Abstract
The principles of mRNA decoding are conserved among all extant life forms. We present an integrative view of all the interaction networks between mRNA, tRNA and rRNA: the intrinsic stability of codon–anticodon duplex, the conformation of the anticodon hairpin, the presence of modified nucleotides, the occurrence of non-Watson–Crick pairs in the codon–anticodon helix and the interactions with bases of rRNA at the A-site decoding site. We derive a more information-rich, alternative representation of the genetic code, that is circular with an unsymmetrical distribution of codons leading to a clear segregation between GC-rich 4-codon boxes and AU-rich 2:2-codon and 3:1-codon boxes. All tRNA sequence variations can be visualized, within an internal structural and energy framework, for each organism, and each anticodon of the sense codons. The multiplicity and complexity of nucleotide modifications at positions 34 and 37 of the anticodon loop segregate meaningfully, and correlate well with the necessity to stabilize AU-rich codon–anticodon pairs and to avoid miscoding in split codon boxes. The evolution and expansion of the genetic code is viewed as being originally based on GC content with progressive introduction of A/U together with tRNA modifications. The representation we present should help the engineering of the genetic code to include non-natural amino acids.
INTRODUCTION
The genetic code is considered as ‘quasi’ universal. Although deviations of the ‘standard’ genetic code have been identified, they occur in only a few organisms and mainly in organelles (1–3). However, as more organisms are being presently studied, new exceptions will certainly be discovered. Nevertheless, because all extant organisms evolved from a small set of ancestral living organisms that, along evolution, continued to exchange DNA information, the basic molecular principles of decoding (including the genetic code) remained essentially the same in all organisms of the three domains of life (Bacteria, Eukarya and Archaea). These principles are anchored in the physics and chemistry of the interplay between the diverse, and often locally neutral, non-covalent and weak molecular interactions engaged, and in the thermodynamics of formation of the several distinct complexes present at the various stages of the multistep translational process. Far from excluding profound improvements, these principles allow for adaptation and even innovations of many of the elements of the modern translation machinery (including the genetic code) according to physiological needs and/or to cellular niches and environments.
The traditional way to represent the 64 codons, as organized in the standard codon table (4–7) took several years to settle down. In the first row of the codon table are indicated all codons starting with a U at the first codon position, with a C in the second row, and a A or a G for the third and fourth rows, respectively. For each column, the same (base) order is used for the second codon base, as well as within each of the 16 decoding boxes for the classification of the 4 codons ending with a different base. This standard and quasi universal codon table is divided into boxes with 4 synonymous codons (hereafter designated as 4-codon boxes or unsplit decoding boxes) where the third base can be either U, C, A or G for coding the same amino acid, and boxes with 2 synonymous codons (hereafter designated as 2-codon boxes or split decoding boxes) where the third base codes for a different amino acid depending on its nature, either pyrimidine (Y) or purine (R). A third category concerns split 3:1-codon boxes where a single codon ending with guanine means methionine or tryptophan (1-codon box). Within these split 2:2- and 3:1-codon boxes are those meaning STOP for opal UGA, ochre UAA and amber UAG. At the time the table was first proposed, knowledge about the detailed molecular mechanisms underlying the decoding process did not exist. A few research groups argued for a different organization of the initial and now standard codon table (8–11). Arguments were based on various criteria or regularities of the code but never with knowledge of all the elements of the whole decoding process, in particular the large diversity of post-transcriptional nucleotide modifications that exist especially in the tRNA.
Recent progress in identifying modified nucleotides and their functions in tRNA (12–14), and information gained from the recent detailed structural, physicochemical and kinetic studies of ribosomes associated with mRNA/aminoacyl-tRNA, has made clear that selection of a given codon by its cognate aminoacyl-tRNA harboring a complementary or nearly complementary anticodon within the ribosomal decoding center is a complex and intricate process (15–22). The ribosome is not a passive machine but an active component (structurally and kinetically speaking) of the codon selection mechanism. Also, the original wobble ‘hypothesis’ (23) has to be broadened in several cases involving non-standard base pairings at the third base pair of the codon/anticodon helix (see for examples: (24–26)). Indeed, it is now becoming clear that some base–base oppositions avoid usual pairings or even H-bonding while still being accommodated within a Watson–Crick-like helix. Indeed, post-transcriptional modifications of nucleotide-34 can change the physicochemical behavior of the base (frequency of tautomerism) and/or the spatial preference of the nucleotide (syn/anti, puckering of the ribose) to precisely allow the third anticodon nucleotide to fit within a mini-helix structure together with the two other base-pairs of the anticodon (15,27–32). In short, wobbling at the third position does occur in various ways, but without movement of the codon base toward the minor groove (see below).
In the present work, we took into consideration all the new elements that we now understand are important for the aminoacyl-tRNA selection at the A site of the decoding center of the ribosome in order to propose a visually useful decoding table embedding multiple structural aspects of translation. This new representation allows one to propose a rationale for the emergence of the contemporary G/C/A/U containing quasi-universal genetic code from an ancient GC-rich code and the multiple constraints on such an evolution.
THE CODON TABLE
We started from an observation, already made at least partly by others (8,9,33,34), on the distribution of codons within the codon table. The codons within the unsplit 4-codon boxes always have one C in the second or either C or G at the first position of codon leading to C = G (G = C) pair in the codon/anticodon triplets. Among these codon boxes, the two conditions are simultaneously satisfied for four amino acids, Ala, Arg, Gly and Pro. In contrast, codons of the split 2:2- and 3:1-codon boxes have, instead, A at the second or either A or U at the first position of the codon leading to a A–U (U–A) pair in the codon/anticodon triplets. Among these, the two conditions are simultaneously satisfied for seven amino acids, Asn, Ile, Leu, Lys, Met, Phe and Tyr. The nine remaining amino acids (but 12 codon boxes because Leu, Ser and Arg each have 2 distinct codon boxes) have a mixture of C = G and U–A pairs, with either a C = G pair at the first position followed by a U–A pair or a U–A pair at the first position followed by a C = G pair. The two additional amino acids that are co-translationally inserted, pyrrolysine and selenocysteine, are recoded to replace stop codons, and will be discussed later in our analysis.
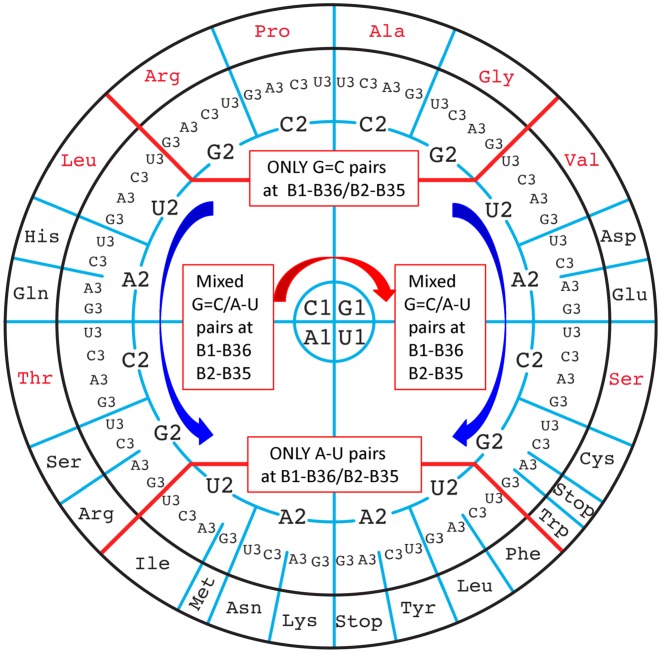
In our new representation, we arrange each codon corresponding to the 20 canonical amino acids on a circle (genetic code wheel) as a function of the nature of the first two base pairs of the codon/anticodon triplet. On the circle, the G = C only codons are disposed at the top, the A–U only codons at the bottom, and the mixed ones in-between on the left and the right. The order of third codon base has been chosen so that proximities of purine- and pyrimidine-ending codons are preserved (Figure 1). Note how the bases C, G, U, A rotate around the circle: for the first position the C1 starts in the top left quadrant, for the second position C2, G2, A2, U2 rotates down in the right part in a right-handed fashion and in the left part in a left-handed fashion. The choice of this unsymmetrical disposition will become more apparent and coherent in the following sections.
Figure 1.
Circular representation of the genetic code emphasizing the inherent regularities of the decoding recognition process. The codons containing solely G = C pairs at the first two positions are at the top, those containing solely A–U pairs at the bottom, and those with mixed pairs of G = C and A–U either at the first or second pair of the codon/anticodon helix in the middle at the right and left. Thick red lines separate the three main regions. The red arrow indicates the direction of rotation for C1, G1, U1, A1 and the blue arrows the direction of rotation for C2, G2, U2, A2 on the right and left parts of the wheel. The amino acids coded by unsplit 4-codon boxes are indicated in red and those by split 2:2- and 3:1-codon boxes, together with the usual stop codons, are indicated in black. Throughout, the codon positions are numbered B1-B2-B3 and the anticodon nucleotides B34-B35-B36, both from 5′ to 3′.
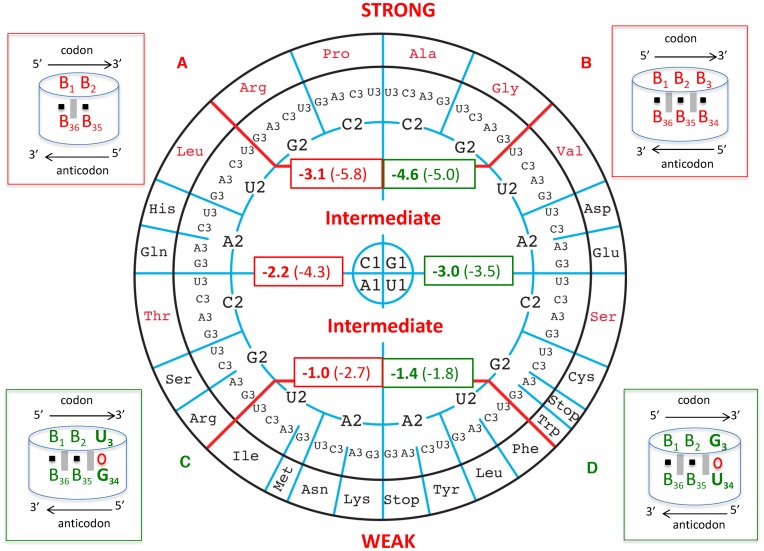
We then computed the energies of the codon/anticodon minihelices (5′-B1-B2-B3-3′ codon paired to 3′-B36-B35-B34-5′ anticodon) following the recent Turner values for dimers (35). First, we considered only the first two base pairs, B1-B2 paired to B36-B35 (Figure 2, box A). The values (indicated as bold red numbers) are distributed on a circular view of the codons. Clearly, from those values, three groups of amino acids can be distinguished. A STRONG (S) group (average −3.1 kcal/mole) with, as above, the four Ala, Arg, Gly, Pro. A WEAK (W) group (average −1.0 kcal/mole) with, again as above, the seven Asn, Ile, Leu, Lys, Met, Phe, Tyr. And finally, an INTERMEDIATE (I) group (average −2.2 kcal/mole) gathers the remaining amino acids (or codon boxes). A similar distribution is observed by computing the triplets of base pairs (B1-B2-B3 with B36-B35-B34) (Figure 2, box B), the corresponding values are in parentheses). Indeed, the distribution from STRONG to WEAK is respected (averages from −5.8, −4.3, −2.7 kcal/mole). Finally, in order to be slightly closer to biological reality, we considered the U3oG34 and G3oU34 Crick-type of wobbles at the third base pair (Figure 2, boxes C and D, respectively). Again, the three groups segregate well with energies, respectively, of −5.0, −3.5 and −1.8 kcal/mole for the U3oG34 wobble and −4.6, −3.0, and −1.4 kcal/mole for the G3oU34 wobble, respectively. Interestingly, the G3oU34 wobble is always less stable than the U3oG34 wobble interactions (see discussion below). There is no overlap between the three categories, in the sense that no INTERMEDIATE codon has a stronger energy than any STRONG codon or a weaker one than any WEAK codon.
Figure 2.
On the circular representation for the code are given the Turner energies calculated for the first two base pairs of the codon/anticodon helix, B1-B2 paired with B36-B35. Most generally, 5′-B1-B2-B3-3′ pairs to 5′-B34-B35-B36-3′, forming pairs B1-B36, B2-B35 and B3-B34. The free energy values for dimers are in kcal/mole and taken from (35). In the scheme (A) at the top left, the black square symbolizes the H-bonds between A–U or G = C and the grey vertical rectangles the stacking interactions between the base pairs. The average values are given in the red rectangles within the codon wheel: it is −3.1 kcal/mole for the top codon boxes, −2.2 kcal/mole for the middle codon boxes and −1.0 kcal/mole for the bottom ones, with therefore 1.0 kcal/mole difference between the three groups. The reported errors on the free energies are less than 0.1 kcal/mole. The top four groups of un-split family codons boxes will be called ‘STRONG’ codon boxes, the middle ones encompassing split and un-split codon boxes are designated ‘INTERMEDIATE’ codon boxes, and the last all split family codon boxes at the bottom as ‘WEAK’ codon boxes. The values in parentheses represent the same calculations with the consideration for the third base pair assumed to be Watson–Crick and without nucleotide modifications (schematized in (B) at the upper right corner). Although such calculations are not biologically meaningful, they may be relevant when considering the evolution of the decoding system. The differences between the averaged values are of the order of −1.5 kcal/mole and around 3.0 kcal/mole between the STRONG and WEAK codon boxes. The same calculations were done with the third base pair either a U3oG34 or a G3oU34 (schemes (C) and (D) at bottom left and right corners respectively). Note that with B2-B35 being either a G = C or C = G pair, the energies are 2.0 kcal/mole (depending on GoU34 or UoG34, respectively) more stable than with B2-B35 being a A–U or U–A pair. Also, whatever the nature of the B2-B35 pair, G3oU34 is always less stable than U3oG34 by about half a kcal/mole. Correct decoding depends on formation of a short double helix-like structure between the three bases of the codon and the anticodon (symbolized by the dashed red cylinder).
Despite the large differences of energy between GC-rich and AU-rich codon–anticodon duplexes, the measured binding constants of natural aminoacyl-tRNAs on 70S ribosomes programmed with mRNA are unexpectedly remarkably similar (20–22,36,37). Likewise the stability of complexes formed between two natural tRNAs harboring complementary anticodons is very much the same whatever the GC/AU composition of the duplex (38,39). Only when non-natural mutant tRNAs are used are large differences of binding constants measured. Obviously, both the ribosome and the built-in features of natural tRNAs have evolved to allow minimization of the differential thermodynamic contributions of the codon–anticodon interactions, allowing tRNA–mRNA interactions to be rather uniform; in other words, to be neither too sticky nor too loose during the process of translation. We will now describe these features, first on the ribosome and then on the transfer RNAs.
THE RIBOSOMAL DECODING SITE AND THE ANTICODON TRIPLET
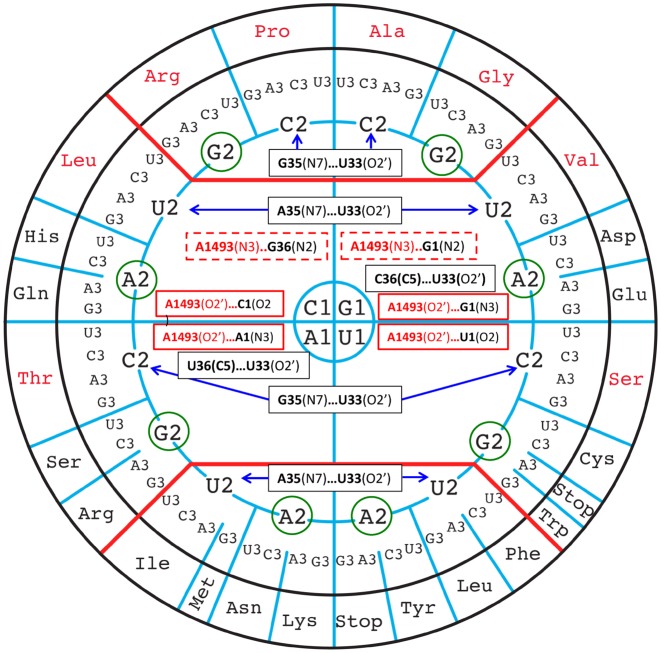
Recent crystallographic data of aminoacyl-tRNAs associated to mRNA programmed ribosomes revealed the contacts made by the ribosomal A-site with the codon/anticodon helix, designated as the ribosomal grip. The two invariant adenine residues A1492 and A1493 of helix 44 of 16S rRNA make A-minor type interactions in the minor groove of the first two base pairs of the codon/anticodon helix (Figure 3) (15–17,27–29,31,32,40–43). Except for those made to the B1-B36 pair, such contacts are almost identical for all types of Watson–Crick pairs and, therefore, can be considered to contribute similarly to the global interaction energies (44) (Table 1). On the circular representation (Figure 3), the A-minor contacts are equally partitioned (listed in plain red boxes), except, importantly, for those between A1493 and B1-B36 (G36 = C1 or G1 = C36, in dashed red boxes), which are specific to the top half of the circle.
Figure 3.
The experimentally identified interactions that are variable between the codon/anticodon base pairs, the ribosomal grip and the anticodon loop nucleotides are shown on the circular representation of the codons. The constant contacts are not shown. The weak contacts involving C–H bonds are also not shown. The rRNA nucleotides are in red and the anticodon loop nucleotides in black. In plain black boxes are the tRNA intra-anticodon loop interactions, in plain red boxes are the interactions of the ribosomal grip with, in dashed red boxes, the contact occurring only in the top half of the wheel. Purine bases at position 2 of codons are circled in green to emphasize the fact that they do not contact the corresponding tRNAs.
Table 1. Regularities in the codon table and the ribosomal grip.
| 4-codons box: always either a C = G pair at 2nd position or a C = G/G = C pair at 1st position (Note that the two states occur in Pro, Ala, Arg, Gly). |
| A1492 … C2 = G35 |
| A1493 … C1 = G36/G36 = C1 |
| 2-codons box: always either a A-U pair at 2nd position or a U-A/A-U pair at 1st position (Note that the two states occur in Phe, Leu, Ile, Met, Asn, Lys, Tyr). |
| A1492 … A2-U35 |
| A1493 … U1-A36/U36-A1 |
Throughout the codon positions are numbered 1-2-3 and the anticodon nucleotides 34-35-36, both from 5′ to 3′. Most generally, they are 5′-B1-B2-B3-3′ pairing to 5′-B34-B35-B36-3′.
In Table 2 and Figure 3 are indicated the constant and variable contacts that occur within the anticodon loop and with the codon/anticodon minihelix (45) (see also Supplementary Figure S1A and B). The contributions of such contacts to aminoacyl-tRNA (aa-tRNA) binding to the ribosome have been established by various methods (46–48). Recent experiments on the effects of 2’O-methyl group in the ribose of mRNA show the strongest reduction in translation efficiency when the 2’O-methyl group is present at the second position, followed by methylation at the third position (25% of unmodified) and the first position (80% of unmodified), confirming further the O2’-hydroxyl group donor contacts at the second and third positions (49). They involve mainly the ribose O2’ of U33 (an almost invariant nucleotide) with the Hoogsteen edge of B35 (where the contact U33(O2’)…R35(N7) is stronger than the U33(O2’)…Y35(C5) contact) (see Tables 1 and 2 and Supplementary Figure S2). Of note is that these contacts therefore slightly disfavor the codon/anticodon interactions with a purine at B2 (Arg, Gly, His/Gln, Asp/Glu, Arg/ser, Cys/Trp) that are circled in green in Figure 3 (discussed below). In short, the ribosomal grip interactions add to the stabilization of mainly codons of the unsplit 4-codon boxes, especially through the A1493(N3) H-bond to the first G = C/C = G pair in the upper part of the circle in Figure 3.
Table 2. Constant and variable or sequence dependent interactions present between the codon/anticodon minihelix and the ribosomal grip or the anticodon loop.
| Constant interactions with rRNA | Variable interactions with rRNA |
|---|---|
| B1-B36 | B1-B36 |
| A1493(N1)…B36(O2’) | A1493(N3)…G36(N2) |
| A1493(O2’)…B1(O2’) | A1493(N3)…G1(N2) |
| A1493(O2’)…Y1(O2) or | |
| A1493(O2’)…R1(N3) | |
| B2-B35 | B2-B35 |
| G530(N3)…B35(O2’) | None observed yet. |
| G530(O2’)…B35(O4’) | |
| G530(N1)…A1492(N1) | |
| A1492(N3)…B2(O2’) | |
| A1492(O2’)…B2(O2’)* | |
| B3-B34 | |
| B3(O2’)…M…G530/C518/S12 | |
| B34 sugar stacking on C1054 | |
| Constant interactions with tRNA | Variable interactions with tRNA |
| B1-B36 | B1-B36 |
| U33(N3)… B36(OR) | U33(O2)….Y36(C5) |
| B2-B35 | B2-B35 |
| U33 Stacking on B35(OR) | U33(O2’)…R35(N7) |
| or weaker U33(O2’)…Y35(C5) | |
Notice that no variable interaction occurs between B2-B35 and the conserved rRNA nucleotides while no interaction occurs between B1-B36 and the other tRNA nucleotides. The contact indicated by * is present in usual A-minor interactions, but presents a long distance in structures for the A-site (3.6 Å or longer). Note also that in standard A-minor contacts, the equivalent C1 = G36 is observed more often than G1 = C36 and C1 = G36 more often than G1 = C36 (44).
In addition, the crystal structures of ribosomal complexes with mRNA and aminoacyl-tRNAs reveal many contacts between a few amino acids of r-proteins and the RNA sugar–phosphate backbones. The majority of these contacts should be general and applicable to any tRNA and mRNA and, thus, should not make a major contribution to the specificity and fidelity of a specific codon–anticodon binding, at least in the decoding A-site of the ribosome. However, r-proteins S4, S5 and S12 have been demonstrated to be involved in the ribosomal grip by controlling the closure of 30S subunit upon correct codon recognition (50–52). The protein S12 especially closely contacts the decoding site with Ser46 binding to the Hoogsteen edge of A1492 (which monitors the second base pair B35-B2) and Pro44 interacting via a solvent molecule with the hydroxyl group of the ribose of B3, together with G530 and C518 (17,53). Among the many conserved and semi-conserved residues present in the rRNA region of the decoding center, a few are post-transcriptionally modified (54,55). However, none of these modified residues is in direct contact with the codon or anticodon in the A-site of the ribosome and therefore should not contribute directly to specific codon selection mechanism. The main functions of these post-transcriptional modifications appear to reside in the modulation of rRNA structure and ligand interactions leading to indirect and general effects on translational fidelity (56). One exception could be the positively charged m7G527 in bacteria. It is located about 5 Å from Pro44 of S12 and the conserved base G530, possibly creating an electrostatic environment that influences the electrostatic state of modified B34, especially with hypermodification at U34.
In summary, the present organization of the genetic code makes a clear energetic segregation between all GC-rich codons belonging to 4-codon family boxes and all AU-rich codons belonging to 2:2 and 3:1 codon family boxes, with stop-codons being among the latter. The codon boxes with a mixture of G = C and A–U pairs in the first and second positions are intermediate in the various energetic parameters considered. Based on the above criteria, codons of the unsplit 4-codon boxes appear to fulfill enough criteria to bind correctly to their cognate aa-tRNAs within the ribosomal A site. They probably possess enough energy from both the internal codon–anticodon mini helix and the additional external stabilizing grip energies from both the ribosome and the tRNA anticodon loop.
Remarkably, in certain ribosomal contexts (Mycoplasmas, mitochondria), the energetics of the first two base pairs of the codon–anticodon mini helix appears to be sufficient; a single tRNA (containing unmodified U34) is able to decode each of the four GC-rich codons of unsplit decoding boxes (57–60), although with different efficiencies. This is known as the 2 out of 3 decoding rule (26,33). In contrast, a minimum of two tRNAs (one with G34, a second one with modified U34—see below—and eventually a third one with C34) is required to read AU-rich codons of each split 2:2-decoding boxes (57–59). Apparently, below a certain energy threshold, the contributions of the first two base pairs are insufficient to sustain translation and a third base pair has to complement the missing energetics. This is probably why the A/U-rich codons of the split 2:2-decoding boxes, where pyrimidine-ending (Y-ending) codons must be distinguished from purine-ending (R-ending) codons, have been selected along evolution for expansion of the genetic code with additional amino acids (see below).
STRUCTURAL CHARACTERISTICS OF THE tRNA ANTICODON HAIRPIN
Transfer RNAs are not passive molecules but active participants during translation. They can modulate their binding and specificity through sequence preferences at key positions, extending beyond the anticodon triplet (61–63), and through very diverse base modifications, particularly in the anticodon loop and stem (14,64). In the following sections, the contributions of the sequence preferences and base modifications to smooth and uniform decoding are analyzed within the framework just described, with a focus restrained to the anticodon stem and loop.
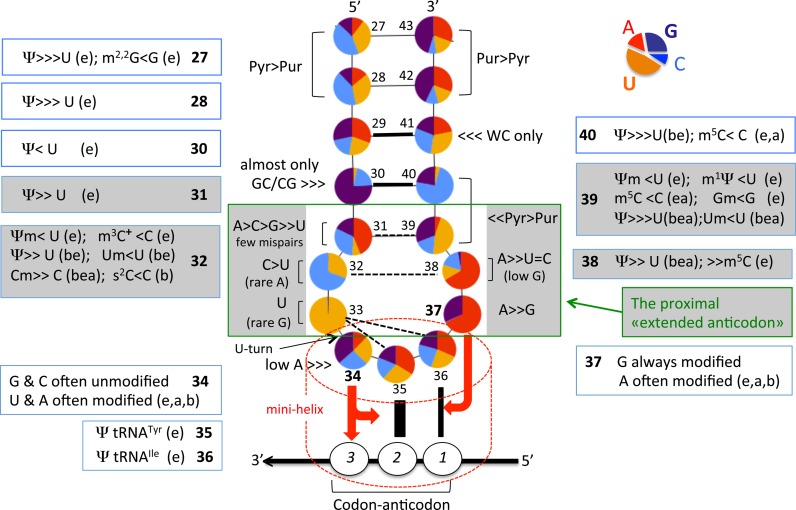
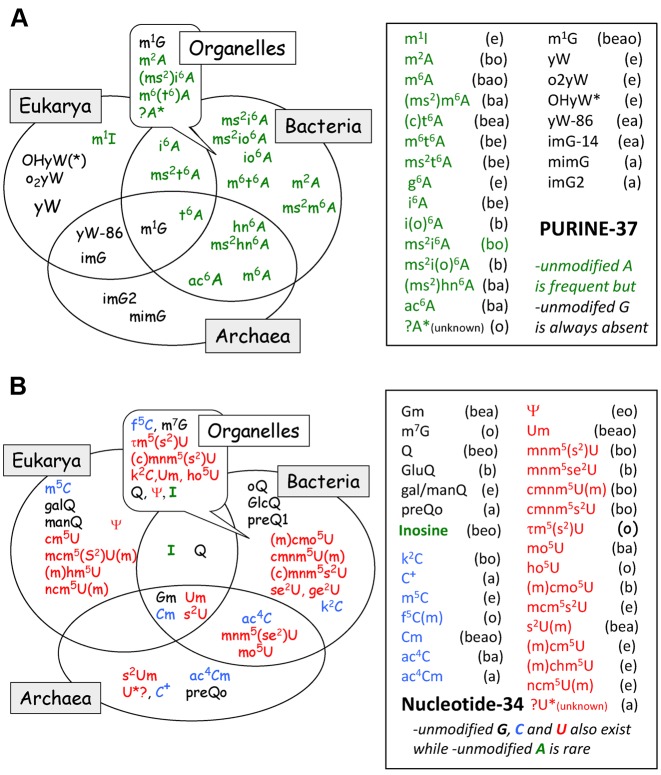
Figure 4 depicts the general distribution of conserved and semi-conserved nucleotides and the presence of modified nucleotides within the anticodon hairpin of 400 native elongator tRNA sequences originating from organisms spanning Bacteria, Eukarya and Archaea (65). Initiator tRNAMet is excluded from the analysis because it participates in a different step of translation and its anticodon hairpin is quite distinct from those observed in elongator tRNAs (66).
Figure 4.
Structural characteristics of anticodon hairpin of tRNA. Identity and relative frequency of a nucleotide (including modified ones) are obtained from compilation of 382 elongator tRNAs belonging to the three domains of life (123 from Eubacteria, 55 from Archaea, 204 from Eukaryota). Initiator tRNAMet, tRNAs coding for Pyl and Sec, all tRNAs from mitochondria/plastids and bacteriophages/virus were excluded from the analysis. The data set comprises tRNA sequences present in the Modomics database (14) that currently contains all sequences available by 2009 in the tRNA database as in ref. (76). Analysis was performed using the software tool tRNAmodviz (http://genesilico.pl/trnamodviz). Distributions of nucleotide residues at each position of the hairpin (positions 27 to 43) are visualized as a pea chart, of which the color code is indicated at the top right corner of the figure. Universal numbering system is used (144). Acronyms of modified nucleotides present in certain isoacceptor tRNA are indicated outside the anticodon hairpin (‘b’ means bacteria, ‘e’, eukaryote and ‘a’, archaea). Those present in the proximal ‘extended anticodon’ (in green square box, see also text) are in grey background. Modified nucleotides present at positions 37 and 34 are listed in Figure 5A and B, respectively. The red arrow pointing from B34 of anticodon to B3 of the codon and the plateau of B35/B2 and the red arrow from B37 to the plateau of B36/B1 symbolize the stabilizing effects of B34/B37 on codon–anticodon pairings.
The distribution of purines and pyrimidines in elongator tRNAs is clearly asymmetric and the presence of a few conserved and semi conserved bases is evident. Some of the bases along the stem are post-transcriptionally modified to Ψ or simple methylated derivatives, which are known to better stack with neighboring bases, thus favoring a locally more rigid conformation (67). In the middle of the stem, base pair B29-B41 is almost always a Watson-Crick pair, and either a G = C or a C = G pair is usually found at positions B30-B40, attesting to the need for a relatively stable helical conformation in this part of the anticodon stem. Depending on the tRNA species, either a C = G or a A–Ψ pair is preferred at B31-B39; thus the nucleotide entering the stem at the 3′-side is generally either a G39 or a Ψ39, two bases which favor the start of a helical stem (35,68). Therefore, all bases of the anticodon stem and particularly the last three base pairs stabilize helical conformations. In the anticodon loop, the first and last residues, B32 and B38, also present a biased distribution that maintains in this case a non-Watson–Crick pair, with B32 almost exclusively a pyrimidine Y (with a preference for a C or a U, again depending on the tRNA species) and B38 mainly a A followed by a less frequently used C (see Table 1 in (69)). Thus, at variance with B31-B39 pair, B32 and B38 tend to form a non-Watson–Crick pair with a single bifurcated contact between exocyclic O2(Y) and exocyclic N6(A) (or N4(C)) (45,70).
At B37 a strictly conserved purine is present (mostly A37 or a modified A37 derivative). When G37 is present, it is invariably modified into N1-methylated G37 or a Wyosine derivative in tRNAPhe of eukaryotes and Archaea (Figure 5A). Modifications at A37 depend on the identity of the adjacent nucleotide at position 36; it is generally modified when B36 is either A or U. The purine residue at 37 stacks over the first codon/anticodon base pair (40,71,72) and propagates the stacking continuity to neighboring residues B38, B39 and the anticodon stem (preferential 3′-stacked conformation). Modifications at the cyclic C2 atom and the exocyclic amine at C6 of A37 (and especially the highly hydrophobic tricyclic derivatives of the Wye-base) reinforce the stacking power of A37 with the neighboring nucleotides (12,72). Likewise, at the first position of the anticodon (B34), a plethora of diverse modified nucleotides are found (Figure 5B, and discussed below). Their functions are mostly related to decoding within the ribosomal decoding site, however some of them, like Cm, Um, s2U, Ψ, ac4C, Gm and Q, also contribute to the pre-structuring of the anticodon loop (31,73,74).
Figure 5.
Phylogenetic distribution of modified and hypermodified nucleosides at (A) B37 and (B) B34 of anticodon hairpin of tRNA from the 3 domains of life. All acronyms are those conventionally used, the corresponding chemical structures, full scientific names and chemical characteristics of most of them can be found in (145) (see also in: Modomics, http://modomics.genesilico.pl/). Only a few tRNA from Archaea have been sequenced so far, therefore information concerning this domain (especially for U34 modifications) is incomplete. Meanings of ‘b’, ‘a’, ‘e’ is the same as above in Figure 4 with that of ‘o’ corresponding to organelles. The data set comprises tRNA sequences present in the Modomics database (14).
THE PROXIMAL ‘EXTENDED ANTICODON’ CONCEPT
More than 30 years ago, Yarus (61) pointed out the strong co-variations between nucleotide B36 of anticodon and several bases of the whole anticodon stem-loop of E. coli tRNAs and introduced the ‘extended anticodon’ concept. Many experiments further confirmed such an apparent but indirect link between the sequence of the anticodon stem and the performance of the experimentally mutated bacterial tRNAs to efficiently translate certain codons both in vivo and in vitro. However, as more tRNA sequences became available from many different organisms, and more experiments were performed, it became evident that these co-variations were particularly relevant for the nucleotides of the anticodon loop and the last base pair B31-B39 of the anticodon loop (indicated in grey background in Figure 4 and thereafter designated as the proximal ‘extended anticodon’).
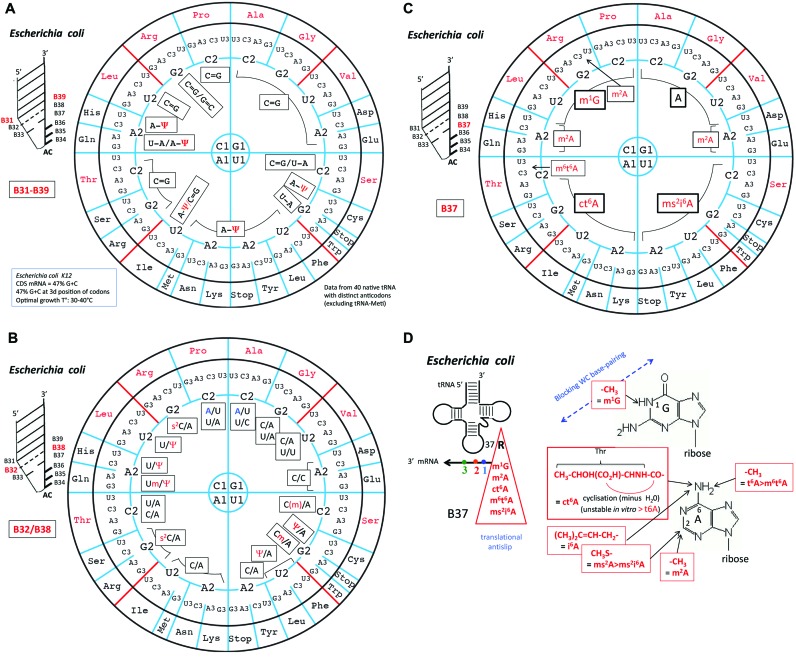
Figure 6A–C corresponds to an update of Yarus’ original data in the form of the ‘wheel of code’ for the 40 elongator tRNAs of the complete E. coli repertoire that read all the 61 codons for the 20 canonical amino acids. In Figure 6A, are displayed the distributions of B31-B39 base pair, B32/B38 in Figure 6B and B37 in Figure 6C (including the acronyms of modified nucleotides). The same analysis has been done for the elongator tRNA repertoire of the archaea Haloferax volcanii and the eukaryote S. cerevisiae and as well as in two extreme situations for which enough sequence information (including for the modified bases) is reasonably well known, Mycoplasma capricolum and human mitochondria (see Supplementary Figure S3). A separate analysis is mandatory for each organism because notable differences exist in the decoding strategies (though not in the genetic codes) of distantly evolutionary related organisms, such as those of Bacteria, Eukarya and Archaea (64,69).
Figure 6.
Architecture of the proximal extended anticodon loop of Escherichia coli tRNAs. On the circular representation for the code are given (A) the base pairs B31-B39, (B) the base opposition B32/B38 and the (C) identity of purine-37 found in the anticodon loop of the various tRNA species corresponding to each of the decoding boxes for the 20 amino acids. Modified nucleotides are indicated in red. Non-random usage of base pairings B31-B39 is apparent, with a frequent use of A31-Ψ39 in tRNA in the ‘weak’ decoding boxes. Likewise, the B32/B38 positions are more variable in the top of the wheel. Modification of B37 is clearly dependent on B1, thus of B36 of anticodon that has to base pair with B1 codon. Only in a limited number of isoacceptor species, another modified B37 (m2A or m6t6A) are found. Same analysis for tRNAs from H. volcanii, S. cerevisiae, M. capricolum and human mitochondria are shown in Supplementary Figure S3. (D) Chemical structures of the modifications found at position B37. Notice the presence of amino acid as part of ct6A modification.
The most frequently found B31-B39 base pair (boxed in black) is C = G, especially in tRNAs of the 4-codon family boxes (Figure 6A, upper part of the wheel). In contrast, a A-Ψ pair is predominantly present in tRNAs of the split 2-codon boxes (lower part of the wheel). Concerning B32/B38, the combination Y32/A38 is the most frequently found one, with both U32 and C32 occasionally modified to Ψ or Um and C32 to Cm or s2C, respectively (Figure 6B). These modifications of the base, especially the pseudouridine and the 2’O-methylation of the ribose, are known to limit significantly the conformational flexibility of the nucleotide and thereby locally rigidify the corresponding portion of the nucleic acid (75).
One remarkable exception is found in tRNAAla (anticodon GGC) and tRNAPro (anticodon GGG) with A32 instead of the quasi universal pyrimidine, but not in the other isoacceptor tRNAAla (anticodon U*GC) and tRNAPro (anticodons UGG and CGG, respectively) in E. coli (65,76). Base-38 is also occasionally a U or C instead of the usual A38, the uridine-38 being often modified to Ψ. This situation is found in majority of tRNA specific for Ala, Leu (of the 4-codon boxes), Val, His, Gln, Asp, and Glu, all belonging to decoding systems of the upper half of the codon wheel. In some instances, B32-B38 can even form a Watson–Crick pair, either U32-A38 (Pro, Gly, Thr) or A32-U38 (Ala, Pro). Again, these tRNAs with a potential B32-B38 Watson–Crick pair all belong to the group of strong 4-codons decoding tRNAs (Ala, Pro) and, surprisingly also to tRNAThr belonging to the intermediate group. The case of tRNAAla has been studied in detail (36,77–79). The take-home lesson is that tRNAs harboring too strong a base-pairing binding capacity have been ‘tuned down’ during evolution for optimal and uniform translation. As discussed above, B32-B38 should not form a Watson–Crick base pair that antagonizes the canonical conformation of the anticodon loop with the anticodon triplet in a correct helical orientation. With complementary bases at B32 and B38, one expect that a Watson–Crick pair is formed in some tRNA population, leading to a less optimal anticodon conformation for pairing with the codon and, thus, possibly rejection from the ribosomal decoding site. Without a selection for the preformed conformation of the anticodon loop, the strong codon/anticodon pairs would allow miscoding by binding to other G/C-rich near- or even non-cognate aa-tRNAs.
The distributions of B37 in Figure 6C (including post-transcriptional modifications) follow each quadrant and thus, strongly depend on the codon base at the first position. This conclusion is valid for the other tRNA repertoires analyzed (Supplementary Figure S3). The presence of stabilizing modifications such as ct6A and ms2i6A in E. coli (or i6A, m1G, yW, m6A in other organisms, Supplementary Figure S3) correlates with the need to stabilize weak neighboring codon–anticodon base pair A1-U36 or U1-A36, respectively. These modifications, as well as the N1-methylation of G37, which is unable to base pair in a Watson–Crick mode, are also required for limiting ribosome frameshifting at specific AU-rich ‘slippery’ sequences of the mRNA (80,81).
In summary, purine-37, together with B31-B39 and B32/B38, and contacts with U33 (grey background in Figure 4), synergistically contribute to maintaining a conserved pre-formed and optimal anticodon conformation, 3′-stacked with both the rest of the anticodon arm and the last two bases B35 and B36 in the anticodon. Supplementary Table S1 shows an approximate partition of these energetic elements that, together with the distribution of key elements on the wheels, helps explain how these apparently disparate interacting elements (including modified bases and ribose methylation) may cooperate to average the binding energies of tRNAs, whatever the GC or AU content of their anticodons to the mRNA-programmed ribosomes. One advantage of such an intricate networking system is the possibility to fine-tune the aminoacyl-tRNA for its ability to read the various cognate codons with appropriate efficiencies while avoiding reading near- and non-cognate codons.
CONTRIBUTION OF NUCLEOTIDE MODIFICATIONS AT THE FIRST tRNA ANTICODON POSITION
A large variety of post-transcriptionally modified nucleotides are found at position 34 of cytoplasmic elongator tRNAs (65,76). Partitioning of these nucleotides according to organisms in the three domains of life is given in Figure 5B. The most frequently and diversely modified nucleotide at position 34 is uracil, followed by cytidine and guanine. The only modified form of the adenine is hypoxanthine (inosine). Methylation of 2’O-ribose also occurs at G34, C34 and U34 (in the latter case, in combination with additional modifications at C5- and/or C2-atom of the base). These chemical adducts to B34 or its ribose alter the nucleotide chemical properties, and thus its polarity, but also the preference for the syn or anti conformer and the base's tautomeric form. They can also control ribose puckering and, in addition, allow for additional interactions with other elements of the translation machinery (see Supplementary Table S2 for an overview).
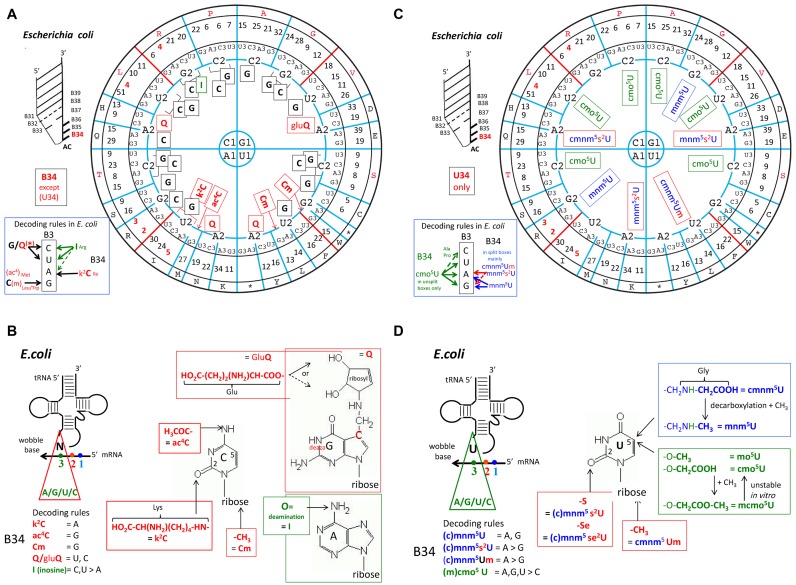
The distribution of modifications at B34 (C34, A34, G34) in elongator tRNAs according to the triplet code is depicted in Figure 7A in the case of E. coli. The same analysis for Haloferax volcanii, S. cerevisiae, Mycoplasma capricolum and human mitochondria are given in Supplementary Figure S4. Again, a clear segregation exists between tRNAs belonging to the ‘strong binding’ class (top part of the wheel) and those belonging to the ‘weak binding’ class (bottom part of the wheel). Except for inosine-34, which is found only in bacterial tRNAArg of the 4-codon boxes, other modified nucleotides at position 34, like ac4C, k2C, Cm, Um, Q and gluQ are found only in tRNAs belonging to the 2:2 and single-codon boxes.
Figure 7.
Identity of nucleotides at the first anticodon position (B34) of E. coli tRNAs. The global codon usage (after (146)) is inserted between the circle for the third base and that for the amino acid type. Conventional one letter code for amino acid is used. For the sake of clarity, the distribution of G34, I34 and C34 derivatives are shown in (A), and U34 derivatives in (B). Modified G/C/I-34 are indicated in red. All Q-containing tRNAs and those containing modified C* are found in tRNA belonging to split 2:2-codon boxes. For the modified U34 (shown in (C)), two chemically distinguishable types of modified residues are found, one harboring an oxyacetic acid group (sometimes methylated) at the C5 atom of uracil (cmo5U or mcmo5U), are indicated as green. These U34 derivatives are found only in isoacceptor tRNA belonging to the unsplit 4-codon family boxes. The second type of modified U34 derivatives harbors a methylaminomethyl (sometimes carboxymethylated) group at the C5 atom of uracil (mnm5U or cmnm5U). They are indicated as blue, some of which are also hypermodified into 2-thiolated derivatives (s2U*) or methylated on the 2′-hydroxyl ribose (U*m). They are found in all split 2:2-codon boxes, and in the Arg/Gly 4-codon boxes (after (88)). Same analysis for tRNAs from H. volcanii, S. cerevisiae, M. capricolum and human mitochondria are shown in Supplementary Figure S4. (D) Chemical structures of the modifications found at positions B34. Notice the presence of amino acid as part of a few modifications.
The situation concerning modified U*34 (*: modified at the C5-atom) and its doubly modified s2U* derivatives constitutes a special case. In the ribosomal grip, the third nucleotide of a codon is constrained by several contacts with the ribosome (specifically with G530, C518 of the 16S rRNA and a residue from RpS12), while the anticodon nucleotide B34 is in a loose stacking contact with C1054. When B34 forms a Watson–Crick pair, this asymmetry in contacts is not discriminatory. However, when forming a GoU wobble pair, the unsymmetrical constraints amplify the non-isostericity of UoG pairs such that G34oU3 wobble pairs can form but U34oG3 pairs form much less readily (for more detail see Supplementary Figure S5). Indeed, in several crystal structures with a U34oG3 opposition, the pair formed is not the standard wobble pair but a Watson–Crick-like pair between tautomeric forms of either G or U (27,29–32). The modifications at U34 are structurally imposed by the constraints on the codon base. Thus, in the non-isosteric GoU pairs, modifications on U34 will be necessary throughout the wheel, especially in the absence of additional C34-containing tRNAs. With C34-containing tRNAs present, the modifications at U34 are less important, since there is no need to form a U34oG3 pair. Despite this, when chemical adducts at the C5-atom of uracil are examined, a clear segregation by type exists depending on whether the modified U34-containing tRNAs belong to exclusively 4-codons (strong) or mainly 2-codons (weak) boxes (Figure 7C indicated in green and in blue, respectively).
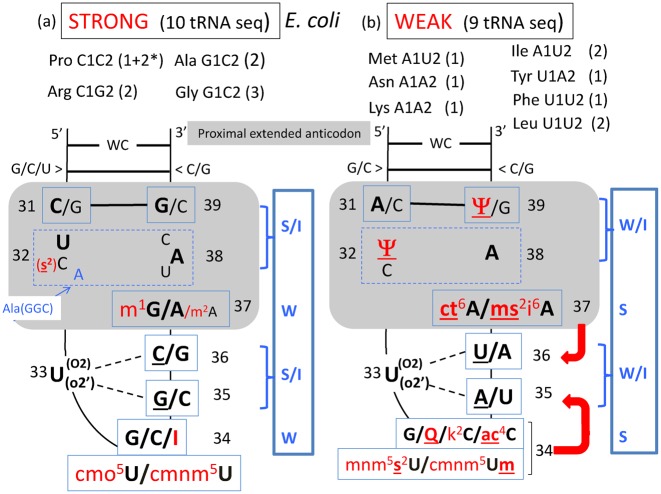
Without exceptions, aminoacyl-tRNAs reading codons of the split 2:2 or 3:1 decoding boxes, as well as those reading codons harboring A and/or U at the B1-B2 codon positions (bottom of the wheel) are the most diversely post-transcriptionally (hyper)-modified at B34, but also at other positions of the anticodon hairpin (compare Figure 6A–C). Evidently, fine-tuning the efficacy and accuracy of mRNA translation is a highly sophisticated process in which the chemistry of each nucleotide (modified or not) within the anticodon hairpin of the various individual natural elongator tRNAs collectively play a pivotal role (61). Figure 8 is a comparison between the general structures of the anticodon loop and proximal stem of the 10 E. coli tRNAs belonging to the STRONG class (panel a) and the 9 other E. coli tRNAs belonging to the WEAK class (panel b). The same figures for the cases of H. volcanii (archaea), S. cerevisiae (eukaryote), M. capricolum (minimalist bacteria) and human mitochondria (organelle) are presented in Supplementary Figure S6. Evidently each tRNA has evolved in such a way that substantial differences exist at almost every position of the molecule, the most characteristic ones being of course B37 and B34, especially for those tRNAs harboring A or U at the two last positions of anticodon (B37 and B38).
Figure 8.
Modulation of codon–anticodon binding according to G+C (STRONG) or A+U (WEAK) binding capability of selected E. coli tRNA species. In both cases, the anticodon hairpin is schematically represented with all the nucleotides of the 5′ branch in continuous stacking up to B34 and the complementary nucleotides of the 3′ branch in continuous stacking up to B32. The U33 turn is indicated with its links to R35 and/or Y36 (underlined). At positions B32 and B38, various combinations of base opposition are found (boxed with dashed lines), the most frequent ones being indicated in bold letters. In red are the modified nucleotides; underlining emphasizes that the chemical adduct reinforces the stacking power of the base with the neighboring nucleotides. Modulation of the strength of codon–anticodon binding occurs by the anticodon loop constraints that mainly depends on the choice of the B32-B38 base opposition, additional interactions with the conserved U33 and the identity of the chemical adducts on B37 and B34 that stabilize the B36-B1 and the B35-B2 interactions, respectively (schematized by red arrows). The number 2 with an asterisk for Pro, means that information came only from tDNA sequence, the corresponding maturated transcripts have not been sequenced yet. On the right of each diagram, an approximate energy scheme is displayed (in a blue rectangle) with S standing for STRONG, I for INTERMEDIATE, and W for WEAK. Same analysis for tRNAs from H. volcanii, S. cerevisiae, M. capricolum and human mitochondria are shown in Supplementary Figure S6.
SHORT SUMMARY OF THE STRUCTURAL ASPECTS
Overall, the three-dimensional structure of the anticodon loop is characterized by two helical stacks interrupted at the phosphate 3′ of the invariant U33 residue: the 5′-stack in continuity with the 5′-strand of the anticodon stem stops at residue U33 (the U-turn), while the 3′-stack extends from residue B34 to B38 in continuity with B39 to B43 in the 3′-strand of the anticodon stem. Although residues B34 to B38 form a helical stack with standard rotation angles between nucleotides, the 3′-strand conformation does not lead to enough rotation to allow B38 to form a Watson–Crick pair with B32 (70). Indeed, formation of a Watson–Crick pair between B32 and B38 would require a change in the conformation of the rest of the loop. B39, however, has rotated enough and is able to form a Watson–Crick pair with B31. Evidently, various combinatorial structural solutions exist to generate a canonical architecture with a balance between flexibility and rigidity of the anticodon hairpin that allows the elongator tRNA to perform a common decoding function on the ribosome. This common functional architecture in elongator tRNAs depends very much on the conserved residue U33. Only in exceptional cases, as in many initiator tRNAMet in eukaryotes (not discussed here) or in the tRNASer of most genius of Candida species harboring a leucine anticodon CUG, have differences at this positions been found (C33 or G33, respectively) (65,76).
Thus, the deciphering of AU-rich codons within the decoding A site of the ribosome requires extra stabilization sources that of course have to arise mainly from the tRNA. Along the course of evolution, various strategies based, as in any complex molecular system, on the interplay between many weak interactions together with a few strong interactions have offered alternatives to overcome this decoding problem. In the proximal ‘extended anticodon’, every single nucleotide between 31 to 39 in the anticodon loop contributes to uniform decoding by adapting its structure to the strength of the codon/anticodon pairings and the contacts within the ribosomal grip. Some combinations of B31-B39 and B32-B38 favor the canonical anticodon triplet conformation, contributing favorably to the free energy of binding, while others can be either too floppy or too rigid (with even the formation of an additional base pair at B32-B38), contributing unfavorably the free energy of binding. The stacking contacts of the modified base B37 sandwiched between B36 and B32-B38 can be extensive or minimal and contribute at the proper interface between the codon/anticodon triplet and the anticodon stem. Finally, the critical role of modifications of B34 for promoting non-standard base pairs has been emphasized.
DEVIATIONS FROM THE STANDARD GENETIC CODE
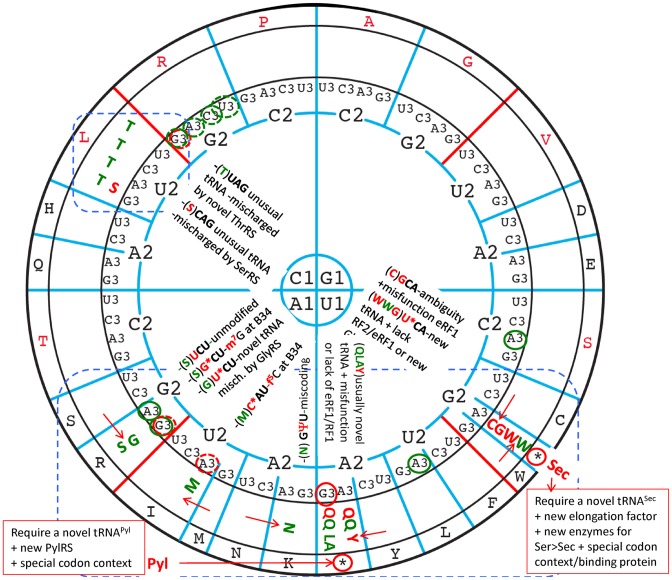
Despite the various decoding strategies existing in Bacteria, Archaea, Eukarya and organelles, the meaning of each of the individual 64 triplets of the genetic code remains remarkably universal. However, the genetic code is not frozen and a few deviations have been identified in the nuclear genomes of a limited number of species (fungi, green algae, ciliates, diplomonads and firmicutes), though mostly in the small A+T rich genomes of mitochondria (metazoan, fungi, green plants and red algae) (1,82,83). Particular codon reassignments appeared to have taken place repeatedly and independently in the different lineages, attesting that certain codons are more prone than others to this evolutionary process. Also, two proteogenic amino acids, selenocysteine (Sec) and pyrrolysine (Pyl), have been added to the standard 20-member amino acid alphabet (natural expansion of the genetic code). These two amino acids are inserted in a few proteins in response to stop codons UGA and UAG, respectively, of cytoplasmic mRNAs of a few bacteria, eukaryotes and even archaea (84,85).
Figure 9 summarizes the best-characterized genetic code deviations identified so far. The most frequently encountered deviations in both nuclear and mitochondrial genomes concern the switch of terminator codons to sense codons (UGA to Trp, Cys, Gly or Sec, UAG to Gln, Leu, Ala or Pyl, and UAA to Gln or Tyr). Identity changes of sense codons are less frequent and found mainly in mitochondrial genomes (except in one case). They are AUA-Ile to Met; AAR-Lys to Asn, AGA/G-Arg to Ser or Gly, all belonging to 2-codons family boxes for which the first base of the codon is either A1 or U1. The few cases where a C1-containing codon is involved are CUY/R-Leu to Thr or Ala in mitochondria of a few filamentous Saccharomycetacea and the CUG-Leu to Ser in the cytoplasm of Candida and Debaryomyces species. In these latter cases, the replacement of one amino acid by another results mainly from mischarging of a tRNA rather than of misreading of a codon during translation. Among the sense codons, those starting with G1 appear most resistant to amino acid reassignment. Also, as a direct consequence of strongly biased codon usage in mitochondria, a few codons are either not used (remain unassigned) or possibly used as alternative stop codons (2).
Figure 9.
Deviations from the standard, almost universal genetic code. The most frequent deviations concern terminator codons unexpectedly efficiently translated as sense codons for amino acids Trp, Gly, Cys, Gln, Tyr, Leu or Ala. Conventional one letter code for amino acid is used. In red are shown amino acid reassignments observed in nuclear genomes while in green are those related to mitochondrial genomes. The less frequently encountered reassignment of sense codons for a non-standard amino acid have been observed essentially in mitochondrial genome (indicated in green), except in one case (a Leu-codon coding for Ser in Candida and Debaryomyces species (83). For more details and references of original papers (in addition to those cited in text), (1,82,147). The codons remaining unassigned and possibly playing the role of occasional alternative stop codon in certain organisms are indicated by a red (nuclear) or green (mitochondria) circle around B3, dotted line means avoided codons, plain lines means unassigned, possibly stop codons. The red arrows between 2 boxes indicate a switches between amino acids within 2:2 decoding boxes. Outside the circle, are indicated the reassignment of stop codon UGA and UAG into Sec (21st proteogenic amino acid) and Pyl (22nd amino acid), respectively (2). The most frequent codons reassignments mostly occur within the blue dotted boxes.
Evidently, the most frequent codon reassignments are found in A/U-rich weaker codon binders and correspond mostly to switch between two amino acids of the same pair of split codon family boxes. These codons are also the most liable to translational miscoding (see for example (86–88)). Similarly, the expansion of the genetic code to additional selenocysteine (Sec) and pyrrolysine (Pyl) occurs mainly, but not exclusively at stop codons, again at the same more frequently AU-rich reassigned codons (85,89).
Changing the meaning of a codon in a genome is challenging. It implies a few coordinated adjustments of various elements of the translation apparatus that necessarily result from a multistep evolutionary process. Among these steps are: (i) mutation in a gene coding for a tRNA (or a duplicated variant) to allow the emergence of a ‘novel’ tRNA able to efficiently read a given codon (or a set of near cognate codons) that is (are) distinct from the original wild type tRNA species; (ii) mutations in the gene coding for an aminoacyl-tRNA synthetase (or a duplicated variant) to allow the mutant enzyme to charge efficiently the newly evolved tRNA with an amino acid distinct to that expected from its anticodon; (iii) mutations in the gene coding for a modification enzyme (or a duplicated variant) to allow (or anneal) specific base modification that can influence the decoding property of the novel tRNA; (iv) mutations in the gene coding for potential competing tRNA (or even its elimination) that may read the same reassigned codon(s); (v) mutations in the termination factor (or even its elimination) to avoid competition with the novel functional suppressor tRNA (discussed in (90)); (vi) sequence rearrangement in mRNA for signaling with stop codon (or sense codon) has to be recoded (or reassigned); (vii) invention of a new elongation factor for selectively bringing the Sec-tRNASec into the decoding site of the translating ribosome; (viii) possibly mutations in the rRNA or r-proteins of the decoding site of the ribosome to allow optimal translation of the newly reassigned codon by its new cognate (or near cognate) aa-tRNA. Not all of these adjustments of the translation apparatus are simultaneously required. However, depending on the case, at least a combination of them has to occur. In this way, each time a reassigned codon, or a stop codon with the appropriate neighboring sequence is encountered in mRNA, the newly assigned amino acid is incorporated following standard decoding mechanism on the ribosome. This is at variance with the phenomena of translational missense error or stop-codon ‘readthrough’ that are mediated by a standard aa-tRNAaa harboring non-cognate anticodon or after occasional mischarging of a tRNA with a wrong amino acid, which in fine give rise to occasional mis-incorporation of an amino acid into the growing polypeptide. The yields of such naturally occurring mischarging and misreading are usually very low (below 10−5 to 10−3), unless, again, mischarging is genetically programmed by multiple sequential mutations in the tRNA, mRNA, various translational proteins and/or even the rRNA or proteins of the ribosome in order to eventually reach the degree of perfection which has been achieved for the reassignment or recoding of codons in a few organisms (91).
Among all the above combinatory strategies for codon reassignments, the more intriguing ones are those involving posttranscriptional modification machineries. For example, in mitochondria of several metazoa and budding yeasts, C34 of the unique Met-tRNAMet is post-transcriptionally modified to 5-formyl-cytidine (f5C34) by a unique mitochondrial 5-formyl-cytosine synthase that allows the mature Met-tRNAMet to translate both codons AUG and AUA codons as Met (92,93). The completeness of such AUA-Ile-to-Met reassignment depends on the absence of a competing lysidine-containing Ile-tRNAIle (anticodon k2CAU). However, in mitochondria of a totally different lineage (Ascidian Halocynthia roretzi), instead of a f5C34, a 5-taurine-2-thio-methyl-2-thiouridine (τm5s2U34) and an exceptionally unmodified U37 were found in Met-tRNAMet (94). In the mitochondria of the squid Loligo bleekeri, an unmodified C34 and a rather normal anticodon loop were found but differences in other parts of the Met-tRNAMet were required for reading the AUA codon as Met (95). Evidently, different independent solutions have been developed along mitochondrial evolution for solving the same problem. Several other examples of this sort have been reported for the reassignements AAA-Lys to Asn (dependence on a peculiar pseudouridine-35 in Asn-tRNAAsn) and AGA/AGG-Arg to Ser or Gly (dependence on a unique m7G34 in Ser-tRNASer or τm5U34 in a B34-mutant of the same Ser-tRNASer) (1). Most of these reassignments depend on base modifications and are prevalent for codons belonging to the A1-quarter of the wheel code and, therefore, frequently depend on the decoding property of a U34-containing tRNAs, for which the panoply of chemical modifications is huge (see discussion above about the non-isostericity of UoG pairs and Figure 5B and Supplementary Figure S5).
The presence of nucleotide modifications, like m6A, m1A, I or Ψ in mRNAs can now be measured and assessed in translation (49,96–98). The presence of m6A has moderate effects on the measured translation efficacy, the effect being strongest with m6A at the first position (49,99). The effects of O6-methylG depend strongly on the codon position, with incorporation at the second position B2 leading surprisingly to stalling of the ribosomes (98). O6-methylG, because of the absence of a hydrogen atom at N1 position of the base, cannot form a Watson–Crick-like base pair with C, but does so with U. Consequently, a decrease in accuracy at the first and third B1 and B3 positions was observed but, unexpectedly, not at the second position. Incorporation of Ψ in bacterial mRNAs led to a 30% repression of translation, with the strongest effect at the B3 position (49). However, in this respect, the conversion of nonsense into sense codons by the presence of Ψ in the mRNA is particularly striking (42,100) (Supplementary Figure S7 shows that the observed miscodings occur at the bottom of the wheel and especially in the U1 quarter). The miscoding effects of Ψ35, in which the absence of a queuosine (Q34) promote misreading of G34oA3 or G34oG3 (101) and have been discussed above. In the published crystal structures, Ψ1 leads to the formation of non-Watson–Crick cis Hoogsteen–Watson–Crick G2oA35 or A2oG35 pairs, with the mRNA base in the syn conformation (42). More crystal structures are necessary to complete our understanding of the molecular basis of these intriguing observations (102).
Because the activity of modification enzymes usually depends on available cofactors of the basic cellular metabolism, we anticipate that the efficiency of such type of codon reassignments depends on the physiological state of the cell and may constitute a regulatory device. They possibly correspond also to early evolutionary steps toward less dependent and more permanent amino acid reassignment.
Briefly, the most frequently encountered code deviations, recoding and codon reassignment, occur in the U1/A1 quadrants, while the G1 quadrant is still unaffected to date. The most frequent miscoding, i.e. decoding errors and code ambiguities also occur in the bottom half of the wheel (together with the Glu/Asp and His/Gln ambiguities) (86,87). Such errors are highly dependent on the tRNA modifications and constitute a rich source of post-transcriptional regulation. Recoding of stop codons (Sec and Pyl) require, not only new enzymes for the new metabolism (4 for Sec and 2 for Pyl), but also several new factors and cofactors and, especially structures in the mRNA for signaling a given stop codon for recoding. Codon reassignments require less specific changes in the translational apparatus (aminoacyl tRNA synthetases, absence of some release factors, etc.) but appear very efficient without competition and should probably be favored for further studies and applications.
THE EVOLUTION OF THE GENETIC CODE
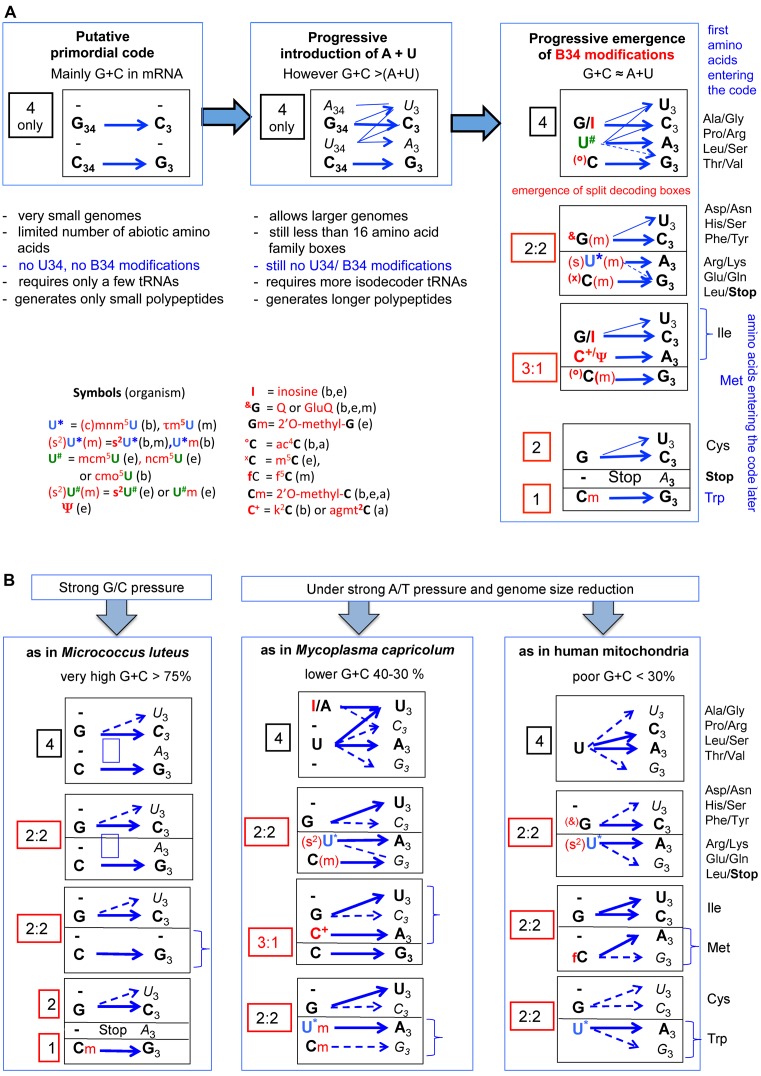
If the organization of the genetic code is, as we suggest, anchored in the structural contacts with the associated energetics correctly integrated within the networks of interactions at the ribosomal decoding grip, it should also reflect historical aspects of the evolution of the genetic code. Nowadays, it is understood that there is not a single explanation for the emergence and expansion of the genetic code as it stands to date. The current most plausible hypothesis is, however, that the earliest proto-biosynthetic system originated from RNA:RNA duplexes with the most stable complementary GC-rich triplets coding for small polypeptides composed of alternate hydrophobic Ala and hydrophilic Gly, the very first amino acids encoded. As more adenine and uracil was introduced into the RNA, this minimalist proto-biosynthetic system later progressively evolved to a more systematic use of repeating GNC, GNY, GNS or SNS triplets (N, Y and S denote respectively: any of the four canonical nucleotides; C or U; and C or G) (103–106). The earliest amino acids used to synthetize ancestral polypeptides were found in the prebiotic soup and selected according their specific interactions with the ancestral codons (the ‘stereochemical hypothesis’: (107,108)). These steps were subsequently expanded through the coevolution with the invention of biosynthetic pathways for new amino acids (the ‘amino acid metabolism hypothesis’, initially pointed out by: (109); reviewed and updated: by (110,111)) and the emergence of the corresponding primordial aminoacyl-tRNA synthetases able to fix these new amino acids on appropriated proto-tRNAs (‘co-evolution with tRNA aminoacylation systems’: (112–114)). Constant refinement at both the replication and translation levels allow to progressively minimize the impact of coding errors and to increase the diversity and functionality of proteins that can be made with a larger amino acid alphabet (the ‘error minimizing code hypothesis’: (107,115,116)). Finally, as mentioned in the previous section, the code can further evolve by reassignment of unused, temporarily ambiguous or less used codons for another canonical or even totally new amino acids (the ‘codon capture theory’: (117,118)). Finally, early horizontal transfer and collective evolution of the code through different subspecies has been emphasized (see for examples (119,120)). In other words, the present day genetic code did not necessarily result solely from divergent evolution, but also from collective evolution via the development of an innovation-sharing process that allows the emergence of a quasi universal genetic code among populations of species ‘speaking the same language’ (121).
The take-home lesson of all the above information is that along the expansion of the genetic code, an optimal stability of complementary codon–anticodon pairs appears to have been the main evolutionary force. Starting with a G/C-rich RNA coding for simple abiotic amino acids like Gly, Ala, Pro and probably a precursor of Arg allows the subsequent development of more complex modern proteosynthetic machineries as we know to date. As argued by Trifonov (122), the temporal order of the sequential acquisition of new amino acids into the code seems to result from a ‘descending’ thermostability of the respective codon:anticodon pairs, with initiation AUG codon among the latest codons to enter the code, together with UAG/UAA/UGA codons which remain unassigned and serve as termination codons. As explained above, this ‘descending’ thermostability was obviously compensated along evolution by a better organization of the tRNA anticodon hairpin and of the ribosomal grip within the ribosome. The main differences in reading the ‘ancient sense strong codons’ of the 4-codon boxes as compared to the subsequently acquired ‘new, weak, sense codons’ of the 2:2-codon and 3:1-codon family boxes is that the former are less dependent than the latter on stabilizing modified nucleotides within the proximal extended anticodon (Figures 6 and 7).
The progressive introduction of A and U into the primordial G/C-rich RNA (both in proto-mRNA and proto-tRNA) was of great advantage for expanding the decoding potentiality of the proteosynthesis machinery (Figure 10A). However, as mentioned in a preceding section, the use of U-rich codons and U34-containing tRNA is promiscuous, both for miscoding and frameshifting of the comma-free mRNA during translation. It was possible to solve this dilemma only by avoiding usage of such U-rich ‘slippery’ codons and by developing specific enzymatic systems for posttranscriptional modifications of base of the anticodon hairpin, mainly at B34 (Figure 7) and B37 (Figure 6C) (64,123–125). Because frequent frameshift events are probably more deleterious to the cell than frequent miscoding of certain codons, it is possible that emergence of B37 modifications systems preceded modifications at B34 (126–128). The advantage of restricting the coding potentiality of certain tRNA by modifications of B34 is that it allows the introduction of new amino acids into the code, which, however, require duplication of a tRNA gene and of the emergence of the corresponding aminoacyl-tRNA synthetases (co-evolution of tRNA genes, tRNA maturation enzymes and tRNA aminoacylation apparatus). The earliest of these modification systems would have appeared before the divergence of Woese's three major domains (129), perhaps very early or at least before completion of the genetic code. These modifications would be universally distributed today (130), while those more recently invented occur either specifically in archaea, bacteria and/or eukaryotes (Figure 5A–B, discussed in (64)). Lateral transfer of genes coding for a given modified enzymes present in bacteria, organelles and/or the cytoplasm of eukaryotes could also have occurred through endosymbiosis or virus infection. Remarkably, the chemical adducts of several modified nucleotides (B34 and B37) contain an amino acid, as in N6-threonylcarbamoyladenosine (t6A37), N6-hydroxynorvalinecarbamoyladenosine (hn6A37), N6-glycinecarbamoyl adenosine (g6A37), 5-taurinomethyl-2-thiouridine (tm5s2U34) and 3-(3-amino-3 carboxypropyl)-uridine (acp3U47), lysidine (k2C34), agmatidine (agm2C34) and glutamyl-queuosine (gluQ34). These modifications might be relics of ancient code, when the RNA hairpin structures harboring the primordial anticodons (the ancestor of tRNA) were strongly associated, and possibly even ‘charged’ with amino acids (131–133).
Figure 10.
Hypothetical stepwise evolution of the genetic code and translation machinery in Bacteria, Eukarya, Archaea and mitochondria. (A) Starting from highly G+C rich small RNA pieces and a few abiotic amino acids, the primordial translation system evolved by extending its coding capacity by progressive introduction of A and U in both the template and decoder RNAs (see also (122)). The final decoding machinery as we know to date (and illustrated on the left panel of the figure) results from a long and complex intricate stepwise coevolution with the emergence of metabolically generated amino acids (up to 20), duplication and speciation of proto-tRNAs with distinct anticodons (up to 40–45 to date), post-transcriptional modification enzymes (up to more than 100 known to date), new amino acid tRNA synthetases (up to 20), complexification of the ribosomal architecture (rRNAs and r-proteins) and the introduction of additional protein factors allowing the extension and ultimate tuning the efficacy and accuracy of the decoding capability of 61/62 sense (cognate and near cognate) codons with in addition 2 to 3 terminators for 20 proteogenomic natural amino acids. Pyrrolysine and selenocysteine have been excluded from our analysis, as well as the situation of the special tRNAMet involved in the initiation of protein synthesis that obviously arose later during cell evolution. Emphasis is given to the importance of B34 modifications (indicated in red) that allows the segregation of the A/U-rich 4-codon boxes into split decoding boxes 2:2 and 3:1 with subsequent additional amino acids to enter the code. (B) Following genomic selection conditions such as directional constraints or mutational pressure on codons (strong G/C as in M. luteus (148) or strong A/T combined with drastic genome size reduction as in mammalian mitochondria (57,60) or the minimalist bacteria M. capricolum (58,59,149), simplifications of the usual translational decoding system are evident, while preserving the split decoding boxes because of the need to encode for 20 amino acids. Less tRNA species with distinct anticodons (isodecoders) are required (22 for human mitochondria, 28 in M. capricolum and 29 in M. luteus, again not including tRNAMet initiator). However, remarkably the only remaining B34 modifications found in natural tRNAs are those related to the split 2:2 and/or 3:1 decoding boxes. More information in relation to the corresponding tRNA modification enzymes of a given tRNA repertoire in the subgroup of Mollicutes (mainly mycoplasmas) can be found in (150). The situation of M. luteus is remarkable for its total absence of U34-containing tRNAs while in mycoplasma and mitochondria U34-containing tRNA are critical. Codon usage correlates with tRNA repertoire and amino acid type (125,137–142). Symbols for each acronym of the most important modified nucleotides are indicated within the figure. In parenthesis b, e, a, m means bacteria, eukaryotes, archaea and mitochondria respectively. More details about their chemical structures are shows in Figure 7 and Supplementary Figure S4. Within each decoding boxes, blue arrows correspond to the most dominant codon:anticodon pairs. Only B34 are indicated. When symbols of B34 modification are in parenthesis, it means that the unmodified version of B34 is used only in certain decoding boxes, while the modified version(s) is (are) used in other decoding boxes (for details see in Figure 7 for E. coli and Supplementary Figure S4 for H. volcanii, S. cerevisiae, M. capricolum and human mitochondria, respectively). When B3 is in bold, it means that the codon usage corresponding to the particular codon is dominant over the other near cognate codons of the same 4- or 2- decoding boxes, while B34 indicated in regular italics correspond to rare codons (for details see as above, Figure 7 for E. coli and Supplementary Figure S4 for the other organisms analyzed).
As a rule, codons ending with pyrimidines U3 or C3 are rarely distinguishable in translation and always behave as synonymous codons, limiting the number of tRNAs able to read four codons to a maximum of three species (Figure 10A). Under special genomic selection conditions, such as strong A/T or G/C pressure, whether coupled or not with genome size reduction, the proteosynthetic machinery can further evolve toward a simpler version than the one found in majority of extant biological cells. In the parasitic Mycoplasmas like M. capricolum and more evidently in mitochondria, both with A/T-rich genome (Figure 10B), fewer isodecoder tRNAs (28 and 35, respectively, instead of 40–45 in most cells) are needed to translate mRNA coding for 20 canonical amino acids. The reduction in the number of distinct tRNAs concerns mainly C34-containing tRNAs in 4-codon boxes and eventually 2:2 decoding boxes with the consequence that now both Met and Trp become encoded by 2 synonymous codons as in mitochondria. More extensive tRNA reduction can occur with A34/G34-containing tRNA as long as the uracil of the remaining U34-containing tRNA is not post-transcriptionally modified (loss of corresponding maturation enzymes). Accordingly, the codon usage in these cells or organelles is strongly biased, with avoidance of NNG codons (see Supplementary Figure S4 and (59,60,117)). Conversely in some Micrococcus strains like M. luteus, with a G/C-rich genome (Figure 10B), reduction of tRNA applies only to U34-containing tRNAs and the usage of codons is almost exclusively NNC and NNG. The main difference between M. luteus (Figure 10B) and primordial cells is that 20 amino acids, and thus 2:2 codon boxes are used, while in the ancestral cells only a few amino acids, corresponding to only a few G/C-containing codons, prevailed.
Briefly, after an initial cooptation of a few abiotic amino acids into the code, further evolution of the primordial RNA with amino acids occurred through intricate but interdependent stepwise expansion of the amino acid biosynthetic pathways, coupled with the evolution of aminoacyl-tRNA synthetases and tRNA modification enzymes. As a result, codon:anticodon pairs with intrinsically weaker interactions provided greater diversity in amino acids (chemical structures and global polarities). These pairs also concentrate a large number of deviations from the genetic code or codon reassignments, and possibly correspond to best candidates for further insertions of additional non-canonical amino acids of special technical interest. The elucidation of mechanisms and the development of tools for the recoding of sense codons are currently actively being researched in order to be able to produce proteins with a variety of non-natural amino acids at several positions (134,135). The rules delineated here should, with further work, help in this development.
CONCLUSIONS
Cells from different organisms, or the same organism but from different tissues, generally have different population of functional tRNAs. Both the total number of tRNA species with distinct anticodons (tRNA repertoire or tRNome) and the relative amounts of each individual isoacceptor species (isodecoders) differ much from one type of cell to another (69,136). Similarly, the frequency with which each codon is translated (codon usage or codon preference) varies depending on the origin of cell and on the levels of gene expression. This is due partly to varying amino acid composition of the proteins synthesized and most importantly to the different use of synonymous codons (codon bias) that is dependent on the global G+C content of the mRNAs (125,137–142).
Inspection and comparison of the mappings of the extended anticodon concept wheels illustrates how, despite the constraining process of decoding and the use of an identical genetic code (except for Met and Trp in certain types of cells like mycoplasma and mitochondria), a large variety of tRNA repertoires and codon usage is still allowed and prevalent. The interpretation of this biological diversity in molecular terms is a daunting task. However, it should be clear that evaluating the frequency of a given codon in individual mRNAs or, as is usually done for statistical reasons, of bulk cellular mRNAs, is not sufficient, and that the exact composition of the tRNA repertoire in a given cell or organism (see below) has also to be determined. Complications arise from the many diverse underlying evolving molecular mechanisms that preside over the biogenesis, stability and functional regulation of each component (tRNAs, mRNAs, snoRNA in Eukarya and Archaea, possibly regulatory RNAs) of the translation machinery.
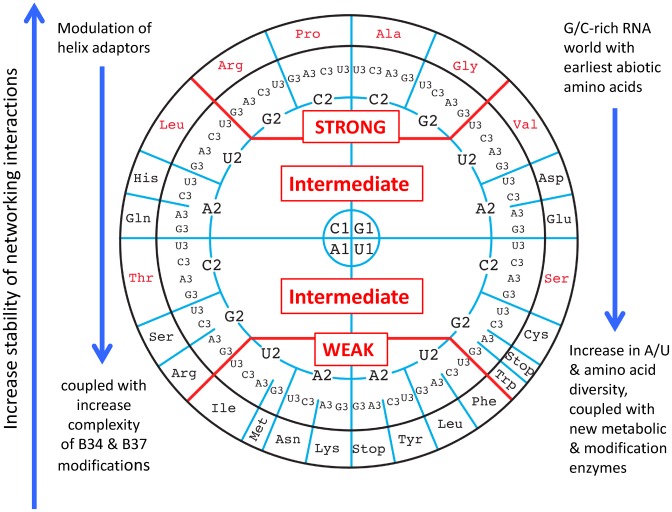
The majority of individual codons in mRNAs can be translated by either a cognate aa-tRNA harboring the three complementary base pairs or by one or several near-cognate aa-tRNA carrying the same amino acid differing by the base-34 or any other important base of the extended anticodon region. The main advantage of representing data related to such complex decoding system in the form of a wheel of genetic code is that all sequence co-variances can be visualized within an internal energy-based logic, for each anticodon of the 61 almost universally used sense codons in a given organism. The concomitant emergence of machineries allowing post-transcriptional enzymatic modifications of bases and ribose at position 34 of especially tRNA species of the split 2:2- and 3:1-codon boxes also play a determinant role, not only to help codon–anticodon stabilization but also to guarantee a better discrimination between the two sets of Y-ending and R-ending codons. Furthermore, for smooth and regular decoding and ribosomal progression along the correct frame of mRNAs, optimal and uniform binding of the various aminoacyl-tRNAs independently of codon types is required (Figure 11).
Figure 11.
Summary scheme illustrating the mapping of the energetics and evolutionary history on the wheel organization of the genetic code. Right vertical arrows: from bottom to top, there is an increase in the strengths of the networking interactions that is coupled with an increase of base modifications at B34 and B37 from top to bottom. Left vertical arrows at the right and left sides: the evolution from the primordial G/C-rich to A/U-rich codon/anticodons triplets required base modifications and the related metabolic enzymatic activities.
The codon usage, the number of tRNAs in each isoacceptor families and the numbers of isodecoder tRNAs all vary with cell type and cellular differentiation state but, despite the overwhelming complexity and potential ambiguity they introduce, as an ensemble they still have to comply with the networks of interactions which act at the decoding A site (Figure 12).
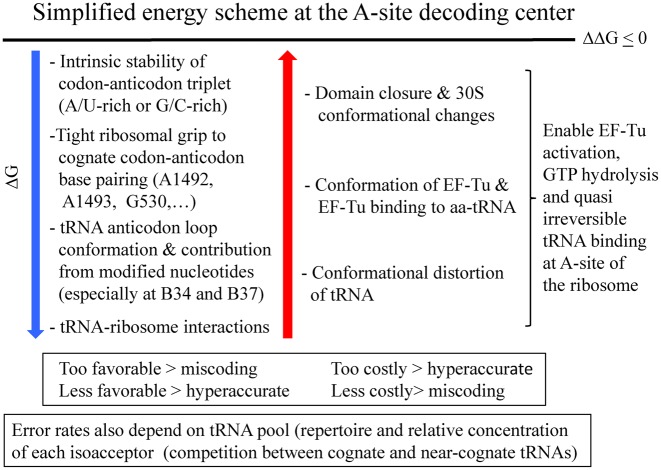
Figure 12.
Summary figure: an approximate and simplified energy scheme at the A site decoding center illustrates how the favorable and costly contributions top the free energies and compensate to maintain a smooth and regular translation process with minor final variations in free energy. On the wheel representation, the energies at the left can be mapped. However, not all energies can be mapped; for example, the interactions between the ribosome and parts other than the anticodon hairpin, the conformational distortions or alternative states of tRNAs, and the energies associated with ribosomal movements.
The multiplicity, diversity and constrained neutrality of the non-covalent and weak molecular interactions between the mRNA and tRNAs within the ribosomal grip allow for such a continuing and divergent evolution toward either complexity or simplification in the genetic code that clearly cannot be viewed as frozen. Changing the meaning of a codon in a genome implies a few coordinated adjustments of various elements of the translation apparatus that necessarily result from a multistep evolutionary process. Thorough rationalization and understanding of the molecular mechanisms underlying the decoding process and the errors in decoding fidelity should contribute to further developments and progress in engineering of the genetic code (85,143).
The main insights gained by the present new organization of the genetic code can be summarized in the following way.
The new organization of the genetic code clearly segregates 4-codon boxes from 2:2- and 3:1-codon boxes.
The 2:2 codon boxes present the weakest intrinsic stabilities and, among them, the STOP codons are the least stable.
The absence of symmetry in the wheel representation is not arbitrary. It correlates with all known parameters acting at the decoding site: the structure of the anticodon loop and stem, the intrinsic stability of the codon/anticodon interactions, the molecular grip exerted by the ribosome at the decoding site and the presence of characteristic modified nucleotides.
The co-variances of base-pair 32–38 and base-opposition 31–39 modulate the adaptation of the codon/anticodon triplet to the anticodon stem and the body of the tRNA.
The complexity of the chemical modifications at base 34 and 37 presents a correlation with AU-richness in the codon/anticodon pairs. The system requires fewer modifications with GC-rich codon/anticodon pairs.
The structurally based network of interactions results in an uniform decoding of all tRNAs that can adapt to the cellular constraints.
The structural and unsymmetrical role is played by residue 34 by exploiting a great diversity of mechanisms: tautomeric or (de)protonation forms, syn/anti conformational changes, and non-Watson–Crick pairs.
Modification of U34 is mandatory for tRNAs belonging to split 2:2-codon boxes: it allows reading only R-ending codons and possibly also the elimination of a C34-containing tRNA that reads the cognate G-ending codon. Conversely, all four codons of an unsplit 4-codon box can be translated by a single tRNA harboring a non-modified U34, but with a preference for A- and U-ending codons (biased codon usage).
Anticodon and codon are two sides of the same coin; the relative usage is necessarily correlated or co-adapted.
In the evolution of the triplet code, a major role was played by thermostability of codon–anticodon interactions. The malleability and, thus, the evolution of the genetic code is primarily based on genomic GC-content, with the progressive introduction of U/A together with tRNA modifications (especially at U34). This leads to a great diversity of number and type of the tRNA repertoire (and thus also of codon usage) depending on GC-content of the genome.
Supplementary Material
Acknowledgments
Drawings of the genetic code as a wheel in various formats are available at http://www-ibmc.u-strasbg.fr/spip-arn/rubrique138?lang=fr or upon requests to the authors. The authors would like to dedicate this article to two friends and colleagues recently deceased, Knut Nierhaus (Berlin) and Mathias Sprinzl (Bayreuth), who both contributed significantly to our understanding of ribosomal translation. The authors wish to thank numerous colleagues for constructive discussions on decoding during the preparation of this article, for help with the drawings, and for critical reading of the MS (J. Chihade, P. Farabaugh, A. Krol, D. Moras, M. Ryckelynck, M. Sissler, G. Yusupova, M. Yusupov).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
LABEX: ANR-10-LABX-0036_NETRNA and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program (to E.W.). Funding for open access charge: LABEX: ANR-10-LABX-0036_NETRNA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Watanabe K., Yokobori S. tRNA modification and genetic code variations in animal mitochondria. J. Nucleic Acids. 2011;2011:623095. doi: 10.4061/2011/623095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling J., O'Donoghue P., Söll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 2015;13:707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezerra A.R., Guimaraes A.R., Santos M.A. Non-standard genetic codes define new concepts for protein engineering. Life (Basel) 2015;5:1610–1628. doi: 10.3390/life5041610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nirenberg M., Leder P., Bernfield M., Brimacombe R., Trupin J., Rottman F., O'Neal C. RNA codewords and protein synthesis, VII. On the general nature of the RNA code. Proc. Natl. Acad. Sci. U.S.A. 1965;53:1161–1168. doi: 10.1073/pnas.53.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 6.Woese C.R., Dugre D.H., Saxinger W.C., Dugre S.A. The molecular basis for the genetic code. Proc. Natl. Acad. Sci. U.S.A. 1966;55:966–974. doi: 10.1073/pnas.55.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agris P.F., Vendeix F.A., Graham W.D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann J. Physico-chemical constraints connected with the coding properties of the genetic system. J. Theor. Biol. 2000;202:129–144. doi: 10.1006/jtbi.1999.1045. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Montano M.A. The fourfold way of the genetic code. Biosystems. 2009;98:105–114. doi: 10.1016/j.biosystems.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Danckwerts H.J., Neubert D. Symmetries of genetic code-doublets. J. Mol. Evol. 1975;5:327–332. doi: 10.1007/BF01732219. [DOI] [PubMed] [Google Scholar]
- 11.Wilhelm T., Nikolajewa S. A new classification scheme of the genetic code. J. Mol. Evol. 2004;59:598–605. doi: 10.1007/s00239-004-2650-7. [DOI] [PubMed] [Google Scholar]
- 12.Agris P.F. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 16.Jenner L., Demeshkina N., Yusupova G., Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 17.Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 18.Ogle J.M., Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 19.Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 20.Wohlgemuth I., Pohl C., Mittelstaet J., Konevega A.L., Rodnina M.V. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R Soc. Lond. B Biol. Sci. 2011;366:2979–2986. doi: 10.1098/rstb.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahlman R.P., Dale T., Uhlenbeck O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Ieong K.W., Johansson M., Ehrenberg M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9602–9607. doi: 10.1073/pnas.1506823112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 24.Nasvall S.J., Chen P., Björk G.R. The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA. 2007;13:2151–2164. doi: 10.1261/rna.731007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada Y., Matsugi J., Ishikura H. tRNA1Ser(G34) with the anticodon GGA can recognize not only UCC and UCU codons but also UCA and UCG codons. Biochim. Biophys. Acta. 2003;1626:75–82. doi: 10.1016/s0167-4781(03)00045-9. [DOI] [PubMed] [Google Scholar]
- 26.Lagerkvist U. Unorthodox codon reading and the evolution of the genetic code. Cell. 1981;23:305–306. doi: 10.1016/0092-8674(81)90124-0. [DOI] [PubMed] [Google Scholar]
- 27.Murphy F.V.t., Ramakrishnan V., Malkiewicz A., Agris P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Na.t Struct. Mol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 28.Murphy F.V.t., Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 29.Weixlbaumer A., Murphy F.V.t., Dziergowska A., Malkiewicz A., Vendeix F.A., Agris P.F., Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurata S., Weixlbaumer A., Ohtsuki T., Shimazaki T., Wada T., Kirino Y., Takai K., Watanabe K., Ramakrishnan V., Suzuki T. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008;283:18801–18811. doi: 10.1074/jbc.M800233200. [DOI] [PubMed] [Google Scholar]
- 31.Vendeix F.A., Murphy F.V.t., Cantara W.A., Leszczynska G., Gustilo E.M., Sproat B., Malkiewicz A., Agris P.F. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol. 2012;416:467–485. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G. Structural insights into the translational infidelity mechanism. Nat. Commun. 2015;6:7251. doi: 10.1038/ncomms8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagerkvist U. ‘Two out of three’: an alternative method for codon reading. Proc. Natl. Acad. Sci. U.S.A. 1978;75:1759–1762. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehmann J., Libchaber A. Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA. 2008;14:1264–1269. doi: 10.1261/rna.1029808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J.L., Dishler A.L., Kennedy S.D., Yildirim I., Liu B., Turner D.H., Serra M.J. Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters. Biochemistry. 2012;51:3508–3522. doi: 10.1021/bi3002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olejniczak M., Uhlenbeck O.C. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006;88:943–950. doi: 10.1016/j.biochi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Wohlgemuth I., Pohl C., Rodnina M.V. Optimization of speed and accuracy of decoding in translation. EMBO J. 2010;29:3701–3709. doi: 10.1038/emboj.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosjean H.J., de Henau S., Crothers D.M. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. U.S.A. 1978;75:610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houssier C., Grosjean H. Temperature jump relaxation studies on the interactions between transfer RNAs with complementary anticodons. The effect of modified bases adjacent to the anticodon triplet. J. Biomol. Struct. Dyn. 1985;3:387–408. doi: 10.1080/07391102.1985.10508425. [DOI] [PubMed] [Google Scholar]
- 40.Jenner L.B., Demeshkina N., Yusupova G., Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 41.Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez I.S., Ng C.L., Kelley A.C., Wu G., Yu Y.T., Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmeing T.M., Voorhees R.M., Kelley A.C., Ramakrishnan V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. 2011;18:432–436. doi: 10.1038/nsmb.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doherty E.A., Batey R.T., Masquida B., Doudna J.A. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 45.Auffinger P., Westhof E. An extended structural signature for the tRNA anticodon loop. RNA. 2001;7:334–341. doi: 10.1017/s1355838201002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Ahsen U., Green R., Schroeder R., Noller H.F. Identification of 2′-hydroxyl groups required for interaction of a tRNA anticodon stem-loop region with the ribosome. RNA. 1997;3:49–56. [PMC free article] [PubMed] [Google Scholar]
- 47.Khade P.K., Shi X., Joseph S. Steric complementarity in the decoding center is important for tRNA selection by the ribosome. J. Mol. Biol. 2013;425:3778–3789. doi: 10.1016/j.jmb.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fahlman R.P., Olejniczak M., Uhlenbeck O.C. Quantitative analysis of deoxynucleotide substitutions in the codon-anticodon helix. J. Mol. Biol. 2006;355:887–892. doi: 10.1016/j.jmb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Hoernes T.P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., Huttenhofer A., Erlacher M.D. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016;44:852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaher H.S., Green R. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell. 2010;39:110–120. doi: 10.1016/j.molcel.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demirci H., Wang L., Murphy F.V.t., Murphy E.L., Carr J.F., Blanchard S.C., Jogl G., Dahlberg A.E., Gregory S.T. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA. 2013;19:1791–1801. doi: 10.1261/rna.040030.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwal D., Gregory S.T., O'Connor M. Error-prone and error-restrictive mutations affecting ribosomal protein S12. J. Mol. Biol. 2011;410:1–9. doi: 10.1016/j.jmb.2011.04.068. [DOI] [PubMed] [Google Scholar]
- 53.Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 54.Polikanov Y.S., Melnikov S.V., Söll D., Steitz T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma S., Lafontaine D.L. 'View From A Bridge': A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015;40:560–575. doi: 10.1016/j.tibs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Jiang J., Seo H., Chow C.S. Post-transcriptional modifications modulate rRNA structure and ligand interactions. Acc. Chem. Res. 2016;49:893–900. doi: 10.1021/acs.accounts.6b00014. [DOI] [PubMed] [Google Scholar]
- 57.Bonitz S.G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F.G., Nobrega M.P., Thalenfeld B.E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 1980;77:3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samuelsson T., Guindy Y.S., Lustig F., Boren T., Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc. Natl. Acad. Sci. U.S.A. 1987;84:3166–3170. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 1989;209:37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki T., Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yarus M. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science. 1982;218:646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]
- 62.Cochella L., Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dale T., Uhlenbeck O.C. Amino acid specificity in translation. Trends Biochem Sci. 2005;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Grosjean H., de Crecy-Lagard V., Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010;584:252–264. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 65.Machnicka M.A., Olchowik A., Grosjean H., Bujnicki J.M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014;11:1619–1629. doi: 10.4161/15476286.2014.992273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barraud P., Schmitt E., Mechulam Y., Dardel F., Tisne C. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008;36:4894–4901. doi: 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Motorin Y., Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 68.Davis D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marck C., Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Auffinger P., Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999;292:467–483. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- 71.Westhof E., Dumas P., Moras D. Loop stereochemistry and dynamics in transfer RNA. J. Biomol. Struct. Dyn. 1983;1:337–355. doi: 10.1080/07391102.1983.10507446. [DOI] [PubMed] [Google Scholar]
- 72.Konevega A.L., Soboleva N.G., Makhno V.I., Semenkov Y.P., Wintermeyer W., Rodnina M.V., Katunin V.I. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 74.Davis D.R., Durant P.C. Nucleoside modifications affect the structure and stability of the anticodon of tRNA(Lys,3) Nucleosides Nucleotides. 1999;18:1579–1581. doi: 10.1080/07328319908044790. [DOI] [PubMed] [Google Scholar]
- 75.Davis D.R. Biophysicalandconformational properties of modified nucleotides in RNA. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. Washingthon D.C.: American Society for Microbiology Press; 1998. pp. 85–102. [Google Scholar]
- 76.Juhling F., Morl M., Hartmann R.K., Sprinzl M., Stadler P.F., Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ledoux S., Olejniczak M., Uhlenbeck O.C. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat. Struct. Mol. Biol. 2009;16:359–364. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murakami H., Ohta A., Suga H. Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat. Struct. Mol. Biol. 2009;16:353–358. doi: 10.1038/nsmb.1580. [DOI] [PubMed] [Google Scholar]
- 79.Shepotinovskaya I., Uhlenbeck O.C. tRNA residues evolved to promote translational accuracy. RNA. 2013;19:510–516. doi: 10.1261/rna.036038.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Curran J.F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- 81.Waas W.F., Druzina Z., Hanan M., Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- 82.Knight R.D., Freeland S.J., Landweber L.F. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 83.Miranda I., Silva R., Santos M.A. Evolution of the genetic code in yeasts. Yeast. 2006;23:203–213. doi: 10.1002/yea.1350. [DOI] [PubMed] [Google Scholar]
- 84.Ambrogelly A., Palioura S., Söll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 85.Mukai T., Englert M., Tripp H.J., Miller C., Ivanova N.N., Rubin E.M., Kyrpides N.C., Söll D. Facile recoding of selenocysteine in nature. Angew. Chem. Int. Ed. Engl. 2016;55:5337–5341. doi: 10.1002/anie.201511657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manickam N., Joshi K., Bhatt M.J., Farabaugh P.J. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res. 2016;44:1871–1881. doi: 10.1093/nar/gkv1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Björk G.R., Hagervall T.G. Transfer RNA modification. EcoSal Plus. 2014;6 doi: 10.1128/ecosalplus.ESP-0007-2013. doi:10.1128/ecosalplus.ESP-0007-2013. [DOI] [PubMed] [Google Scholar]
- 89.Brocker M.J., Ho J.M., Church G.M., Söll D., O'Donoghue P. Recoding the genetic code with selenocysteine. Angew. Chem. Int. Ed. Engl. 2014;53:319–323. doi: 10.1002/anie.201308584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lehman N. Molecular evolution: Please release me, genetic code. Curr. Biol. 2001;11:R63–66. doi: 10.1016/s0960-9822(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 91.Namy O., Rousset J.P., Napthine S., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 92.Takemoto C., Spremulli L.L., Benkowski L.A., Ueda T., Yokogawa T., Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616–1627. doi: 10.1093/nar/gkp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cantara W.A., Murphy F.V.t., Demirci H., Agris P.F. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. U.S.A. 2013;110:10964–10969. doi: 10.1073/pnas.1222641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suzuki T., Miyauchi K., Suzuki T., Yokobori S., Shigi N., Kondow A., Takeuchi N., Yamagishi A., Watanabe K. Taurine-containing uridine modifications in tRNA anticodons are required to decipher non-universal genetic codes in ascidian mitochondria. J. Biol. Chem. 2011;286:35494–35498. doi: 10.1074/jbc.M111.279810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ohira T., Suzuki T., Miyauchi K., Suzuki T., Yokobori S., Yamagishi A., Watanabe K. Decoding mechanism of non-universal genetic codes in Loligo bleekeri mitochondria. J. Biol. Chem. 2013;288:7645–7652. doi: 10.1074/jbc.M112.439554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., Leon-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 98.Hudson B.H., Zaher H.S. O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity. RNA. 2015;21:1648–1659. doi: 10.1261/rna.052464.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hudson G.A., Bloomingdale R.J., Znosko B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA. 2013;19:1474–1482. doi: 10.1261/rna.039610.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karijolich J., Yu Y.T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tomita K., Ueda T., Watanabe K. The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res. 1999;27:1683–1689. doi: 10.1093/nar/27.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Svidritskiy E., Madireddy R., Korostelev A.A. Structural basis for translation termination on a pseudouridylated stop codon. J. Mol. Biol. 2016;428:2228–2236. doi: 10.1016/j.jmb.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eigen M., Winkler-Oswatitsch R. Transfer-RNA, an early gene. Naturwissenschaften. 1981;68:282–292. doi: 10.1007/BF01047470. [DOI] [PubMed] [Google Scholar]
- 104.Eigen M., Winkler-Oswatitsch R. Transfer-RNA: the early adaptor. Naturwissenschaften. 1981;68:217–228. doi: 10.1007/BF01047323. [DOI] [PubMed] [Google Scholar]
- 105.Ikehara K. Origins of gene, genetic code, protein and life: comprehensive view of life systems from a GNC-SNS primitive genetic code hypothesis. J. Biosci. 2002;27:165–186. doi: 10.1007/BF02703773. [DOI] [PubMed] [Google Scholar]
- 106.Trifonov E., Berezovsky I. Molecular evolution from abiotic scratch. FEBS Lett. 2002;527:1–4. doi: 10.1016/s0014-5793(02)03165-4. [DOI] [PubMed] [Google Scholar]
- 107.Yarus M., Caporaso J.G., Knight R. Origins of the genetic code: the escaped triplet theory. Annu. Rev. Biochem. 2005;74:179–198. doi: 10.1146/annurev.biochem.74.082803.133119. [DOI] [PubMed] [Google Scholar]
- 108.Yarus M., Widmann J.J., Knight R. RNA-amino acid binding: a stereochemical era for the genetic code. J. Mol. Evol. 2009;69:406–429. doi: 10.1007/s00239-009-9270-1. [DOI] [PubMed] [Google Scholar]
- 109.Wong J.T. A co-evolution theory of the genetic code. Proc. Natl. Acad. Sci. U.S.A. 1975;72:1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ronneberg T.A., Landweber L.F., Freeland S.J. Testing a biosynthetic theory of the genetic code: fact or artifact? Proc. Natl. Acad. Sci. U.S.A. 2000;97:13690–13695. doi: 10.1073/pnas.250403097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Di Giulio M. An extension of the coevolution theory of the origin of the genetic code. Biol. Direct. 2008;3:37. doi: 10.1186/1745-6150-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Klipcan L., Safro M. Amino acid biogenesis, evolution of the genetic code and aminoacyl-tRNA synthetases. J. Theor. Biol. 2004;228:389–396. doi: 10.1016/j.jtbi.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 113.Delarue M. An asymmetric underlying rule in the assignment of codons: possible clue to a quick early evolution of the genetic code via successive binary choices. RNA. 2007;13:161–169. doi: 10.1261/rna.257607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith T.F., Hartman H. The evolution of Class II Aminoacyl-tRNA synthetases and the first code. FEBS Lett. 2015;589:3499–3507. doi: 10.1016/j.febslet.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 115.Freeland S.J., Wu T., Keulmann N. The case for an error minimizing standard genetic code. Orig. Life Evol. Biosph. 2003;33:457–477. doi: 10.1023/a:1025771327614. [DOI] [PubMed] [Google Scholar]
- 116.Buhrman H., van der Gulik P.T., Klau G.W., Schaffner C., Speijer D., Stougie L. A realistic model under which the genetic code is optimal. J. Mol. Evol. 2013;77:170–184. doi: 10.1007/s00239-013-9571-2. [DOI] [PubMed] [Google Scholar]
- 117.Osawa S., Jukes T.H., Watanabe K., Muto A. Recent evidence for evolution of the genetic code. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schultz D.W., Yarus M. On malleability in the genetic code. J. Mol. Evol. 1996;42:597–601. doi: 10.1007/BF02352290. [DOI] [PubMed] [Google Scholar]
- 119.Smets B.F., Barkay T. Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat. Rev. Microbiol. 2005;3:675–678. doi: 10.1038/nrmicro1253. [DOI] [PubMed] [Google Scholar]
- 120.Shackelton L.A., Holmes E.C. The role of alternative genetic codes in viral evolution and emergence. J. Theor. Biol. 2008;254:128–134. doi: 10.1016/j.jtbi.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 121.Vetsigian K., Woese C., Goldenfeld N. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trifonov E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene. 2000;261:139–151. doi: 10.1016/s0378-1119(00)00476-5. [DOI] [PubMed] [Google Scholar]
- 123.Atkins J.F., Baranov P.V. The distinction between recoding and codon reassignment. Genetics. 2010;185:1535–1536. doi: 10.1534/genetics.110.119016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van der Gulik P.T., Hoff W.D. Unassigned codons, nonsense suppression, and anticodon modifications in the evolution of the genetic code. J. Mol. Evol. 2011;73:59–69. doi: 10.1007/s00239-011-9470-3. [DOI] [PubMed] [Google Scholar]
- 125.Novoa E.M., Pavon-Eternod M., Pan T., Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 126.Björk G.R., Jacobsson K., Nilsson K., Johansson M.J., Bÿstrom A.S., Persson O.P. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thiaville P.C., El Yacoubi B., Kohrer C., Thiaville J.J., Deutsch C., Iwata-Reuyl D., Bacusmo J.M., Armengaud J., Bessho Y., Wetzel C., et al. Essentiality of threonylcarbamoyladenosine (t(6) A), a universal tRNA modification, in bacteria. Mol. Microbiol. 2015;98:1199–1221. doi: 10.1111/mmi.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sample P.J., Koreny L., Paris Z., Gaston K.W., Rubio M.A., Fleming I.M., Hinger S., Horakova E., Limbach P.A., Lukes J., et al. A common tRNA modification at an unusual location: the discovery of wyosine biosynthesis in mitochondria. Nucleic Acids Res. 2015;43:4262–4273. doi: 10.1093/nar/gkv286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McKenney K.M., Alfonzo J.D. From prebiotics to probiotics: The evolution and functions of tRNA modifications. Life (Basel) 2016;6 doi: 10.3390/life6010013. doi:10.3390/life6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Di Giulio M. Reflections on the origin of the genetic code: a hypothesis. J. Theor. Biol. 1998;191:191–196. doi: 10.1006/jtbi.1997.0580. [DOI] [PubMed] [Google Scholar]
- 132.Grosjean H., de Crecy-Lagard V., Björk G.R. Aminoacylation of the anticodon stem by a tRNA-synthetase paralog: relic of an ancient code? Trends Biochem. Sci. 2004;29:519–522. doi: 10.1016/j.tibs.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 133.Szathmary E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet. 1999;15:223–229. doi: 10.1016/s0168-9525(99)01730-8. [DOI] [PubMed] [Google Scholar]
- 134.Liu C.C., Schultz P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 135.Chin J.W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 136.Goodenbour J.M., Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 138.Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982;18:199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- 139.Rocha E.P. Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ran W., Higgs P.G. The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol. Biol. Evol. 2010;27:2129–2140. doi: 10.1093/molbev/msq102. [DOI] [PubMed] [Google Scholar]
- 141.Quax T.E., Claassens N.J., Söll D., van der Oost J. Codon bias as a means to fine-tune gene expression. Mol. Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wald N., Alroy M., Botzman M., Margalit H. Codon usage bias in prokaryotic pyrimidine-ending codons is associated with the degeneracy of the encoded amino acids. Nucleic Acids Res. 2012;40:7074–7083. doi: 10.1093/nar/gks348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Iwane Y., Hitomi A., Murakami H., Katoh T., Goto Y., Suga H. Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes. Nat. Chem. 2016;8:317–325. doi: 10.1038/nchem.2446. [DOI] [PubMed] [Google Scholar]
- 144.Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Limbach P.A., Crain P.F., McCloskey J.A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nakamura Y., Gojobori T., Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Beier H., Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kano A., Andachi Y., Ohama T., Osawa S. Novel anticodon composition of transfer RNAs in Micrococcus luteus, a bacterium with a high genomic G + C content. Correlation with codon usage. J. Mol. Biol. 1991;221:387–401. doi: 10.1016/0022-2836(91)80061-x. [DOI] [PubMed] [Google Scholar]
- 149.de Crecy-Lagard V., Marck C., Brochier-Armanet C., Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 150.Grosjean H., Breton M., Sirand-Pugnet P., Tardy F., Thiaucourt F., Citti C., Barre A., Yoshizawa S., Fourmy D., de Crecy-Lagard V., et al. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet. 2014;10:e1004363. doi: 10.1371/journal.pgen.1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.