Abstract
Using a combination of array comparative genomic hybridization, mate pair and cloned sequences, and FISH analyses, we have identified in multiple myeloma cell lines and tumors a novel and recurrent type of genomic rearrangement, i.e. interchromosomal rearrangements (translocations or insertions) and intrachromosomal inversions that contain long (1–4000 kb; median ∼100 kb) identical sequences adjacent to both reciprocal breakpoint junctions. These duplicated sequences were generated from sequences immediately adjacent to the breakpoint from at least one—but sometimes both—chromosomal donor site(s). Tandem duplications had a similar size distribution suggesting the possibility of a shared mechanism for generating duplicated sequences at breakpoints. Although about 25% of apparent secondary rearrangements contained these duplications, primary IGH translocations rarely, if ever, had large duplications at breakpoint junctions. Significantly, these duplications often contain super-enhancers and/or oncogenes (e.g. MYC) that are dysregulated by rearrangements during tumor progression. We also found that long identical sequences often were identified at both reciprocal breakpoint junctions in six of eight other tumor types. Finally, we have been unable to find reports of similar kinds of rearrangements in wild-type or mutant prokaryotes or lower eukaryotes such as yeast.
INTRODUCTION
Multiple myeloma (MM), an age-dependent monoclonal bone marrow plasma cell tumor, mostly—if not always—is preceded by a premalignant monoclonal gammopathy of uncertain significance (MGUS) tumor (1). The sporadic progression of MGUS to MM occurs at an average rate of ∼1% per year. This transition is associated with increased expression of MYC, which sometimes is a consequence of secondary genomic rearrangements that reposition MYC near one of a promiscuous array of super-enhancers (SE) (2–7).
MM and MGUS tumor cells are similar to long-lived, post-germinal center bone marrow plasma cells, which have undergone three kinds of B cell-specific DNA modification events: V(D)J recombination (VDJR), somatic hypermutation (SHM) and IGH class switch recombination (CSR) (8). The pathogenesis of MGUS and MM appears to involve two major pathways: hyperdiploidy or primary IGH translocations (9,10). Hyperdipoid (HRD) tumors typically have 48–60 chromosomes, with multiple trisomies or tetrasomies involving eight chromosomes (3,5,7,9,11,15,19,21). Primary IGH translocations, which mostly are present in non-hyperdiploid (NHRD) tumors, simultaneously dysregulate MMSET (WHSCI) and FGFR3, or one of three CYCLIN D genes or one of three MAF genes (1,9). These mostly balanced translocations, which position an oncogene under control of IGH intronic (Emu) and/or 3′ IGH (Eα1; Eα2) SE, are thought to occur mostly as a result of errors in one of the three B cell-specific DNA modifications (11–13).
Secondary rearrangements, which have a similar prevalence in HRD and NHRD tumors, represent progression events that do not involve the B cell-specific DNA modification mechanisms, which seem to be inactive in MGUS and MM tumors (5). Secondary rearrangements include: IGH rearrangements (rarely involving the primary IGH translocation partners), most if not all IGK and IGL rearrangements, MYC rearrangements with or without IG involvement, and a promiscuous array of other chromosomal regions (5,14–16). In contrast to primary IGH translocations, secondary rearrangements usually are complex translocations or insertions, which often are unbalanced and can involve three or more chromosomes. Moreover, the secondary rearrangements in MM have structural features that are similar to the complex unbalanced rearrangements that have been identified in many kinds of solid tumors (17–19). The mechanisms generating these rearrangements are poorly understood
In the present manuscript we show the frequent occurrence of translocations, insertions and inversions that contain long identical sequences adjacent to both reciprocal breakpoint junctions in MM cell lines and primary tumors. The duplicated sequences were generated from sequences located at the breakpoint from at least one—but often both—chromosomal donor sites. We also found that this novel category of rearrangements were present in six of eight other tumor types that have reciprocal rearrangements.
MATERIALS AND METHODS
Mate pair libraries
The Illumina Mate Pair Library Preparation kit was used for library construction following the manufacturer's instructions and two samples were run on one lane of an Illumina HiSeq2000 with 50bp reads. The sequences were aligned to hg19 using BWA and a BAM file containing only the clustered discordant reads with clustered mates was created. Breakpoints with more then five discordant reads were identified, as described previously (4).
Analysis of mate pair libraries
Samtools was used to convert BAM files to SAM files. Breakpoints were divided into four groups, corresponding to four different kinds of rearrangements: (i) deletions, (ii) tandem duplications, (iii) intrachromosomal inversions, (iv) interchromosomal rearrangements. For the last two groups all non-reciprocal rearrangements were excluded. The remaining rearrangements were compared to 69 normal individual Whole Genome Sequences (CGI) to eliminate germline structural variations (www.completegenomics.com/sequence-data/download-data/). In addition, reciprocal rearrangements that were present in at least one other mate pair library prepared for 22 MMCL were also excluded. The remaining reciprocal rearrangements were analyzed to identify the presence of duplicated sequences at both breakpoint junctions (see Figure 1 for an example).
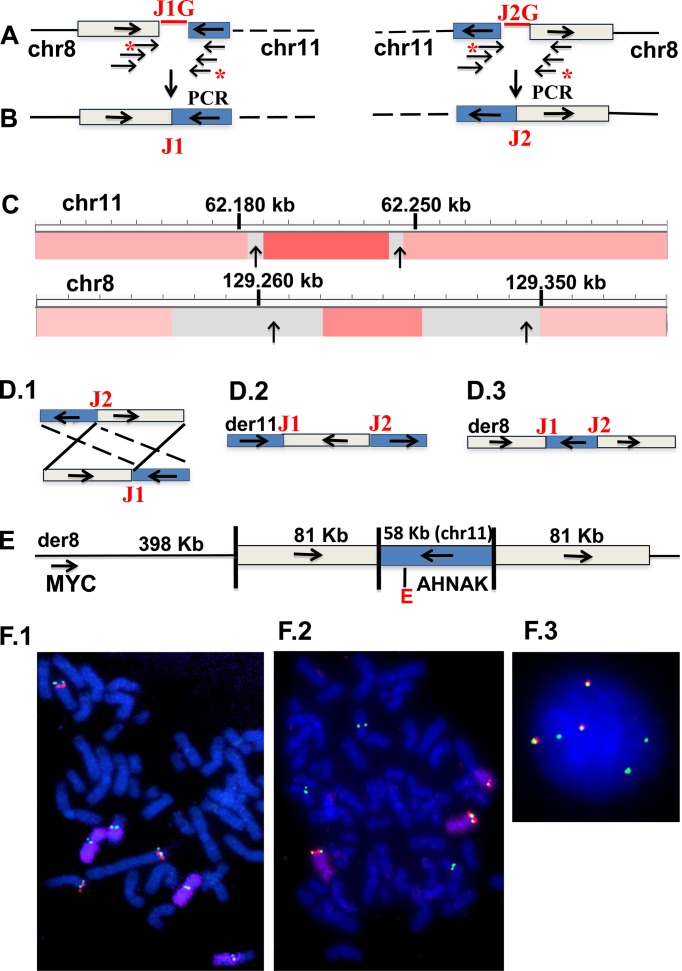
Figure 1.
Characterization of interchromosomal MYC locus rearrangement in EJM MMCL. Horizontal arrows indicate reference genome orientations of chromosomal regions. (A) Paired mate pair sequences are read from opposite ends of ∼2.5 kb fragments that encompass the breakpoint junctions. Discordant mate pair sequences identify a reciprocal chr8:chr11 rearrangement, with the sequences marked by asterisks closest to the breakpoint, but with gaps (J1G, J2G) of uncertain size; (B) oligonucleotides flanking the two sequences with asterisks generated a PCR product that defines the precise breakpoint junctions (J1, J2) and sizes of apparent duplicated sequences (boxed); (C) agilent 244K CGH with copy number and cloned breakpoints (vertical arrows) shown in the Integrative Genome Viewer (IGV) for chr8 and chr11 (note that gray indicates regions deficient in probes); (D) D.1, 2, 3 depict three possible structures: reciprocal translocation, insertion of chr8 into chr11, or insertion of chr11 into chr8; (E) FISH results (F) confirm the D.3 structure, as schematically summarized; (F.1) metaphase FISH (MYC, green; chr11 probe (G248P8668H1) [Supplementary Table S5], red; chr11 whole chromosome paint (wcp), purple; F.2 metaphase FISH (same chr 11 probe, green; chr8 duplication probe (G248P89052F10), red; chr8 wcp, purple; F.3 interphase FISH with same probes as in F.2 but without chr8wcp.
Identifying potential copy number gains at chromosomal breakpoints
Apparent reciprocal breakpoint junctions involving two chromosomes or distant sites on one chromosome were identified. For each chromosome or chromosomal site in these junctions, the breakpoint position on the antisense strand (−) is subtracted from the breakpoint position on the sense strand (+). Negative and positive results, respectively, indicate a possible copy number loss (CNL) or a possible copy number gain (CNG), which in some cases actually represents an inserted sequence. An example of this analysis is shown in Figure 1. We have defined CNG or CNL as each being >1 kb and <4 Mb (Tables 1–3). Note that breakpoints with CNG (CNL) determined from mate pair libraries can be significantly larger (smaller) than the calculated size, so that the number of breakpoints with CNG (CNL) is an underestimate (overestimate), e.g. Tables 1 and 2, and corresponding text.
Table 1. Tandem duplications, reciprocal interchromosomal rearrangements and reciprocal inversions in eight MMCLs.
| MMCL | TD | Interchromosomal rearrangements | Inversions | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | CNG × 2 | CNG × 1 | CNG + CNL | CNL × 2 | Total | CNG × 2 | CNG × 1 | ||
| AMO | 9 | 10 | 1 | ||||||
| INA6 | 5 | 1 | 1 | ||||||
| JIM3 | 72 | 17 | 1 | 5 | 1 | ||||
| JJN3 | 29 | 7 | 4 | 1 | 1 | 2 | 1 | ||
| KP6 | 8 | 4 | 4 | ||||||
| MOLP8 | 12 | 10 | 2 | 1 | 1 | ||||
| XG2 | 21 | 7 | 3 | 3 | 1 | 1 | |||
| XG6 | 6 | 5 | 1 | 2 | |||||
| TOTAL | 162 | 61 | 14 | 2 | 2 | 2 | 13 | 3 | 1 |
TD = tandem duplication.
CNG = copy number gain; CNL = copy number loss.
CNG × 2 = insertion with flanking duplicated sequences OR translocation with duplicated sequences from both donor breakpoints at both breakpoint junctions.
Table 3. Reciprocal interchromosomal breakpoints in 140 tumorsa.
| Tumor | # | Recipr rearr | Small CNAb | Large CNAc | CNG × 1 | CNG × 2 | CNG + CNL | CNL × 1 | CNL × 2 |
|---|---|---|---|---|---|---|---|---|---|
| BRCA | 35 | 68 | 10 | 6 | 17 | 22 | 2 | 7 | 4 |
| CRC | 14 | 8 | 2 | 2 | 1 | 1 | 1 | 1 | |
| GBM | 16 | 14 | 4 | 3 | 5 | 2 | |||
| HCC | 19 | 16 | 1 | 3 | 3 | 9 | |||
| KIRC | 4 | ||||||||
| LUSC | 19 | 21 | 6 | 5 | 3 | 5 | 2 | ||
| MM | 7 | 6 | 3 | 1 | 2 | ||||
| OV | 9 | 13 | 1 | 1 | 4 | 4 | 1 | 1 | 1 |
| PR | 7 | 9 | 3 | 1 | 1 | 3 | 1 | ||
| UCEC | 10 | 12 | 1 | 1 | 2 | 8 | |||
| ALL | 140 | 167 | 28 | 25 | 31 | 55 | 7 | 12 | 9 |
aAnalysis of data in Supplementary Table S3, Yang et al., Cell 153(2013), p.919.
bLess than 1 kb CNG or CNL at breakpoints for both chromosomes.
cGreater than 4 Mb CNG or CNL at breakpoints for at least one chromosome.
CNG or CNL in other five groups are >1 kb and <4 Mb.
Table 2. Reciprocal interchromosomal rearrangements in 25 MM tumors.
| Tumor | Primary IGH | Other interchromosomal rearrangements | |||||
|---|---|---|---|---|---|---|---|
| samples | Total | CNG × 2 | CNG × 1 | CNG + CNL | CNL × 1 | CNL × 2 | |
| A1855 | 11;14 | 0 | |||||
| A1864 | 11:14(der14) | 0 | |||||
| A1868 | 8 | 1 | 1 | 1 | 4 | ||
| A1886 | 1 | 1 | |||||
| A1892 | 1 | 1 | |||||
| A1898 | 4 | (2)a | 1 | 1 | |||
| A1904 | 2 | 2 | |||||
| A1910 | 2 | 2 | |||||
| A1916 | 2 | 1 | 1 | ||||
| A1922 | 1 | 1 | |||||
| A1928 | 6 | 2 | 1 | 3 | |||
| A1934 | 11;14(der14) | 0 | |||||
| A1940 | 2 | 2 | |||||
| A1946 | 3 | 1 | 2 | ||||
| A1952 | 11;14(der14) | 0 | |||||
| A1958 | 14;16 | 2 | |||||
| A1964 | 11;14 | 0 | |||||
| A1970 | 1 | 1 | |||||
| A1976 | 4;14 | 1 | 1 | ||||
| A1982 | 11;14(der14) | 0 | |||||
| A1988 | 1 | 1 | |||||
| A1994 | 11;14 | 0 | |||||
| A2066 | 11;14 | 2 | 2 | ||||
| A2070 | 11;14 | 2 | 1 | 1 | |||
| A2079 | 4;14 | 1 | 1 | ||||
| TOTAL | 42 | 13(15) | 5 | 13 | 6 | 0 | |
CNG (CNL) = copy number gain (loss) >1 kb and <4 Mb.
CNGX2 = insertion with flanking duplicated sequences OR translocation with sequences from both chromosome breakpoints at both breakpoint junctions.
aTwo rearrangements with CNG > 4 Mb included in parentheses.
Cell culture, FISH analyses, comparative genomic hybridization and enhancer predictions have been described previously, with references and additional details provided in Supplementary Methods.
RESULTS
MYC locus gains often have duplicated sequences at rearrangement breakpoints
Figure 1 shows how mate pair analyses, polymerase chain reaction (PCR) product breakpoint sequences, comparative genomic hybridization (CGH) and FISH experiments were used to determine the structure of an 8;11 interchromosomal rearrangement in the EJM MM cell line (MMCL). Mate pair sequences identified the approximate locations of two breakpoints (JG1, JG2) (Figure 1A). Flanking oligonucleotides enabled PCR products to identify the precise breakpoints (Figure 1B): J1, chr8:129,346,425(+);chr11:62,245,125(+) and J2, chr8:129,265,218(−);chr11:62,187,330(−) [(+) and (−) indicate, respectively, forward and reverse orientations (compared to the reference genome) of sequences going toward the breakpoint, as indicated by the horizontal arrows on the chromosomes]. CGH analyses (Figure 1C) showed CNG on chr8 and on chr11, which are consistent with CNG predicted from the breakpoint sequences. These results suggested three possible structures (Figure 1D): D.1, t(8;11) reciprocal translocation; D.2, insertion of chr8 fragment into chr11; D.3, insertion of chr11 fragment into chr8. However, FISH analyses (Figure 1F) showed that the chr11 and chr8 probes from the regions of CNG co-localized on chr8, with no detectable chr11 whole chromosome paint (wcp) sequences near the co-localized probe. All together these results are consistent only with D.3, i.e. insertion of a 58-kb chr11 (AHNAK enhancer) fragment between flanking 81-kb chr8 duplicated sequences located ∼500 kb downstream of MYC (Figure 1E).
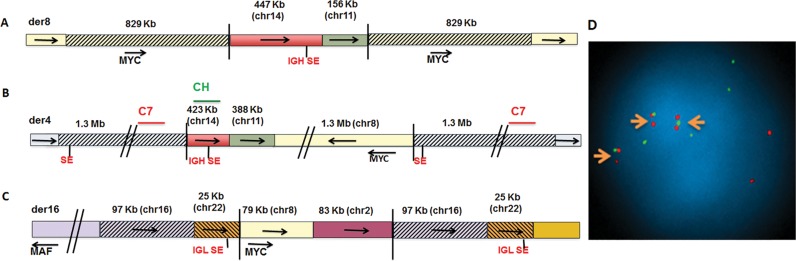
Six additional examples of MYC locus rearrangements (four more complex insertions, one translocation and one inversion), all of which have duplicated sequences at breakpoint junctions, are shown in Figure 2 and Supplementary Figure S1. The characterization of these rearrangements was similar to the results described above for EJM, with FISH data having been reported previously in some cases (5,20). The KMS-12 MMCL has an insertion of 447-kb chr14 (containing an 3′IGH/Ealpha2 SE) and 156-kb chr11 sequences (not including CYCLIN D1) between flanking 829-kb duplicated chr8 sequences that include MYC (Figure 2A). There was a CNG not only of the flanking regions on chr8, but also of the inserted sequences from both chr11 and chr14 (Supplementary Figure S2 and Table S1). Given that KMS-12 has a balanced primary t(11;14) and that mate pair analysis showed only one chr14;chr11 breakpoint (Supplementary Table S1), these results indicate that the inserted chr14;chr11 breakpoint had been derived from a der14 t(11;14) chromosome, which had CYCLIN D1 positioned about 370-kb downstream of an 3′ IGH SE. The MM-M1 MMCL is similar to KMS-12 in having 229-kb chr11 and 652-kb chr14 (IGH SE) sequences inserted between flanking ∼1 Mb chr8 (MYC) duplicated sequences (Supplementary Figures S1A and 2; Table S1). The MOLP8 MMCL also is similar to KMS-12 but more complex with 423-kb chr14 (IGH SE), 388-kb chr11 sequences and 1.3 Mb chr8 (MYC) sequences inserted between flanking 1.3 Mb chr4 duplicated sequences (Figure 2B and D; Supplementary Figure S2 and Table S1). Curiously there is an SE on chr4 between the TBC1DA and KLF3 genes that is closer to MYC than the IGH SE (Supplementary Table S2). The RPMI 8226 MMCL (Figure 2C and Supplementary Table S1) has an insertion of 79-kb chr8 (MYC) and 83-kb chr2 sequences between flanking duplicated sequences containing 97 kb of chr16 and 25 kb of chr22 (3′IGL SE) on der16 t(16;22), so that MYC and MAF can be dysregulated by separate 3′ IGL SE (5,20). Presumably the rearrangement involving MYC occurred on a der16 t(16;22) on which MAF was dysregulated by the IGL SE.
Figure 2.
Examples of complex MYC locus rearrangements in three MMCL. (A) Insertion of chr11 (not including CCND1) and chr14 (IG SE) sequences between flanking duplicated sequences (cross-hatched) on chr8 in KMS-12 MMCL; (B) Insertion of chr8, 11 and 14 sequences between flanking duplicated sequences on chr4 in MOLP-8 MMCL; (C) Insertion of chr8 and chr2 sequences between flanking duplicated sequences containing chr22 and chr16 sequences from a der(16)t(16;22) translocation in 8226 MMCL; (D) Interphase FISH showing insertion of chr14 CH (IGH SE) probe (green) between duplicated chr4 sequences (C7 probe [Supplementary Table S5], red).
Complex translocation products in the Karpas 620 MMCL (Supplementary Figure S1B and Table S1) appear to be the result of a translocation between chr8 and der14 of a t(11;14) translocation (21). The resultant der14 and der8 each contain 828-kb chr8 fragments (MYC), and also 475-kb chr14 (IGH SE) and 229-kb chr11 fragments that appear to have been contributed by the der14 from a primary t(11;14) translocation.
Previously, we found that nearly one third of MYC rearrangements in MM involve large deletions that reposition MYC near the NSMCE2 intron 4 SE, which is ∼2.3 Mb centromeric to MYC, e.g. H929 MMCL and MMRC0156 MM tumor shown in Supplementary Figure S2 (4). However, the KMS-34 MMCL has a 2.3 Mb inversion that repositions MYC near the NSMCE2 SE (Supplementary Figure S1C). This inversion included duplicated sequences shared by the two breakpoints, 24 kb from sequences telomeric to the NSMCE SE and 18 kb from sequences telomeric to MYC (Supplementary Table S1). The duplicated sequences at the inversion breakpoints are too small to be detected by 244K array CGH (Supplementary Figure S2). However, this result suggests that segmental CNGs of sequences including the NSMCE2 SE and sequences centromeric to MYC, which were identified by CGH in the MMRC0167 tumor, may be the consequence of an inversion similar to that seen in KMS34, but with the duplicated sequences large enough to be detected by CGH (Supplementary Figure S2).
Duplications also occur at rearrangement breakpoints that do not involve MYC
We analyzed the results of mate pair sequences and CGH for eight MMCLs, and did FISH or SKY analyses for some rearrangements since mate pair and CGH data cannot distinguish insertions and translocations. This enabled us to identify and characterize interchromosomal insertions and translocations, and also inversions that did not involve MYC but that contained large duplicated sequences at breakpoint junctions (Figure 3; Supplementary Figure S3 and Table S2).
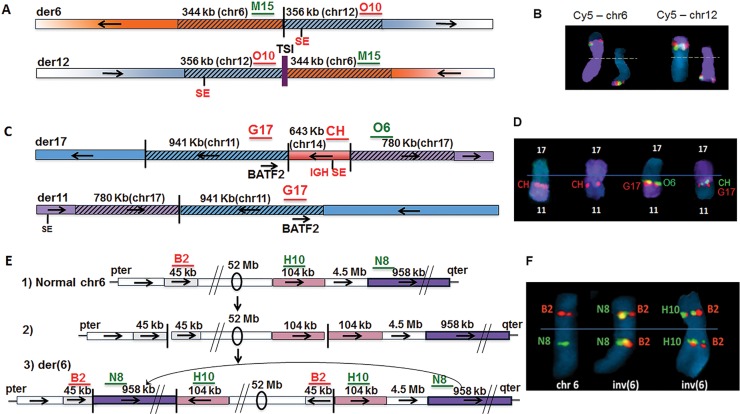
Figure 3.
Complex interchromosomal non-MYC locus rearrangements in three MMCL. (A) t(6;12) with duplications on both derivative chromosomes and insertion of chr1 TSI on der(12) in KP6 MMCL; (B) Metaphase FISH of t(6;12) in KP6, with localization of chr6 probe (M15, green signal) and chr12 probe (O10, red signal) on der(6) and der (12), with chr6 and chr12WCP probes (purple), respectively, on the left and right; (C) t(11;17) with duplicated sequences from chr11 (including BATF2) and chr17 breakpoints at der11 and der17 breakpoint junctions, and insertion of chr14 segment including 3′ IGH SE on der17 in MOLP8 MMCL; (D) Metaphase FISH showing localization of probes on der17; data not shown for der11; (E) A total of 52 Mb chr6 inversion with duplicated breakpoint sequences, and insertion of distant sequences from 6q at the 6p breakpoint junction in JJN3 MMCL: (1) anatomy of normal chr6, with B2, N8, H10 indicating the approximate position of FISH probes; (2) hypothetical inversion precursor without insertion; (3) observed inversion with insertion; (F) Metaphase FISH showing chr6 and inv(6) with probes for relevant sequences (B2, H10, N8). Probe details in Supplementary Table S5. Horizontal arrows indicate orientation of chromosomal segments.
The XG-2 MMCL had two insertions with large duplicated sequences flanking the insertions: (i) a 40-kb chr20 fragment containing the CD40 gene is inserted between flanking 25-kb chr22 duplicated sequences, both of which include the IGL SE (Supplementary Figure S3A) (15); and (ii) a 906-kb chr22 fragment (IGL SE) was inserted between flanking 594-kb chr20 duplicated sequences, both of which contain the MAFB gene (Supplementary Figure S3B) (5).
We identified three MMCLs with translocations not involving MYC that had large duplicated sequences from both chromosomes at the breakpoints. First, the XG-2 MMCL had a t(12;14) translocation, with both der12 and der14 containing a 34-kb fragment from chr12 and a 37-kb fragment including the IGH SE from chr14 (Supplementary Figure S3C) (5). Somewhat surprisingly, we have been unable to identify dysregulation of a gene on chr12 as a consequence of the presence of the IGH SE at both breakpoints. Second, the KP6 MMCL has a t(6;12) translocation, with both der6 and der12 containing a 344-kb chr6 fragment and a 356-kb chr12 fragment at the translocation breakpoint junctions (Figure 3A and B). As reported previously (22), der12 also had an 85 bp chr1 ‘templated sequence insertion’ (TSI), which was hypothesized to have been derived from an RNA template, located between the chr6 and chr12 breakpoints. Both duplicated regions contain SEs, and most notably the TXNDC5 associated SEs that recurrently are involved in MYC rearrangements in MM (4). However, we have been unable to identify any target genes that are dysregulated by these SEs. Finally, the MOLP8 MMCL has a complex rearrangement, which includes a t (11;17) translocation with both der11 and der17 containing a 941-kb chr11 fragment (BATF2 gene) and a 780-kb chr17 fragment at the respective breakpoints; there is also insertion of a 643-kb chr14 fragment (IGH SE) at the 11;17 junction on der 17 (Figure 3C and D). The expression of BATF2 RNA was much higher in MOLP8 compared to most MMCL (data not shown), which seems likely to be a consequence of the IGH SE localized near BATF2 on der17.
Two MMCLs had inversions that did not involve MYC but had duplicated sequences at both breakpoints. First, the JIM3 MMCL (Supplementary Figure S3D and Table S2) had ∼3.6 Mb chr20 inversion, which included duplicated sequences shared by the two breakpoints, i.e. 2788 and 106 kb, respectively, from the centromeric and telomeric ends of the inversion. Second, the JJN3 MMCL (Figure 3E and F) had an ∼59 Mb inversion on chr6, which included duplicated sequences shared by the two breakpoints, i.e. 45 kb from the p-arm and 104 kb from the q-arm. This complex inversion also included insertion of a 958-kb fragment, which was derived from sequences distal to the 6q breakpoint, between the inverted ends on the p-arm.
Prevalence of breakpoint junctions with duplicated sequences in eight MMCL
To better define the number and fraction of interchromosomal rearrangements and inversions that had large duplicated sequences at breakpoint junctions, we analyzed mate pair sequence data for eight MMCLs (Table 1). The data include non-polymorphic reciprocal breakpoint junctions, which identified apparent CNG that could represent either inserted sequences or duplicated breakpoint sequences. Among the eight MMCLs, there were a total of 62 reciprocal interchromosomal rearrangements, with 14 having apparent CNG for both donor chromosomes (CNG × 2) at the breakpoint junctions. Therefore, at least 23% of these interchromosomal rearrangements appear to be either: (i) translocations containing sequences adjacent to the breakpoints on both donor chromosomes that are present at the breakpoint junctions on both derivative chromosomes; or (ii) insertions that are flanked by duplicated sequences on the recipient chromosome. There were a total of 13 inversions, with four (31%) having duplicated sequences at one or both breakpoint junctions, so that the relative frequency of duplicated breakpoint junction sequences is similar for both interchromosomal rearrangements and intrachromosomal inversions. These same eight MMCLs also have a total of 162 large non-polymorphic tandem duplications, which were identified from the mate pair sequence data and confirmed by the 244K CGH analyses.
Prevalence of interchromosomal breakpoint junctions with duplicated sequences in 25 MM tumors
Analysis of 2.5 kb mate pair libraries for 25 primary MM tumors from untreated patients identified 42 (54 including 12 primary IGH translocations) non-polymorphic reciprocal interchromosomal breakpoint junctions (Table 2 and Supplementary Table S3). Thirteen (∼30%) of the 42 rearrangements, which were present in 10 of the 25 MM tumors, had large sequences (1–4000 kb) of apparent CNG for both chromosomes at the breakpoint junctions. Therefore, the occurrence of duplications at interchromosomal breakpoint junctions appears to have a similar prevalence in MMCL and untreated primary MM tumors.
Identification of interchromosomal breakpoint junction with duplicated sequences in other tumors
Published whole genome sequence data for 140 tumors representing ten different types of tumors were analyzed in order to identify apparent CNG and CNL at interchromosomal breakpoint junctions (Table 3 and Supplementary Table S4A and B). About 55 of 167 (33%) reciprocal interchromosomal breakpoint junctions, had an apparent CNG (1–4000 kb) from both parental chromosomes at both breakpoint junctions, including all six non-MM tumor types that had ten or more reciprocal interchromosomal rearrangements. To confirm the predicted CNGs from breakpoints (‘Materials and Methods’ section), we generated copy number variation from the WGS data (Supplementary Methods) for 35 breast cancer tumors. This data confirmed that most predicted CNGs >50 kb do have the expected CNG (Supplementary Table S4A).
DISCUSSION
For MMCL and untreated primary MM tumors, we found that approximately one-third of secondary interchromosomal rearrangements (translocations or insertions) and inversions had reciprocal breakpoint junctions containing large CNG (1–4000 kb; average ∼400 kb; median ∼100 kb) contributed by at least one—but sometimes both—donor chromosome breakpoint(s) (Tables 1 and 2; Supplementary Tables S2 and 3). Tandem duplications in MMCL had a similar size distribution (Supplementary Figure S4), with two specific examples involving the MYC locus in the KP6 and XG2 MMCL (Supplementary Figure S2 and Table S1). In addition, we identified more complex rearrangements involving the insertion of two independent fragments between flanking duplicated sequences, or insertion of a fragment into inversion or interchromosomal translocation breakpoint junctions that contained large duplicated sequences. Given that a similar fraction of interchromosomal rearrangements in six other tumor types examined had similarly large identical sequences at reciprocal breakpoint junctions (Table 3; Supplementary Table S4A and B), it appears that the mechanism(s) generating the breakpoint duplications are active in a significant fraction of most kinds of malignant tumors.
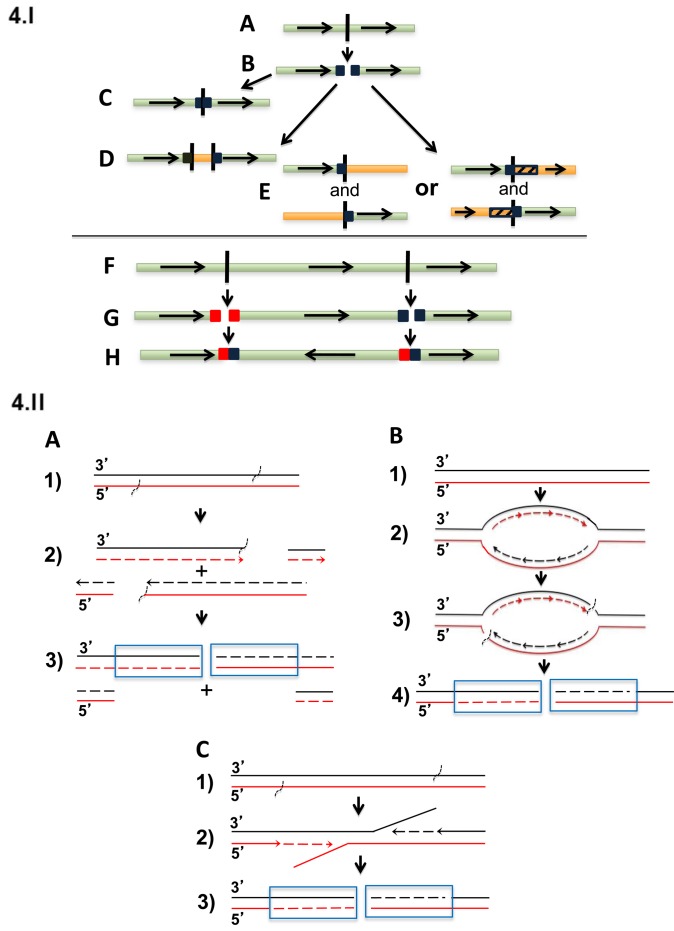
One possible model for generating large duplicated sequences at the various types of rearrangement junctions is shown in Figure 4.I. This model suggests that large duplicated sequences sometimes are present on both sides of a double-stranded breakpoint (see below for possible mechanisms). The break could then be repaired to generate: (i) a head-to-tail tandem duplication; (ii) insertion of an unrelated DNA segment between the duplicated sequences; (iii) a reciprocal translocation involving another chromosome or chromosome region that may or may not also have duplicated sequences at both ends of its breakpoint; or (iv) an inversion.
Figure 4.
Model and possible mechanisms for generation of breakpoint duplications. (I) Four possible consequences of double stranded break with duplicated sequences on both sides of the breakpoint. (A) germline sequence; (B) generation of duplicated sequences on both sides of a breakpoint; repair generating (C) tandem head-to-tail duplication; (D) insertion of an unrelated DNA segment between the duplicated sequences; (E) translocation involving another chromosome or chromosome region that may also have duplicated sequences at both ends of its breakpoint; (F) second double strand break on same chromosome; (G) generation of duplicated sequences on both sides of the two breakpoints; (H) inversion of segment between two breakpoints, with duplicated sequences at both breakpoint junctions. (II) Possible mechanisms that might generate duplications at the ends of breakpoints. Duplicated sequences are boxed. (A) Replication bubbles as a source of duplications and deletions at reciprocal breakpoints; (1) unreplicated DNA with vertical marks indicating location of future breakpoints; (2) new strands (dashed lines) and with the single strand breaks shown; (3) replication completed and the chromatids separate to daughter cells: Outcome 1: Daughter cell 1 inherits a chromosome with loss of the segment of chromosome between the two breakpoints. Daughter cell 2 inherits two copies of this segment, to give duplication at the breakpoint. Outcome 2: both cells receive exactly balanced products. (B) Re-replication bubbles as an origin of duplications at reciprocal breakpoints. (1) Unreplicated DNA; (2) re-replication of a chromosomal segment due to dysregulation of repair and/or replication controls; (3) re-replication generates slowed or stalled forks and DNA damage; (4) outcome: DNA with duplications at double strand break. (C) Errors in repair of single stranded breaks as origin of duplications at reciprocal breakpoints. (1) DNA with staggered single strand breaks; (2) strand displacement DNA synthesis without repair of single strand breaks; (3) DNA with duplications at double strand break.
The combination of CGH and mate pair or cloned breakpoint sequences could not distinguish translocations and insertions, but FISH studies were required to make this distinction. Among the rearrangements associated with duplicated sequences at the breakpoint, we characterized 11 insertions: eight in Figures 1 and 2, Supplementary Figures S1 and 3, but also VPC6 in Supplementary Figure S1, Table S1 and two not described here; four translocations (two of which included an insertion at the breakpoint on one derivative chromosome) (Figure 3, Supplementary Figures S1 and 3); and three inversions (one of which included an insertion at one of the two breakpoints) (Figure 3; Supplementary Figures S1 and 3). Given the limited number of rearrangements that we characterized, we do not know if the observed distribution is an accurate representation of the different types of rearrangements that have large duplicated sequences at breakpoint junctions. However, our data suggest that insertions are more prevalent than translocations or inversions. For most rearrangements with duplicated breakpoint sequences >40 kb we were able to verify a CNG from CGH data (Figure 1; Supplementary Figure S1 and Tables S2 and 4A). In addition, for all insertions >40 kb we identified a CNG of the inserted sequences (e.g. Figure 1), which could be a result of incorporation of an un-degraded DNA fragment or a Fork Stalling Template Switch or Microhomology-Mediated Break-Incuced Replication (MMBIR) mechanism (23,24). We identified no insertions that did not have large duplications flanking the inserted sequences, which suggests that this is a preferred mechanism for insertions in MMCL.
What mechanisms might be responsible for generating large duplications at the ends of breakpoints? Since breakpoint junctions for secondary rearrangements in MM often have short microhomologies of 1–9 bases (4), it is possible that an MMBIR mechanism (25,26) might be involved in this process. Consequently, duplication might arise from replication (Figure 4.IIA) (27) or re-replication bubbles (Figure 4.IIB) (28,29). Although many of these duplications are >100 kb and therefore larger than the ∼100 kb distance between neighboring replication origins, larger bubbles could be created when adjacent bubbles coalesce (30). Alternatively, duplications might be generated by staggered single strand breaks and delayed repair (Figure 4.IIC) (21).
The first mechanism proposed replication bubbles as generating two ends containing duplicated sequence and two ends containing a corresponding deletion (Figure 4.IA) (27). If the two copies with the duplication end up in one daughter cell, this cell has duplicated sequences at both ends, while the other daughter cell will have a corresponding loss of the duplicated sequences. Analysis of translocations in three breast cancer cell lines showed a similar prevalence of duplications and deletions at translocation breakpoints, providing some support for this model (27). Although we see duplications more often than deletions at breakpoints, the possibility of selection against deletions and the small size of our dataset does not rule out this model for MM.
The second mechanism proposed re-replication bubbles as an origin of duplications (Figure 4.IIB). It has been demonstrated that re-replication of a chromosomal segment due to dysregulation of replication controls can efficiently induce non-allelic homologous recombination-mediated tandem duplication/amplification of that segment in the budding yeast Saccharomyces cerevisiae (29,31). Re-replication generates slowed or stalled forks and DNA damage (28,32,33). Other studies showed that in contrast to stalled replication forks in S phase, where breakage among thousands of replication forks is a rare accident, during re-replication, isolated re-replication forks are unlikely to be rescued by converging forks from neighboring origins and fork breakage might be the rule rather than the exception (29). Given that dysregulation of a CYCLIN D gene is present in virtually all MGUS and MM tumors (1,34), it notable that a high level CYCLIN D1 expression has been shown to be associated with re-replication (35).
The third mechanism (Figure 4.IC) proposes that delayed repair of staggered single strand breaks could generate a double stranded breakpoint with the sequences between single strand breaks present on both ends of the double stranded breakpoint (21).
We have characterized breakpoints in four MMCL that have a primary t(11;14) translocation and also an MYC locus rearrangement involving a 3′ IGH SE. Remarkably, all four MMCL had either an insertion (KMS-12, MM-M1, MOLP8) or a translocation (Karpas 620), which repositioned an 11;14 fragment containing a 3′ IGH SE (but not CCND1) into the MYC locus. These results suggest that MYC has captured an 11;14 fragment containing a 3′ IGH SE from der14 of a t(11;14) translocation. We also note that others have used an IGH capture procedure that identified 8;11 breakpoints in two primary MM tumors with t(11;14) translocations (6). We do not understand why MYC seems to preferentially capture the 3′IGH SE from der14 t(11;14) instead of from a normal copy of chr14.
For MM, all of our examples of rearrangements associated with large duplicated sequences appeared to be secondary genomic rearrangements. Does this phenomenon also occur with primary IGH translocations? We had cloned sequences or mate pair sequences that provided reciprocal breakpoints for presumptive primary IGH translocations in 13 MMCL and eight untreated MM tumors (Table 3), but none had large duplicated sequences at breakpoint junctions (11). We also examined recently published reciprocal breakpoints for presumptive primary IGH translocations in 32 MM tumors, and found that only three tumors had large duplicated sequences from the non-14 partner chromosome (12). Together, these data show that 3 of 53 (∼6%) of presumptive primary IGH translocations are associated with large duplicated sequences. Although most MM tumors have primary IGH translocation breakpoints suggesting a role for IGH switch recombination or less often VDJ joining, the three tumors with large duplications did not have breakpoints suggesting a role for IGH switch recombination or VDJ joining. Two of these tumors had t(14;16) translocations and the other had a t(6;14) translocation. As we proposed previously, all of the primary IGH translocation partners except for 4p16 can be involved in secondary rearrangements (1,5). Therefore, it is possible that the presumptive primary IGH translocations that are associated with large duplications are actually secondary rearrangements.
It is interesting that the duplicated regions often contain SE but sometimes conventional enhancers (Supplementary Table S2), perhaps indicative of a higher probability of these rearrangements occurring in regions of open chromatin and/or selection for dysregulation of a target gene. In addition, the duplications may have novel functional consequences. One example is the insertion of chr8 sequences (including MYC) between flanking duplicated sequences, which include the IGL SE from a t(16;22) breakpoint junction (Figure 2C). This complex rearrangement results in the presumptive dysregulation of MAF (chr16) and MYC (chr8) by separate copies of the IGL SE. There also are a number of insertions containing a SE or oncogene, with the duplicated flanking sequences containing, respectively, an oncogene or SE, which presumably increases the probability of a functional configuration of the oncogene with the SE (Figures 1–3, Supplementary Figures S1 and 3; Table S2). Finally, it is not clear if the association of duplications with breakpoint junctions occurs only in tumor cells but also in normal wild-type or mutant prokaryotic cells or lower eukaryotes.
ACCESSION NUMBERS
The mate pair sequence data on MMCL and 25 primary MM tumors have been published under SRA accession number SRP064107.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (to W.M.K); National Institutes of Health Grants [CA186781, CA195688 to P.L.B.]; the Predolin Foundation (to P.L.B). Funding for open access charge: Intramural Research Funds of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kuehl W.M., Bergsagel P.L. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J. Clin. Invest. 2012;122:3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesi M., Robbiani D.F., Sebag M., Chng W.J., Affer M., Tiedemann R., Valdez R., Palmer S.E., Haas S.S., Stewart A.K., et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chng W.J., Huang G.F., Chung T.H., Ng S.B., Gonzalez-Paz N., Troska-Price T., Mulligan G., Chesi M., Bergsagel P.L., Fonseca R. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Affer M., Chesi M., Chen W.D., Keats J.J., Demchenko Y.N., Tamizhmani K., Garbitt V.M., Riggs D.L., Brents L.A., Roschke A.V., et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014;28:1725–1735. doi: 10.1038/leu.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabrea A., Martelli M.L., Qi Y., Roschke A., Barlogie B., Shaughnessy J.D., Jr, Sawyer J.R., Kuehl W.M. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes Chromosomes Cancer. 2008;47:573–590. doi: 10.1002/gcc.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker B.A., Wardell C.P., Brioli A., Boyle E., Kaiser M.F., Begum D.B., Dahir N.B., Johnson D.C., Ross F.M., Davies F.E., et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014;4:e191. doi: 10.1038/bcj.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Max E.E. Immunoglobulins: Molecular Genetics. In: Paul WE, editor. Fundamental Immunology. 7th edn. Philadelphia: Lippincott Williams and Wilkins; 2013. pp. 150–182. [Google Scholar]
- 9.Fonseca R., Bergsagel P.L., Drach J., Shaughnessy J., Gutierrez N., Stewart A.K., Morgan G., Van Ness B., Chesi M., Minvielle S., et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smadja N.V., Fruchart C., Isnard F., Louvet C., Dutel J.L., Cheron N., Grange M.J., Monconduit M., Bastard C. Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia. 1998;12:960–969. doi: 10.1038/sj.leu.2401041. [DOI] [PubMed] [Google Scholar]
- 11.Bergsagel P.L., Kuehl W.M. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 12.Walker B.A., Wardell C.P., Johnson D.C., Kaiser M.F., Begum D.B., Dahir N.B., Ross F.M., Davies F.E., Gonzalez D., Morgan G.J. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood. 2013;121:3413–3419. doi: 10.1182/blood-2012-12-471888. [DOI] [PubMed] [Google Scholar]
- 13.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 14.Annunziata C.M., Davis R.E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keats J.J., Fonseca R., Chesi M., Schop R., Baker A., Chng W.J., Van Wier S., Tiedemann R., Shi C.X., Sebag M., et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawyer J.R., Lukacs J.L., Thomas E.L., Swanson C.M., Goosen L.S., Sammartino G., Gilliland J.C., Munshi N.C., Tricot G., Shaughnessy J.D., Jr, et al. Multicolour spectral karyotyping identifies new translocations and a recurring pathway for chromosome loss in multiple myeloma. Br. J. Haematol. 2001;112:167–174. doi: 10.1046/j.1365-2141.2001.02546.x. [DOI] [PubMed] [Google Scholar]
- 17.Roschke A.V., Tonon G., Gehlhaus K.S., McTyre N., Bussey K.J., Lababidi S., Scudiero D.A., Weinstein J.N., Kirsch I.R. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 18.Stephens P.J., McBride D.J., Lin M.L., Varela I., Pleasance E.D., Simpson J.T., Stebbings L.A., Leroy C., Edkins S., Mudie L.J., et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L., Luquette L.J., Gehlenborg N., Xi R., Haseley P.S., Hsieh C.H., Zhang C., Ren X., Protopopov A., Chin L., et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shou Y., Martelli M.L., Gabrea A., Qi Y., Brents L.A., Roschke A., Dewald G., Kirsch I.R., Bergsagel P.L., Kuehl W.M. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc. Natl. Acad. Sci. U.S.A. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dib A., Glebov O.K., Shou Y., Singer R.H., Kuehl W.M. A der(8)t(8;11) chromosome in the Karpas-620 myeloma cell line expresses only cyclin D1: yet both cyclin D1 and MYC are repositioned in close proximity to the 3′IGH enhancer. DNA Repair (Amst) 2009;8:330–335. doi: 10.1016/j.dnarep.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onozawa M., Zhang Z., Kim Y.J., Goldberg L., Varga T., Bergsagel P.L., Kuehl W.M., Aplan P.D. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7729–7734. doi: 10.1073/pnas.1321889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colnaghi R., Carpenter G., Volker M., O'Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin. Cell Dev. Biol. 2011;22:875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 26.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howarth K.D., Pole J.C., Beavis J.C., Batty E.M., Newman S., Bignell G.R., Edwards P.A. Large duplications at reciprocal translocation breakpoints that might be the counterpart of large deletions and could arise from stalled replication bubbles. Genome Res. 2011;21:525–534. doi: 10.1101/gr.114116.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archambault V., Ikui A.E., Drapkin B.J., Cross F.R. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 2005;25:6707–6721. doi: 10.1128/MCB.25.15.6707-6721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn K.J., Li J.J. Single-stranded annealing induced by re-initiation of replication origins provides a novel and efficient mechanism for generating copy number expansion via non-allelic homologous recombination. PLoS Genet. 2013;9:e1003192. doi: 10.1371/journal.pgen.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berezney R., Dubey D.D., Huberman J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 31.Green B.M., Finn K.J., Li J.J. Loss of DNA replication control is a potent inducer of gene amplification. Science. 2010;329:943–946. doi: 10.1126/science.1190966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green B.M., Li J.J. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell. 2005;16:421–432. doi: 10.1091/mbc.E04-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melixetian M., Ballabeni A., Masiero L., Gasparini P., Zamponi R., Bartek J., Lukas J., Helin K. Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 2004;165:473–482. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergsagel P.L., Kuehl W.M., Zhan F., Sawyer J., Barlogie B., Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal P., Lessie M.D., Lin D.I., Pontano L., Gladden A.B., Nuskey B., Goradia A., Wasik M.A., Klein-Szanto A.J., Rustgi A.K., et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.