated 300,400 new cases of and 145,400 deaths from oral cancer were reported (2). Two-thirds of all cases are recorded in developing countries; the Indian subcontinent alone accounts for one-third of the global burden (2). Oral cancer has a predilection for males and socially disadvantaged population groups in developing countries as well as minority ethnic groups in developed countries (1, 2). Although the oral cavity is accessible to direct examination, oral cancer is usually detected at late stages (3). Despite advances in surgery, radiation and chemotherapy, the 5-year survival rates remain below 50%, one of the lowest for major cancers (4, 5). Furthermore, patients who survive a first cancer of the oral cavity are at very high risk of developing a recurrent or second primary oral cancer (4, 6).
Risk factors for oral cancer
Oral cancer is of multifactorial origin; several risk factors act individually or in combinations in the pathogenesis of the disease, and the mix varies from one population to another. The major risk factors of OSCC, the most extensively studied, include smoking of cigarettes, cigars, beedies and pipes, smokeless tobacco usage, betel quid (with or without tobacco) chewing, other forms of areca nut usage, heavy alcohol consumption and human papillomavirus (HPV) infection (7–10). Tobacco smoking and alcohol consumption alone account for a population attributable risk of 74% (11). Viral infections particularly explain the elevated incidence of cancer among the young population (12, 13) and non-smoking females (14). Genetic susceptibility of the individual arising from polymorphisms in carcinogen metabolising enzymes and DNA repair mechanisms (15–17), often in a background of dietary micronutrient deficiencies, is another factor associated with oral carcinogenesis (13). Lesser known risk factors include exposure to excessive solar radiation/UV light, sulphur dioxide, pesticides, mists from strong inorganic acids and burning of fossil fuels (18, 19). Periodontitis has also been shown in several studies to be associated with increased risk of OSCC (20), with a possible underlying mechanism being long-term chronic inflammation (21).
Can bacteria be a risk factor of oral cancer?
Approximately 15% of oral cancers cannot be explained by the aforementioned major risk factors (22), which has evoked the need to explore for other potential risk factors. In the last two decades, significant evidence has emerged implicating bacteria in the aetiology of some cancer types such as Helicobacter pylori in gastric cancer and mucosa-associated lymphoid tissue (MALT) lymphomas (23), Chlamydia trachomatis in cervical cancer (24), Salmonella typhi in gallbladder cancer (25) and both Bacteroides fragilis and Fusobacterium nucleatum in colon cancer (26, 27). This has recently inspired and triggered a considerable amount of research into the possible role of bacteria in oral oncogenesis, which has indeed provided some evidence to support such a role. Relevant work has been already reviewed in a number of publications (28–30). However, the overwhelmingly growing literature keeps calling for new, updated reviews.
Therefore, this narrative review aims to dissect existing literature on the association between bacteria and oral cancer and to provide an in-depth update about the underlying mechanisms, with particular focus on the work done on Porphyromonas gingivalis and F. nucleatum.
The healthy oral microbiome
Bacteria living within the body of an average healthy adult human outnumber the human cells by at least 10-folds (31, 32). Furthermore, the collective genome of these bacteria (metagenome) surpasses the human genome by orders of magnitude (>150-folds in terms of gene content) (33, 34). Therefore, humans can be thought of as ‘Superorganisms whose metabolism represents an amalgamation of microbial and human attributes’ (35). The ‘human microbiome’ is a term used to define all microorganisms in the human body and their collective genomes (36). Microbial communities found in the different body habitats (e.g. upper respiratory tract, mouth, skin, gut and urogenital tract) contribute to the overall microbiome.
The oral cavity has several distinct niches that provide unique conditions and nutrients for populating microbes, predominately bacteria (37). Each niche (e.g. hard palate, soft palate, lateral and dorsal surfaces of the tongue, and tooth surfaces above and below the gingival margin) displays site specificity and distinct bacterial profile (38). Once established, the oral microbial communities maintain a stable composition ‘microbial homeostasis’ and exhibit commensal and mutualistic relation with the host (39). The host provides its microbial communities with an environment where they can flourish; in return, microbes protect the host as they colonise specific surfaces and prevent adherence and/or hinder growth of pathogenic bacteria (40).
The human mouth harbours one of the most diverse microbiota; at least 687 species/phylotypes have been identified, for each of which a reference 16S rRNA sequence is available in the Human Oral Microbiome Database (HOMD 14.5; www.homd.org) (41). These species belong to 185 genera and 12 phyla, namely Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Spirochaetes, SR1, Synergistetes, Saccharibacteria (TM7) and Gracilibacteria (GN02). Thirty-two percent of these species remain uncultivated and 14% have not been named.
Prior to 2000, the composition of the oral microbiome had been assessed via culture methods or close-ended molecular techniques, mainly checkerboard DNA–DNA hybridisation and polymerase chain reaction (PCR). After 2000, clonal analysis of 16S rRNA gene with Sanger sequencing was employed to identify uncultivable species in oral samples and assess diversity of the oral microbiota (42). Recently, the advent of high throughput, next-generation sequencing (NGS) has enabled analysis of the microbial communities at unprecedented depth and breadth, providing unmatched opportunity for profiling the oral microbiome in health and identifying microbial shifts associated with disease (43).
Oral microbiome dysbiosis and disease
Under certain circumstances, the homeostatic state of oral bacterial flora can be lost, resulting in an ‘ecological shift’ or ‘dysbiosis’ characterised by increased abundance of pathogenic bacteria and expression of virulence properties (39, 40). Subsequently, these pathogens become capable of causing diseases within and beyond the oral cavity, reverting the oral microbiota relationship with the host from mutualistic to parasitic (44). A consortium of microbes rather than one species is usually involved in causing disease (45). Classic examples of oral microbial dysbiosis include dental caries and periodontal infections. In caries, the ecological shift favours growth of acidogenic and aciduric species, namely mutans streptococci, lactobacilli and Bifidobacteria (46). In periodontal diseases, proteolytic bacteria that challenge the host inflammatory response are in play (47). The leading bacteria at periodontal destruction sites include members of the so-called red complex, namely P. gingivalis, Tannerella forsythia and Treponema denticola (48), as well as a number of new taxa such as oral Synergistetes and Saccharibacteria (TM7) (49). Currently, there is increasing interest in the possible role of microbial dysbiosis in cancers.
Oral microbial dysbiosis in oral cancer
The relationship between bacterial profiles and OSCC has been thoroughly studied (Table 1). In the first association study, Nagy et al. (50) performed culture-based analysis of surface swabs and found the levels of Porphyromonas spp., Fusobacterium spp. and other bacterial species to be significantly higher on OSCC tissue compared with adjacent healthy mucosa. In a subsequent study using immunohistochemistry for detection of P. gingivalis, Katz et al. (51) found that sections of gingival squamous cell carcinoma displayed higher staining intensity than those of healthy gingival tissue samples, indicating a higher colonisation of cancerous tissues by this bacterium. Interestingly and consistent with the above findings, both P. gingivalis and F. nucleatum have been linked recently to pancreatic and colorectal cancers (CRC), respectively (26, 52–54).
Table 1.
Summary of epidemiological studies that assessed the association between bacteria and oral cancer
| Study | N | Technology used | Case sample | Control sample | Taxa associated with oral cancer |
|---|---|---|---|---|---|
| Nagy et al., 1998 (50) | 21 | Cultivation; biochemical identification | Tumour surface swabs | Contagious mucosa surface swabs | Fusobacterium, Porphyromonas, Actinomyces, Propionibacterium spp. and Candida albicans |
| Katz et al., 2011 (51) | 15 | Immunohistochemical staining | FFPE gingival carcinoma tissue | FFPE normal tissue | Porphyromonas gingivalis |
| Tateda et al., 2000 (55) | 270 | Cultivation, PCR and Southern-blot | Tumour tissue, gingival smears & oropharyngeal swabs | None | Streptococcus anginosus |
| Sasaki et al., 2005 (56) | 49 | PCR | Fresh tumour tissue, dental plaque & saliva | Fresh tissue, leukoplakia, lymphoma & rhabdomyosarcoma | S. anginosus |
| Morita et al., 2003 (57) | 63 | Real-time PCR | Fresh tumour tissue | Fresh non-cancerous tissue | Association with S. anginosus ruled out |
| Mager et al., 2005 (60) | 274 | Checkerboard DNA–DNA hybridisation | Unstimulated saliva | Unstimulated saliva | Capnocytophaga gingivalis, Prevotella melaninogenica and S. mitis |
| Hooper et al., 2006 (61) | 51 | Cultivation; 16S rRNA gene sequencing | Fresh tumour tissue | Fresh contagious tissue | Micrococcus luteus, P. melaninogenica, Exiguobacterium oxidotolerans, Fusobacterium naviforme, Staphylococcus aureus and Veillonella parvula |
| Hooper et al., 2007 (59) | 20 | 16S rRNA metagenomics (Sanger sequencing) | Fresh tumour tissue | Fresh contagious tissue | Ralstonia insidiosa, Fusobacterium naviforme, Peptostreptococcus micros, Clavibacter michiganensis subsp. tessellarius, Capnocytophaga sp. oral strain S3 and Prevotella sp. oral clone BE073 |
| Pushalkar et al., 2012 (58) | 20 | 16S rRNA metagenomics (Sanger sequencing) & DGGE | Fresh tumour tissue | Fresh contagious tissue | Parvimonas sp. oral taxon 110, Eubacterium infirmum, Eubacterium brachy, Gemella haemolysans, Gemella morbillorum, Gemella sanguinis, Johnsonella ignava, Peptostreptococcus stomatis, S. gordonii, S. parasanguinis I and S. salivarius |
| Pushalkar et al., 2011 (62) | 05 | 16S rRNA metagenomics (NGS; Roche's 454) & DGGE | Stimulated saliva | Stimulated saliva | Genera Streptococcus, Rothia, Gemella, Peptostreptococcus, Lactobacillus, Micromonas and Porphyromonas |
| Schmidt et al., 2014 (63) | 94 | 16S rRNA metagenomics (NGS; Illumina) | Tumour surface swabs | Surface swabs: contra-lateral normal; healthy and pre-cancer subjects | Genus Fusobacterium and phylum Bacteriodetes (Streptococcus and Rothia showed inverse association) |
| Al-Hebshi et al., 2015 (64) | 03 | 16S rRNA metagenomics (NGS; Roche's 454) | Fresh tumour tissue | None | Bacteroides fragilis |
PCR: polymerase chain reaction; FFPE: formalin-fixed, paraffin-embedded; DGGE: denaturing gradient gel electrophoresis; NGS: next-generation sequencing.
Streptococcus anginosus has also been implicated in OSCC. In one study, S. anginosus was detected by PCR in all head and neck squamous cell carcinoma, including OSCC, samples studied (55). Consistently, Sasaki et al. (56) detected S. anginosus in 19/42 (45%) OSCC samples, but in none of the leukoplakia, lymphoma or rhabdomyosarcoma samples tested. However, Morita et al. (57) identified S. anginosus in only 5/38 (13%) of their OSCC samples. In addition, more recent studies have identified S. anginosus in non-tumorous oral tissue at equal and even higher frequency compared with tissue from the tumours, suggesting that S. anginosus is a normal coloniser of the oral mucosa (58, 59).
Mager et al. (60) studied the differences in salivary counts of 40 common oral bacteria between 45 OSCC cases and 229 cancer-free controls using checkerboard DNA–DNA hybridisation. They found Capnocytophaga gingivalis, Prevotella melaninogenica and S. mitis to be significantly elevated in the saliva of cases. When used as diagnostic markers, these three bacterial species were found to differentiate between cases and controls with 80% sensitivity and 83% specificity. However, these results have not been replicated in any subsequent study.
In 2006, Hooper et al. (61), using culture methods, isolated 80 bacterial species from within the tissue of 20 OSCC biopsies. Further analysis of 10 specimens using 16S rRNA Sanger sequencing identified an additional 28 bacterial species (59). These studies provided some evidence for tumour specificity of bacteria, as some species were exclusively found in either the cancerous or non-cancerous tissues. However, the majority of bacteria identified in the tumours were saccharolytic and aciduric (Table 1), suggesting a selection process by the tumour microenvironment rather than a potential association with carcinogenesis. In 2012, Pushalkar et al. (58), also using 16S rRNA Sanger sequencing, detected 80 bacterial species in 10 specimens of OSCC, 35 species of which reported for the first time in OSCC, thus expanding the bacterial diversity within OSCC tissues to 140 species. In this study, however, a totally different panel of species was found to be associated with the tumours (Table 1).
A caveat of conventional culture methods and Sanger sequencing is low analysis depth, that is, the limited number of strains/clones that can be affordably analysed, which hampers reproducible detection of possibly relevant species, particularly those with relatively low abundance. This limitation has been surpassed by the advent of NGS, which allows the study of microbial communities at unprecedented depth and breadth (43). To date, three studies have employed NGS to characterise the oral bacteriome associated OSCC. In one study, Pushalkar et al. (62) identified in salivary samples from three OSCC cases and two healthy controls 860 operational taxonomic units (OTUs) matching known species; 244 and 398 of these were exclusively present in the OSCC and control samples, respectively. However, the comparison between the two groups was limited to the genus level; the genera Streptococcus, Rothia, Gemella, Peptostreptococcus, Porphyromonas, Micromonas and Lactobacillus were found to be more abundant in the OSCC cases, whereas Prevotella, Neisseria, Leptotrichia, Capnocytophaga, Actinobacillus and Oribacterium were higher in the controls. In a larger-scale study, Schmidt et al. (63) analysed swabs of lesion surface and contra-lateral normal mucosa from 18 OSCC patients, eight pre-cancer cases and nine healthy controls. In contrast to the study by Pushalkar et al., the abundance of genera Streptococcus and Rothia was significantly lower in the tumour samples compared with contra-lateral normal as well as to the pre-cancer samples; instead, the tumours were associated with significantly higher proportions of the genus Fusobacterium. In addition, the phylum Bacteroidetes was notably more abundant in both cancerous and normal tissues of OSCC patients compared with pre-cancer and healthy control subjects, suggesting that higher colonisation with this phylum may be associated with increased risk of OSCC, and thus serve as a biomarker.
A limitation to the studies by Pushalkar et al. (62) and Schmidt et al. (63), as the case with most oral microbiome studies employing NGS, was the low taxonomic resolution, that is, failure to reliably classify NGS sequences beyond the genus level, which hinders accurate assessment of the possible association between bacteria and oral cancer. This limitation been overcome in a third study by Al-hebshi et al. (64) utilising a novel bioinformatic algorithm that exploits well-curated databases of 16S rRNA reference sequences, including the HOMD, for the classification of individual NGS reads to the species level. By applying the algorithm to three samples of OSCC DNA, the study revealed the presence of 228 bacterial species, of which 35 species were present in all samples. Two of the samples contained B. fragilis, a bacterium seldom detected in the oral cavity. Interestingly, six proteins from this species had been identified in saliva of OSCC in an earlier study (65). Furthermore, B. fragilis has been recently implicated in colon cancer (27). Together, these findings are suggestive of a role of B. fragilis in oral carcinogenesis, a possibility worth further investigation.
It is evident, therefore, that there is currently no consensus among studies on which bacterial species are linked to oral cancer. In addition, due to the cross-sectional nature of these studies, it is not possible to tell whether any microbial dysbiosis identified is involved in the aetiology of oral cancer or just a consequence of it. Nevertheless, and despite the lack of strong epidemiological evidence to support a role for P. gingivalis and F. nucleatum in oral cancer, the carcinogenic properties of these two species, particularly P. gingivalis, have been extensively demonstrated in vitro and in experimental animals as detailed in the section below.
Possible mechanisms by which oral bacteria contribute to oral carcinogenesis
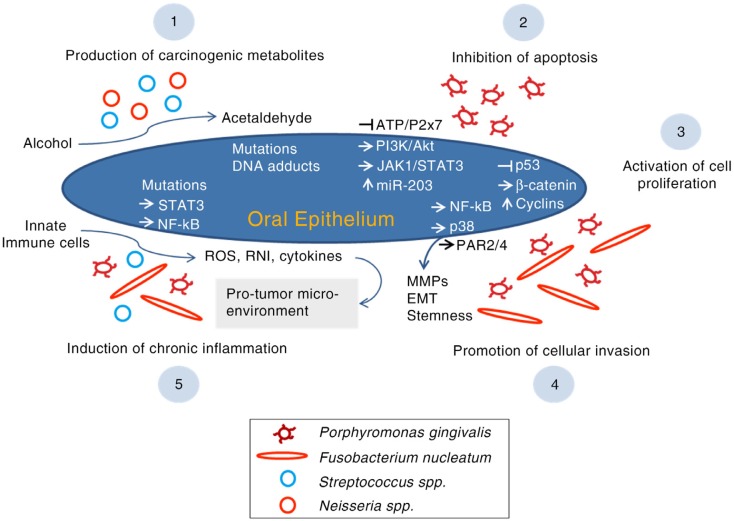
Oral bacteria possibly contribute to oral carcinogenesis via a number of mechanisms including inhibition of apoptosis, activation of cell proliferation, promotion of cellular invasion, induction of chronic inflammation and production of carcinogens (Fig. 1).
Fig. 1.
The possible mechanisms by which oral bacteria contribute to oral carcinogenesis. ROS, reactive oxygen species; RNI, reactive nitrogen intermediates, MMPs, matrix metalloproteinases; PAR, protease-associated receptor; EMT, epithelial to mesenchymal transition.
Inhibition of apoptosis
P. gingivalis represses chemically induced apoptosis in primary cultures of gingival epithelial cells (GECs) (66), which seems to be mediated by various mechanisms. P. gingivalis stimulates JAK1/STAT3 and PI3K/Akt signalling, which controls intrinsic mitochondrial apoptosis pathways (67, 68). At the mitochondrial membrane, the activity of proapoptotic BAD (BCL-2-associated death promoter) is suppressed resulting in elevated BCL2 (anti-apoptotic): BAX (proapoptotic) ratio, which in turn deceases the release of the apoptosis effector cytochrome c (69). Downstream, activity of both caspase-9 and the executioner caspase-3 is blocked (68, 69). In a different mechanism, P. gingivalis has been shown to upregulate microRNA-203 in GECs, which through downregulation of SOCS3 (suppressor of cytokine signalling 3) increases the activity of STAT3 (signal transducer and activator of transcription 3) and, in turn, inhibits apoptosis (70). Furthermore, P. gingivalis secretes a nucleoside diphosphate kinase (NDK), which prevents ATP-dependent apoptosis mediated through purinergic receptor P2X7 on GECs (71); NDK might also interfere with an anticancer immune response mediated by ATP activation of P2X7 receptors on dendritic cells (29, 72). In 2015, Binder Gallimidi et al. (73) reported that chronic coinfection with P. gingivalis and F. nucleatum promotes progression of chemically induced oral cancer in a murine model via activation of the IL6/STAT3 axis.
Activation of cell proliferation
In addition to its anti-apoptotic properties, P. gingivalis accelerates progression of GECs through the S and G2 phases of the cell cycle via upregulation of cyclins (A, D and E), activation (phosphorylation) of cyclin-dependent kinases (CDKs) and diminishing the level of p53 tumour suppressor (74, 75). These effects are dependent on the possession of fimbriae (FimA adhesin). Since bacterial lipopolysaccharide (LPS) has been reported to dysregulate p53, a similar role by P. gingivalis LPS is possible (29). P. gingivalis may also contribute to a proliferative phenotype in GECs through activation of β-catenin via a gingipain-dependent proteolytic process (76).
F. nucleatum also promotes cell proliferation. In human epithelial cells, infection by F. nucleatum results in upregulation of 12 kinases, the majority of which are involved in cell proliferation and cell survival signalling as well as DNA repair (77). A strong relationship between F. nucleatum and CRC is particularly evident. In vitro, fusobacterial adhesion FadA has been found to bind to E-cadherin on CRC cells and in turn activate the β-catenin signalling pathway (78). Downstream, this results in elevated transcriptional activity of oncogenes and pro-inflammatory cytokines, and subsequently enhanced CRC cell proliferation. Consistently, expression levels of the FadA gene in colon tissue from patients with CRC have been shown to be >10-fold higher than those in normal individuals (78).
Promoting cellular migration and invasion
Both P. gingivalis and F. nucleatum promote cellular invasion in OSCC. Using an OSCC cell line, P. gingivalis infection was demonstrated to upregulate expression of pro-matrix metalloproteinase-9 (pro-MMP-9) by the activation of the ERK1/2-ETS1, p38/HSP27 and PAR/NF-kB pathways; gingipains then cleave the proenzyme into active MMP-9, enhancing cellular invasion (79, 80). Repeated exposure to P. gingivalis can also increase invasiveness of OSCC cells by triggering epithelial to mesenchymal transition (EMT), acquisition of stemness and enhanced production of MMP-1 and MMP-10 (81). In a similar fashion, the infection of human epithelial cells by F. nucleatum increases the production of MMP-13 (collagenase 3) through the activation of mitogen-activated protein kinase p38 and promotes cellular migration, possibly via stimulation of Etk/BMX, S6 kinase p70 and RhoA kinase (77).
Induction of inflammation
Chronic inflammation, triggered by infections or environmental exposures, plays a pivotal role in all stages of carcinogenesis including induction, progression, invasion and metastasis (82). Reactive oxygen species (ROS), reactive nitrogen intermediates (RNI) and cytokines produced by inflammatory cells are believed to contribute to initiation of cancer by inducing mutations, genomic instability and epigenetic alterations. Inflammatory cytokines then activate key transcription factors such as STAT3 and NF-kB within the premalignant cells; this in turn supports pro-malignant processes including proliferation, angiogenesis, and invasion and metastasis, and most importantly, results in a sustained tumour-promoting inflammation within the tumour microenvironment (82, 83).
Chronic inflammation has, therefore, been anticipated as one of the potential pathways by which bacteria contribute to oral carcinogenesis (30). Indeed, this provides a plausible explanation for the strong association between periodontitis and higher risk of OSCC (20). A pro-inflammatory potential is documented for some oral bacterial species. F. nucleatum has been found to be associated with high cytokine levels in CRC and to create an inflammatory microenvironment supportive of tumour progression (84, 85). In GECs as well as OSCC cell lines, P. gingivalis upregulates B7-H1 and B7-DC receptors, both of which are known to contribute to chronic inflammation (72). Increased production of IL-1, IL-6, IL-8 and TNF-α in response to P. gingivalis infection has been documented in engineered human oral mucosa (86). Similar pro-inflammatory properties have been reported for other oral bacteria including Eikenella corrodens, S. anginosus and S. mitis (87, 88).
Production of carcinogens
Ethanol itself is not a carcinogen but its metabolites acetaldehyde, hydroxyl ethyl radicals and hydroxyl radicals are carcinogenic (89, 90). The International Agency for Research on Cancer classified acetaldehyde associated with alcohol consumption as a Group 1 carcinogen in humans, with the capability to cause sister chromatid exchanges, point mutations, DNA adducts and hyperproliferation of epithelium (8, 91). It is well established that certain bacteria and Candida spp. in the oral cavity possess the enzyme alcohol dehydrogenase (ADH), which catalyses the production of mutagenic amounts of acetaldehyde under aerobic or microaerophilic conditions (30, 92–94). Examples of such oral bacteria include S. salivarius, S. intermedius, S. mitis (95) and non-pathogenic Neisseria spp. (96).
Conclusion and future directions
There is an increased interest in the potential role of bacteria in oral cancer as evident from the increasing number of publications addressing the topic. However, published studies disagree on which specific bacteria or patterns of oral microbial dysbiosis to associate with oral cancer. This is probably due to the significant methodological variation between studies with respect to sampling (e.g. deep tissue biopsy vs. surface swab), selection of control tissues and microbial profiling technology. 16S rRNA metagenomics with NGS has great potential in exploring the oral microbiome shifts associated with oral cancer, but currently lacks adequate standardisation, particularly in terms of DNA extraction, selection of primers and hypervariable region for amplification, and bioinformatics. In addition, 16S rRNA microbial profiling does not inform about microbial gene expression and microbial function. Therefore, future work should, in addition to methodological standardisation, embody full metagenomics or metatranscriptomics analysis into case-control studies to fully explain the role of the oral microbiome in oral cancer and generate information of translational value, for example, identify diagnostic and prognostic markers.
The oral carcinogenic properties of P. gingivalis and F. nucleatum are well documented in vitro as well as in experimental animals; many of the mechanisms underlying these have been elucidated. It has been suggested, therefore, that the detection of P. gingivalis and F. nucleatum in oral potentially malignant conditions could be used as a marker of risk of malignant transformation. Virulence factors such as FimA of P. gingivalis and FadA of F. nucleatum may also serve as novel targets for therapeutic intervention of oral cancer. However, it is important to note that a strong epidemiological evidence for the association of P. gingivalis and F. nucleatum with oral cancer is still lacking. Large-scale, well-designed longitudinal studies that involve follow-up of microbiota associated with precancerous lesions while adequately control for the confounding effects of established risk factors are required to demonstrate if these two or other species contribute or even have an exclusive role in oral carcinogenesis in a subset of patients. Such an approach will also help resolve the legitimate hen and egg question: which species, if any, is involved in carcinogenesis (driver) and which is simply an adapter to the cancer tissue (passenger)?
There are a number of well-established risk factors for oral cancers and a presumed multifactorial aetiology. The balance of risk factors varies from individual to individual. In the age of personalised medicine, it is important to isolate causes and the molecular damage caused, in order to plan personalised therapy. Further work on the oral bacteriome associated with oral cancers has an exciting future and can generate hypotheses for exploring mechanisms in cell and animal models.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors. Perera M is supported by a Griffith University Postgraduate Research Scholarship.
References
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; [Google Scholar]
- 3.Silverman S. Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J Am Dent Assoc. 2001;132:7S–11S. doi: 10.14219/jada.archive.2001.0382. [DOI] [PubMed] [Google Scholar]
- 4.Alfano MC, Horowitz AM. Professional and community efforts to prevent morbidity and mortality from oral cancer. J Am Dent Assoc. 2001;132:24S–9S. doi: 10.14219/jada.archive.2001.0385. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Zhang S, Yue K, Wang XD. The recurrence and survival of oral squamous cell carcinoma: a report of 275 cases. Chin J Cancer. 2013;32:614–18. doi: 10.5732/cjc.012.10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez-Mahia I, Seoane J, Varela-Centelles P, Tomas I, Alvarez Garcia A, Lopez Cedrun JL. Predictors for tumor recurrence after primary definitive surgery for oral cancer. J Oral Maxillofac Surg. 2012;70:1724–32. doi: 10.1016/j.joms.2011.06.228. [DOI] [PubMed] [Google Scholar]
- 7.Petti S. Lifestyle risk factors for oral cancer. Oral Oncol. 2009;45:340–50. doi: 10.1016/j.oraloncology.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V, et al. A review of human carcinogens – part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033–4. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 9.Syrjänen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32:59–66. doi: 10.1016/j.jcv.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Gupta B, Johnson NW. Systematic review and meta-analysis of association of smokeless tobacco and of betel quid without tobacco with incidence of oral cancer in South Asia and the Pacific. PLoS One. 2014;9:e113385. doi: 10.1371/journal.pone.0113385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen PE. Oral cancer prevention and control – the approach of the World Health Organization. Oral Oncol. 2009;45:454–60. doi: 10.1016/j.oraloncology.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Llewellyn CD, Linklater K, Bell J, Johnson NW, Warnakulasuriya S. An analysis of risk factors for oral cancer in young people: a case-control study. Oral oncol. 2004;40:304–13. doi: 10.1016/j.oraloncology.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Llewellyn C, Johnson NW, Warnakulasuriya K. Risk factors for oral cancer in newly diagnosed patients aged 45 years and younger: a case–control study in Southern England. J Oral Pathol Med. 2004;33:525–32. doi: 10.1111/j.1600-0714.2004.00222.x. [DOI] [PubMed] [Google Scholar]
- 14.Bleyer A, editor. Cancer of the oral cavity and pharynx in young females: increasing incidence, role of human papilloma virus, and lack of survival improvement. Semin Oncol. 2009;36:451–9. doi: 10.1053/j.seminoncol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Kietthubthew S, Sriplung H, Au WW, Ishida T. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health. 2006;209:21–9. doi: 10.1016/j.ijheh.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Bau D, Tsai M, Huang C, Lee C, Tseng H, Lo Y, et al. Relationship between polymorphisms of nucleotide excision repair genes and oral cancer risk in Taiwan: evidence for modification of smoking habit. Chin J Physiol. 2007;50:294. [PubMed] [Google Scholar]
- 17.Hatagima A, Costa EC, Marques CF, Koifman RJ, Boffetta P, Koifman S. Glutathione S-transferase polymorphisms and oral cancer: a case-control study in Rio de Janeiro, Brazil. Oral Oncol. 2008;44:200–7. doi: 10.1016/j.oraloncology.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Thornhill MH. The sun, the ozone layer and the skin: the role of ultraviolet light in lip and skin cancer. Dent Update. 1993;20:236–40. [PubMed] [Google Scholar]
- 19.Kumar SKS, Zain RB. Aetiology and risk factors for oral cancer – a brief overview. Annal Dent Univ Malaya. 2004;11:41–50. [Google Scholar]
- 20.Meyer MS, Joshipura K, Giovannucci E, Michaud DS. A review of the relationship between tooth loss, periodontal disease, and cancer. Cancer Causes Control. 2008;19:895–907. doi: 10.1007/s10552-008-9163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi C, Gudino C, Gibson F, III, Genco C. Review: pathogen-induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell-specific innate immune inflammatory pathways. Mol Oral Microbiol. 2010;25:305–16. doi: 10.1111/j.2041-1014.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chocolatewala N, Chaturvedi P, Desale R. The role of bacteria in oral cancer. Indian J Med Paediatr Oncol. 2010;31:126. doi: 10.4103/0971-5851.76195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mager D. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. doi: 10.1186/1479-5876-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowska J, Fischer N, Markowski M, Nalewaj J. The role of Chlamydia trachomatis infection in the development of cervical neoplasia and carcinoma. Med Wieku Rozwoj. 2005;9:83–6. [PubMed] [Google Scholar]
- 25.Nagaraja V, Eslick G. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther. 2014;39:745–50. doi: 10.1111/apt.12655. doi: http://dx.doi.org/10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 26.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–8. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–6. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 28.Meurman JH. Oral microbiota and cancer. J Oral Microbiol. 2010;2:5195. doi: 10.3402/jom.v2i0.5195. doi: http://dx.doi.org/10.3402/jom.v2i0.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10:e1003933. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooper SJ, Wilson MJ, Crean SJ. Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head Neck. 2009;31:1228–39. doi: 10.1002/hed.21140. [DOI] [PubMed] [Google Scholar]
- 31.Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35:249–55. doi: 10.1093/carcin/bgt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–35. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27:1012–22. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Do T, Devine D, Marsh PD. Oral biofilms: molecular analysis, challenges, and future prospects in dental diagnostics. Clin Cosmet Investig Dent. 2013;5:11–19. doi: 10.2147/CCIDE.S31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18:109–20. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen T, Yu W-H, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakamoto M, Umeda M, Benno Y. Molecular analysis of human oral microbiota. J Periodontal Res. 2005;40:277–85. doi: 10.1111/j.1600-0765.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- 43.Siqueira JF, Jr, Fouad AF, Rocas IN. Pyrosequencing as a tool for better understanding of human microbiomes. J Oral Microbiol. 2012;4:10743. doi: 10.3402/jom.v4i0.10743. doi: http://dx.doi.org/10.3402/jom.v4i0.10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parahitiyawa N, Scully C, Leung W, Yam W, Jin L, Samaranayake L. Exploring the oral bacterial flora: current status and future directions. Oral Dis. 2010;16:136–45. doi: 10.1111/j.1601-0825.2009.01607.x. [DOI] [PubMed] [Google Scholar]
- 45.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–90. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 47.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Microbiol. 2010;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 48.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 49.Al-hebshi NN, Al-Alimi A, Taiyeb-Ali T, Jaafar N. Quantitative analysis of classical and new putative periodontal pathogens in subgingival biofilm: a case-control study. J Periodontal Res. 2015;50:320–9. doi: 10.1111/jre.12210. [DOI] [PubMed] [Google Scholar]
- 50.Nagy K, Sonkodi I, Szöke I, Nagy E, Newman H. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34:304–8. [PubMed] [Google Scholar]
- 51.Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3:209–15. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–8. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michaud DS, Izard J. Microbiota, oral microbiome, and pancreatic cancer. Cancer J. 2014;20:203–6. doi: 10.1097/PPO.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tateda M, Shiga K, Saijo S, Sone M, Hori T, Yokoyama J, et al. Streptococcus anginosus in head and neck squamous cell carcinoma: implication in carcinogenesis. Int J Mol Med. 2000;6:699–703. doi: 10.3892/ijmm.6.6.699. [DOI] [PubMed] [Google Scholar]
- 56.Sasaki M, Yamaura C, Ohara-Nemoto Y, Tajika S, Kodama Y, Ohya T, et al. Streptococcus anginosus infection in oral cancer and its infection route. Oral Dis. 2005;11:151–6. doi: 10.1111/j.1601-0825.2005.01051.x. [DOI] [PubMed] [Google Scholar]
- 57.Morita E, Narikiyo M, Yano A, Nishimura E, Igaki H, Sasaki H, et al. Different frequencies of Streptococcus anginosusinfection in oral cancer and esophageal cancer. Cancer Sci. 2003;94:492–6. doi: 10.1111/j.1349-7006.2003.tb01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pushalkar S, Ji X, Li Y, Estilo C, Yegnanarayana R, Singh B, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hooper SJ, Crean S-J, Fardy MJ, Lewis MA, Spratt DA, Wade WG, et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. 2007;56:1651–9. doi: 10.1099/jmm.0.46918-0. [DOI] [PubMed] [Google Scholar]
- 60.Mager D, Haffajee A, Devlin P, Norris C, Posner M, Goodson J. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hooper SJ, Crean SJ, Lewis MA, Spratt DA, Wade WG, Wilson MJ. Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol. 2006;44:1719–25. doi: 10.1128/JCM.44.5.1719-1725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pushalkar S, Mane SP, Ji X, Li Y, Evans C, Crasta OR, et al. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol. 2011;61:269–77. doi: 10.1111/j.1574-695X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz EL, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9:e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Hebshi NN, Nasher AT, Idris AM, Chen T. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol. 2015;7:28934. doi: 10.3402/jom.v7.28934. doi: http://dx.doi.org/10.3402/jom.v7.28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie H, Onsongo G, Popko J, de Jong EP, Cao J, Carlis JV, et al. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:486–98. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, Watanabe K, El-Sabaeny A, et al. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol Lett. 2001;200:145–9. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 67.Yilmaz Ö, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72:3743–51. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao L, Jermanus C, Barbetta B, Choi C, Verbeke P, Ojcius D, et al. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Mol Oral Microbiol. 2010;25:89–101. doi: 10.1111/j.2041-1014.2010.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moffatt CE, Lamont RJ. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect Immun. 2011;79:2632–7. doi: 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yilmaz Ö, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, et al. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 2008;10:863–75. doi: 10.1111/j.1462-5822.2007.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology. 2011;216:1302–10. doi: 10.1016/j.imbio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 73.Binder Gallimidi A, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6:22613–23. doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, et al. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–8. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan C, Xu X, Tan L, Lin L, Pan Y. The effects of Porphyromonas gingivalis on the cell cycle progression of human gingival epithelial cells. Oral Dis. 2014;20:100–8. doi: 10.1111/odi.12081. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Y, Sztukowska M, Wang Q, Inaba H, Potempa J, Scott DA, et al. Noncanonical activation of beta-catenin by Porphyromonas gingivalis. Infect Immun. 2015;83:3195–203. doi: 10.1128/IAI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uitto V-J, Baillie D, Wu Q, Gendron R, Grenier D, Putnins EE, et al. Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells. Infect Immun. 2005;73:1171–9. doi: 10.1128/IAI.73.2.1171-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inaba H, Sugita H, Kuboniwa M, Iwai S, Hamada M, Noda T, et al. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014;16:131–45. doi: 10.1111/cmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inaba H, Amano A, Lamont RJ, Murakami Y. Involvement of protease-activated receptor 4 in over-expression of matrix metalloproteinase 9 induced by Porphyromonas gingivalis. Med Microbiol Immunol. 2015;204:605–12. doi: 10.1007/s00430-015-0389-y. [DOI] [PubMed] [Google Scholar]
- 81.Ha NH, Woo BH, Kim da J, Ha ES, Choi JI, Kim SJ, et al. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. 2015;36:9947–60. doi: 10.1007/s13277-015-3764-9. [DOI] [PubMed] [Google Scholar]
- 82.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andrian E, Grenier D, Rouabhia M. In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infect Immun. 2004;72:4689–98. doi: 10.1128/IAI.72.8.4689-4698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y, Kato H, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95:569–74. doi: 10.1111/j.1349-7006.2004.tb02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yumoto H, Nakae H, Yamada M, Fujinaka K, Shinohara C, Ebisu S, et al. Soluble products from Eikenella corrodens stimulate oral epithelial cells to induce inflammatory mediators. Oral Microbiol Immunol. 2001;16:296–305. doi: 10.1034/j.1399-302x.2001.016005296.x. [DOI] [PubMed] [Google Scholar]
- 89.Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol Res Health. 2006;29:245–54. [PMC free article] [PubMed] [Google Scholar]
- 90.Seitz H, Gärtner U, Egerer G, Simanowski UA. Ethanol metabolism in the gastrointestinal tract and its possible consequences. Alcohol Alcohol Suppl. 1994;2:157–62. [PubMed] [Google Scholar]
- 91.Seitz HK, Cho CH. Contribution of alcohol and tobacco use in gastrointestinal cancer development. Methods Mol Biol. 2009;472:217–41. doi: 10.1007/978-1-60327-492-0_9. [DOI] [PubMed] [Google Scholar]
- 92.Homann N, Tillonen J, Meurman JH, Rintamäki H, Lindqvist C, Rautio M, et al. Increased salivary acetaldehyde levels in heavy drinkers and smokers: a microbiological approach to oral cavity cancer. Carcinogenesis. 2000;21:663–8. doi: 10.1093/carcin/21.4.663. [DOI] [PubMed] [Google Scholar]
- 93.Väkeväinen S, Mentula S, Nuutinen H, Salmela K, Jousimies-Somer H, Färkkilä M, et al. Ethanol-derived microbial production of carcinogenic acetaldehyde in achlorhydric atrophic gastritis. Scand J Gastroenterol. 2002;37:648–55. doi: 10.1080/00365520212500. [DOI] [PubMed] [Google Scholar]
- 94.Salaspuro MP. Acetaldehyde, microbes, and cancer of the digestive tract. Crit Rev Clin Lab Sci. 2003;40:183–208. doi: 10.1080/713609333. [DOI] [PubMed] [Google Scholar]
- 95.Kurkivuori J, Salaspuro V, Kaihovaara P, Kari K, Rautemaa R, Grönroos L, et al. Acetaldehyde production from ethanol by oral streptococci. Oral Oncol. 2007;43:181–6. doi: 10.1016/j.oraloncology.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 96.Muto M, Hitomi Y, Ohtsu A, Shimada H, Kashiwase Y, Sasaki H, et al. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: implications for carcinogenesis in upper aerodigestive tract. Int J Cancer. 2000;88:342–50. [PubMed] [Google Scholar]