Graphical abstract
Keywords: Single domain antibodies, Rhizavidin, Quantum dots, Surface plasmon resonance
Highlights
-
•
Single domain antibody—rhizavidin fusion bioconjugated biotin coated quantum dots.
-
•
Provided facile and effective method to orient antibodies on QD surface.
-
•
Accomplished improved production of His-tagged rhizavidin (RZh) in E. coli.
-
•
Demonstrated utility of RZh as a replacement for tetrameric biotin binders.
Abstract
Straightforward and effective methods are required for the bioconjugation of proteins to surfaces and particles. Previously we demonstrated that the fusion of a single domain antibody with the biotin binding molecule rhizavidin provided a facile method to coat biotin-modified surfaces with a highly active and oriented antibody. Here, we constructed similar single domain antibody—rhizavidin fusions as well as unfused rhizavidin with a His-tag. The unfused rhizavidin produced efficiently and its utility for assay development was demonstrated in surface plasmon resonance experiments. The single domain antibody-rhizavidin fusions were utilized to coat quantum dots that had been prepared with surface biotins. Preparation of antibody coated quantum dots by this means was found to be both easy and effective. The prepared single domain antibody-quantum dot reagent was characterized by surface plasmon resonance and applied to toxin detection in a fluoroimmunoassay sensing format.
1. Introduction
Single domain antibodies (sdAbs), derived from heavy chain only antibodies found in camelids (i.e. camels, llamas, and alpacas), often possess high affinity and the ability to refold and bind antigen after heat denaturation [1], [2], [3], [4]. Because these binding elements are produced recombinantly, they can be tailored through protein engineering for specific applications. SdAbs, in general, produce very well in Escherichia coli; sdAb fusion constructs are also produced more readily than traditional antibody binding fragments that include both heavy and light chains. For example, expression of sdAbs as genetic fusions with peptides, or high-melting temperature proteins is a proven route towards generating highly stable molecules with enhanced utility [5], [6], [7], [8], [9], [10]. Fusions have been used for a variety of purposes including: providing improved cytoplasmic production of stable sdAbs, increasing solubility of sdAbs, as well as providing oriented capture reagents [5], [7], [11], [12].
We recently described the production and characterization of a genetic fusion between a sdAb and rhizavidin (RZ) [10], a biotin binding protein (derived from the symbiotic nitrogen-fixing bacterium Rhizobium etli CFN42) [13]. Unlike streptavidin, RZ putatively forms a homodimer instead of a tetramer [13], [14]. RZ has a melting temperature of over 100 °C in the presence of biotin, and ∼75 °C in its absence. The sdAb-RZ fusion provided the same desirable characteristics, such as providing oriented capture, as fusions with the streptavidin core protein [11]. Importantly, protein production was improved by approximately 20-fold compared to the sdAb-streptavidin core fusion [10].
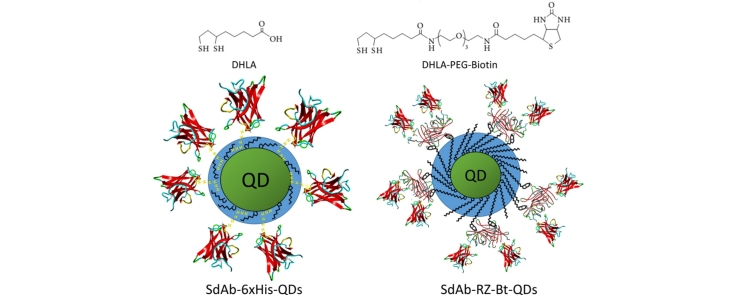
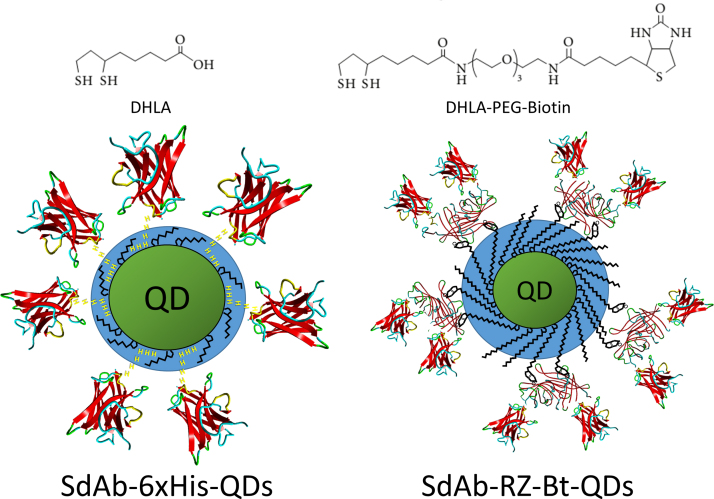
Luminescent quantum dots (QDs) provide robust fluorophores that have been incorporated for applications including biosensing and imaging [15], [16]. Conjugates of sdAbs with QDs, which couple stable recognition elements with robust fluorophores, have been described for detection, imaging and diagnostic applications [17], [18], [19], [20], [21], [22], [23]. Several methods for bioconjugation of proteins to QDs have been described; for instance, we have previously utilized directional conjugation of sdAbs to QDs through an extended poly histidine tail [17], [20], [24]. One of the previous generation of sdAb-QD reagents we tested was based on QDs made water compatible through capping with dihydrolipoic acid (DHLA). QDs functionalized with DHLA-PEG based-ligands are not as amenable to conjugation through an extended histidine tail, however they offer functionality and stability over a wider pH range [17], [25]. An advantage of sdAbs is their ability to function over a wide range of conditions [26], [27] including intracellular [28]. Therefore it is desirable to have a facile system for the directional conjugation of sdAbs to QDs functionalized with DHLA-PEG ligands that provide an increased biocompatibility. The recent development of DHLA-PEG capped QDs with a portion of the cap functionalized with biotin [29], [30], in conjunction with fusions of sdAbs with RZ provides an alternate route for directional conjugates of sdAbs on QDs. A schematic illustrating both a sdAb-QD conjugate formed using DHLA-capped QDs with attachment of the sdAb through an extended histidine tail and a sdAb-QD conjugate utilizing the DHLA-PEG biotinylated QDs and a sdAb-RZ genetic fusion is shown in Fig. 1. Having a wide variety of methods to form effective sdAb-QD conjugates is advantageous as it provides researchers the ability to choose the conjugation method most appropriate for their assay or imaging conditions.
Fig. 1.
Schematic of sdAb-QDs prepared previously, through an extended histidine tail on the sdAb and through the current method utilizing biotinylated QDs and sdAb-RZ. The left side shows a DHLA-capped QD onto which sdAb have been conjugated through an extended histidine tail. The right side shows a QD capped with 80% DHLA-PEG550-OMe and 20% DHLA-PEG400-biotin onto which sdAb-RZ are conjugated through the RZ-biotin interaction. The structure of the sdAb is from PDB:4W70 [40] and the RZ structure from PBD:3EW2 [14]. The components are not drawn to scale.
This current work focuses on ricin detection. Ricin is a 60–65 kDa highly potent toxin which consists of an A and B subunit. The A subunit is the enzymatic portion responsible for ribosome inactivation, while the B subunit binds the cell to facilitate entry of the toxin [31]. To detect ricin the sdAb, D12f, which has both high affinity and good thermal stability (Tm = 78 °C) [32], was produced as a fusion with RZ. D12f better complements the high stability of RZ than the original C8 anti-ricin sdAb used as a fusion partner with RZ, which binds the same epitope and has a high affinity for ricin, but melts ∼60 °C. In addition, because we had observed sporadic degradation of constructs that utilized the llama heavy chain antibody’s upper hinge as a linker, we switched to a generic10-amino acid Gly-Ser linker to join D12f to RZ. We also prepared the unfused RZ with a C-terminal hexa histidine tag (RZh), evaluated its biophysical characteristics and demonstrated its utility for use as a regenerable ligand via surface plasmon resonance (SPR) using HTE (6x-His binding) sensor chips. Nevertheless, the main objective was demonstrating the utility of the sdAb-RZ fusion by formation of a bioconjugate between the D12f-RZ and QDs that have biotins incorporated on a portion of their capping ligands. The oriented immobilization provided by the RZ on the QDs yielded a highly active sdAb that binds target effectively.
2. Materials and methods
2.1. Construction SdAb-RZ fusions with Gly-Ser linker
The D12f-L10-RZ was constructed by first inserting the RZ into the XhoI site of a pET22b expression vector in which the D12f sdAb sequence had been cloned into NcoI-NotI sites (D12f-pET22b); this vector includes a C-terminal 6xHis tag [32]. The RZ fragments flanked with a XhoI site at both ends were amplified from the original vector using PCR and inserted to the XhoI site within D12f-pET22b. D12f-RZ [33] then served as a template to insert a 10 amino acid Gly-Ser linker (L10, GGGGSGGGGS) using the Quikchange II mutagenesis kit and minor modifications to the manufacturer’s protocol (Agilent Technologies; Santa Clara, CA). Mutagenesis was achieved using the forward primer, 5′-GCGGCCGCACTCGAGGGCGGTGGCGGTAGCGGCGGTGGCGGTTCTTTTGATGCGTCCAATTTTAAA-3′, and its reverse complement sequence as the reverse primer. In brief, low annealing temperature (47 °C) and 18 amplification cycles were employed and the rest of procedures were unchanged from the manufacturer’s protocol, to obtain the mutated plasmid. All the clones were confirmed by sequencing (Euofins Genomics; Huntsville, AL). The mutant protein is termed D12f-L10-RZ. All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs, Inc. (Ipswich, MA).
2.2. Construction of unfused RZ with a C-terminal hexa histidine tag (RZh) in pET22b(+)
The rhizavidin fragment was excised from D12f-RZ using the restriction enzyme XhoI. Gel purification of the excised fragment was performed with QIAquick Gel Extraction Kit (QIAGEN). Using the rhizavidin fragment and the pET22b (+) Vector, a 3:1 ligation at 16 °C was performed overnight. Correct sequences were identified by DNA sequencing (Euofins Genomics).
2.3. Expression, production, and purification of SdAb-RZ and RZh proteins
Protein expression and purification were performed similar to methods described previously [7]. Briefly, a single colony of Rosetta (DE3) cells harboring the expression plasmid was used to inoculate a 50 mL overnight culture grown at 30 °C in Luria broth (LB) in the presence of 100 μg/mL ampicillin and 35 μg/mL chloramphenicol. The following day, a 500 mL culture in LB was inoculated using the overnight culture and grown at 30 °C for 3 h. The temperature was then reduced to 25 °C and protein expression was induced with the addition of 0.5 mM IPTG (isopropyl β-d-1-thiogalactopyranoside). After 3 h, protein was harvested by osmotic shock. Briefly, first cell pellets from 500 mL of culture were homogenized in 14 mL cold sucrose-tris (750 mM sucrose, 100 mM Tris pH 7.5). Next 28 mL of 1 mM ethylenediaminetetraaceticacid (EDTA; pH 8) was added drop-by-drop to each sample and the samples were shaken for 15 min on ice. Finally 1 mL of 500 mM MgCl2 was added, the samples were incubated a further 10 min and the cells were pelleted. 5 mL of 10 × IMAC buffer (0.2 M Na2HPO4, 4 M NaCl, 0.2 M imidazole, pH 7.5) and 0.5 mL of Ni Separose (GE Healthcare) were added to the supernatant and the sample tumbled overnight at 4 °C on a rotisserie. Next, the resin was washed with 1 × IMAC buffer and eluted with IMAC buffer containing 500 mM imidazole. Samples were purified by immobilized metal affinity chromatography (IMAC) and size-exclusion chromatography using the BioLogic Duo-flow chromatography system (Bio-Rad, Hercules, CA) and either an ENrich SEC 70 10 × 300 mm column or a Superdex G75 10 × 300 mm column (GE Healthcare, Piscataway, NJ) equilibrated in phosphate buffer saline (PBS) +0.02% azide and run at 0.5 mL/min.
Yields of RZh from the periplasmic production were determined to be 13.5 ± 2.5 mg/L by measuring the absorbance at 280 nm using a NanoDrop spectrophotometer. Previous reports detailed the recombinant production of unfused RZ from the cytoplasm with an average yield of 4 mg/L [13], [14]. Thus, periplasmic production of the RZh with the tag was significantly improved from the previously reported RZ production [13], [14], even though production into the periplasmic space is generally considered a limiting factor [34], [35]. In part this may be due to improved recovery from the IMAC versus the 2-iminobiotin column.
To evaluate the quaternary state (monomer, dimer, or tetramer) of the fusion constructs, 0.5 mL samples of the proteins either with or without excess biotin (20 μM) added were rerun on the ENrich SEC 70 column (Fig. S1). Unexpectedly, size exclusion chromatography of the IMAC purified RZh, in the absence of biotin eluted as a tetramer. However, upon additional size exclusion separations of dilutions of the RZh protein, a concentration dependent tetramer to dimer transition was observed. In the presence of excess biotin we observed that a concentration independent dimer form predominated. On denaturing gels, following the standard protocol to denature the protein first, RZh ran as a 29.1 kDa dimer both in the presence and absence of biotin, attesting to the high stability of the dimer interaction (Fig. S2). In general, it appears that the biotin bound form of RZh is not only more thermostable, but is also more soluble as it was observed to consistently elute from the gel filtration column with a more narrow half-height bandwidth than was observe for biotin free RZh (Fig. S1B and S1C). Perhaps the biotin binding pocket shows some propensity to interact weakly with the column matrix or weak tetramer to dimer transitions are occurring.
2.4. Biotin functionalized QDs
Luminescent CdSe-CdZnS-ZnS core-shell-shell quantum dots were prepared as described previously [36]. The size of the CdSe-CdZnS-ZnS core-shell-shell QDs utilized in this work was 4.6 ± 0.44 nm as measured by transmission electron microscopy (TEM); the QDs had an emission centered at 540 nm and were capped with a combination of 80% DHLA-PEG550-OMe and 20% DHLA-PEG400-biotin. There are about 100–200 ligands per QD and thus there are estimated to be 20–40 biotins per QD [30].
The biotin-functionalized QDs (Bt-QDs), as well as DHLA-PEG capped QDs with an emission at 540 nm, were diluted in PBS and applied in triplicate at concentrations ranging from 100 nM down to 0.8 nM (5-fold dilution series) to wells of a NeutrAvidin coated 96-well plate that had been washed three times with PBS containing 0.05% tween (PBST). After incubating for ∼2 h the photoluminescence (PL) was measured and then the QD solutions were removed and wells were washed 3 times with PBS and final PL spectra were collected. The measurement taken before washing the wells confirmed that the spectra of the QD dilutions were as expected; the measurement after washing the wells indicates how the QDs bound to the plate. All fluorescence emission spectra were measured from each well using a Tecan Infinite M1000. The excitation wavelength was 300 nm. Data was collected every 5 nm from 450 to 600 nm. Each QD sample was examined in triplicate.
2.5. Conjugation of sdAb-RZ to biotin functionalized QDs
Biotin functionalized QDs (∼200 pmol; 10 μL of a 19.8 μM solution) and D12f-L10-RZ (∼4000 pM; 77 μL of a 52 μM solution) were mixed, made up to 1 mL in PBS and incubated for approximately a half hour on ice. This gave a molar ratio of 20:1 sdAb-RZ to QDs, a ratio chosen to saturate the QD surface. The sdAb-RZ-QD mixture was applied to a float-a-lyzer with a 100 kDa cut off (Spectra/Por) and dialyzed against PBS with three buffer changes with the final dialysis overnight against fresh buffer. The next day the sdAb-RZ-QDs were removed from the dialysis cup and the cup was rinsed with 1 mL PBS to give a final volume of 2 mL with a concentration of ∼100 nM QD reagent, assuming no loss on dialysis.
2.6. Binding determinations by surface plasmon resonance (SPR)
All SPR experiments were performed using a Bio-Rad ProteOn XPR36 and the data were analyzed using the accompanying ProteOn Manager 3.1software using a global analysis Langmuir fit. Raw data is shown for the immobilization steps, for the ricin binding and subsequent amplification with QDs. The data is corrected for interspot signal and zero concentration, when appropriate. For the evaluation of the QD bioconjugates three different anti-ricin antibodies were utilized: D12f and H1W are both sdAb developed by us [32], and mAb 5F4, which was the kind gift of Dr. Thomas O’Brien, Tetracore (Rockville, MD). These three antibodies all bind to different epitopes on the ricin A chain and were immobilized using a concentration of 20 μg/mL onto a GLC chip, which has a compact carboxyl modified polymer layer with a binding capacity of approximately one protein monolayer, using 10 mM acetate buffer pH 5.0 by the standard EDC amine coupling chemistry recommended by Bio-Rad. The immobilized antibodies were then allowed to bind ricin for 120 s, 100 μL/min, at a range of concentrations from 100 to 0 nM, then biotin capped QDs (Bt-QDs; 10 nM) that had been saturated with the D12f-L10-RZ fusion (prepared as described above) were flowed over the chip for 120 s, followed by 240 s of buffer (PBS +0.005% Tween-20). Following regeneration of the same chip with 0.085% phosphoric acid the antibodies were allowed to bind 100 nM ricin on all lanes for 120 s. Then to evaluate the dose response of D12f-L10-RZ coated Bt-QDs, concentrations of 10, 3.3, 1.1, 0.37, 0.124, and 0 nM QD conjugates were each flowed over one lane for 120 s followed by 240 s of buffer.
The RZh was tested on a Bio-Rad HTE sensor chip, which has a high density of tris-NTA complexes for improved capturing of histidine-tagged protein providing excellent binding stability and regeneration capability in the capture of histidine-tagged proteins. The HTE chip was first exposed to 10 mM Ni sulfate for 300 s at 30 μL/min (not shown). Lanes 1–5 were then coated with RZh (only 1 lane is shown, however similar results were obtained on all five lane) or D12f-L10-RZ (only a single lane was used as a control) via their 6xHis-tags by flowing for 20 μg/mL of each over the respective lane at 30 μL/min for 300 s. The RZh was then allowed to bind Bt-D12f (20 μg/mL) for 300 s. Afterwards the chip was turned 90° and lanes were challenged with a range of ricin concentrations (100–0 nM) for 90 s at 100 μL/min, the off rate was followed for 600 s. While the HTE chip has less disassociation of the His-tagged protein than Bio-Rad’s lower density HTG chips, as RZh is a dimer and can bind with two His tails, its off rate is further suppressed. In addition, the interspot correction capability of the ProteOn allows for correction of background drift.
2.7. Experion automated electrophoresis system
To further evaluate the quaternary structure of the proteins and their stability samples either native or denatured were analyzed on the Bio-Rad Experion system following provided instructions, see Bio-Rad’s quick guide bulletin 10004490. Protein samples were examined in native and reduced conditions, both with and without added biotin (20 μM). (Fig. S2).
2.8. SdAb-QD bioconjugate use in fluoroimmunoassays
Fluoroimmunoassays were carried out similar to the previous sdAb-QD assays [20]. Wells of a 96-well plate (Nunc, maxisorb) were coated overnight at 4 °C with capture antibody using the monoclonal antibody 5F4 at a concentration of 3 μg/mL in PBS. The remaining incubation steps were all carried out at room temperature. The next morning, the coating liquid was dumped out and the wells blocked for an hour with PBS containing 4% powdered milk. Wells were washed two times with PBST and then dilutions of ricin, made up in PBS containing 1 mg/mL BSA (PBS-B) were applied to the wells (4-fold dilutions starting at 10 μg/mL). After ∼1.5 h wells were washed with PBST. The sdAb-RZ-QD reagent was diluted to ∼14 nM in PBS-B and 75 μL was added per well and incubated for ∼1.5 h. The sdAb-RZ-QD solution was removed and the wells washed with PBST. The plate was slapped on paper towels to remove remaining wash buffer from wells and fluorescence at 545 nm was measured on a Tecan Infinite M1000 using an excitation of 300 nm. Measurements were done in triplicate; controls in which there was no coating antibody, no ricin, or no sdAb-RZ-QDs were also performed.
3. Results and discussion
3.1. Production of sdAb-RZ genetic fusion
Recently, we published the first report of genetic fusions between sdAbs and the biotin binding protein RZ [10]. In that work we showed that the sdAb-RZ conjugates appeared to have the same ability to provide oriented capture surfaces as we had found with sdAb fusions to a streptavidin core [11], with the advantage of 20-fold better protein production of ∼14 mg/L of sdAb-RZ fusions. All these constructs included a sequence from the natural llama upper hinge linking the two proteins, and used a pECAN45-based expression vector.
For the production of sdAbs, we have found our best protein expression when the sdAb sequence was inserted into the pET22b expression vector without including any sequence from the upper hinge [37]. Thus, for second-generation sdAb-RZ reagents we switched to the pET22b expression vector, and constructed a version of the sdAb-RZ that replaced the upper hinge linker with a more generic one that contained the amino acid sequence comprising the restriction sites used in cloning and an additional 10 amino acid Gly-Ser linker (AAALEGGGGSGGGGS) between the two domains. In addition, for this work we utilized the D12f ricin binding sdAb which shows excellent affinity with a high melting temperature (78 °C) to provide a reagent with improved ruggedness compared to the original RZ fusion with the anti-ricin C8 sdAb that melts ∼60 °C [32]. When purified the fusion protein, D12f-L10-RZ, eluted from the ENrich SEC 70 as a ∼60 kDa dimer, with biotin having no effect on the elution volume, indicating the fusion construct always exists as a dimer (Fig. S1A). On the Experion, D12f-L10-RZ ran as ∼63 kDa dimer when in the native state, but as ∼30.3 kDa monomer when denatured (Fig. S2).
3.2. Demonstration of the utility of unfused RZh
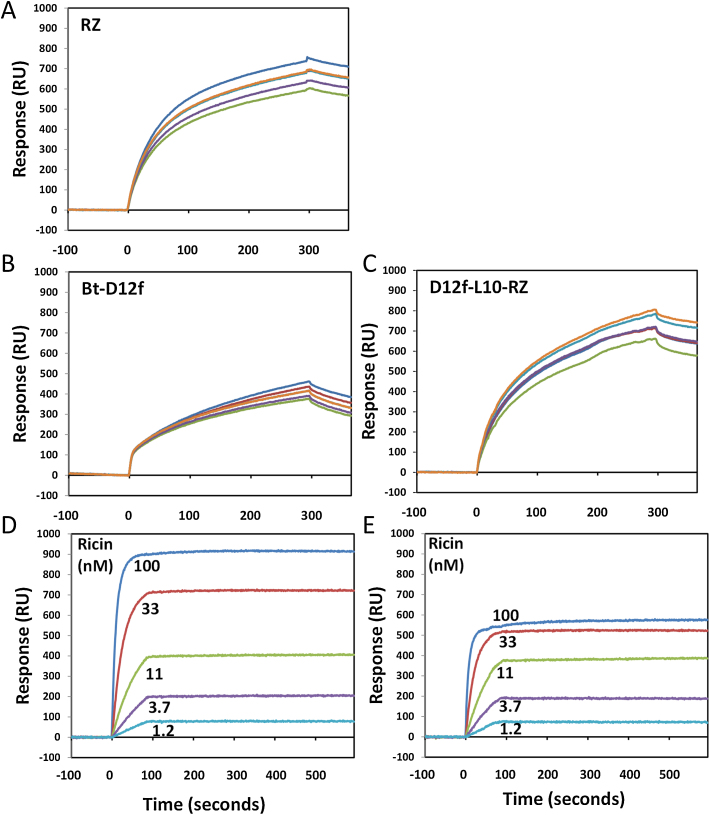
In addition to our prime objective, we also prepared and evaluated the utility of the RZh protein by performing a test similar to that described by Zhou et al. [38], see Fig. 2. The RZh and the D12f-L10-RZ were both captured via their 6xHis tags onto the surface of a HTE chip. RZh being a dimer, the two His tags form a more stable interaction to the tris-NTA surface than monomeric proteins with a single His tag. The RZh was then allowed to bind a biotinylated D12f (Bt-D12f). After the capture reagents were immobilized, the chip was turned 90° and various concentration of ricin (100–0 nM) were then flowed over the chip. The quantity of ricin bound by the two surfaces was very comparable and the affinity observed was the same for both forms of D12f (KD = 86 ± 24 pM). The oriented RZh were calculated to bind ∼1.3 Bt-D12f for each RZh dimer and the Bt-D12f bind ∼1 ricin for every 2 Bt-D12f immobilized. Thus, an excellent level of binding activity was obtained. These experiments show that RZh can be used in place of the other His-tagged biotin binding proteins for a wide array of applications, with the added advantage that RZh produced at least a 10-fold better yield in E. coli than the strep-core biotin binding protein (not shown).
Fig. 2.
Demonstrating the effectiveness of rhizavidin (RZh) using HTE chip on the ProteOn SPR. Panel A shows the rhizavidin (20 μg/mL) binding to all 6 spots on one lane of the HTE chip via the 6xHis tags. Panel B shows the same spots as in panel A being coated with a biotinylated anti-ricin sdAb, D12f (Bt-D12f). Panel C shows six spots from another lane being coated with the D12f-L10-RZ via the His tags. After the sensor chip is rotated 90°, the ability of both lanes so prepared (panels B and C) to bind to ricin was demonstrated as shown in panels D and E, for the Bt-D12f on the RZh alone and the D12-L10-RZ respectively.
3.3. Testing biotin-functionalized QDs
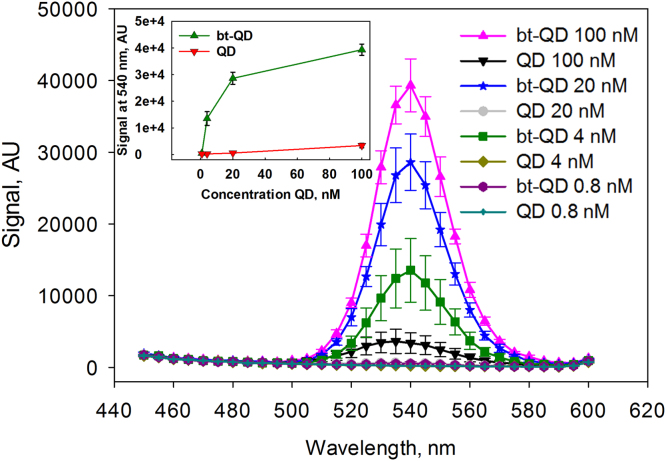
Functionality of the QDs capped with 20% biotinylated ligand (Bt-QDs) was first assessed by examining their ability to bind a NeutrAvidin coated plate. Dilution series of QDs, with and without biotin functionalized ligand, were applied to wells of a NeutrAvidin coated plate. The biotin-functionalized QDs showed binding to the NeutrAvidin, in contrast similar QDs that were only coated with DHLA-PEG-OMe showed only minimal non-specific binding to the NeutrAvidin (Fig. 3). This experiment verified the availability of the biotin on the QDs for binding to NeutrAvidin and indicated that the Bt-QDs would specifically bind to NeutrAvidin.
Fig. 3.
Binding of QDs capped with DHLA PEG and 20% biotin reagent (Bt-QD), and QDs capped only with DHLA PEG to NeutrAvidin coated wells. The QD dilutions (100 nM, 20 nM, 4 nM and 0.8 nM) were first applied to NeutrAvidin coated wells and allowed to bind, before the wells were washed and fluorescence spectra were measured. Spectra are color coded; the key on the right side of the plot indicates which colors and symbols correspond to which type of QD and which dilution. The inset on the left side of the plot shows the values of the fluorescence emission at 540 nm for both the Bt-QDs (green triangles) and the QDs capped only with DHLA PEG (red triangles) plotted against QD concentration. Excitation was at 300 nm. This plot indicates that the Bt-QDs were specifically binding to the NeutrAvidin. Data points represent the average fluorescence from three wells; error bars are the standard deviation.
3.4. Evaluation of sdAb-RZ-QDs by SPR
The Bt-QDs were conjugated to D12f-L10-RZ and their binding to ricin evaluated by SPR to determine the binding kinetics of the QD reagents and evaluate them for the detection of ricin. Previously we had observed that sdAb coupled to DHLA capped QDs through an extended His tail provided a >2-fold amplification over unconjugated sdAbs for the detection of ricin, while commercial streptavidin functionalized QDs provided less of an amplification [20]. In this work the D12f-L10-RZ provided the recognition element that was conjugated to the QDs. The high affinity RZ-biotin interaction provided oriented immobilization of D12f on the Bt-QD surface. A schematic of two types of sdAb-QD conjugates is shown in Fig. 1.
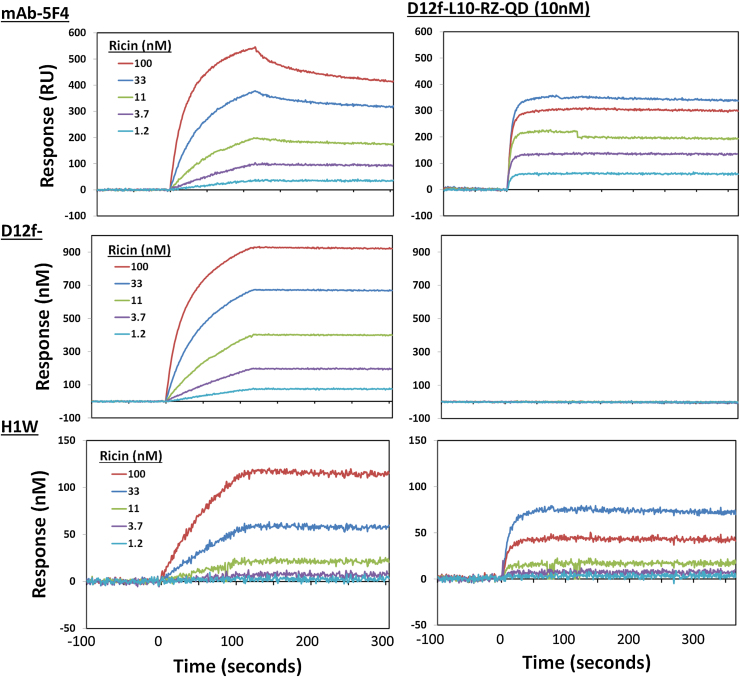
The sdAb-RZ-QDs were examined to determine their binding kinetics and their ability to amplify signals in sandwich assays for ricin. Ricin was bound by three different immobilized capture antibodies: a monoclonal antibody (mAb 5F4), the D12f sdAb, and the H1W sdAb that recognizes a different epitope on ricin than D12f (Fig. 4 left panels) [32]. The sdAb-RZ-QDs were then applied to the antibody-captured ricin (Fig. 4 right panels). Binding kinetics for the capture reagents, as well as the sdAb-RZ-QDs are shown in Table 1. The interaction of the D12f-L10-RZ-QD with ricin is multivalent, and is reflected in the slow off rates which are beyond the limit of our ability to accurately measure. When off rates become very low the ability to differentiate becomes more challenging as the value can easily be affected by drift. As expected, no binding of the D12f-L10-RZ-QDs was recorded when ricin was captured by the immobilized D12f. This is because ricin is a simple heterodimeric toxin without repeating epitopes. In order to form a successful sandwich binding, reagents must recognize different portions of the toxin.
Fig. 4.
Effectiveness of Biotin—QDs coated with D12f-L10-RZ to function as an amplifier in SPR sandwich assays. A GLC SPR chip was coated with anti-ricin binders [mAb-5F4 (top), D12f (middle), and H1W (bottom)]. The chip was turned 90°, and allowed to bind ricin for 120 s at a range of concentrations from 100 to 0 nM as indicated, left column. The left side of the figure shows the binding of ricin dilutions to the three capture antibodies. Then sdAb-RZ-QD complexes at ∼10 nM were flowed over the surfaces for 120 s followed by buffer to monitor for dissociation as is shown on the right side of the figure. The sdAb-RZ-QD complexes bound rapidly in a ricin dose dependent manner.
Table 1.
Binding constants determined for the three capture antibodies and apparent values determined for the D12f-RZ-QDs using (1) mAb 5F4 and (2&3) H1W as capture molecule for ricin.
| ka M−1 s−1 | kd 1 s−1 | KD M | |
|---|---|---|---|
| mAb 5F4 | 6.2 × 105 | 4.1 × 10−4 | 6.7 × 10−10 |
| D12f | 6.6 × 105 | 4.3 × 10−5 | 6.5 × 10−11 |
| H1W | 3.6 × 105 | 2.4 × 10−4 | 7.5 × 10−10 |
| 1D12f-L10-RZ-QD | 2.0 × 107 | 3.1 × 10−18 | 6.7 × 10−25 |
| 2D12f-L10-RZ-QD | 1.0 × 107 | 2.3 × 10−17 | 6.7 × 10−24 |
| 3C8-QD | 6.4 × 106 | 2.2 × 10−4 | 3.5 × 10−11 |
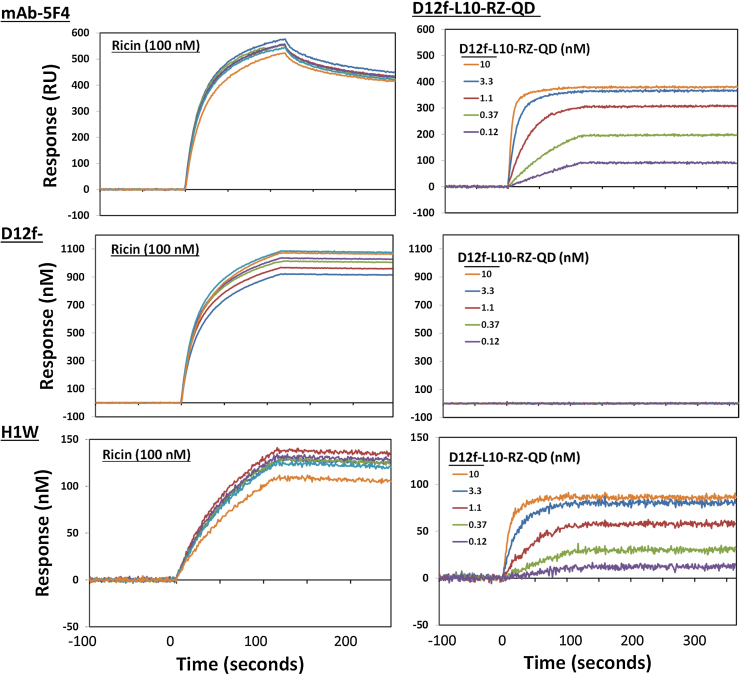
An evaluation of the dose response of the D12f-L10-RZ-QD was carried out, looking at the signal generated by dilutions of the sdAb-RZ-QDs on a captured ricin surface. The three anti-ricin antibodies (mAb 5F4, H1W and D12f) were exposed to 100 nM ricin; the left panel of Fig. 5 shows that for each of the three capture reagents, the signal generated by applying ricin was roughly equivalent across the 6 lanes. Applying a dilution series of the sdAb-RZ-QDs showed a dose-dependent on rate as is expected for a typical analyte (Fig. 5 right side). As before no binding was observed with the D12f capture reagent.
Fig. 5.
Evaluation of dose response of QD conjugates. A GLC SPR chip was coated with anti-ricin binders [mAb-5F4 (top), D12f (middle), and H1W (bottom)], the same as in Fig. 5. Each lane was exposed to ricin 100 nM for 90 s (left column). As can be seen on the left side of the figure, for each of the three capture antibodies, this generated approximately the same signal in each of the six lanes. Next the lanes were exposed to the SdAb-RZ-QD conjugates at range of dilutions, starting at 10 to 0 nM (as indicated in the legend). Data is shown on the right side of the figure. A dose dependent on rate was observed, similar as one would see for any typical analyte.
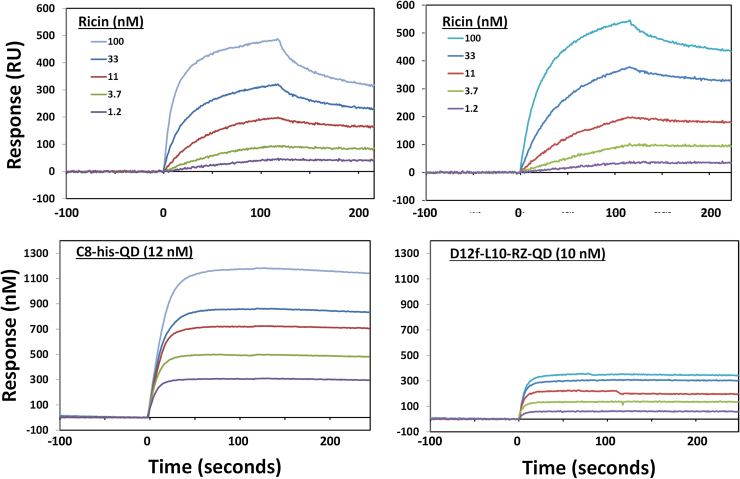
Fig. 6 shows the comparison of the DHLA capped QDs coated with C8, a sdAb highly homologous to D12f, via its extended His-tag and the sdAb-RZ-QD conjugates prepared for this work. The SPR binding studies that utilize the sdAb-QDs enable us to compare on rates; as this is a multi-valent system, the on rates are relative. Our current QDs displayed faster on rate binding kinetics by ∼3.1 times than those used previously (Table 1); thus, at least 3 fold lower concentrations of current QDs can be used. This is a useful advantage when dealing with expensive reagents such QDs. Fig. 6 also shows a comparison of the amplification observed, previously greater than 2-fold was obtained versus the less than 1-fold increase seen with our current QDs. It is unclear, why these QD bioconjugates provide less amplification, even though they appear just as active as the previously utilized QDs. Previously we had also observed less amplification when examining sdAb-QD conjugates prepared utilizing commercial streptavidin coated QDs [20]; potentially the ligands used to make the QDs biocompatible are a factor in amplification. While the greater thickness of Bt-PEG layer on the QDs, as well as the increased thickness of the sdAb-RZ conjugate, increase the distance of the QDs from the surface, additional experimentation is required to determine if this change was responsible for the reduced amplification. The D12f-L10-RZ-QDs bound with high specificity, as no binding to ricin that had been captured using covalently immobilized D12f-sdAb was observed. SPR testing of QD bioconjugates, however, was envisioned more as a means to evaluate the nature of the QD bioconjugates rather than as a method for SPR amplification. Alternatives such as gold nanoparticles are likely superior for that application [39].
Fig. 6.
Comparison of DHLA capped QDs prepared coated with the anti-ricin sdAb C8 via its His-tag (left panel) [20] with the current Bt-QDs conjugated with D12f-L10-RZ (right panel). For both sets of data ricin at various concentration was captured via mAb 5F4 immobilized on the surface of the SPR sensor chip (top panels). The DHLA-QDs gave an amplification >3-fold ricin alone, however the Bt-QDs appear to have a superior on rate >2-fold.
3.5. Fluroimmunoassays utilizing sdAb-RZ-QDs
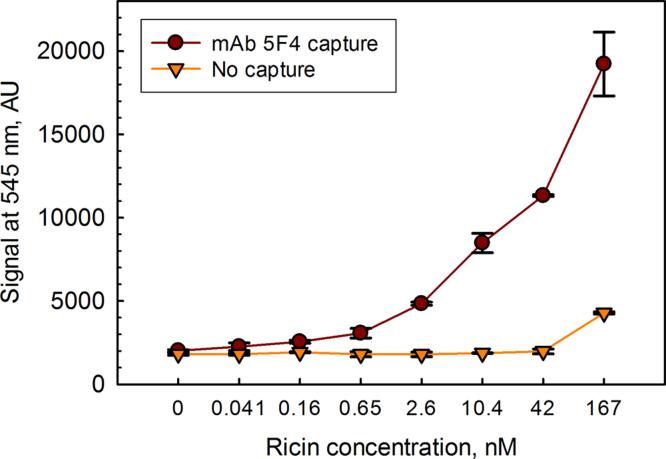
We had previously shown that sdAbs directionally conjugated to DHLA-capped CdSe–ZnS core–shell QDs could be successfully utilized in fluoroimmunoassays for the detection of ricin [20]. Fig. 7 shows a fluoroimmunoassay utilizing the sdAb-RZ-QD reagent for ricin detection. Examination of binding to wells that had no capture antibody show non-specific binding only at the highest ricin concentration; this signal is likely due to non-specific binding of the ricin toxin, as there was no binding of the sdAb-RZ-QD reagent to similar wells having lower amounts of ricin. Using the sdAb-RZ-QD reporter reagent, detection of ricin was essentially identical to what we had observed previously using sdAbs conjugated to QDs through an extended His tail as the reporter and the same monoclonal antibody as a capture [20]. In both cases we demonstrated detection of toxin down to ∼0.16 nM (∼10 ng/mL). These sdAb-RZ reagents should enable oriented immobilization on any biotinylated surface, including commercial preparations of biotinylated QDs.
Fig. 7.
Fluoroimmunoassay, utilizing sdAb-RZ-QD reagents for the detection of ricin. In these assays, a capture antibody was adsorbed onto wells and exposed to dilutions of ricin. Excess toxin was washed and then the sdAb-RZ-QD reagent allowed to bind to captured ricin. Finally wells were washed and fluorescence measured from each well at 545 nm (300 nm excitation). Dark brown circles represent signal generated by a sandwich assay with capture monoclonal antibody 5F4. Light brown triangles are the signal in the absence of capture antibody and represent a combination of non-specific binding by both the ricin antigen and the fluorescent sdAb-RZ-QD tracer. Ricin concentrations are given in nM; for reference 100 nM ricin ∼6 μg/mL. Each ricin concentration was assayed in triplicate, the average value is plotted; error bars represent the standard deviation.
4. Conclusions
This work detailed the production and characterization of sdAb-RZ fusions with a generic Gly-Ser linker in place of the hinge region as well as RZh (RZ with a His tag). The proteins produced well (at least 10 mg/L), and were fully functional, with both the RZ portion showing binding to biotin, and the sdAb portion of the fusions recognizing ricin. The focus however was demonstrating the bioconjugation of sdAb-RZ to Bt −QDs. The antigen recognition function was provided by a previously described high-affinity ricin binding sdAb. The advantages of QD bioconjugation using a sdAb-RZ fusion include the ability to prepare a wide range of sdAb-RZ fusions that can be conjugated to stable bio-compatible biotin coated QDs as desired. The dimeric nature of RZ makes the attachment bivalent and thus makes for an extremely stable noncolavent interaction. Just as important, the sdAb-RZ coats the BT-QDs in an oriented manner, providing a highly active surface. As the RZ monomer is of similar size as an sdAb molecule, they pack efficiently onto the surface of the QD. This format provides a useful alternative to preparation of streptavidin-QDs that are coated with biotin tagged ligands. The use of sdAb-RZ to bioconjuagate QDs or any biotinylated nano or microparticle hold great promise as diagnostic and imaging reagents as well as for detection applications.
Acknowledgements
Funding for this project was provided by the Defense Threat Reduction AgencyCBCALL12-DIAGB5-2-0037 and CBCALL12-LS6-2-0036. Other support for this work was provided by ONR/NRL 6.1 and 6.2 Base funds. KBT was an American Society for Engineering Education postdoctoral fellow.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2016.03.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Eyer L., Hruska K. Single-domain antibody fragments derived from heavy-chain antibodies: a review. Vet. Med. 2012;57:439–513. [Google Scholar]
- 2.de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb. Cell Fact. 2011;10 doi: 10.1186/1475-2859-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wesolowski J., Alzogaray V., Reyelt J., Unger M., Juarez K., Urrutia M., Cauerhff A., Danquah W., Rissiek B., Scheuplein F., Schwarz N., Adriouch S., Boyer O., Seman M., Licea A., Serreze D.V., Goldbaum F.A., Haag F., Koch-Nolte F. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med. Microbiol. Immunol. 2009;198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 5.Walper S.A., Battle S.R., Lee P.A.B., Zabetakis D., Turner K.B., Buckley P.E., Calm A.M., Welsh H.S., Warner C.R., Zacharko M.A., Goldman E.R., Anderson G.P. Thermostable single domain antibody-maltose binding protein fusion for Bacillus anthracis spore protein BclA detection. Anal. Biochem. 2014;447:64–73. doi: 10.1016/j.ab.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Sherwood L.J., Osborn L.E., Carrion R., Patterson J.L., Hayhurst A. Rapid assembly of sensitive antigen-capture assays for Marburg virus using in vitro selection of llama single-domain antibodies, at biosafety level 4. J. Infect. Dis. 2007;196:S213–S219. doi: 10.1086/520586. [DOI] [PubMed] [Google Scholar]
- 7.Goldman E.R., Brozozog-Lee P.A., Zabetakis D., Turner K.B., Walper S.A., Liu J.L., Anderson G.P. Negative tail fusions can improve ruggedness of single domain antibodies. Protein Expr. Purif. 2014;95:226–232. doi: 10.1016/j.pep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Hussack G., Luo Y., Veldhuis L., Hall J.C., Tanha J., MacKenzie R. Multivalent anchoring and oriented display of single-domain antibodies on cellulose. Sensors. 2009;9:5351–5367. doi: 10.3390/s90705351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherwood L.J., Hayhurst A. Hapten mediated display and pairing of recombinant antibodies accelerates assay assembly for biothreat countermeasures. Sci. Rep. 2012;2 doi: 10.1038/srep00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J.L., Zabetakis D., Walper S.A., Goldman E.R., Anderson G.P. Bioconjugates of rhizavidin with single domain antibodies as bifunctional immunoreagents. J. Immunol. Methods. 2014;411:37–42. doi: 10.1016/j.jim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Walper S.A., Brozozog Lee P.A., Goldman E.R., Anderson G.P. Comparison of single domain antibody immobilization strategies evaluated by surface plasmon resonance. J. Immunol. Methods. 2013;388:68–77. doi: 10.1016/j.jim.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Pleschberger M., Saerens D., Weigert S., Sleytr U.B., Muyldermans S., Sara M., Egelseer E.M. An S-layer heavy chain camel antibody fusion protein for generation of a nanopatterned sensing layer to detect the prostate-specific antigen by surface plasmon resonance technology. Bioconjug. Chem. 2004;15:664–671. doi: 10.1021/bc049964w. [DOI] [PubMed] [Google Scholar]
- 13.Helppolainen S.H., Nurminen K.P., Maatta J.A.E., Halling K.K., Slotte J.P., Huhtala T., Liimatainen T., Yla-Herttuala S., Airenne K.J., Narvanen A., Janis J., Vainiotalo P., Valjakka J., Kulomaa M.S., Nordlund H.R. Rhizavidin from Rhizohium etli: the first natural dimer in the avidin protein family. Biochem. J. 2007;405:397–405. doi: 10.1042/BJ20070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meir A., Helppolainen S.H., Podoly E., Nordlund H.R., Hytonen V.P., Maatta J.A., Wilchek M., Bayer E.A., Kulomaa M.S., Livnah O. Crystal structure of rhizavidin: insights into the enigmatic high-affinity interaction of an innate biotin-binding protein dimer. J. Mol. Biol. 2009;386:379–390. doi: 10.1016/j.jmb.2008.11.061. [DOI] [PubMed] [Google Scholar]
- 15.Petryayeva E., Algar W.R., Medintz I.L. Quantum dots in bioanalysis: a review of applications across various platforms for fluorescence spectroscopy and imaging. Appl. Spectrosc. 2013;67:215–252. doi: 10.1366/12-06948. [DOI] [PubMed] [Google Scholar]
- 16.Algar W.R., Kim H., Medintz I.L., Hildebrandt N. Emerging non-traditional Forster resonance energy transfer configurations with semiconductor quantum dots: investigations and applications. Coord. Chem. Rev. 2014;263:65–85. [Google Scholar]
- 17.Gemmill K.B., Deschamps J.R., Delehanty J.B., Susumu K., Stewart M.H., Glaven R.H., Anderson G.P., Goldman E.R., Huston A.L., Medintz I.L. Optimizing protein coordination to quantum dots with designer peptidyl linkers. Bioconjug. Chem. 2013;24:269–281. doi: 10.1021/bc300644p. [DOI] [PubMed] [Google Scholar]
- 18.Sukhanova A., Even-Desrumeaux K., Kisserli A., Tabary T., Reveil B., Millot J.M., Chames P., Baty D., Artemyev M., Oleinikov V., Pluot M., Cohen J.H.M., Nabiev I. Oriented conjugates of single-domain antibodies and quantum dots: toward a new generation of ultrasmall diagnostic nanoprobes. Nanomed. Nanotechnol. Biol. Med. 2012;8:516–525. doi: 10.1016/j.nano.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Fatehi D., Baral T.N., Abulrob A. In vivo imaging of brain cancer using epidermal growth factor single domain antibody bioconjugated to near-Infrared quantum dots. J. Nanosci. Nanotechnol. 2014;14:5355–5362. doi: 10.1166/jnn.2014.9076. [DOI] [PubMed] [Google Scholar]
- 20.Anderson G.P., Glaven R.H., Algar W.R., Susumu K., Stewart M.H., Medintz I.L., Goldman E.R. Single domain antibody-quantum dot conjugates for ricin detection by both fluoroimmunoassay and surface plasmon resonance. Anal. Chim. Acta. 2013;786:132–138. doi: 10.1016/j.aca.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Wegner K.D., Linden S., Jin Z.W., Jennings T.L., el Khoulati R., Henegouwen P., Hildebrandt N. Nanobodies and nanocrystals: highly sensitive quantum dot-based homogeneous FRET immunoassay for serum-based EGFR detection. Small. 2014;10:734–740. doi: 10.1002/smll.201302383. [DOI] [PubMed] [Google Scholar]
- 22.Zaman M.B., Baral T.N., Zhang J., Whitfield D., Yu K. Single-domain antibody functionalized CdSe/ZnS quantum dots for cellular imaging of cancer cells. J. Phys. Chem. C. 2009;113:496–499. [Google Scholar]
- 23.Zaman M.B., Baral T.N., Jakubek Z.J., Zhang J., Wu X., Lai E., Whitfield D., Yu K. Single-domain antibody bioconjugated near-IR quantum dots for targeted cellular imaging of pancreatic cancer. J. Nanosci. Nanotechnol. 2011;11:3757–3763. doi: 10.1166/jnn.2011.4167. [DOI] [PubMed] [Google Scholar]
- 24.Blanco-Canosa J.B., Wu M., Susumu K., Petryayeva E., Jennings T.L., Dawson P.E., Algar W.R., Medintz I.L. Recent progress in the bioconjugation of quantum dots. Coord. Chem. Rev. 2014;263:101–137. [Google Scholar]
- 25.Mei B.C., Susumu K., Medintz I.L., Delehanty J.B., Mountziaris T.J., Mattoussi H. Modular poly(ethylene glycol) ligands for biocompatible semiconductor and gold nanocrystals with extended pH and ionic stability. J. Mater. Chem. 2008;18:4949–4958. [Google Scholar]
- 26.Dona V., Urrutia M., Bayardo M., Alzogaray V., Goldbaum F.A., Chirdo F.G. Single domain antibodies are specially suited for quantitative determination of gliadins under denaturing conditions. J. Agric. Food Chem. 2010;58:918–926. doi: 10.1021/jf902973c. [DOI] [PubMed] [Google Scholar]
- 27.Harmsen M.M., van Solt C.B., van Bemmel A.M.V., Niewold T.A., van Zijderveld F.G. Selection and optimization of proteolytically stable llama single-domain antibody fragments for oral immunotherapy. Appl. Microbiol. Biotechnol. 2006;72:544–551. doi: 10.1007/s00253-005-0300-7. [DOI] [PubMed] [Google Scholar]
- 28.Alzogaray V., Danquah W., Aguirre A., Urrutia M., Berguer P., Vescovi E.G., Haag F., Koch-Nolte F., Goldbaum F.A. Single-domain llama antibodies as specific intracellular inhibitors of SpvB, the actin ADP-ribosylating toxin of Salmonella typhimurium. FASEB J. 2011;25:526–534. doi: 10.1096/fj.10-162958. [DOI] [PubMed] [Google Scholar]
- 29.Susumu K., Uyeda H.T., Medintz I.L., Mattoussi H. Design of biotin-functionalized luminescent quantum dots. J. Biomed. Biotechnol. 2007 doi: 10.1155/2007/90651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susumu K., Uyeda H.T., Medintz I.L., Pons T., Delehanty J.B., Mattoussi H. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J. Am. Chem. Soc. 2007;129:13987–13996. doi: 10.1021/ja0749744. 7 pages. [DOI] [PubMed] [Google Scholar]
- 31.Kozlov Y.V., Sudarkina O.Y., Kurmanova A.G. Ribosome-inactivating lectins of plants. Mol. Biol. 2006;40:711–723. [PubMed] [Google Scholar]
- 32.Turner K.B., Liu J.L., Zabetakis D., Lee A.B., Anderson G.P., Goldman E.R. Improving the biophysical properties of anti-ricin single-domain antibodies. Biotechnol. Rep. 2015;6:27–35. doi: 10.1016/j.btre.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raphael M., Christodoulides J., Byers J., Anderson G., Liu J., Turner K., Goldman E., Delehanty J. Optimizing nanoplasmonic biosensor sensitivity with orientated single domain antibodies. Plasmonics. 2015:1–7. doi: 10.1007/s11468-015-9969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riggs P. Expression and purification of recombinant proteins by fusion to maltose-binding protein. Mol. Biotechnol. 2000;15:51–63. doi: 10.1385/MB:15:1:51. [DOI] [PubMed] [Google Scholar]
- 35.Georgiou G., Segatori L. Preparative expression of secreted proteins in bacteria: status report and future prospects. Curr. Opin. Biotechnol. 2005;16:538–545. doi: 10.1016/j.copbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Susumu K., Oh E., Delehanty J.B., Pinaud F., Gemmill K.B., Walper S., Breger J., Schroeder M.J., Stewart M.H., Jain V., Whitaker C.M., Huston A.L., Medintz I.L. A new family of pyridine-appended multidentate polymers as hydrophilic surface ligands for preparing stable biocompatible quantum dots. Chem. Mater. 2014;26:5327–5344. [Google Scholar]
- 37.Turner K.B., Zabetakis D., Legler P.M., Goldman E.R., Anderson G.P. Isolation and epitope mapping of staphylococcal enterotoxin B single-domain antibodies. Sensors. 2014;14:10846–10863. doi: 10.3390/s140610846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M., Luo R., Shezifi D., Nimri S. A novel biotinylated ligand capture method with surface regeneration capability for label-free biomolecular interaction analysis. Bioradiations. 2013;(February(12)) Protocols and tips. [Google Scholar]
- 39.Szunerits S., Spadavecchia J., Boukherroub R. Surface plasmon resonance: signal amplification using colloidal gold nanoparticles for enhanced sensitivity. Rev. Anal. Chem. 2014;33:153–164. [Google Scholar]
- 40.George J., Compton J.R., Leary D.H., Olson M.A., Legler P.M. Structural and mutational analysis of a monomeric and dimeric form of a single domain antibody with implications for protein misfolding. Proteins. 2014;82:3101–3116. doi: 10.1002/prot.24671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.