Abstract
Hemagglutinin (HA) is a protein located on the surface of the influenza virus that mediates viral fusion to the host cellular membrane. During the fusion process the HA fusion peptide (HAfp), formed by the first 23 N-terminal residues of HA and structurally characterized by two alpha helices (Helix A and Helix B) tightly packed in a hairpin-like arrangement, is the only part of the virus in direct contact with the host membrane. After encountering the host cell HAfp is believed to insert into the membrane, thereby initiating the fusion of the viral and host membranes. Detailed characterization of the interactions between the HAfp and cellular membrane is therefore of high relevance to the mechanism of viral entry into the host cell. Employing HMMM membrane representation with enhanced lipid mobility, we have performed a large set of independent simulations of unbiased membrane binding of HAfp. We have been able to capture spontaneous binding and insertion of HAfp consistently in nearly all the simulations. A reproducible membrane-bound configuration emerges from these simulations, despite employing a diverse set of initial configurations. Extension of several of the simulations into full membrane systems confirms the stability of the membrane-bound form obtained from HMMM binding simulations. The resulting model allows for the characterization of important interactions between the peptide and the membrane, and the details of the binding process of the peptide for the first time. Upon membrane binding, Helix A inserts much deeper into the membrane than Helix B, suggesting that the former is responsible for hydrophobic anchoring of the peptide into the membrane. Helix B, in contrast, is found to establish major amphipathic interactions at the interfacial region thereby contributing to binding strength of HAfp.
Introduction
Viral infection starts with the fusion of the viral and host membranes,1 a process mediated by hemagglutinin (HA), a homotrimeric glycoprotein located on the surface of the virus.2 HA monomers are each composed of two major domains: HA1, a globular domain anchored in the viral membrane, and HA2, the domain that mediates attachment to the host membrane. During the fusion process, the N-terminal 23 residues of HA2, known as the HA fusion peptide (HAfp), are the only part of the virus in direct contact with the host membrane.3 At high or normal pH HAfp is buried within the core of HA,4 but lowering the pH, e.g. in the endosomes, induces a significant conformational change in HA that results in the exposure of HAfp,5,6 so that it can target and insert into the host cellular membrane, thereby initiating the fusion of the viral and host membranes. Although the detailed mechanism by which HA mediates membrane fusion remains elusive, it is well established that the role of HAfp in this process hinges on its direct interaction with and insertion into the host membrane.7,8 In addition to its anchoring role, it has been proposed that upon membrane insertion, HAfp promotes significant changes in the lipid bilayer structure, including membrane curvature9,10 and lipid protrusion,11 that can further facilitate membrane fusion.
Due to its importance in the fusion process, and aiming at providing a detailed picture of the underlying molecular events for its function, a large number of biophysical studies have investigated HAfp in its isolated form, i.e., in the absence of the remainder of HA2. At low or neutral pH, binding experiments have shown that different variants of HAfp are able to insert into lipid bilayers, thus promoting membrane fusion.12–14 The most recent structure of HAfp is the NMR structure of full length HAfp (subtype H1) in micelles, consisting of 23 residues.15 This structure contains three additional C-terminal residues (Trp-21, Tyr-22 and Gly-23) which are strictly conserved among all the known serotypes of HA, but were not resolved in the previous HAfp structures.14 The structure of HAfp in micelles15 is characterized by two alpha helices tightly packed in a hairpin-like arrangement, with the hydrophobic side chains facing the same side in both helices while the polar residues are mostly located at the interhelical interface. Subsequent NMR studies aimed at characterizing the interaction of HAfp with micelles revealed the pH- and length-dependence of both the conformation and fusogenic activity of the peptide,16,17 suggesting an important role for Trp-21, Tyr-22 and Gly-23 in interaction with the membrane.
Because of the highly dynamic nature of the peptide–membrane interactions, the details of the membrane binding mechanism of HAfp are difficult to elucidate experimentally. To overcome this, computational approaches employing molecular dynamics (MD) simulations have been employed to elucidate the role of HAfp in membrane fusion by characterizing its interactions with artificial lipid bilayers.9,11,18–25 These studies have employed a variety of models, including all-atom representations,9,11,18–22,24,25 and coarse-grained models.23 Simulations in membranes of different lipid compositions have suggested that wild-type HAfp induces significant structural changes in the lipids, resulting in thinning of the bilayer,9,18–21 lipid protrusion,11,24 and in positive curvature of the membrane,21,23 while mutations of HAfp (e.g., E11A and W14A) greatly reduce thinning and curvature-inducing effects.21 The induced disruption of the membrane by HAfp is suggested to promote the formation of a fusion stalk where lipids in the outer leaflets of both the host and viral membranes make contacts,11 which could eventually lead to pore formation and irreversible fusion between the viral and host cell membranes.7,8
Understanding the interaction of the HAfp with the membrane is a crucial step towards elucidating the role of the peptide in membrane fusion. The majority of the previous computational studies have employed a model peptide containing only the first 20 N-terminal residues of HAfp,14 with only one using on the recently resolved full-length, 23-residue HAfp.25 Although the first 20 residues of HAfp are sufficient to maintain fusogenic activity,12,13,16 the full length sequence of the peptide has been suggested to regulate conformational changes of HAfp which might be relevant to membrane fusion.16 More importantly, previous MD studies have employed a predefined placement of HAfp in the membrane, based on EPR spectroscopy.14 Since membrane relaxation is a slow process, placing the peptide in a predefined position could potentially bias the results of the simulations, specifically the resulting insertion depth and the orientation of peptide in the membrane. On the other hand, capturing the spontaneous association and insertion of proteins with membranes is challenging as the process is primarily hampered by the slow dynamics of membrane lipids and their insufficient sampling on the timescales currently accessible by atomistic MD simulations. Recently, an alternative approach to study peripheral membrane proteins based on a highly mobile membrane-mimetic (HMMM) model has been developed.26 This membrane model employs a biphasic solvent system27 together with short-tailed lipids located at the solvent/water interface. The design of the HMMM model allows for an increase of 1-2 orders in the lipid diffusion, accelerating the spontaneous association of proteins with the bilayer.26 The HMMM model has been successfully employed to capture the membrane binding and insertion of a variety of proteins, such as the GLA domain of the human coagulation factor VII,26 cytochrome P450,28 talin,29 α–synuclein,30 synaptotagmin,31 and synaptobrevin,32 while preserving the atomistic detail of the interactions between the protein and lipid head groups.33
In this study, we employed the HMMM model to study the process of spontaneous membrane binding of the full-length HAfp. The HMMM model is particularly suitable to study HAfp–membrane interactions, given the expected partitioning of the peptide at the interfacial region as suggested by the micelle-derived structure and subsequent NMR studies,15,34 since the short-tailed lipids accurately reproduce relevant interactions and free energies of amino acid insertion into this region of the membrane with great detail.33 The size of the system together with the accelerated lipid dynamics of the HMMM membrane allow for an increased sampling of membrane binding events. Spontaneous binding and insertion of the peptide into the membrane were captured in 18 independent simulations, resulting in a convergent model of the membrane-bound HAfp, despite the designed scatter of the initial configurations in different simulations. The insertion depth and the orientation of the membrane-bound HAfp are preserved after converting the HMMM model to a full lipid bilayer. These simulations provide a convergent membrane-bound model for influenza HAfp resulting from spontaneous binding of the peptide, that is not biased by initial placement of the peptide in the membrane.
Methods
Membrane preparation
The methods used in this work closely follow the protocol described in References 26 and 28 for HMMM membrane preparation and simulations.26,28 The HMMM membrane was constructed by placing two leaflets of divaleryl-phosphatidylcholine (DVPC) at the interface of water and 1,1-dichloroethane (DCLE), as described in detail elsewhere.26 The DVPC molecules were generated by shortening the lipid tails of a palmitoyl-oleoyl-phosphatidylcholine (POPC) molecule to only 5 carbons.26 The HMMM membrane was assembled with the Packmol software,35 employing a DCLE box with dimensions of 65×65×10 Å3 and containing 308 molecules of the organic solvent, and placing 128 short-tailed lipids at each water/DCLE interface, with 64 lipids on each side. The resulting structure was then solvated using the SOLVATE plugin of VMD,36 resulting in a system with ~23,000 atoms. The solvated membrane system was energy minimized for 10,000 steps and simulated for 2 ns, using an NPnAT ensemble with constant area, with a target pressure and temperature of 1.0 atm and 310 K, respectively. A constant area of 4,356 Å2 (66×66 Å2) was employed, yielding an area per lipid (AL) of ~68 Å2, which closely matches the experimental AL for POPC.37,38 The resulting membrane was employed in the subsequent membrane-binding simulations.
Modeling the fusion peptide
The NMR structure of subtype H1 HAfp was obtained from the RCSB Protein Data Bank39 (PDB entry 2KXA).15 This PDB entry contains 10 conformations of HAfp obtained in DPC micelles at pH 7.4, all presenting very similar structures. Given the high structural similarity, only one conformer (the first one) was used for the simulations. Glu and Asp residues of the peptide were modeled in their unprotonated (charged) forms, in order to model the charge of the peptide at neutral pH. HAfp was placed in a box with dimensions of 60×60×60 Å3 and containing ~20,000 water molecules, generated with the SOLVATE plugin of VMD,36 and neutralized with 100 mM NaCl ions using the AUTOIONIZE plugin. The solvated peptide system was then energy minimized for 10,000 steps and simulated for 50 ps with the Cα atoms of the peptide harmonically restrained (k = 5 kcal mol−1 Å−2), and a 20 ns run with no restraints followed. The resulting equilibrated structure of HAfp in solution was used as the initial structure for the membrane-binding simulations with the HMMM membrane model.
Membrane-binding simulations
The equilibrated HAfp structure was added to the membrane model by placing its center-of-mass (COM) at a position 7-10 Å above the PO4 plane of the cis leaflet, without contacting the membrane. In order to test the convergence of the membrane-binding simulations, seven different starting orientations of HAfp were employed (Fig. 1). An additional layer (10 Å) of water was added using the SOLVATE plugin to the resulting structures to ensure sufficient hydration. To neutralize the systems, 100 mM NaCl was added using the AUTOIONIZE plugin of VMD.36 These initial peptide-membrane systems consisted of a box with dimensions of 65×65×72 Å3containing 29,622 atoms.
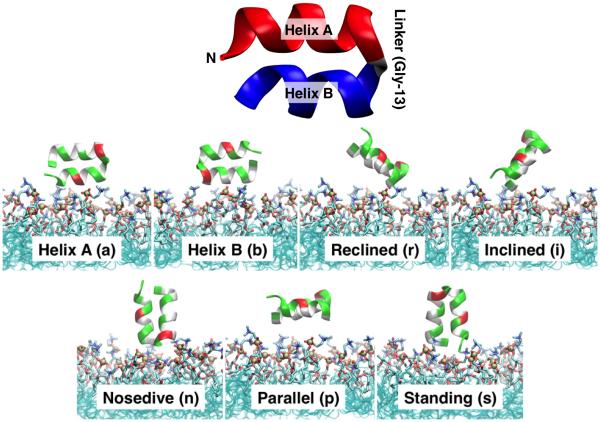
Figure 1. Initial configurations of HAfp in membrane-binding simulations.
(Top) Cartoon representation of important structural details of HAfp. Helix A (red) is composed of residues 3 to 12, and Helix B (blue) is composed of residues 14 to 22, brought together by a small linker region (Gly-13). (Bottom) Seven different initial positions were used for membrane-binding simulations. Different orientations and heights of the peptide with respect to the membrane plane (z-axis) were employed in order to test the convergence of the membrane-bound form of HAfp. A set of 3 independent 100–ns simulations were performed for each initial configuration. The peptide is shown in cartoon representation, with polar residues shown in green, hydrophobic residues in white, and acidic residues in red. Different configurations were labeled according to their orientation with respect to the membrane normal (z-axis) and the part of the peptide initially facing the membrane: (a) Helix A and (b) Helix B are placed closer to the membrane surface, respectively; and the linker region between Helices A and B placed at a (r) reclined, (i) inclined, (n) nosedive, (p) parallel, and (s) standing positions with respect to the membrane.
The systems were energy minimized for 10,000 steps and simulated for 50 ps with the Cα atoms of the peptide harmonically restrained (k = 5 kcal mol−1 Å−2), followed by a production run of 100 ns. Three independent runs for each starting position were performed, resulting in a total of 21 independent membrane–binding simulations, from which a set of 18 yielded membrane insertion of HAfp within 100 ns (Table 1). A harmonic restraint along the z-axis, with a force constant k = 0.05 kcal mol−1 Å−2, was applied to the carbonyl carbon atoms of the DVPC lipids to mimic the atomic distributions of a full lipid bilayer more closely, and to prevent the occasional lipid diffusion into the aqueous phase, expected for these surfactant-like molecules.26
Table 1.
Summary of the simulations. The systems are labeled according to the initial orientation a, b, r, i, n, p, s of HAfp (see Fig. 1). The suffix (1, 2 or 3) indicates the replicate number for the respective system. A letter f at the end of the system name indicates the extension of the membrane-bound system fo a full POPC membrane. The simulations in which insertion of the peptide was observed (18 out of 21) are indicated in the left column.
| System name | Simulation time | Membrane insertion | |
|---|---|---|---|
| Helix A |
a1
a2 a3 |
100 ns 100 ns 100 ns |
− + + |
| Helix B |
b1
b2 b3 |
100 ns 100 ns 100 ns |
+ + + |
| Reclined |
r1
r2 r3 r3f |
100 ns 100 ns 100 ns 30ns (POPC) |
− − + already membrane-bound |
| Inclined |
i1
i2 i3 i2f |
100 ns 100 ns 100 ns 30ns (POPC) |
+ + + already membrane-bound |
| Nosedive |
n1
n2 n3 |
100 ns 100 ns 100 ns |
+ + + |
| Parallel |
p1
p2 p3 |
100 ns 100 ns 100 ns |
+ + + |
| Standing |
s1
s2 s3 s1f |
100 ns 100 ns 100 ns 30ns (POPC). |
+ + + already membrane-bound |
In addition to the HMMM systems, and to further examine the stability of the resulting membrane-bound HAfp, simulations employing a full POPC lipid bilayer were also performed. These simulations used the final, membrane-bound peptides produced by the HMMM simulations as their starting configuration, and followed the membrane preparation protocol described in Reference 28. Three membrane-bound models of HAfp were adopted from the last frames of simulations r3, i2, and s1 (Table 1). For these systems, the DVPC bilayer was transformed into a POPC bilayer by removing the DCLE molecules and adding the missing carbons of the lipid tails, while preserving the positions of the lipid atoms already present in the HMMM model (head groups and initial few carbons of the lipid tails). The positions of the newly added lipid tail atoms were then optimized by adopting the coordinates of the corresponding atoms obtained from randomly selected POPC molecules from a membrane previously equilibrated. During these steps, the original contacts established between the lipids and HAfp in the HMMM membrane-binding simulations were preserved. The resulting POPC-membrane systems, hereby referred to as r3f, i2f and s1f, were then minimized and equilibrated for 100 ps while restraining (k = 1 kcal mol−1 Å−2) the heavy atoms of the peptide and the short-tailed lipids already present before extending the tails to allow for the newly added atoms to relax. Following this step, the systems were equilibrated without restraints for 30 ns, with the last 20 ns used for structural analysis.
Simulation protocol
Simulations were performed using NAMD2,40 utilizing the CHARMM27 force field with CMAP 41 corrections for proteins and CHARMM3642 parameters for lipids. The TIP3P model was used for water.43 All the HMMM simulations were performed in NPnAT ensembles at 1.0 atm and 310 K, and with a time step of 2 fs. The POPC systems were simulated as an NPT ensemble with similar parameters. Constant pressure was maintained using the Nosé-Hoover Langevin piston method,44,45 and constant temperature was maintained by Langevin dynamics with a damping coefficient γ of 0.5 ps−1applied to all atoms. Nonbonded interactions were cut off after 12 Å with a smoothing function applied after 10 Å. The particle mesh Ewald (PME) method46 was used for long-range electrostatic calculations with a grid density of > 1 Å−3.
Analysis
The position of HAfp with respect to the membrane was characterized by the height (depth of insertion) along the membrane normal (z-axis) of the center of mass (COM) of the heavy atoms of the hydrophobic side chains of HAfp (Leu-2, Phe-3, Ile-6, Phe-9, Ile-10, Trp-14, Met-17, Ile-18, and Trp-21), in order to monitor the insertion of the peptide during the simulations. Membrane insertion was assumed when the COM of the hydrophobic side chains of the peptide was located around or below the PO4 level (see Fig. 2). After insertion, the profile of HAfp in the membrane was characterized by the average distance along the z-axis between a selected set of the side chains without the backbone (e.g., Helix A and Helix B) and the PO4 plane of the cis leaflet. The average was taken over the last 10 ns of the simulation (i.e., after insertion was completed). The effect of the membrane on the structure of the peptide was quantified by measuring the backbone root mean square deviation (RMSD) with respect to the NMR structure.15 In addition, to quantify the structural variability of the peptide among different simulations, for each pair of the simulation systems, backbone RMSD was also calculated. For each pair this was done by generating an ensemble of 100 peptide backbone structures from the last 1 ns of the first simulation (10 ps interval), calculating RMSDs of all 100 frames to those obtained from the second simulation, and reporting the average of all the 100 × 100 RMSD values.
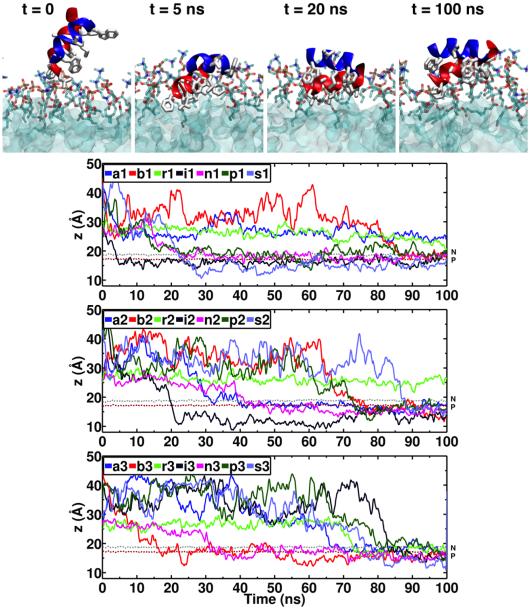
Figure 2. Spontaneous membrane binding and insertion of HAfp.
(Top) Snapshots taken at different times from System i1, representing a typical example of spontaneous membrane insertion observed in HMMM simulations. Helix A and Helix B are shown in red and blue, respectively; hydrophobic side chains are shown in white. (Bottom) Time evolution of the position along the membrane normal (z-axis) of the center of mass (COM) of the heavy atoms of hydrophobic side chains of HAfp (Leu-2, Phe-3, Ile-6, Phe-9, Ile-10, Trp-14, Met-17, Ile-18 and Trp-21) for all the 21 simulation systems. Membrane insertion was assumed when the COM of the hydrophobic side chains was located at or below the PO4 level. The average positions of the phosphorus (PO4) and the nitrogen (choline) atoms of the lipid head groups are shown using brown and gray dotted lines, respectively.
During the simulations, the orientation of the peptide was characterized by the orientation of its two helices (Helices A and B) individually. The helix axis was defined by the average backbone N-H bond vector of residues 3 to 12 for Helix A, and residues 14 to 22 for Helix B. The orientation of each helix was then characterized by the tilt angle between its main axis and the membrane normal (the z-axis). The tilt angle for each helix was averaged over the last 10 ns of each simulation. Rotation of the peptide around the helical hairpin principal axes was calculated for the last 10 ns of simulation for each system, employing the angles defined by Chen et al.47 to characterize protein orientation.
The interaction of HAfp with the membrane was also characterized by the number of hydrogen bonds. The criteria used to determine the presence of a hydrogen bond was a distance less than 3 Å between donor and acceptor heavy atoms, and a donor-H-acceptor angle of at least 150 °.
Results and Discussion
Membrane binding and insertion of HAfp were captured in multiple independent simulations, permitting detailed characterization of the binding steps of the peptide for the first time. The peptide was modeled at neutral pH), with charged Glu-11 and Asp-19 side chains. This condition was chosen to preserve the helical structure of the peptide, which has been shown to be disrupted at low pH.17 The resulting membrane-bound form of the peptide also allowed for the characterization of important structural features of the peptide, such as depth of insertion and membrane orientation. Although the interaction of HAfp with a membrane has been studied previously with MD simulations using a wide variety of approaches,9,18–24,48–52 the process leading to peptide insertion has not been described at an atomic level. Previous MD studies have used a predefined placement of HAfp in the membrane, e.g., based on EPR measurements.14 To our knowledge, this is the first study reporting spontaneous insertion of HAfp to a membrane.
Spontaneous Membrane Binding and Insertion of HAfp
In order to study the binding of HAfp to the membrane, seven completely different initial configurations (i.e., positions and orientations of the peptide above the membrane) were tested (Fig. 1), with each configuration simulated in 3 independent 100–ns runs. In all cases the peptide was initially at least 7 Å away from the surface of the membrane, ensuring that the process of binding is not biased by initial placement of the peptide inside the membrane, which is a common practice when employing conventional atomistic membranes in simulation studies. Spontaneous binding of the HAfp to the membrane within 100 ns was observed in a total of 18 out of 21 systems (1–3 for each initial configuration). Membrane insertion was assumed when the COM of the hydrophobic side chains of the peptide was located at or below the PO4 level (Fig. 2). In the cases where membrane insertion was not observed in 100 ns, the peptide either interacted with the membrane through some of its side chains (Phe-9 and Trp-14) without completing insertion or it simply diffused into the aqueous phase without encountering the membrane (Fig. 2).
The first step of insertion occurs when bulky aromatic side chains (Phe-9, Trp-14, or Trp-21) encounter the membrane and start to insert (Fig. 2). After this initial encounter, the rest of the hydrophobic and aromatic side chains begin to penetrate the head group region of the membrane, while the polar residues remain exposed to the aqueous solution, achieving a partitioning at the lipid-water interface, also reported as the peptide configuration in micelles based on NMR measurements.15 After binding, insertion and stabilization of HAfp over the lipid layer is achieved during the remainder of the simulation, as characterized by the peptide insertion depth measured along the membrane normal (Fig. 2). The time required for the peptide to encounter the membrane varies greatly as it diffuses freely in water and does not depend on the initial configuration; for example, in the i1 and i2 cases binding occurs around 5 and 22 ns, respectively.
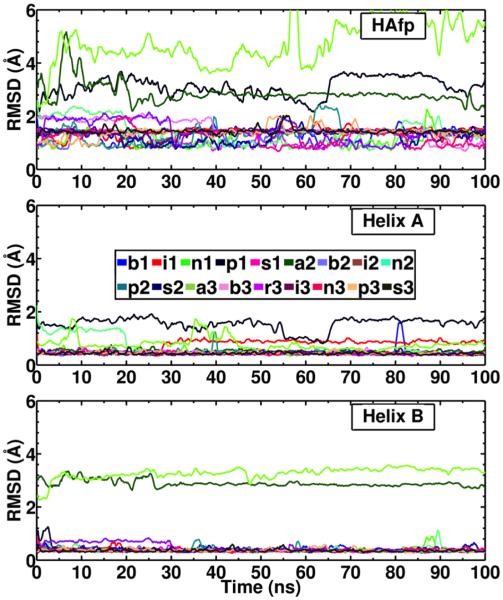
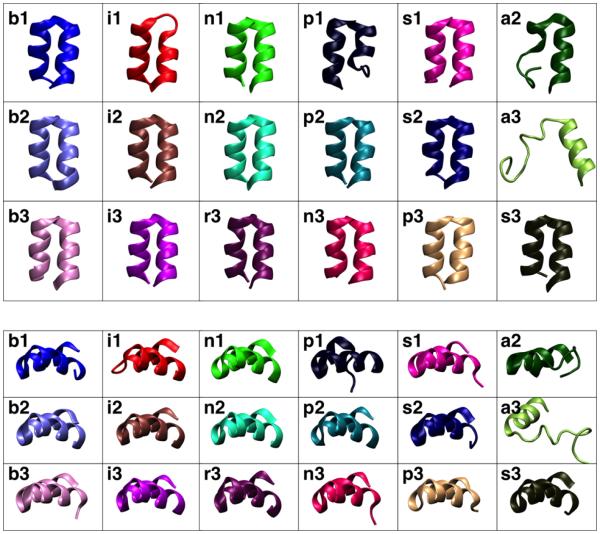
In general, the structure of the peptide is largely unaffected by the membrane after insertion, as measured by the backbone RMSD with respect to the NMR structure (Fig. 3). Stabilization of the overall structure of HAfp after membrane binding is expected since the NMR structure was obtained in DPC micelles.15 For the majority of the systems, the helical hairpin structure of the peptide is preserved upon membrane binding (backbone RMSD < 2 Å with respect to the NMR structure), as observed in the final structures of the peptide obtained from each simulation (Fig. 4). This is also supported by the low mutual RMSD (< 2 Å) obtained between all the membrane-binding simulations (Table 2). However, in some systems (p1, a2 and a3) the hairpin conformation is distorted. The RMSD of the individual helices reveals that the major disruption to the hairpin conformation in these systems occurs in Helix A (Fig. 4). The RMSD of the helices in each case decreases after membrane binding is completed, e.g., Helix A in system n2, indicating that the membrane stabilizes the structure of the peptide, an expected outcome which has also been observed in MD simulations of subtype H3 HAfp.20
Figure 3. Structural stability of HAfp during membrane-binding simulations.
Time evolution of the backbone RMSD of HAfp (Top), Helix A (Middle) and Helix B (Bottom) of HAfp.
Figure 4. Membrane-bound structures of HAfp.
Final structures of the membrane-bound HAfp obtained from each simulation. The top (Top) and side (Bottom) views of the peptide are shown for each system. The coloring of each system corresponds to that used in Fig. 3.
Table 2.
Pairwsie backbone RMSD between the final states of the 18 membrane–binding simulations (see Methods). The average RMSD is presented along with the standard deviation in parentheses.
| b1 | i1 | n1 | p1 | s1 | a2 | b2 | i2 | n2 | |
|---|---|---|---|---|---|---|---|---|---|
| b1 | 0.8 (0.1) | 0.8 (0.2) | 3.1 (0.2) | 1.3 (0.2) | 2.5 (0.2) | 0.6 (0.1) | 0.7 (0.1) | 0.7 (0.1) | |
| i1 | 0.8 (0.1) | 0.8 (0.1) | 3.1 (0.2) | 1.4 (0.2) | 2.5 (0.1) | 0.8 (0.1) | 0.8 (0.1) | 0.8 (0.1) | |
| n1 | 0.8 (0.2) | 0.8 (0.1) | 3.0 (0.2) | 1.4 (0.2) | 2.5 (0.1) | 0.7 (0.1) | 0.7 (0.1) | 0.7 (0.1) | |
| p1 | 3.1 (0.2) | 3.1 (0.2) | 3.0 (0.2) | 3.3 (0.2) | 3.5 (0.2) | 3.0 (0.2) | 3.0 (0.2) | 3.0 (0.2) | |
| s1 | 1.3 (0.2) | 1.4 (0.2) | 1.4 (0.2) | 3.3 (0.2) | 2.3 (0.2) | 1.3 (0.2) | 1.3 (0.2) | 1.3 (0.2) | |
| a2 | 2.5 (0.2) | 2.5 (0.1) | 2.5 (0.1) | 3.5 (0.2) | 2.3 (0.2) | 2.4 (0.1) | 2.5 (0.2) | 2.4 (0.2) | |
| b2 | 0.6 (0.1) | 0.8 (0.1) | 0.7 (0.1) | 3.0 (0.2) | 1.3 (0.2) | 2.4 (0.1) | 0.6 (0.1) | 0.6 (0.1) | |
| i2 | 0.7 (0.1) | 0.8 (0.1) | 0.7 (0.1) | 3.0 (0.2) | 1.3 (0.2) | 2.5 (0.2) | 0.6 (0.1) | 0.7 (0.1) | |
| n2 | 0.7 (0.1) | 0.8 (0.1) | 0.7 (0.1) | 3.0 (0.2) | 1.3 (0.2) | 2.4 (0.2) | 0.6 (0.1) | 0.7 (0.1) | |
| p2 | 0.6 (0.1) | 0.8 (0.1) | 0.7 (0.1) | 3.0 (0.2) | 1.3 (0.2) | 2.5 (0.2) | 0.6 (0.1) | 0.6 (0.1) | 0.6 (0.1) |
| s2 | 0.7 (0.2) | 0.9 (0.2) | 0.8 (0.2) | 3.1 (0.2) | 1.3 (0.2) | 2.5 (0.2) | 0.7 (0.2) | 0.7 (0.2) | 0.7 (0.2) |
| a3 | 5.9 (0.3) | 5.8 (0.3) | 5.7 (0.3) | 6.2 (0.3) | 5.6 (0.2) | 5.3 (0.2) | 5.8 (0.2) | 5.8 (0.3) | 5.8 (0.3) |
| b3 | 1.5 (0.2) | 1.6 (0.1) | 1.6 (0.1) | 3.4 (0.2) | 1.0 (0.3) | 2.3 (0.1) | 1.5 (0.2) | 1.5 (0.1) | 1.5 (0.2) |
| d3 | 0.7 (0.1) | 0.8 (0.1) | 0.7 (0.1) | 3.0 (0.2) | 1.3 (0.2) | 2.5 (0.1) | 0.6 (0.1) | 0.6 (0.1) | 0.7 (0.1) |
| i3 | 0.6 (0.1) | 0.8 (0.1) | 0.7 (0.1) | 3.0 (0.2) | 1.3 (0.2) | 2.4 (0.2) | 0.6 (0.1) | 0.6 (0.1) | 0.6 (0.1) |
| n3 | 1.5 (0.2) | 1.6 (0.1) | 1.5 (0.1) | 3.3 (0.2) | 1.0 (0.3) | 2.3 (0.1) | 1.5 (0.1) | 1.4 (0.1) | 1.4 (0.1) |
| p3 | 1.0 (0.2) | 1.2 (0.2) | 1.1 (0.2) | 3.2 (0.2) | 1.2 (0.3) | 2.5 (0.2) | 1.0 (0.2) | 1.0 (0.2) | 1.0 (0.2) |
| s3 | 1.0 (0.1) | 1.2 (0.1) | 1.1 (0.2) | 3.3 (0.2) | 1.1 (0.2) | 2.5 (0.2) | 1.0 (0.1) | 1.0 (0.1) | 1.1 (0.1) |
| p2 | s2 | a3 | b3 | r3 | i3 | n3 | p3 | s3 | |
|---|---|---|---|---|---|---|---|---|---|
| b1 | 0.6 (0.1) | 0.7 (0.2) | 5.9 (0.3) | 1.5 (0.2) | 0.7 (0.1) | 0.6 (0.1) | 1.5 (0.2) | 1.0 (0.2) | 1.0 (0.1) |
| i1 | 0.8 (0.1) | 0.9 (0.2) | 5.8 (0.3) | 1.6 (0.1) | 0.8 (0.1) | 0.8 (0.1) | 1.6 (0.1) | 1.2 (0.2) | 1.2 (0.1) |
| n1 | 0.7 (0.1) | 0.8 (0.2) | 5.7 (0.3) | 1.6 (0.1) | 0.7 (0.1) | 0.7 (0.1) | 1.5 (0.1) | 1.1 (0.2) | 1.1 (0.2) |
| p1 | 3.0 (0.2) | 3.1 (0.2) | 6.2 (0.3) | 3.4 (0.2) | 3.0 (0.2) | 3.0 (0.2) | 3.3 (0.2) | 3.2 (0.2) | 3.3 (0.2) |
| s1 | 1.3 (0.2) | 1.3 (0.2) | 5.6 (0.2) | 1.0 (0.3) | 1.3 (0.2) | 1.3 (0.2) | 1.0 (0.3) | 1.2 (0.3) | 1.1 (0.2) |
| a2 | 2.5 (0.2) | 2.5 (0.2) | 5.3 (0.2) | 2.3 (0.1) | 2.5 (0.1) | 2.4 (0.2) | 2.3 (0.1) | 2.5 (0.2) | 2.5 (0.2) |
| b2 | 0.6 (0.1) | 0.7 (0.2) | 5.8 (0.2) | 1.5 (0.2) | 0.6 (0.1) | 0.6 (0.1) | 1.5 (0.1) | 1.0 (0.2) | 1.0 (0.1) |
| i2 | 0.6 (0.1) | 0.7 (0.2) | 5.8 (0.3) | 1.5 (0.1) | 0.6 (0.1) | 0.6 (0.1) | 1.4 (0.1) | 1.0 (0.2) | 1.0 (0.1) |
| n2 | 0.6 (0.1) | 0.7 (0.2) | 5.8 (0.3) | 1.5 (0.2) | 0.7 (0.1) | 0.6 (0.1) | 1.4 (0.1) | 1.0 (0.2) | 1.1 (0.1) |
| p2 | 0.7 (0.2) | 5.8 (0.2) | 1.5 (0.1) | 0.7 (0.1) | 0.6 (0.1) | 1.5 (0.1) | 1.0 (0.2) | 1.1 (0.1) | |
| s2 | 0.7 (0.2) | 5.9 (0.3) | 1.5 (0.2) | 0.7 (0.2) | 0.7 (0.2) | 1.5 (0.2) | 1.0 (0.2) | 1.0 (0.1) | |
| a3 | 5.8 (0.2) | 5.9 (0.3) | 5.5 (0.2) | 5.8 (0.3) | 5.8 (0.2) | 5.4 (0.3) | 5.8 (0.3) | 5.8 (0.2) | |
| b3 | 1.5 (0.1) | 1.5 (0.2) | 5.5 (0.2) | 1.5 (0.1) | 1.5 (0.1) | 0.9 (0.2) | 1.4 (0.2) | 1.3 (0.2) | |
| d3 | 0.7 (0.1) | 0.7 (0.2) | 5.8 (0.3) | 1.5 (0.1) | 0.6 (0.1) | 1.5 (0.1) | 1.0 (0.2) | 1.1 (0.1) | |
| i3 | 0.6 (0.1) | 0.7 (0.2) | 5.8 (0.2) | 1.5 (0.1) | 0.6 (0.1) | 1.5 (0.1) | 1.0 (0.2) | 1.1 (0.1) | |
| n3 | 1.5 (0.1) | 1.5 (0.2) | 5.4 (0.3) | 0.9 (0.2) | 1.5 (0.1) | 1.5 (0.1) | 1.3 (0.2) | 1.2 (0.2) | |
| p3 | 1.0 (0.2) | 1.0 (0.2) | 5.8 (0.3) | 1.4 (0.2) | 1.0 (0.2) | 1.0 (0.2) | 1.3 (0.2) | 0.8 (0.2) | |
| s3 | 1.1 (0.1) | 1.0 (0.1) | 5.8 (0.2) | 1.3 (0.2) | 1.1 (0.1) | 1.1 (0.1) | 1.2 (0.2) | 0.8 (0.2) |
Membrane-Bound Configuration of HAfp
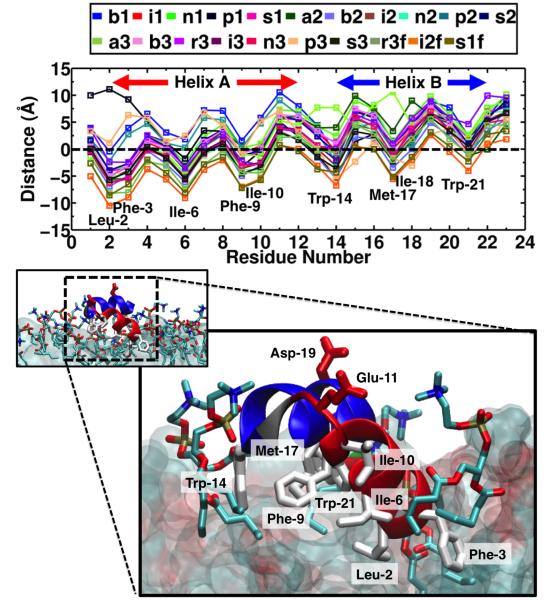
To further characterize the membrane-bound form of HAfp, the average insertion depth of each side chain was calculated (see Methods). For all membrane-bound systems, the peptide adopts a profile in which Helix A is more inserted than Helix B (Fig. 5). After insertion, hydrophobic residues Leu-2, Phe-3, Ile-6, Phe-9 and Ile-10 in Helix A are buried (i.e., below the PO4 groups, with a COM side chain z-position < 0 in Fig. 5) for most of the simulations. NMR measurements of the interactions of HAfp with a DPC micelle15 have suggested that Ile-6, Phe-9 and Ile-10 are buried in the hydrophobic region of the micelle, consistent with the observed insertion of these side chains in our membrane-binding simulations. In Helix B, amphipathic side chain Trp-14 and Met-17 are buried for most simulations (relative z-position < 0 in Fig. 5), whereas Trp-21 is mostly located at the head group region (relative z-position > 0 in Fig. 5). The insertion profile reveals that the insertion of Helix A is deeper than Helix B, indicating that Helix A establishes core hydrophobic interactions, through residues Leu-2, Phe-3, Ile-6, Phe-9 and Ile-10 with the membrane. The insertion depth of Helix A is in close agreement with the EPR data of membrane insertion at pH 7.4 reported for the first 11 residues of HAfp subtype H3 (i.e., GLFGAIAGFIE),14 which are shared with the peptide studied here (HAfp subtype 1)".
Figure 5. Membrane insertion profile of HAfp.
(Top) Average distance along the z-axis of the side chains of the inserted HAfp to the phosphorous (PO4) level of the lipids. The average was taken over the last 10 ns of each simulation. (Bottom) Representative membrane-bound structure of HAfp, taken from the final snapshot of System i1. Helices A and B are shown in red and blue, respectively, and Gly-13 (linker) is shown in gray. Side chains of hydrophobic residues Leu-2, Phe-3, Ile-6, Phe-9, Ile-10, Trp-14, Met-17 and Trp-21 (white), acidic residues Glu-11 and Asp-19 (red), and the lipids interacting with the peptide (heavy atom distance < 2.5 Å) are shown in stick representation. The rest of the membrane is shown in surface representation.
We observed that by increasing the realism of the membrane (i.e., extending the short-tailed lipids to POPC lipids, see Methods) the insertion profile is preserved, as shown in Systems r3f, i2f and s1f (Fig. 5). This indicates that the HMMM simulations have been able to achieve a stable configuration (in terms of insertion depth and orientation) of the membrane-bound form of the peptide, while capturing the process of spontaneous binding without any initial bias, such as the placement of the peptide in the membrane. The resulting membrane–bound HAfp indicates that the peptide interaction with the cis leaflet (i.e., head groups and first few carbons of tails) has been captured by the HMMM model.
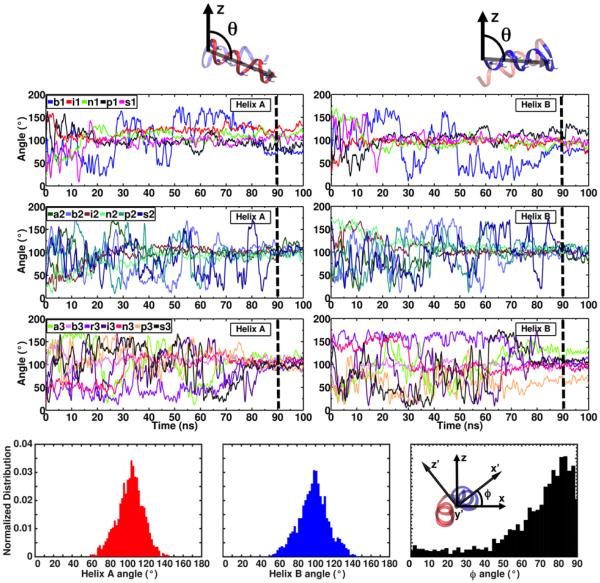
To characterize the orientation of the peptide after insertion, the tilt angle between individual helices of the fusion peptide and the membrane normal was calculated (see Methods). Despite the wildly different initial orientations of the peptide (Table 3), a convergent orientation is achieved during the simulations for all the membrane-bound systems (Fig. 6). The tilt angle calculated for Helix A for membrane-bound HAfp ranges from 76.0 ° to 126.0 °, with an average of 102.8 ° over the 18 simulations. The tilt angle for helix B ranges from 66.0 ° to 126.4 °, with an average of 97.9 °. The range of tilt angles that are sampled in these simulations are in close agreement with the recent NMR data showing that the HAfp is able to undergo rocking motions when bound to a bicelle, with an amplitude of about 20 ° around the helix axes in the range of tens of nanoseconds.34 Moreover, these tilt angles indicate that the peptide adopts an almost parallel orientation with respect to the membrane plane after insertion, which is in agreement with what has been suggested in NMR studies.15,34 The orientation of the peptide was further characterized by the rotation around the helical hairpin axis (Fig. 6), following the scheme proposed by Chen et al. to characterize protein orientations.47 This rotation ranges between 50° to 80°, resulting from a deeper insertion of Helix A compared to Helix B, consistent with the observed insertion profile of the peptide (Fig. 5). The orientation of HAfp after insertion is preserved when the short-tailed lipids are extended to full POPC lipids in Systems r3f, i2f and s1f (Table 4), with average angles of 105.1 ° and 97.9 ° for Helices A and B, respectively. This suggests that the peptide–lipid interactions that determine the insertion depth and orientation of the peptide upon membrane binding are successfully sampled with the HMMM model. It is important to note however that the membrane curvature and lipid protrusion-based mechanisms in which HAfp binding has been suggested to directly contribute to the process of membrane fusion9,24 can not be captured with the HMMM model membrane, due to the short-tailed lipid representation, and decoupling of the two lipid leaflets. The full POPC membrane simulations performed here are likely too short to capture any associated membrane curvature.
Table 3.
Initial (solution) and membrane-bound (averaged over the last 10ns) tilt angles of Helices A and B of HAfp in membrane-binding simulations. The average over all the membrane-bound systems is also presented. The standard deviations are in parentheses.
| System | Helix A (°) | Helix B (°) | ||
|---|---|---|---|---|
| Initial tilt angle (solution) |
Membrane-bound tilt angle |
Initial tilt angle (solution) |
Membrane-bound tilt angle |
|
| b1 | 78.3 | 76.0 (4.4) | 100.5 | 75.7 (3.7) |
| i1 | 143.9 | 126.0 (6.5) | 38.1 | 90.4 (8.1) |
| n1 | 20.7 | 112.8 (4.7) | 171.5 | 91.8 (4.6) |
| p1 | 102.8 | 83.4 (6.5) | 86.3 | 122.7 (6.7) |
| s1 | 51.1 | 106.0 (6.0) | 140.1 | 93.9 (7.6) |
| a2 | 99.1 | 116.3 (5.3) | 63.7 | 86.7 (5.8) |
| b2 | 78.7 | 101.5 (6.1) | 110.0 | 105.0 (5.9) |
| i2 | 30.5 | 102.8 (3.9) | 129.5 | 101.7 (4.9) |
| n2 | 22.9 | 96.7 (7.1) | 168.0 | 104.4 (7.1) |
| p2 | 88.0 | 106.7 (9.9) | 72.1 | 102.2 (9.7) |
| s2 | 27.1 | 105.4 (5.9) | 162.9 | 97.6 (7.3) |
| a3 | 98.3 | 113.1 (7.7) | 64.2 | 126.4 (6.2) |
| b3 | 89.3 | 112.3 (5.7) | 87.2 | 88.6 (7.4) |
| r3 | 65.0 | 89.6 (5.1) | 139.7 | 109.6 (5.1) |
| i3 | 161.2 | 106.5 (4.8) | 56.0 | 97.0 (4.9) |
| n3 | 13.8 | 105.6 (5.9) | 170.5 | 94.0 (5.6) |
| p3 | 82.5 | 92.2 (5.5) | 87.8 | 66.0 (5.6) |
| s3 | 14.9 | 97.6 (4.4) | 170.7 | 107.7 (3.1) |
| Average | 102.8 (12.2) | 97.9 (14.6) | ||
Figure 6. Orientation of HAfp during membrane binding.
(Top) Time series of the tilt angle of individual helices of HAfp with respect to the membrane normal (z-axis). Each tilt angle is defined as the angle between the average N-H bond vectors (shown in stick representation) of Helix A (residues 3-12) or Helix B (residues 14-22) and the z-axis, respectively (see Methods). Dashed lines show the section of the simulations (last 10 ns) that was used to calculated the average tilt angles, reported in Table 2. (Bottom) Distributions of Helix A tilt angle (left), Helix B tilt angle (middle), and the rotation around the helical hairpin axis (right), obtained for the last 10 ns of the simulation for all the systems. The helical hairpin axis (y′-axis) is perpendicular to the other principal axes of the peptide (x′-axis and z′-axis), going into the page.
Table 4.
Tilt angles of Helices A and B of membrane-bound HAfp in a full POPC membrane. The angles are averaged for each system over the last 10 ns of simulation. The average over all the systems is also presented. The standard deviations are in parentheses.
| System | Helix A (°) | Helix B (°) |
|---|---|---|
| s1f | 98.2 (5.2) | 106.8 (5.3) |
| i2f | 108.3 (5.7) | 96.6 (6.5) |
| r3f | 108.9 (5.5) | 90.9 (5.6) |
| Average | 105.1 (6.0) | 97.9 (8.1) |
Interaction of HAfp with the Membrane
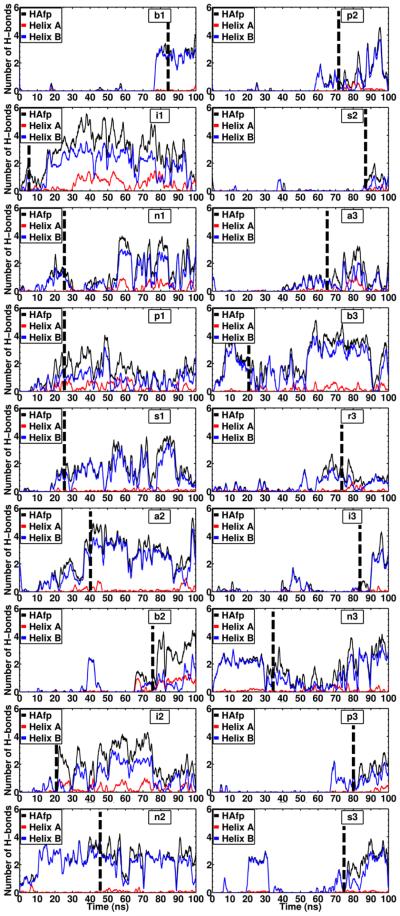
In order to characterize the interaction of the peptide and the membrane, the pattern of H–bonds between the two was calculated for all membrane-bound systems (Fig. 7). These calculations were done by taking into account the H-bond contribution of both the backbone and the side chain of each residue of HAfp. Overall, Helix B contributes to the majority of the H-bonds formed with the phospholipid head groups. Because Helix B contains five H–bond forming side chains, namely Trp-14, Thr-15, Asp-19, Trp-21, and Tyr-22, a larger contribution to the H–bond formation from this helix is expected. In contrast, Helix A only contains one H–bond forming side-chain (Glu-11), therefore its overall contribution is significantly smaller than Helix B. In the membrane–bound form of HAfp at pH 7.4 modeled in this work, charged residues Glu-11 and Asp-19 do not contribute to any charge interactions between the peptide and the membrane, and due to the helicity of the peptide the side chains point away from the membrane (Fig. 5).
Figure 7. Hydrogen bonds between HAfp helices and the membrane.
Time series of the number of hydrogen bonds formed between HAfp and the membrane in the systems where membrane insertion was observed (18 out of 21 simulations). In each plot the dashed line indicates the time at which peptide insertion occured.
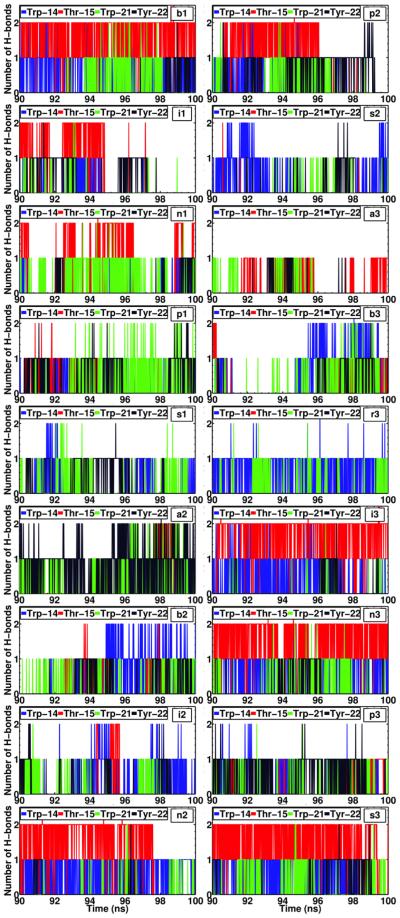
Since the helical structure of the peptide remains largely unaltered during the course of membrane-binding, it is expected that most of the H–bonds between the peptide and the membrane involve the side chains. This suggests that H-bond donor side-chains stabilize the peptide after binding with the membrane (Fig. 8). Trp-14, Thr-15, Trp-21 and Tyr-22 account for approximately 99% of all the H–bonds formed between Helix B and the lipid head groups, while the side chains of Thr-15 and Tyr-22 account for the majority of the total contribution (Fig. 8). The observation that tryptophan and tyrosine residues contribute to the H–bonding of the peptide within the lipid head groups is in line with the well-known role of these residues in anchoring membrane proteins to the interfacial region of the membrane.53,54 Moreover, this observation suggests the importance of residues Trp-21 and Tyr-22 in the stabilization of the peptide at the membrane–water interface after spontaneous binding occurs, something that, to our best knowledge, has not been reported before, since the previously available structures of HAfp did not include these highly conserved residues.10,14,55 Recent NMR experiments have shown that these residues are important to stabilize the helical-hairpin structure of the peptide.16 Moreover, the larger contribution of Helix B to membrane–biding observed in out simulations is supported by experimental evidence showing that the truncated sequence of HAfp, containing only the first 14 residues, is void of fusogenic activity.16 This result, taken together with the observed deeper insertion of Helix A in our simulations, suggests that Helix A contributes to peptide insertion by establishing the majority of hydrophobic interactions after binding to the membrane, while Helix B increases the affinity by providing amphipathic interactions at the membrane interface.
Figure 8. HAfp residues contributing to hydrogen bonding with the membrane.
Time series for the last 10 ns (90–100 ns) of simulation (i.e., after the peptide is inserted) of the hydrogen bonds formed by residues Trp-14, Thr-15, Trp-21, and Tyr-22 with the membrane in the systems where membrane insertion was observed. Both contributions from the backbone and side chains to H–bonds were accounted for in the calculation.
Conclusions
In this study, we have employed the HMMM membrane model to investigate the binding and insertion of the hemagglutinin fusion peptide of the influenza virus. The HMMM membrane model provides an enhanced lipid mobility, allowing to capture the spontaneous insertion of the peptide in atomistic simulations. The HMMM membrane model, together with the small size of the system, allowed a greater sampling of the peptide-membrane systems. The spontaneous binding and insertion of the peptide into the membrane were captured consistently in 18 out of the 21 independent simulations performed. Despite employing broadly different initial configurations for the simulations, a reproducible membrane-bound state of the peptide was observed in the simulations. The insertion profile and the orientation of the membrane-bound HAfp are preserved after converting the HMMM model to a full lipid bilayer. To our knowledge, this is the first study reporting spontaneous insertion of HAfp to a membrane.
It was observed that the helical hairpin structure of the peptide is preserved upon membrane binding. In the membrane–bound form, Helix A is inserted in the membrane deeper than Helix B. The observed deeper insertion of Helix A in our simulations, suggests that Helix A contributes to peptide insertion by establishing the majority of hydrophobic interactions after binding to the membrane, while Helix B increases the affinity by providing amphipathic interactions at the interfacial region. The membrane-bound model of HAfp, as well as the details of membrane-binding captured in the simulations could be expanded in a subsequent work to study the mechanism of pore formation induced by HAfp. It has been suggested that multiple copies of the peptide are necessary for pore formation,56,57 while the peptide undergoes a pH-induced conformational change.17 This mechanism could be further explored by employing our membrane-bound model as well as the enhanced lipid dynamics of the HMMM membrane to study the peptide–peptide interactions that preclude pore formation.
Acknowledgement
This work was supported in part by the National Institutes of Health (grants R01-GM101048, R01-GM086749, U54-GM087519, and P41-GM104601). All simulations have been performed using XSEDE resources (grant No. MCA06N060).
References
- (1).Luo M. Influenza Virus Entry. Adv. Exp. Med. Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wiley DC, Skehel JJ. The Structure and Function of the Haemagglutinin Membrane Glycoprotein of Influenza Virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- (3).Durrer P, Galli C, Hoenke S, Corti C, Glück R, Vorherr T, Brunner J. H+–induced Membrane Insertion of Influenza Virus Hemagglutinin Involves the HA2 Amino-terminal Fusion Peptide but not the Coiled Coil Region. J. Biol. Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- (4).Wilson IA, Skehel JJ, Wiley DC. Structure of the Haemagglutinin Membrane Glyco-protein of Influenza Virus at 3 Å Resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- (5).Stegmann T, Delfinogll JM, Richards FM, Helenius A. The HA2 Subunit of Influenza Hemagglutinin Inserts into the Target Membrane Prior to Fusion. J. Biol. Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- (6).Tsurudome M, Glück R, Graf R, Falchetto R, Schaller U, Brunne J. Lipid Interactions of the Hemagglutinin HA2 NH2–Terminal Segment During Influenza Virus-Induced Membrane Fusion. J. Biol. Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- (7).Gruenke JA, Armstrong RT, Newcomb WW, Brown JC, White JM. New Insights into the Spring-Loaded Conformational Change of Influenza Virus Hemagglutinin. J. Virol. 2002;76:4456–4466. doi: 10.1128/JVI.76.9.4456-4466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tamm LK. Hypothesis: Spring-Loaded Boomerang Mechanism of Influenza Hemagglutinin-Mediated Membrane Fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- (9).Lagüe P, Roux B, Pastor RW. Molecular Dynamics Simulations of the Influenza Hemagglutinin Fusion Peptide in Micelles and Bilayers: Conformational Analysis of Peptide and Lipids. J. Mol. Biol. 2005;354:1129–1141. doi: 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
- (10).Lai AL, Park H, White JM, Tamm LK. Fusion Peptide of Influenza Hemagglutinin Requires a Fixed Angle Boomerang Structure for Activity. J. Biol. Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- (11).Kasson PM, Lindahl E, Pande VS. Atomic-Resolution Simulations Predict a Transition State for Vesicle Fusion Defined by Contact of a Few Lipid Tails. PLoS Comput. Biol. 2010;6:e1000829. doi: 10.1371/journal.pcbi.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Han X, Tamm LK. A Host-Guest System to Study Structure-Function Relationships of Membrane Fusion Peptides. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Han X, Tamm LK. pH-Dependent Self-Association of Influenza Hemagglutinin Fusion Peptides in Lipid Bilayers. J. Mol. Biol. 2000;304:953–965. doi: 10.1006/jmbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- (14).Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane Structure and Fusion-Triggering Conformational Change of the Fusion Domain from Influenza Hemagglutinin. Nat. Struct. Mol. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- (15).Lorieau JL, Louis JM, Bax A. The Complete Influenza Hemagglutinin Fusion Peptide Adopts a Tight Helical Hairpin Arrangement at the Lipid:Water Interface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lorieau JL, Louis JM, Bax A. The Impact of Influenza Hemagglutinin Fusion Peptide Length and Viral Subtype in its Structure and Dynamics. J. Am. Chem. Soc. 2012;99:189–195. doi: 10.1002/bip.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lorieau JL, Louis JM, Schwieters CD, Bax A. pH-Triggered, Activated-State Conformations of the Influenza Hemagglutinin Fusion Peptide Revealed by NMR. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19994–19999. doi: 10.1073/pnas.1213801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Huang Q, Chen C-L, Herrmann A. Bilayer Conformation of Fusion Peptide of Influenza Virus Hemagglutin: A Molecular Dynamics Simulation Study. Biophys. J. 2004;87:14–22. doi: 10.1529/biophysj.103.024562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Vaccaro L, Cross KJ, Kleinjung J, Straus SK, Thomas DJ, Wharton SA, Skehel JJ, Franternali F. Plasticity of Influenza Haemagglutinin Fusion Peptides and Their Interaction with Lipid Bilayers. Biophys. J. 2005;88:25–36. doi: 10.1529/biophysj.104.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Volynsky PE, Polyansky AA, Simakov NA, Arseniev AS, Efremov RG. Effect of Lipid Composition on the "Membrane Response" Induced by a Fusion Peptide. Biochemistry. 2005;44:14626–14637. doi: 10.1021/bi0514562. [DOI] [PubMed] [Google Scholar]
- (21).Li J, Das P, Zhou R. Single Mutation Effects on Conformational Change and Membrane Deformation of Influenza Hemagglutinin Fusion Peptides. J. Phys. Chem. B. 2010;114:8799–8806. doi: 10.1021/jp1029163. [DOI] [PubMed] [Google Scholar]
- (22).Legare S, Lagüe P. The Influenza Fusion Peptide Adopts a Flexible Flat V Conformation in Membranes. Biophys. J. 2012;102:2270–2278. doi: 10.1016/j.bpj.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fuhrmans M, Marrink SJ. Molecular View of the Role of Fusion Peptides in Promoting Positive Membrane Curvature. J. Am. Chem. Soc. 2012;134:1543–1552. doi: 10.1021/ja207290b. [DOI] [PubMed] [Google Scholar]
- (24).Larsson P, Kasson PM. Lipid Tail Protrusion in Simulations Predicts Fusogenic Activity of Influenza Fusion Peptide Mutants and Conformational Models. PLoS Comput. Biol. 2013;9:e1002950. doi: 10.1371/journal.pcbi.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Brice AR, Lazaridis T. Structure and Dynamics of a Fusion Peptide Helical Hairpin on the Membrane Surface: Comparison of Molecular Simulations and NMR. J. Phys. Chem. B. 2014;118:4461–4470. doi: 10.1021/jp409412g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ohkubo YZ, Pogorelov TV, Arcario MJ, Christensen GA, Tajkhorshid E. Accelerating Membrane Insertion of Peripheral Proteins with a Novel Membrane Mimetic Model. Biophys. J. 2012;102:2130–2139. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Arcario MJ, Ohkubo YZ, Tajkhorshid E. Capturing Spontaneous Partitioning of Peripheral Proteins using a Biphasic Membrane-Mimetic Model. J. Phys. Chem. B. 2011;115:7029–7037. doi: 10.1021/jp109631y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. Characterizing the Membrane-Bound State of Cytochrome P450 3A4: Structure, Depth of Insertion, and Orientation. J. Am. Chem. Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Arcario MJ, Tajkhorshid E. Membrane-Induced Structural Rearrangement and Identification of a Novel Membrane Anchor in Talin F2F3. Biophys. J. 2014;107:2059–2069. doi: 10.1016/j.bpj.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Vermaas JV, Tajkhorshid E. Conformational Heterogeneity of α-Synuclein in Membrane. Biochim. Biophys. Acta Biomembr. 2014;1838:3107–3117. doi: 10.1016/j.bbamem.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wu Z, Schulten K. Synaptotagmin’s Role in Neurotransmitter Release Likely Involves Ca2+-induced Conformational Transition. Biophys. J. 2014;107:1156–1166. doi: 10.1016/j.bpj.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Blanchard AE, Arcario MJ, Schulten K, Tajkhorshid E. A Highly Tilted Membrane Configuration for the Pre-Fusion State of Synaptobrevin. Biophys. J. 2014;107:2112–2121. doi: 10.1016/j.bpj.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pogorelov TV, Vermaas JV, Arcario MJ, Tajkhorshid E. Partitioning of Amino Acids into a Model Membrane: Capturing the Interface. J. Phys. Chem. B. 2014;118:1481–1492. doi: 10.1021/jp4089113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lorieau JL, Louis JM, Bax A. Whole-Body Rocking Motion of a Fusion Peptide in Lipid Bilayers from Size-Dispersed 15N NMR Relaxation. J. Am. Chem. Soc. 2011;133:14184–14187. doi: 10.1021/ja2045309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Martínez L, Andrade R, Birgin EG, Martínez JM. PACKMOL: A Package for Building Intial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009;30:2157–2164. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- (36).Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (37).Armen RS, Uitto OD, Feller SE. Phospholipid Component Volumes: Determination and Application to Bilayer Structure Calculations. Biophys. J. 1998;75:734–744. doi: 10.1016/S0006-3495(98)77563-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kučerka N, Tristram-Nagle S, Nagle JF. Structure of Fully Hydrated Fluid Phase Lipid Bilayers with Monounsaturated Chains. J. Membr. Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- (39).The RCSB Protein Data Bank. http://www.rcsb.org/pdb, (accessed May 15, 2015)
- (40).Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).MacKerell AD, Jr., Feig M, Brooks CL., III Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comp. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- (42).Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD., Jr. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comp. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- (44).Feller SE, Zhang Y, Pastor RW. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- (45).Martyna GJ, Tobias DJ, Klein ML. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- (46).Darden T, York D, Pedersen LG. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- (47).Chen C-H, Málková Šárka, Pingali SV, Long F, Garde S, Cho W, Schlossman ML. Configuration of PKCα-C2 Domain Bound to Mixed SOPC/SOPS Lipid Monolayers. Biophys. J. 2009;97:2794–2802. doi: 10.1016/j.bpj.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bechor D, Ben-Tal N. Implicit Solvent Model Studies of the Interactions of the Influenza Hemagglutinin Fusion Peptide with Lipid Bilayers. Biophys. J. 2001;80:643–655. doi: 10.1016/S0006-3495(01)76045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Spassov VZ, Yan L, Szalma S. Introducing an Implicit Membrane in Generalized Born/Solvent Accessibility Continuum Solvent Models. J. Phys. Chem. B. 2002;106:8726–8738. [Google Scholar]
- (50).Sammalkorpi M, Lizaridis T. Configuration of Influenza Hemagglutinin Fusion Peptide Monomers and Oligomers in Membranes. Biochim. Biophys. Acta. 2007:30–38. doi: 10.1016/j.bbamem.2006.08.008. [DOI] [PubMed] [Google Scholar]
- (51).Sammalkorpi M, Lazaridis T. Modeling a Spin-Labeled Fusion Peptide in a Membrane: Implications for the Interpretation of EPR Experiments. Biophys. J. 2007;92:10–22. doi: 10.1529/biophysj.106.092809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Panahi A, Feig M. Conformational Sampling of Influenza Fusion Peptide in Membrane Bilayers as a Function of Termini and Protonation States. J. Phys. Chem. B. 2010;114:1407–1416. doi: 10.1021/jp907366g. [DOI] [PubMed] [Google Scholar]
- (53).Stopar D, Spruijt RB, Hemminga MA. Anchoring Mechanisms of Membrane-Associated M13 Major Coat Protein. Chem. Phys. Lipids. 2006;141:83–93. doi: 10.1016/j.chemphyslip.2006.02.023. [DOI] [PubMed] [Google Scholar]
- (54).Schiffer M, Chang CH, Stevens FJ. The Functions of Tryptophan Residues in Membrane Proteins. Protein Eng., Des. Sel. 1992;5:213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- (55).Lai AL, Tamm LK. Locking the Kink in the Influenza Hemagglutinin Fusion Domain Structure. J. Biol. Chem. 2007;282:23946–23956. doi: 10.1074/jbc.M704008200. [DOI] [PubMed] [Google Scholar]
- (56).Danieli T, Pelletier SL, Henis YI, White JM. Membrane Fusion Mediated by the Influenza Virus Hemagglutinin Requires the Concerted Action of at Least Three Hemagglutinin Trimers. J. Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Blumenthal R, Sarkar DP, Durell S, Howard DE, Morris SJ. Dilation of the Influenza Hemagglutinin Fusion Pore Revealed by the Kinetics of Individual Cell-Cell Fusion Events. J. Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]