Abstract
Background
Pathogenic mutations in the granulin gene (GRN) are causative in 5-10% of patients with frontotemporal dementia (FTD), mostly leading to reduced progranulin protein (PGRN) levels. Upcoming therapeutic trials focus on enhancing PGRN levels.
Methods
Fluctuations in plasma PGRN (n = 41) and its relationship with cerebrospinal fluid (CSF, n = 32) and specific single nucleotide polymorphisms were investigated in pre- and symptomatic GRN mutation carriers and controls.
Results
Plasma PGRN levels were lower in carriers than in controls and showed a mean coefficient of variation of 5.3% in carriers over 1 week. Although plasma PGRN correlated with CSF PGRN in carriers (r = 0.54, p = 0.02), plasma only explained 29% of the variability in CSF PGRN. rs5848, rs646776 and rs1990622 genotypes only partly explained the variability of PGRN levels between subjects.
Conclusions
Plasma PGRN is relatively stable over 1 week and therefore seems suitable for treatment monitoring of PGRN-enhancing agents. Since plasma PGRN only moderately correlated with CSF PGRN, CSF sampling will additionally be needed in therapeutic trials.
Key Words: Frontotemporal dementia, Progranulin protein, Granulin gene (GRN), Plasma, Cerebrospinal fluid, Biomarker, Single nucleotide polymorphism
Introduction
Frontotemporal dementia (FTD), a common type of presenile dementia, shows an autosomal dominant inheritance in 20-30% [1]. Pathogenic mutations in granulin(GRN) are a major cause of heritable FTD and mostly reduce progranulin protein (PGRN) levels in blood and cerebrospinal fluid (CSF) by haploinsufficiency [2,3,4,5,6]. PGRN plays an important role in neurite outgrowth and inflammation, which may be the link to neurodegeneration [7].
As PGRN levels vary greatly between individuals, various genetic and environmental regulators may play a role [5,8,9,10]. A number of single nucleotide polymorphisms (SNPs) have been associated with altered CSF or plasma PGRN levels: rs5848 (GRN),rs646776 [near sortilin 1(SORT1)] and rs1990622 [near transmembrane protein 106B(TMEM106B)] [10,11,12,13,14].
Current FTD research is shifting towards disease-modifying agents, and sensitive biomarkers are essential to evaluate these potential agents in the clinic. Histone deacetylase inhibitors, alkalizing reagents and inhibitors of vacuolar ATPase, have been shown to enhance PGRN expression in carrier-derived cells and might therefore inhibit the disease process [15,16]. Although blood PGRN poorly correlates to CSF PGRN in healthy controls and Alzheimer's disease, sporadic FTD and amyotrophic lateral sclerosis [10,17,18], this information is lacking in GRN mutation carriers. To use PGRN levels as biomarkers in therapeutic trials on PGRN-enhancing agents, a better understanding in GRN mutation carriers is needed on correlations between levels in blood and CSF, fluctuations over time and variability between subjects.
In this study, patients and presymptomatic carriers of pathogenic GRN mutations were studied to investigate (1) the correlation between plasma and CSF levels in GRNmutation carriers, (2) the fluctuations of plasma PGRN over time, and (3) the associations between three SNPs (rs5848, rs646776 and rs1990622) and PGRN levels in plasma and CSF.
Methods
Subjects
A group of 57 (37 women, 20 men) asymptomatic first-degree relatives of patients with FTD caused by a pathogenic GRN mutation (at-risk group) was selected from our longitudinal neuropsychological and MRI study in genetic FTD [19]. Participants were selected depending on the availability of plasma and/or CSF (no biosample available: n = 10). The participants originate from three different families with GRNmutations (p.Ser82Valfs, p.Gln125* or p.Val411Serfs mutation). After screening of these GRN mutations [20], participants were divided into those with (presymptomatic carriers, n = 28) and those without a pathogenic GRN mutation (controls, n = 29); investigators and at-risk individuals remained blinded to the individual carrier status. Plasma was available from all 57 at-risk individuals; a lumbar puncture was carried out in 28 of them (16 presymptomatic GRNcarriers and 12 controls, see online suppl. fig. 1; see www.karger.com/doi/10.1159/000447738 for all online suppl. material). Longitudinal blood samples for the determination of variability of plasma PGRN were collected in 37 at-risk subjects (18 presymptomatic GRNcarriers and 19 controls), and the other 20 subjects did not participate, mainly because of logistical reasons (e.g. long travel distance) or a lack of motivation.
Additionally, 10 patients with a pathogenic GRNmutation (p.Ser82Valfs, p.Gln125*, p.Val411Serfs and p.Gln130Serfs) were included from our previously described cohorts [21,22]. Plasma was available from 7 patients (longitudinal sampling in 4) and CSF from 7 (plasma also available in 4).
The study was approved by the medical ethics committee. All participants or legal representatives provided written informed consent for the blood and/or CSF collections.
Plasma and CSF Collection
Blood collections were performed according to standard procedures. For longitudinal analyses, blood was collected in a local hospital or nursing home at 5 time points during 1 week: 0 h, 6 h, 12 h, 24 h and 7 days. The collections at the time points 0 h, 24 h and 7 days were performed in the morning, while the 6-hour time point was performed after noon and the 12-hour time point in the evening. To diminish the burden for FTD patients resident in a nursing home, the venipuncture at 12 h was omitted (n = 3). To assess the influence of fasting on plasma PGRN, participants fasted for a minimum of 8 h before the venipuncture at 24 h. Plasma was isolated from K2EDTA-coated tubes (Becton Dickinson) by direct centrifugation at 1,300 relative centrifugal force for 20 min at room temperature, aliquoted and directly stored at −80°C.
CSF was collected using standard procedures into polypropylene tubes. The first 1-3 ml were discarded and samples were centrifuged in the polypropylene tubes at 2,000 relative centrifugal force for 10 min at +4°C. CSF was aliquoted into polypropylene vials and immediately stored at −80°C.
Laboratory Methods
All measurements were performed blinded to the mutation status and clinical stage. PGRN concentrations in plasma and CSF samples were determined using a qualified immunoassay based on an enzyme-linked immunosorbent assay (ELISA) kit from BioVendor (Brno, Czech Republic) following kit instructions. Low, medium and high PGRN concentration quality control samples were run on each assay plate with an acceptance coefficient of variation (CV) cutoff of ≤20%. PGRN replicates with a CV of >20% were excluded from the analyses. CSF was diluted 1:2 and plasma 1:40. All reported concentrations fell within the qualified range of the assays. PGRN concentrations were determined on a standard curve by plotting optical density versus concentration, using four-parameter logistic curve-fitting.
DNA was extracted from whole blood following standard procedures. SNPs rs5848 [NM_ 002087.2(GRN):c.*78C>T], rs646776 [NM_001408.2(CELSR2):c.*1859C>T, near SORT1] and rs1990622 [NM_000007.13:g.12283787A>G, near TMEM106B] were genotyped using Taqman assays (respective assay numbers C_7452046_20, C_3160062_10 and C_11171598_10; Life Technologies, Carlsbad, Calif., USA) on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, Calif., USA). Genotypes were assigned using SDS v3.1 software (Applied Biosystems).
Statistical Analysis
Statistical analyses were performed using SPSS 21.0 for Windows (Chicago, Ill., USA) and GraphPad Prism 6 (La Jolla, Calif., USA), applying a significance level of p < 0.05. Comparisons between two groups were made by Mann-Whitney U tests because of non-normally distributed data; multiple groups were compared by Kruskal-Wallis tests. Pearson's correlation coefficient (r) was used to correlate two normally distributed variables; otherwise, Spearman's correlation coefficient (rs) was used. To assess the variability (variance) in CSF PGRN levels accounted for by plasma PGRN, the coefficient of determination (R2 = r2) was used [10]. Age at onset (AAO) was defined as the age of first symptoms noted by a caregiver. For the longitudinal blood withdrawals, repeated-measures ANOVAs were used and variability per individual was assessed by the CV (ratio of the standard deviation to the mean) and compared between the groups with t tests. In the case of longitudinal withdrawals, the median of each individual was used for comparisons of single plasma PGRN values; for plasma-CSF correlations, we used the plasma sample closest in time to the lumbar puncture. Associations between SNP genotypes and PGRN levels were analyzed by multivariate regression with GRNmutation status, age and gender as covariates.
Results
Cohort Characteristics
The baseline characteristics of the cohort are displayed in table 1. Presymptomatic carriers, controls and patients did not differ in age (p = 0.08). Mini-Mental State Examination scores were lower in patients than in at-risk individuals (p < 0.01); no differences were found between controls and presymptomatic carriers (p = 0.51).
Table 1.
Subject characteristics
| Plasma available (n = 64) |
CSF available (n = 35) |
|||
|---|---|---|---|---|
| controls | GRN mutation carriers | controls | GRN mutation carriers | |
| Number (male) | 29 (11) | 35 (12)a | 12 (4) | 23 (9) |
| Number of symptomatic subjects | – | 7 | – | 7 |
| MMSE (range) | 30 (25–30)c | 30 (0–30)b | 30 (28–30) | 29 (18–30) |
| AAO, years (IQR) | – | 59.1 (57.6–65.8)c | – | 57.6 (55.1–65.5)c |
| Age at collection, years (IQR) | 58.6 (51.8–63.5) | 58.1 (53.4–64.3) | 58.1 (50.8–64.0) | 57.4 (54.7–61.0) |
| Time between onset and collection, years (range) | – | 2.3 (–1.2 to 5.3) | – | 2.0 (–1.2 to 5.0) |
| Time between plasma and CSF collection, days (range) | 35 (0–265) | 19 (0–597) | 35 (0–265) | 19 (0–597) |
| PGRN level, ng/ml (range) | 28.5 (21.5–39.2) | 8.0 (5.2–11.3) | 0.76 (0.60–1.25) | 0.29 (0.15–0.46) |
Values are displayed as medians. MMSE = Mini-Mental State Examination; IQR = interquartile range.
Mutations: p.Ser82Valfs, n = 23; p.Gln125*, n = 9; p.Val411Serfs, n = 2; p.Gln130Serfs, n = 1.
Presymptomatic GRN mutation carriers median MMSE score = 30 (range 26–30), symptomatic carriers median MMSE score = 18 (range 0–26).
Only known AAO in 7 patients and 1 presymptomatic carrier who converted after collection.
Plasma PGRN Levels
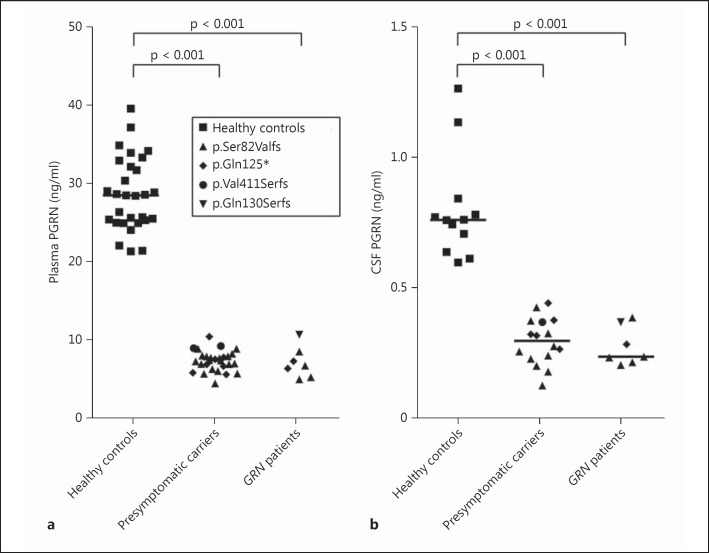
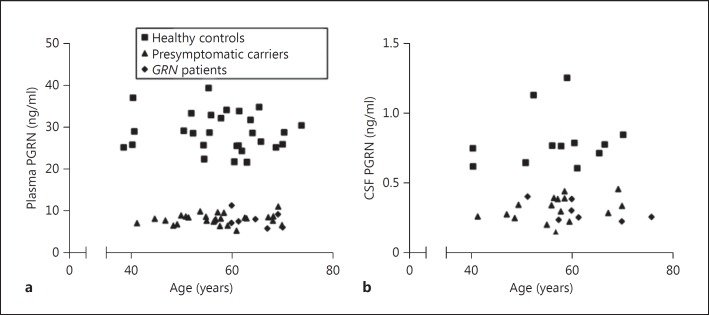
GRNmutation carriers had lower plasma PGRN levels than controls, without any overlap between the groups (p < 0.001; table 1; fig. 1a). Median level in carriers was 28% of that in the controls. Maximum variation between subjects was a factor of 1.8 in controls and 2.2 in carriers (ratio of the highest to the lowest expression per group). PGRN levels did not differ between presymptomatic carriers and patients (p = 0.51). Differences between various mutations did not reach significance (p = 0.06), and limited numbers across the mutations did not allow for post hoc testing. Plasma PGRN did not correlate with age (fig. 2a), gender or AAO.
Fig. 1.
PGRN levels in plasma and CSF. PGRN in plasma from healthy controls (n = 29), presymptomatic GRN mutation carriers (n = 28) and GRNpatients (n = 7) (a) and in CSF from healthy controls (n = 12), presymptomatic GRN mutation carriers (n = 16) and GRN patients (n = 7) (b). Each individual is represented as a data point and labeled by type of mutation. Horizontal lines are median plasma levels per group.
Fig. 2.
PGRN levels as a function of age. Scatterplots of PGRN levels in plasma (a) and CSF (b) by age at collection in healthy controls (squares), presymptomatic GRNmutation carriers (triangles) and GRN patients (diamonds). To prevent disclosure of genetic status, a 23-year-old subject was excluded from the graph (but not from the analyses).
CSF PGRN Levels
Median CSF PGRN in carriers was 39% of that in controls, without overlap of the levels between the groups (p < 0.001; table 1; fig. 1b). In carriers, the lowest expression level of the CSF PGRN was 3.1 times lower than that of the highest expression level; in controls this was a 2.1-fold difference. CSF PGRN did not differ between presymptomatic carriers and patients (p = 0.58), or between various GRN mutations (p = 0.16). CSF PGRN did not correlate with age (fig. 2b), gender or with AAO.
Correlation between Plasma and CSF
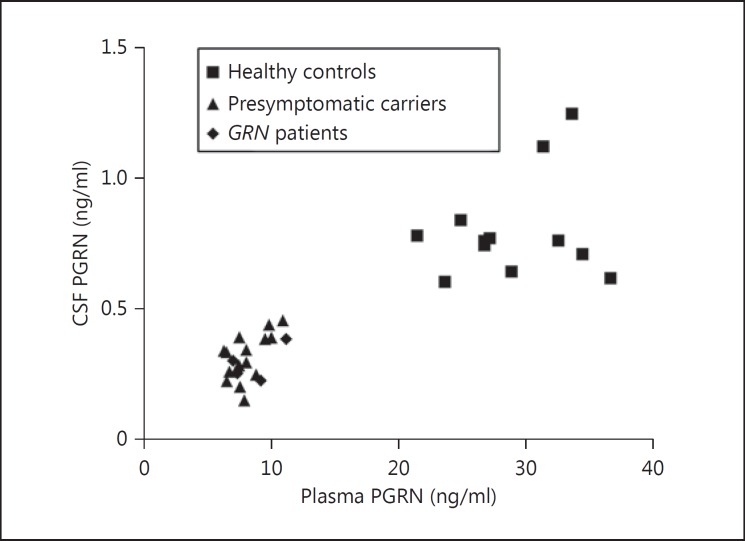
Within the group of subjects with both plasma and CSF samples (16 presymptomatic carriers, 4 GRN patients and 12 controls), PGRN plasma levels correlated significantly with CSF levels (rs = 0.80, p < 0.001; fig. 3). The median interval between blood and CSF collection was 32 days (range 0-597; see table 1). The correlation between PGRN levels in CSF and plasma collected on the same day in 13 subjects (6 presymptomatic carriers, 3 GRNpatients and 4 controls) was similar to the total group (rs = 0.78, p = 0.002). The correlation between PGRN levels in plasma and CSF was weaker when both subgroups were separately analyzed: r = 0.54 (p = 0.02) in carriers and r = 0.21 (p = 0.51) in controls. The coefficient of determination, R2, was 0.29 in carriers; this means that in our group of carriers, 29% of the variability in CSF PGRN levels is explained by the variability in plasma PGRN levels and the remaining 71% are still unaccounted for.
Fig. 3.
Correlation plasma versus CSF PGRN. Scatterplot of the correlation (rs = 0.80, p <0.001) between PGRN levels in plasma and CSF in 12 healthy controls (squares), 16 presymptomatic GRNmutation carriers (triangles) and 4 GRNpatients (diamonds).
Longitudinal Plasma PGRN Levels
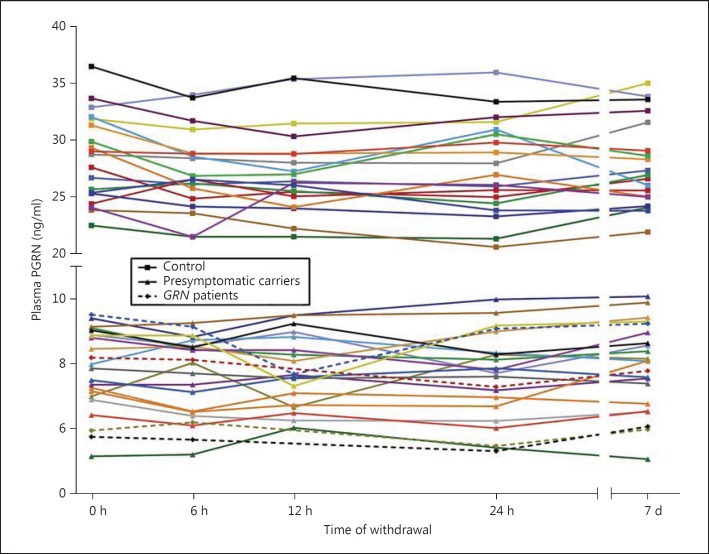
Plasma PGRN levels were found to be stable over 1 week in carriers and controls (fig. 4), with a mean CV in carriers of 5.3% (range 2.3-9.7) and in controls of 4.7% (range 1.4-8.6; not statistically different, p = 0.32). Repeated-measures analyses showed no significant differences over the five time points (p = 0.13 in carriers and p = 0.053 in controls); the trend for a difference in controls was caused by a nonsignificant lower plasma PGRN at 6 versus 0 h (p = 0.09, Bonferroni post hoc test). The fasting PGRN plasma levels (24-hour time point) were not significantly different from other time points (e.g. fasting PGRN level vs. time point 0 h in carriers, p = 0.18, paired t test).
Fig. 4.
Longitudinal plasma PGRN. PGRN levels over time in plasma of healthy controls (n = 19, squares), presymptomatic GRN mutation carriers (n = 18, triangles) and GRN patients (n = 4, diamonds). Each withdrawal is represented with a data point; lines connect data points of each individual. At the 12-hour time point, 3 data points are missing (all GRN patients).
Associations between SNPs and PGRN Levels
The associations of the SNPs with PGRN levels are displayed in online supplementary table 1 and online supplementary figure 2. GRNmutation carriers had lower PGRN plasma levels with each additional minor allele (A) of rs5848 (GRN) without an effect in CSF. In contrast, in the control group an effect was found in the CSF only. The minor allele (G) of rs646776 (near SORT1) was associated with lower PGRN levels in plasma of GRNmutation carriers, without a significant effect in controls or in CSF. For rs1990622 (near TMEM106B), the minor allele (C) was associated with lower plasma PGRN levels in the entire group of subjects; no significant associations were observed for CSF.
Discussion
This study showed that CSF PGRN levels in a large series of presymptomatic GRNmutation carriers were already lower than in age-matched controls. Plasma PGRN levels strongly correlated with CSF levels; however, this was largely explained by the difference in GRNcarrier status and in carriers, only 29% of the variability in CSF PGRN was explained by plasma PGRN. Plasma PGRN levels within subjects fluctuated by 5% over a 1-week period, which has major implications for the clinical trial design aiming at PGRN restoration. Known SNPs only partly explained variation in plasma and CSF PGRN levels between subjects.
The significantly lower plasma PGRN levels in GRNmutation carriers than in healthy controls found in this study, and the similar levels in presymptomatic carriers and patients are in line with previous reports [4,5,8,9,11]. The gap between plasma PGRN levels in our carriers versus noncarriers was even larger and the spread of PGRN levels was smaller than in previous studies [4,5,8]. This might be explained by a standardized collection of samples and the use of a new qualified ELISA assay with strict performance acceptance criteria and quality control samples on each plate. As previously suggested, plasma PGRN can serve as a reliable screening tool for pathogenic GRNmutations in patients with seemingly sporadic FTD or with an unspecified early-onset dementia [4,5,9]. Plasma PGRN can also serve as a rapid ex vivo screening tool in patients with variants of unknown significance in GRN. This identification of pathogenic GRNmutations becomes even more important with forthcoming potential treatments increasing PGRN expression.
PGRN levels in CSF of presymptomatic carriers and patients did not show overlap with controls at all ages, which is in line with earlier reports in small series of GRN patients [4,6]. With the development of PGRN-enhancing therapies, a crucial question remains whether and at what age enhancing PGRN will have an effect on disease course. As this study showed for the first time that CSF PGRN levels are already reduced in the presymptomatic stage, additional biomarkers (clinical, neuroimaging and/or biochemical) are needed to determine disease onset and to track disease progression in therapeutic trials.
The strong correlation between peripheral and CSF PGRN levels contrasts with a weaker correlation found in studies in healthy controls (partial r = 0.17 and r = 0.36) and no significant correlation in sporadic FTD [10,17,18]. This could be explained by the clear dichotomy in PGRN levels between GRNmutation carriers and noncarriers; our observed correlation was mainly driven by GRN genotype, as subgroup analyses by carrier status showed lower correlations. In our opinion, plasma PGRN levels can serve as an easily accessible biomarker to assess target engagement for potentially disease-modifying agents. However, since plasma PGRN levels in carriers explained only 29% of the variability of CSF PGRN levels, plasma PGRN cannot predict CSF PGRN. Moreover, these and previous data suggest a differential regulation of PGRN in plasma/serum versus CSF, and likely the majority of CSF PGRN is synthesized in the central nervous system [10,18,23]; therefore, it could also be that PGRN-enhancing agents have differential effects on different tissues. Although it is unknown whether CSF indeed reflects PGRN levels in the brain and/or interstitial space [18], drug effects could be missed in the worst case if solely relying on plasma PGRN, and CSF sampling will additionally be needed to evaluate effects of pharmacological interventions. The performance of plasma versus CSF PGRN as a surrogate biomarker depends on the place and size of effect of potential PGRN-restoring agents, and remains to be investigated in longitudinal trials.
This first observation of stable PGRN levels over a 1-week period in carriers has important implications for therapeutic trials of pharmacological agents aiming to restore PGRN levels. It is in line with the findings of no significant differences over a longer time period in mainly noncarriers [23]. The mean observed CV over a week of 5% can be technically explained by the assay variability. Plasma PGRN did not alter under fasting conditions, which additionally indicates that plasma PGRN levels can be a suitable biomarker of target engagement. Furthermore, our data can support sample size estimations, greatly facilitating clinical trial design: using intrasubject over intersubject variability significantly reduces trial size in this rare disease.
It was recently published that more commonly used PGRN ELISA kits (Adipogen and R&D) detected CSF PGRN in non-GRNcarriers only at the lower detection range of the kits [24]. Because CSF PGRN of GRN carriers is at least 50% lower, we opted for the more sensitive protocol of Biovendor. R&D and Biovendor yielded similar PGRN concentrations; additionally, Biovendor was qualified to detect low ranges of CSF PGRN (qualified range: 0.018-2.1 ng/ml, data not shown). A limitation is that comparison with studies that used Adipogen and meta-analyses for cutoff levels would be complicated, due to different normality values among the kits.
PGRN levels are known to vary widely among subjects and several factors have been found which partly explain this variability. In this study, neither age at collection, nor gender were correlated with PGRN levels, in contrast to some earlier reports in noncarriers [4,5,10,12,13]. This might be explained by the limited age range and the small sample size. However, in carriers, previous studies did not detect a correlation with age, similar to our result [8,9]. The same holds true for AAO, for which conflicting results have been reported and this study did not find a correlation [5,8,9].
Several SNPs have been found to influence PGRN levels: rs5848 in GRN, rs646776 near SORT1,and rs1990622 near TMEM106B. The correlation between lower PGRN levels and the minor allele of rs5848 in our series is in line with findings from previous studies [10,12,25]. Probably microRNA-659 binds more efficiently to this minor allele, resulting in translational suppression of PGRN [26]. The significantly lower plasma PGRN levels with each minor allele in our GRN mutation carriers suggest that the translational suppression also takes place when there is only one functional GRN allele.
In our GRN mutation carriers and in previous studies, rs646776 (near SORT1) correlated significantly with plasma PGRN levels, but not with PGRN CSF levels [10,13,27]. This SNP is probably a liver-specific regulator of SORT1 and therefore only a peripheral modifier [10,28].
The minor allele (C) in rs1990622 (TMEM106B)was not associated with higher plasma PGRN levels, in contrast to the strong association in GRN patients and healthy controls found in previous studies [11,14]. Such an association was supported by a proposed functional link between TMEM106B and PGRN and by a delay in AAO with each minor allele [11,14,29,30,31]. Our findings might be explained by analysis in a few families resulting in genetic bias. Larger cohorts are needed in order to investigate the exact role of TMEM106B on PGRN and AAO in various genetic backgrounds.
Major strengths of our study include longitudinal plasma collections over a week within the same individuals, uniformly performed by a single investigator and collection protocol. Furthermore, presymptomatic carriers and controls were well matched since they originate from the same families. A methodological weakness of this study is the interval of more than 1 day between plasma and CSF collection in half of the cases. However, a subgroup analysis of samples collected on the same day showed a correlation comparable to that in the entire group. This might be expected given the demonstrated low variability in plasma PGRN levels over time in this study and in CSF PGRN levels recently reported [23]. Future studies are warranted to analyze variations between plasma sampled more than 1 week apart and fluctuations in PGRN CSF over time in GRN mutation carriers. Additionally, the population was too small to get robust conclusions on the effect of the studied SNPs on PGRN levels in GRN mutation carriers and should be studied in larger cohorts.
To conclude, PGRN levels in plasma and CSF were already low in presymptomatic GRNmutation carriers and separated completely from noncarriers. Although PGRN levels in plasma and CSF strongly correlated, plasma PGRN levels only explain 29% of the variability of CSF PGRN levels in GRNmutation carriers, and therefore both blood and CSF sampling is needed in PGRN-enhancing trials. Plasma PGRN levels can serve as biomarker of target engagement for potentially disease-modifying agents addressing PGRN steady state, as they were relatively stable over 1 week. Further research is required to elucidate which other factors are associated with PGRN regulation, with emphasis on the differences between the regulation in plasma and CSF.
Disclosure Statement
H.H. Meeter received support from Alzheimer Nederland (grant No. WE.09-2014-04). H. Patzke and G. Loewen are employees of FORUM Pharmaceuticals Inc. E.G.P. Dopper received support from Dioraphte Foundation grant No. 09-02-03-00, the Association for Frontotemporal Dementias Research Grant 2009, The Netherlands Organization for Scientific Research (NWO) grant HCMI 056-13-018 and Netherlands Alzheimer Foundation. Y.A.L. Pijnenburg, R. van Minkelen and J.C. van Swieten report no disclosures.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
This project was supported by FORUM Pharmaceuticals Inc. (who is developing a PGRN-enhancing therapy) and grants from ‘Nationaal Initiatief Hersenen en Cognitie’, Alzheimer Nederland (grant No. 056-13-018) and Van Leersumfonds (grant No. VLF2013220). We are greatly indebted to Dorothy Flood (FORUM Pharmaceuticals Inc., USA) for critically reviewing the manuscript. We would like to thank all study participants for taking part in this study.
References
- 1.Seelaar H, Rohrer JD, Pijnenburg YAL, Fox NC, Van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J Neurol Neurosurg Psychiatry. 2011;82:476–486. doi: 10.1136/jnnp.2010.212225. [DOI] [PubMed] [Google Scholar]
- 2.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 3.Eriksen JL, Mackenzie IR. Progranulin: normal function and role in neurodegeneration. J Neurochem. 2008;104:287–297. doi: 10.1111/j.1471-4159.2007.04968.x. [DOI] [PubMed] [Google Scholar]
- 4.Ghidoni R, Benussi L, Glionna M, Franzoni M, Binetti G. Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology. 2008;71:1235–1239. doi: 10.1212/01.wnl.0000325058.10218.fc. [DOI] [PubMed] [Google Scholar]
- 5.Finch N, Baker M, Crook R, Swanson K, Kuntz K, Surtees R, et al. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009;132:583–591. doi: 10.1093/brain/awn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme P, Van Hoecke A, Lambrechts D, Vanacker P, Bogaert E, Van Swieten J, et al. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol. 2008;181:37–41. doi: 10.1083/jcb.200712039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci. 2014;37:388–398. doi: 10.1016/j.tins.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Sleegers K, Brouwers N, Van Damme P, Engelborghs S, Gijselinck I, Van der Zee J, et al. Serum biomarker for progranulin-associated frontotemporal lobar degeneration. Ann Neurol. 2009;65:603–609. doi: 10.1002/ana.21621. [DOI] [PubMed] [Google Scholar]
- 9.Ghidoni R, Stoppani E, Rossi G, Piccoli E, Albertini V, Paterlini A, et al. Optimal plasma progranulin cutoff value for predicting null progranulin mutations in neurodegenerative diseases: a multicenter Italian study. Neurodegener Dis. 2012;9:121–127. doi: 10.1159/000333132. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson AM, Finch NA, Thomas CS, Wojtas A, Rutherford NJ, Mielke MM, et al. Progranulin protein levels are differently regulated in plasma and CSF. Neurology. 2014;82:1871–1878. doi: 10.1212/WNL.0000000000000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruchaga C, Graff C, Chiang H, Wang J, Hinrichs AL, Spiegel N, et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiung GYR, Fok A, Feldman HH, Rademakers R, MacKenzie IRA. rs5848 polymorphism and serum progranulin level. J Neurol Sci. 2011;300:28–32. doi: 10.1016/j.jns.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, et al. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87:890–897. doi: 10.1016/j.ajhg.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cenik B, Sephton CF, Dewey CM, Xian X, Wei S, Yu K, et al. Suberoylanilide hydroxamic acid (vorinostat) up-regulates progranulin transcription: rational therapeutic approach to frontotemporal dementia. J Biol Chem. 2011;286:16101–16108. doi: 10.1074/jbc.M110.193433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capell A, Liebscher S, Fellerer K, Brouwers N, Willem M, Lammich S, et al. Rescue of progranulin deficiency associated with frontotemporal lobar degeneration by alkalizing reagents and inhibition of vacuolar ATPase. J Neurosci. 2011;31:1885–1894. doi: 10.1523/JNEUROSCI.5757-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morenas-Rodríguez E, Cervera-Carles L, Vilaplana E, Alcolea D, Carmona-Iragui M, Dols-Icardo O, et al. Progranulin protein levels in cerebrospinal fluid in primary neurodegenerative dementias. J Alzheimers Dis. 2015;50:1–8. doi: 10.3233/JAD-150746. [DOI] [PubMed] [Google Scholar]
- 18.Wilke C, Gillardon F, Deuschle C, Dubois E, Hobert MA, Müller J, et al. Serum levels of progranulin do not reflect cerebrospinal fluid levels in neurodegenerative disease. Curr Alzheimer Res. 2016;13:654–662. doi: 10.2174/1567205013666160314151247. [DOI] [PubMed] [Google Scholar]
- 19.Dopper EGP, Rombouts SARB, Jiskoot LC, den Heijer T, De Graaf JR, de Koning I, et al. Structural and functional brain connectivity in presymptomatic familial frontotemporal dementia. Neurology. 2014;83:e19–e26. doi: 10.1212/WNL.0000000000000583. [DOI] [PubMed] [Google Scholar]
- 20.Seelaar H, Kamphorst W, Rosso SM, Azmani A, Masdjedi R, de Koning I, et al. Distinct genetic forms of frontotemporal dementia. Neurology. 2008;71:1220–1226. doi: 10.1212/01.wnl.0000319702.37497.72. [DOI] [PubMed] [Google Scholar]
- 21.Rosso SM, Kaat LD, Baks T, Joosse M, de Koning I, Pijnenburg Y, et al. Frontotemporal dementia in the Netherlands: patient characteristics and prevalence estimates from a population-based study. Brain. 2003;126:2016–2022. doi: 10.1093/brain/awg204. [DOI] [PubMed] [Google Scholar]
- 22.Van Der Flier WM, Pijnenburg YAL, Prins N, Lemstra AW, Bouwman FH, Teunissen CE, et al. Optimizing patient care and research: the Amsterdam dementia cohort. J Alzheimers Dis. 2014;41:313–327. doi: 10.3233/JAD-132306. [DOI] [PubMed] [Google Scholar]
- 23.Feneberg E, Steinacker P, Volk AE, Weishaupt JH, Wollmer MA, Boxer A, et al. Progranulin as a candidate biomarker for therapeutic trial in patients with ALS and FTLD. J Neural Transm. 2016;123:289–296. doi: 10.1007/s00702-015-1486-1. [DOI] [PubMed] [Google Scholar]
- 24.Willemse EAJ, Durieux-Lu S, Van der Flier WM, Pijnenburg YAL, de Jonge R, Teunissen CE. Stability of progranulin under pre-analytical conditions in serum and cerebrospinal fluid. J Alzheimers Dis. 2016;53:107–116. doi: 10.3233/JAD-160061. [DOI] [PubMed] [Google Scholar]
- 25.Vercellino M, Fenoglio C, Galimberti D, Mattioda A, Chiavazza C, Binello E, et al. Progranulin genetic polymorphisms influence progression of disability and relapse recovery in multiple sclerosis. Mult Scler. 2015 doi: 10.1177/1352458515610646. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 26.Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu F, Padukkavidana T, Vægter CB, Brady OA, Zheng Y, Mackenzie IR, et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brady OA, Zheng Y, Murphy K, Huang M, Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet. 2013;22:685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB, Castanedes-Casey M, et al. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013;126:781–791. doi: 10.1111/jnc.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data