Abstract
The RNA world hypothesis refers to the early period on earth in which RNA was central in assuring both genetic continuity and catalysis. The end of this era coincided with the development of the genetic code and protein synthesis, symbolized by the apparition of the first non-random messenger RNA (mRNA). Modern transfer-messenger RNA (tmRNA) is a unique hybrid molecule which has the properties of both mRNA and transfer RNA (tRNA). It acts as a key molecule during trans-translation, a major quality control pathway of modern bacterial protein synthesis. tmRNA shares many common characteristics with ancestral RNA. Here, we present a model in which proto-tmRNAs were the first molecules on earth to support non-random protein synthesis, explaining the emergence of early genetic code. In this way, proto-tmRNA could be the missing link between the first mRNA and tRNA molecules and modern ribosome-mediated protein synthesis.
INTRODUCTION
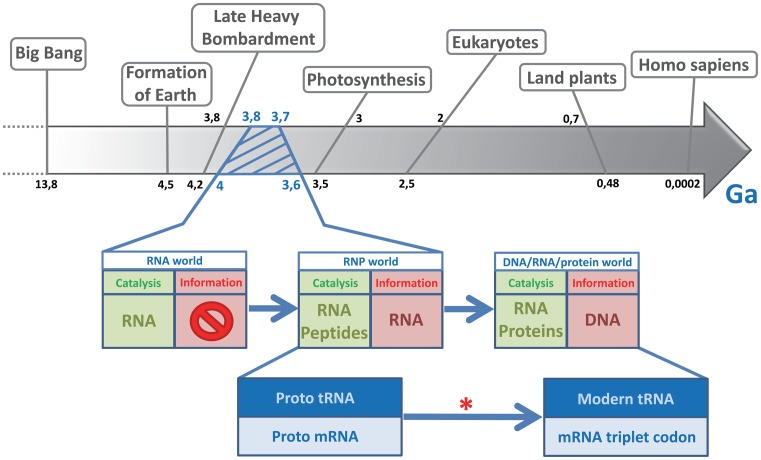
The RNA world hypothesis was first proposed over forty years ago as a major and early step in the evolution of life, at a time when there was no protein synthesis mechanism as it exists now (1–3). The theory is based on the capacity of RNA to simultaneously catalyze enzymatic reactions and store genetic information, as now done by proteins and DNA, respectively. RNA's intrinsic weaknesses support such a slow shift to modern molecular biology, in which genetic information passes from DNA to RNA and possibly to proteins. Indeed, despite the versatility of RNA, DNA has a higher molecular stability for carrying genetic information, and proteins have higher catalytic abilities. At some point during the RNA world, an evolutionary leap took place between the first system able to replicate molecules responsible for biochemical reactions (i.e. self-replicating RNA), and the cell that replicates a whole genome encoding for these biochemical activities. This evolutionary leap was embodied by the emergence of non-random coding RNA, which served as the first medium for genetic information. From there, RNA and peptides had to co-evolve through a ribonucleoprotein (RNP) world (Figure 1). This step would have been fundamental for the development of the code of life. At that time (Figure 1, red star), the world must have consisted of: a network of small RNAs sufficiently evolved as to have different catalytic and self-replicating properties; some primitive amino acids such as alanine, glycine, and aspartic acid; and the minimal prerequisites for translation, such as the first proto-ribosomes. Coding RNA may then have evolved to become non-random and specifically recognized as messenger RNA rather than another RNA type. The evolution of the translation mechanism will have been the result of ‘molecular Darwinism’, or in other words a random phenomenon leading progressively to a selective advantage.
Figure 1.
Timeline of early events, emphasizing the transition from an RNA world to modern life. During the history of life on earth, the RNA world lasted from the first appearance of short catalytic RNAs right up to the transition to a modern period in which genetic information carried by DNA and RNA became translated into proteins. The RNP (ribonucleoprotein) world was the intermediate period in which RNA and the first random peptides coexisted as informational and catalytic molecules. The red star indicates when the genetic medium stopped being random. Ga, giga-annum, or 109 (1 000 000 000) years.
Many molecular fossils of the RNA world are still present and even active in modern organisms. Candidates must be either catalytic, ubiquitous, and/or central to some aspect of metabolism (4). Transfer-messenger RNA (tmRNA) is a hybrid molecule present in all bacteria. It exhibits properties of both transfer and messenger RNA, and permits the rescue of ribosomes arrested during translation. So not only does tmRNA play a key cellular role in modern bacteria, but in a single molecule it also has two major and ancient functions that were necessary for the transition from an RNA world to the modern protein synthesis pathways.
ORIGINS OF THE GENETIC CODE
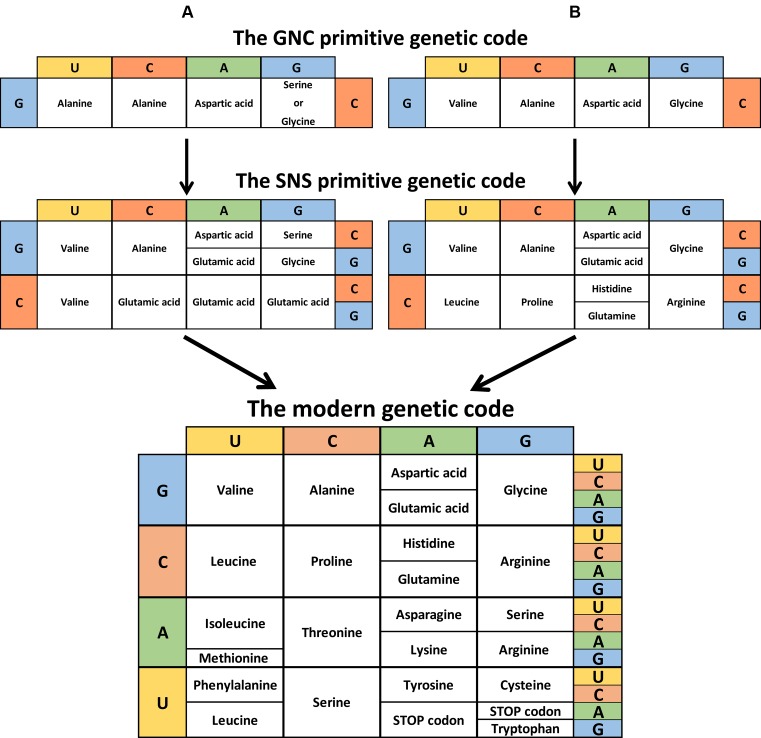
The genetic code is the set of rules by which the information encoded within DNA and messenger RNA (mRNA) is translated into proteins. The information is contained in the mRNA sequence, combined into codons or nucleotide triplets. Each codon corresponds to either a specific amino acid or to a stop signal which terminates protein synthesis. With four different nucleotides and a code made of nucleotide triplets, there are 34 = 64 possibilities to code 20 amino acids and three stop codons. Consequently, modern genetic code is degenerated, or in other words, most amino acids are encoded by more than one codon. Although we may not know what led to the current distribution of codons and their corresponding amino acids, these distributions are not random. For example, amino acids that share the same biosynthetic pathway or similar polarities and/or side-chain sizes tend to be close to each other on the genetic code table (Figure 2) (5).
Figure 2.
Theory of the genetic code evolution. This shows the evolutionary pathway going from the GNC code (4 codons) to the SNS code (16 codons) to the universal genetic code (64 codons). (A) Adapted from Massimo Di Giulio (72). (B) Adapted from Kenji Ikehara (10). (C) Instead of the conventional representation, the modern genetic code is shown reflecting the order of codon occurrence (columns G and U inverted).
Many theories have been put forward about the origins of the genetic code. Francis Crick first described merely a ‘frozen accident’ (1). This would mean that the system of twenty amino acids and their designated codons was good enough to work, but too resistant to change to improve. Other possibilities are: that the current shape of the code depends greatly on specific primordial biochemical interactions (such as those between RNA and amino acids) (6); that modern genetic code grew from an earlier and simpler code through a ‘biosynthetic expansion’ process (7); or that it resulted from information channels (8). Though the different forces that led to the evolution of the genetic code are unknown, there is a generally accepted model describing its appearance. In this, an RNA charged with an amino acid (proto-tRNA) targeted a proto-mRNA by complementary interactions. The evolution of interactions between the proto-tRNA and the proto-mRNA then gave birth to the first mRNAs coding for non-random short peptides (9). This reaction was probably promoted by a proto-ribosome. Today, based on the biosynthetic pathway of each amino acid and on the coevolution of transfer RNA (tRNA) and aminoacyl-tRNA synthetases (aaRS), we can reliably predict the order in which the codons appeared. The first generation of codons were of the GNS type (G = G; N = A, U, C, or G; and S = C or G), while the second generation were SNS (10). This early genetic code continued to evolve, maximizing its efficiency, until it became as it is today (Figure 2).
MODERN RIBOSOMES AND PROTO-RIBOSOMES
Nowadays, the information carried by DNA is first transcribed into mRNA, which in turn can be translated into proteins by the ribosome. Ribosomes are made of two subunits, themselves made up of ribosomal RNA (rRNA) and proteins. The ribosomes translate mRNA codons into one of the twenty amino acids that make up proteins. In a broad outline of the process (see (11) for a complete review), the small subunit permits the decoding of genetic information carried by mRNA, while the large subunit catalyzes the link between amino acids (peptide bonds). The first atomic resolution structures of the ribosome allowed for a precise description of its catalytic heart, the Peptidyl Transfer Center (PTC). Strikingly, the ribosome is a ribozyme, since only RNA performs its key role of peptide bond formation (12,13). The PTC's highly conserved structure is symmetrical, made up of a ‘stem–elbow–stem’ RNA of approximately 180 nucleotides. Its structure probably originated from the dimerization of two stem–elbow–stem motifs forming a pocket. This form evolved because it offers the best substrate orientation for peptide bond catalysis. The PTC is currently considered to be the first proto-ribosome dating from the RNA world (14–16).
ORIGINS OF TRANSFER RNA: BRIDGING TWO GENETIC CODES
Transfer RNA (tRNA) molecules have a major role to link the genetic information carried by mRNA codons and the corresponding amino acids necessary for protein synthesis. Because of their central role, tRNA have two distinct characteristics corresponding to two different genetic codes (17). First, a tRNA carries the anticodon, a specific nucleotide triplet which corresponds to the mRNA codon. Secondly, each tRNA also binds with high specificity to the amino acid corresponding to its anticodon, in a reaction catalyzed by a specific aminoacyl-tRNA synthetase (aaRS). In this sense, tRNA is a key molecule for combining ribonucleotide information (ancient RNA world) and peptide information (modern protein world). The specific attachment of a particular amino acid to its corresponding tRNA is referred to as the ‘second genetic code’. In fact, this second code must have appeared first (17) then evolved together with the aminoacyl-tRNA synthetases, a family of enzymes believed to be among the oldest proteins on earth (18). Today, aaRS discriminates between different tRNAs by recognizing elements in both the anticodon loop and acceptor stem of the RNA (19). The tRNA molecule is present in all organisms and its secondary structure is among the most evolutionarily conserved. Consequently, it is commonly accepted that at least from the Last Universal Common Ancestor (LUCA), its origin is monophyletic.
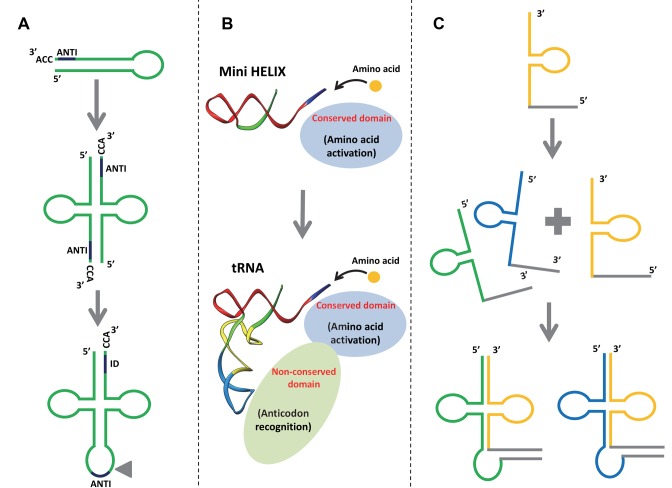
Several models propose explanations for the molecular mechanisms leading to the formation of modern tRNA (Figure 3). Ancestral tRNA could have been encoded by split genes, which later were merged to encode modern tRNA (Figure 3A) (20). Modern tRNAs could instead be the result of a fusion between two ancient RNA minihelices (Figure 3B) (21). A third model is based on the sequence analysis of archaeal tRNA genes, focusing on the presence of introns for clues (Figure 3C) (22). Indeed, during archaeal evolution the nucleotide sequences of the 3′-end (CCA sequence) were more conserved than those of the 5′-halves. Since the 3′-end is required for adding the amino acid residue to tRNA, it would seem that the 3’-arm evolved first. A variety of tRNA species would have been generated at a later stage through asymmetric combination with different 5′-tRNA halves. Accordingly, most tRNA sequences have vestiges of double hairpin folding, suggesting that the structure of tRNA molecules could have been the result of double hairpin formation in the ancient prebiotic world (23).
Figure 3.
Different models explaining the origins of tRNA. (A) tRNA may originate from the dimerization of two hairpin structures. ANTI, anticodon; ID, the discrimination region for identifying tRNA. The triangle represents the position where the intron is found in tRNA genes (73). (B) tRNA may originate from the late fusion between two RNA minihelices. A new aminoacyl tRNA synthetase (aaRS) domain links the operational RNA code to the trinucleotides of the genetic code (21). (C) tRNA may originate from the fusion of split genes of non-contiguous tRNAs (22).
Whatever the scenario, tRNA evolution is closely linked to aminoacylation. Although it is not precisely known how it operated in the RNA world, aminoacylation can be achieved through spontaneous chemical reactions. Indeed, it is possible to aminoacylate an RNA minihelix with an aminoacyl phosphate oligonucleotide (24), which would explain the importance of amino acid chirality in this mechanism. In another study it was shown that a simple pocket formed by a complex of four nucleotides (the C4N RNA hairpin) can be aminoacylated with specificity at its 3′-end by a simple Val–Asp dipeptide (25). The emergence of aaRS-like ribozymes certainly increased reaction specificity. Such ribozymes, referred to as flexizymes, have not yet identified in vivo, but they were isolated by means of in vitro selection (26). In the early stages of the translational system's development, the discrimination was made close to the CCA 3′-end (27). During evolution, it moved away from the acceptor stem loop until it arrived to the current situation, with aaRS recognition performed in both the anticodon and acceptor stem loops (28).
FUNCTION AND STRUCTURE OF TRANSFER-MESSENGER RNA
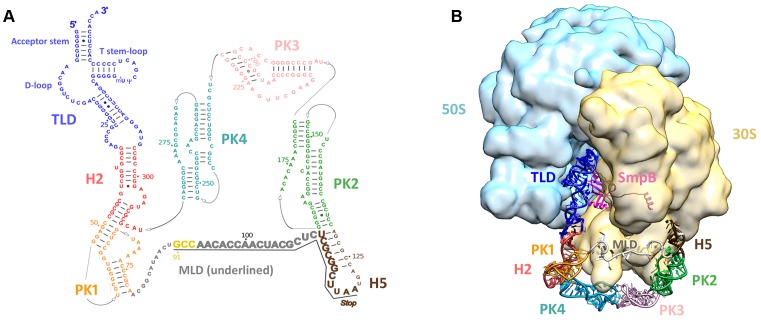
In all bacteria, trans-translation is the primary rescue system that permits the release of stalled ribosomes as well as the elimination of the related incomplete proteins and mRNA. A particular RNA performs this process: tmRNA associated with Small protein B (SmpB). tmRNA is a hybrid molecule carrying out both transfer and messenger RNA activities, and its total length varies between about 260 and 430 nucleotides, depending on the cell species (Figure 4). It is always aminoacylated by alanine. As for SmpB, its topology makes it similar to several other modern RNA‐binding proteins associated with translation, such as ribosomal protein s17, aspartyl tRNA synthetase, or the prokaryotic translation initiation factor IF1 (29). This structural link not only suggests that they may share a common ancestor or be linked by an evolutionary relationship, but also that they emerged late, when modern translation was already set up. Thanks to SmpB, the tRNA-like domain (TLD) of tmRNA recognizes and enters the vacant decoding site of ribosomes stalled at the 3’-end of truncated mRNAs, thus restarting translation. In a sophisticated ballet, protein synthesis then switches on the mRNA part of tmRNA. This allows the truncated protein to be extended by a short sequence of amino acids that tags it for destruction by cellular proteases, while the incomplete mRNA is released to be destroyed by ribonucleases (for a full review see (30)). Trans-translation occurs frequently, accounting for as much as 2–4% of translation reactions in Escherichia coli (31). It is found almost exclusively in the bacterial world (32), with a few exceptions. Trans-translation is also found in the plastomes of some primitive algae (33–35); in some rare diatoms (Stramenopila) that acquired genes from marine bacteria (36); and in the Jakobids, sub-lineages of the very distant organismal group Ecavates (37). The Jakobids are interesting due to their uniquely bacterial-like mitochondrial genomes (38). They are considered to be some of the most ancient living eukaryotes, which would explain the presence of tmRNA in their mitochondrial genome. Although faced with the same need for protein synthesis quality control, eukaryotic and archaeal cells have distinct rescue systems that have evolved differently. Although faced with the same need for quality control of protein synthesis, eukaryotic and archaeal cells have distinct rescue systems that have evolved differently. These include nonsense-mediated decay (NMD), no-go decay (NGD), and nonstop decay (NSD) in eukaryotes (39,40); and NGD in archaea (41). At the beginning of the DNA–RNA–protein era, translation errors and (proto)-ribosome stalling must have been far more frequent, and the need for an unlocking system such as trans-translation was even more necessary. Therefore, one can assume that a selective pressure led to the appearance of tmRNA then to its high conservation during evolution, an assumption which seems confirmed by the fact that tmRNA is now ubiquitous in all bacteria. On the other hand, SmpB is likely to have arised as modern tmRNA needed a more accurate way to recognize and rescue stalled ribosomes. tmRNA might therefore be the molecular fossil that coevolved with the translational system up to the present day.
Figure 4.
tmRNA secondary and tertiary structures. (A) Secondary structure diagram of Thermus thermophilus tmRNA. Watson–Crick base pairs are connected by lines and GU pairs are represented by dots. Domains are in color: the tRNA-like domain (TLD) is blue; helix 2 (H2) is red; pseudoknot 1 (PK1) is orange; helix 5 (H5) is brown; pseudoknot 2 (PK2) is green; pseudoknot 3 (PK3) is pink; and pseudoknot 4 (PK4) is light blue. The nucleotides within the internal open reading frame (ORF) are underlined and shown in a larger font. The resume codon is yellow and the STOP codon is indicated. (B) Cryo-EM map of the alanyl-tmRNA-SmpB complex bound to a stalled ribosome. 3D molecular model of tmRNA based on the homology modeling of each independent domain followed by flexible fitting into the cryo-EM density map of the accommodated step (74). EMDB entry: EMD-5188. Color code: tmRNA domains are the same as in (A); SmpB is magenta; the small 30S subunit is yellow; the large 50S subunit is light blue.
ORIGINS OF TRANSFER-MESSENGER RNA
Introns: common features in tmRNA and tRNA?
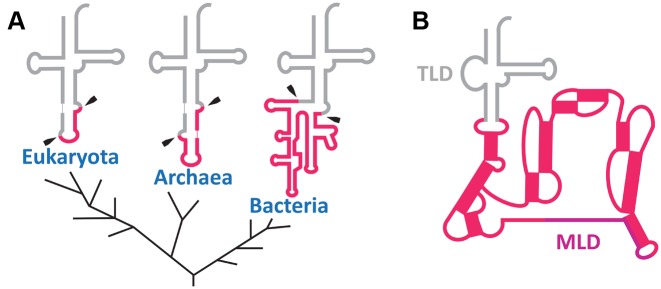
In the three domains of life, some rare tRNA molecules still undergo splicing of non-coding sequences (introns) located within the anticodon stem–loop (Figure 5A) (42). In bacteria, these introns are eliminated by self-splicing, while in archaea and eukaryotes they are taken care of by endonucleases. The origin of these introns is still being debated. In an ‘introns-early’ scenario (27,43), all essential tRNA genes had an intermediate block in the anticodon loop that had to be removed, and large parts of these introns were lost during evolution. A second scenario is ‘introns-late’ (44), and it theorizes that introns were inserted into genes after the emergence of tRNA. Although there is no evidence for a biological role for these non-coding intron regions, it must be noted that they are present in all three domains of life. The selective pressures that allowed the conservation of some of them up to the present day remain unclear, but their presence seems to be a signature of tRNA evolution. Remarkably, a simple comparison of the secondary structures shows that the positioning of the introns in the anticodon stem–loop of modern tRNA is the same as the large and structured ring abutting the TLD of tmRNA (Figure 5B). Other similarities—such as the length of these extra sequences and the presence of highly structured stem–loops—reinforce the idea that there is a link between modern tmRNA and tRNA. In bacteria, these introns belong to self-splicing group I introns which must be ancestral as they have characteristics that are omnipresent in different bacterial phyla and as they have self-catalytic activity reminiscent of the RNA world (45). The typical secondary structure of a group I intron consists of approximately ten paired elements organized into three domains (46). Self-splicing group I introns use a mechanism based on two distinct guanines as a cofactor (Figure 6A). The first guanine is located at the beginning of the intron in the Internal Guide Sequence (IGS), a highly conserved element in a stem–loop structure. An interesting particularity of this stem–loop is that the guanine used for the self-splicing reaction is associated with a uracil (G:U) (Figure 6A). This mismatch is conserved in all pre-tRNA molecules having a group I intron (47). Strikingly this mismatch characteristic is also found in the TLD domain of tmRNA, at the position G24:U328 ± 1 (Figure 6B) which would correspond to the start of a pre-tRNA intron (Figure 6C). The possible reason for having an intron in the tRNA anticodon is so strange today that neither its origins nor any evolutionary advantages that may have led to its preservation are known (48).
Figure 5.
Position and secondary structure similarities between the tRNA intron and tmRNA pseudoknots. (A) The tRNA intron in the three major kingdoms (Bacteria, Archaea and Eukaryota), adapted from Akio Kanai (22). Introns are framboise and mature tRNA is gray. Intron clipping sites are indicated with black arrows. (B) Secondary structure of tmRNA. The tRNA-like (TLD) and mRNA-like (MLD) domains are indicated, and the pseudoknots are framboise. Note the similar positioning of the tRNA introns in the three domains of life and in the other tmRNA domains.
Figure 6.
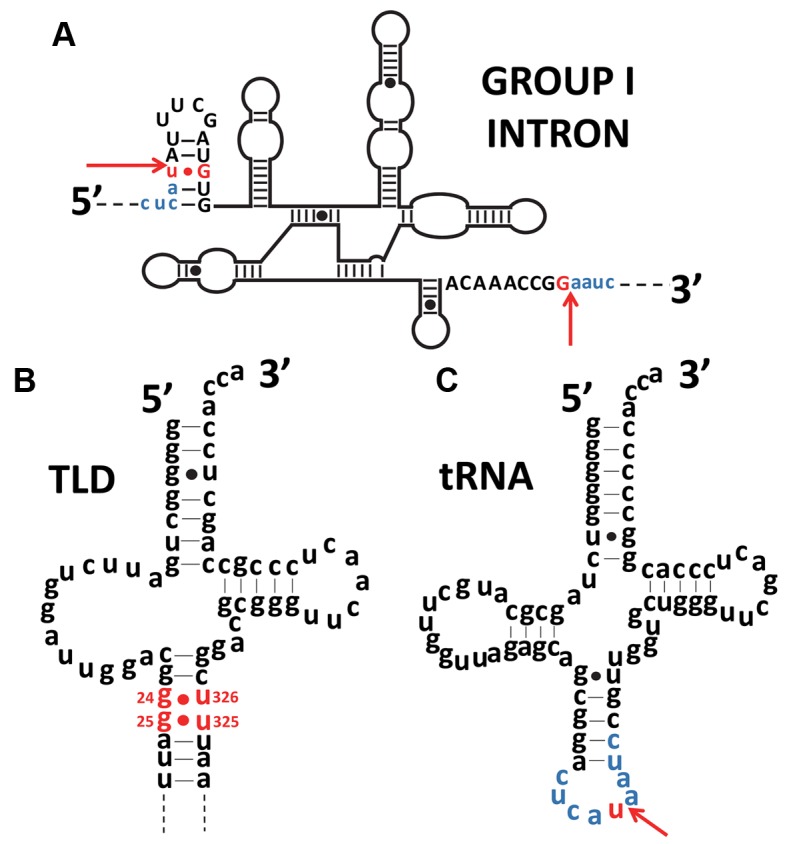
Similarity of the positions of G:U mismatches in the tRNA intron and in tmRNA. (A) Secondary structure of the Azoarcus group I intron. Exon sequences are in lower case and blue, while introns are in upper case letters, with red arrows indicating the splice boundaries. The conserved G-U mismatch necessary for self-splicing and the guanine partner are red. (B) Secondary structure of the Escherichia coli tRNA-like domain (TLD) of tmRNA. The conserved G-U mismatches in the TLD are red. A similarity in position between the G-U mismatch of the tRNA intron and the TLD is noticeable. (C) Secondary structure of a tRNAIle (CAU) from Azoarcus. The red arrow indicates the insertion site for the introns shown in (C). The exon sequence common to figures (A) and (C) is blue. Mismatches are signaled by dots.
Although it is difficult to predict whether tmRNA derives from a tRNA carrying a type I intron or vice versa, one could imagine a scenario in line with the introns-early hypothesis (27). In this variation, a universal proto-tmRNA with a large intron could be the common ancestor of both tRNA and tmRNA molecules. Gradually, some portions of this loop would have been lost, finally giving birth to the modern panel of tRNAs (49). tmRNA could result from the evolution of this loop into an internal open reading frame, encoding several codons. tmRNA probably arose very early, evolving from a proto-tmRNA that was aminoacylated by alanine only (see below).
Alanines at the crossroads of tmRNA-based aminoacylation and tagging events
Alanine is the simplest chiral amino acid, and along with glycine and aspartic acid the first to be present on earth (50–52). Accordingly, alanine was one of the first amino acids encoded by the first generation of codons (Figure 2). Remarkably, tmRNA is always charged by alanyl-tRNA synthetase (AlaRS), a class II tRNA synthetase that also catalyzes the esterification of alanine to tRNAAla. Contrary to recognition of most aminoacyl-tRNA synthetases, AlaRS does not require a specific signature on the anticodon moiety of the corresponding tRNA. Instead, it depends mainly on the presence of both a conserved G3:U70 base pair in the acceptor stem of the tRNAAla isoacceptor, and an adenosine at the discriminator position adjacent to the 3’-terminal CCA (53). This exception is extremely conserved within the three branches of life, and without a doubt reflects the ancient recognition of RNA minihelices by the first enzymes (54). Strikingly, this age-old signature is always present in tmRNA sequences, supporting its early apparition as well as the maintenance through the ages of specific aminoacylation of tmRNA, even in the absence of an anticodon (18,55).
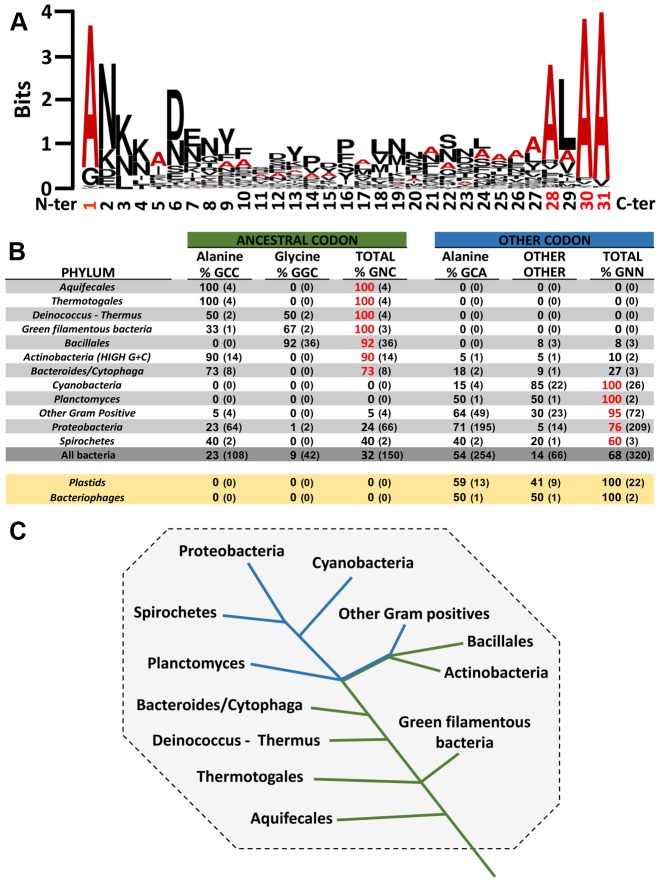
Similarly, four alanines are also encoded by the mRNA-like domain (MLD) that has the following consensus sequence: A*AN—-ALAA (e.g A*ANDENYALAA in E.coli, with the first A* carried by the tmRNA TLD) (Figure 7A). The first alanine codon is essential to trans-translation. Indeed, this region is important because it ensures the proper placement of the reading frame which allows resumption of translation in the absence of a start codon (56). Remarkably, when comparing the sequences of the resume codon among the various bacterial species, the more primitive the bacteria, the more ancestral GNC codons are recovered (Figure 7B–C). The three conserved tmRNA alanine codons (——–A-AA) that follow are also crucial, allowing specific recognition of the tagged protein by proteases. Indeed, most tmRNA-tagged proteins are degraded by ClpXP or less frequently by ClpAP proteases. Within the typical tagging sequence, ClpX binds the C-terminal residues (———LAA), whereas ClpA recognizes both C- and N-terminal tag residues (AA—–ALA-) (57,58). Degradation is also influenced by the adaptor protein SspB that binds to the tag's N-terminal portion to deliver the tagged peptide tagged to ClpXP (59).
Figure 7.
Consensus of amino acids and repartition of first codon of mRNA-like domain in bacteria phyla. (A) Sequence of the consensus sequence and diversity of the amino acids in the MLD. Sequence conservation is measured in bits, going from weakly conserved (0) to highly conserved (4). Alanine is in red and the other amino acids are in black. The alignment was obtained using 708 tag sequences from the UTHSCSA RNP database (http://rnp.uthscsa.edu/rnp/tmRDB/peptide/peptide.html) (66). The consensus logo was created using WebLogo (http://weblogo.berkeley.edu/logo.cgi) (75). (B) Analysis of the first MLD codon in different bacteria phyla. The first codon of the MLD is indexed by the ancestral codons (GNC) and the others. The percentage of each is bold and the sequence number is in parentheses. If the percentage of a codon in a phylum is superior to 50%, it is red. Non-bacterial tmRNA is yellow. Sequence alignments are presented for 940 different tmRNAs. They were taken from the ‘tmRNA website’ (76). Briefly, the first codon is identified thanks to the highly-conserved upstream sequence determinants. Of course, the resume codon sequence itself is another likely determinant. (C) Variations in the first MLD codon in the different bacteria phyla. The green line indicates ancestral codons for the first codon of the MLD, and the blue line indicates other codons. Adapted from the phylogenetic tree of all extant organisms based on 16S rRNA gene sequence data, as originally proposed by Woese.
tmRNA-mediated initiation and termination
In the RNA world, for an RNA to be considered a messenger molecule it had to be recognized through specific signatures. Today, this recognition is complex and involves many protein partners such as initiation and release factors. In the RNA world, the mechanism would have been simpler since only the PTC catalytic domain was present. Consequently, the first mRNAs with a non-random coding sequence needed an initiation signal for starting on the correct codon and accurately stopping translation. In fact, tmRNA fulfills all of these conditions. Indeed, the TLD domain of tmRNA can resume translation on its internal coding sequence, even in the absence of any canonical initiation factors. The presence of structured RNA regions upstream of a coding sequence is one of the simplest mechanisms known for initiating translation, as observed today in many tRNA-like structures (60,61). In the same way, tmRNA also carries a signal for terminating translation on its internal reading frame. All modern tmRNAs carry this stop codon within a stem–loop (Figure 4), known to be a basic signal to stop translation (62). Since in primordial times there were no release factors, stop codons, nor ribosomes with helicase activity, the primitive stop signal was probably a stem–loop RNA structure. In the absence of a whole ribosome, the mRNA could not anchor to maintain interactions with the PTC catalytic domain. Yet these interactions were certainly necessary to encourage the translation's evolution in the RNA world. Again, tmRNA possesses the ability to host these interactions via its non-coding domain. The large pseudoknot ring of tmRNA can perform many interactions with the translational complex and may have been used for anchoring the mRNA part to the first proto-ribosomes.
A UNIVERSAL PROTO-tmRNA AT THE ORIGINS OF MODERN tRNAs AND mRNAs?
We suggest that tmRNA fulfills all the criteria for being the missing link between an ancient RNA-driven world and the apparition of modern protein synthesis, which is organized around tRNA, mRNA and ribosomes. The model is based on an early prebiotic world in which various sets of short RNA molecules were first synthesized by the non-enzymatic bonding between free-floating nucleotides present in the primordial soup (Figure 8). Driven by evolutionary forces, these short RNAs would then have tended to pair with each other and/or fold into minihelices, forming hairpin structures. This stage likely promoted further interactions and permitted the first forms of the genetic code to emerge, while new RNA catalytic properties generated self-replicating molecules and the first aminoacylated RNA. We propose that in the second stage of an intriguing ‘intron scenario’, a primitive proto-tmRNA carrying a tRNA acceptor stem with a large intron was obtained by fusing two separate hairpin RNA. Such an ancestral molecule would possess both proto-tRNA and proto-mRNA functions (archaic forms of peptidylation and coding, respectively) within a single molecule. This would in fact explain the emergence of early genetic code, the two parts having evolved very closely. At this stage, proto-tmRNA could contain different acceptor branches charged by their specific early amino acids (e.g. alanine, see above). The large intronic loop abutted to the acceptor branch would contain several primitive codons (e.g. alanine codons). Indeed, according to a recent model by Di Giulio, the first mRNAs on earth might have also been peptidated, making tmRNA a perfect candidate to be one of the first mRNAs (63). However, it would also act as a strand of primitive anticodons (49) interacting with the primitive codons of other proto-tmRNAs simply by creating antiparallel duplexes as in modern anticodon-codon interactions. Even if we cannot rule out the theory that the first interactions between proto-tmRNAs occurred in the absence of any other partners (64), these proto-tmRNAs might have rapidly interacted with the first forms of the early peptidyl transferase center (PTC) while the first genetic code was evolving. Indeed, we can assume that the proto-PTC might have emerged in the same way, by self-folding and dimerization of RNA chains, thus providing the first framework for stereochemistry favoring for peptide bond formation and substrate-mediated catalysis (16,65). At this stage, protein synthesis was surely limited to quite short peptides. Co-evolution between these peptides and RNAs might have created a ‘virtuous circle’, meaning that RNA persisted thanks to peptide protection, the increased RNAs produced more peptides, this led to longer RNAs and peptides, and so on (66). We propose that in a subsequent stage, proto-tmRNAs eventually gave birth to tRNA and mRNA, and evolved into modern tmRNA molecules. In this stage, tRNA was made by variable intronic processing, resulting in the placement of a particular anticodon in the contemporary anticodon position (49). Relics of such a transition can be found today in tRNA variable Ioops and/or introns, which may correspond to the intron loop's unprocessed excess nucleotides. As for tmRNA, it would have emerged from one of the proto-tmRNAs that had an ancestral tRNAAla acceptor branch (i.e. having a G:U base pair in the acceptor stem). Interestingly, AlaRS is one of the rare enzymes that continues to recognize only the tRNA acceptor stem (early ‘second genetic code’) rather than the tRNAAla anticodon. It is possible that, despite its lack of anticodon, the need for tmRNA to be correctly aminoacylated is part of the evolutionary force that drove AlaRS to not incorporate the anticodon into its recognition process (67). Due to its essential role in cells, tmRNA is present in all bacteria, reinforcing the idea that it arose before bacterial phylogenic divergence (68). Three simple criteria can be used to establish RNA ‘antiquity’: catalysis, ubiquity, and central placement in the cellular metabolism (4). Many modern RNAs, particularly non-coding ones (ncRNAs), can thus be considered as relics of an ancient RNA world. However, during the slow and gradual replacement of RNAs by ribonucleoproteins (RNPs) and proteins in catalysis, there must have been a crucial step that allowed for the switchover to ribosome-mediated translation. While many RNAs meet the three criteria, introducing a central role in protein synthesis as a fourth criterion makes the game more complicated. Indeed, unlike tmRNA, only few modern RNAs carry specific features related to translational events. Among these, tRNA-like structures (TLSs) are thought to be molecular fossils of the original RNA world. These structures are often located at the 3’-end of the single-stranded RNA genomes of many bacterial and plant viruses, allowing both for tRNA mimicry (including aminoacylation) and regulation of RNA genome replication (61). In an appealing ‘genomic tag model’, Weiner and Maizels (69) suggested that these 3′-terminal TLSs were molecular fossils that originally identified genomic RNA molecules for replication and also functioned as primitive telomeres. In this case, aminoacylation would have come along later, fortuitously via an aminoacylating replicase. Such a scenario does not however contradict our model. It could even explain how the first aminoacylated proto-tmRNAs arose after the cleavage of aminoacylated TLS-tagged genomic RNAs.
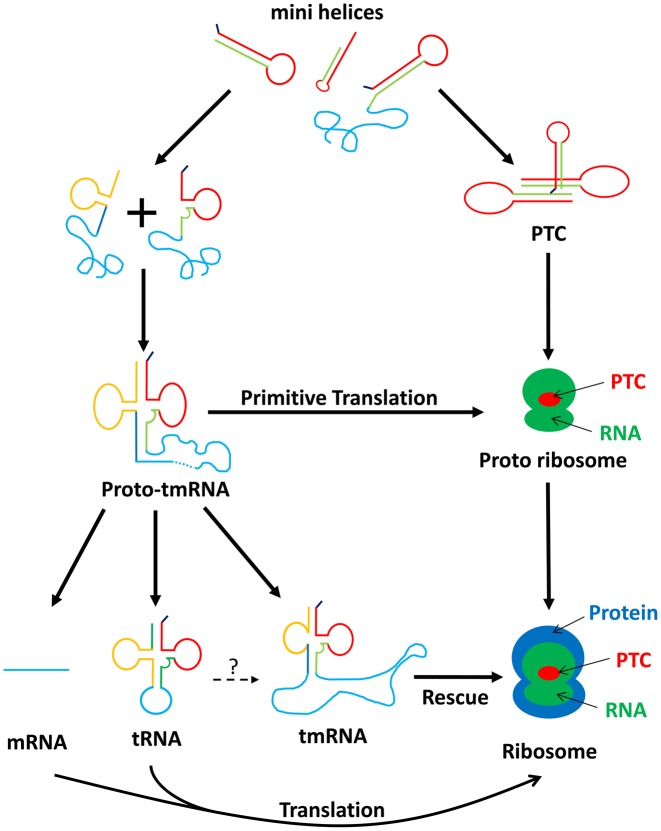
Figure 8.
Schematic model representing the different possibilities for the origin of tmRNA. In the beginning of the RNA world, there was a mix of different minihelices. A homodimerisation between two of these would have generated the first proto-ribosome, the Peptidyl Transferase Center (PTC). With aleatory amino acids or aminoacylated minihelices, the PTC created the first random peptide syntheses. A heterodimerization between two minihelices (plus sign) then surely generated a proto-tmRNA. In this step, the proto-tmRNA would have been used as a proto-mRNA. At the same time, the PTC must have evolved into a proto-ribosome by acquiring new RNA that improved its activity. The proto-ribosome would then have continued to evolve via the new synthesized proteins, finally ending up as the ribosomal complex seen today. In contrast to this ribosomal development, the proto-tmRNA took varying evolutionary paths. Through self-splicing, it produced a proto-tRNA that evolved into modern tRNA. It also led to modern tmRNA, which now serves as a rescue system in all bacteria. In addition, proto-tmRNA provided the first non-random genetic medium, which evolved into the RNA genome then into modern mRNA. However, we cannot exclude the theory that a proto-tRNAAla grew into modern tmRNA, through the insertion of nucleotides into its gene (question mark) (77).
Modern tmRNA has two main functions (embodied in mRNA and tRNA) that are indispensable to the emergence of protein synthesis. But contrary to isolated mRNA or tRNA, it is only found in bacteria and therefore does not fulfill the second criterion of being ubiquitous. Nevertheless, it is interesting to note that overall prokaryotic and eukaryotic/archaea share several common features, including the addition of a degradation signal to incomplete peptides, and specific degradation of problematic mRNAs. However, in eukaryotes and archaea, the degradation pathway mainly involves the sophisticated ubiquitin proteasome system (70,71), which suggests that tmRNA was lost with ubiquitin's appearance. Accordingly, the Dom34p-mediated no-go decay (NGD) protein is universal in eukaryotes and archaea, suggesting that NGD is probably an ancient mechanism already present in their last common ancestor (41). Evolution of mRNA decay systems in eukaryotes might then have been driven by eRF1 and eRF3 gene duplications. We can therefore assume that tmRNA was lost very early, replaced by more elaborate protein-based quality control mechanisms (i.e. NGD, NSD and NMD as discussed above) in response to the greater diversity of potential clients in eukaryotes and archaea.
In conclusion, we believe that a proto-tmRNA dating from the RNA world is the common ancestor of both modern tRNA and mRNA. This early RNA might have been the first player in protein synthesis and the evolution of the genetic code, making it one of the oldest known fossils of the RNA world.
Acknowledgments
We wish to thank Emmanuel Giudice for generously helping to create the figures used here, and Juliana Berland for insightful comments on the manuscript.
FUNDING
Agence Nationale pour la Recherche and Direction Générale de l'Armement (ANR-14-ASTR-0001); Institut Universitaire de France. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 2.Orgel L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 3.Woese C.R. Harper & Row. 1967. The genetic code the molecular basis for genetic expression. ASIN: B000I3M2KK. [Google Scholar]
- 4.Jeffares D.C., Poole A.M., Penny D. Relics from the RNA world. J. Mol. Evol. 1998;46:18–36. doi: 10.1007/pl00006280. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg A.L., Wittes R.E. Genetic code: aspects of organization. Science. 1966;153:420–424. doi: 10.1126/science.153.3734.420. [DOI] [PubMed] [Google Scholar]
- 6.Yarus M., Widmann J.J., Knight R. RNA-amino acid binding: a stereochemical era for the genetic code. J. Mol. Evol. 2009;69:406–429. doi: 10.1007/s00239-009-9270-1. [DOI] [PubMed] [Google Scholar]
- 7.Ronneberg T.A., Landweber L.F., Freeland S.J. Testing a biosynthetic theory of the genetic code: fact or artifact. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13690–13695. doi: 10.1073/pnas.250403097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tlusty T. A colorful origin for the genetic code: information theory, statistical mechanics and the emergence of molecular codes. Phys. Life Rev. 2010;7:362–376. doi: 10.1016/j.plrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Foltan J.S. tRNA genes and the genetic code. J. Theor. Biol. 2008;253:469–482. doi: 10.1016/j.jtbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Ikehara K. Origins of gene, genetic code, protein and life: comprehensive view of life systems from a GNC-SNS primitive genetic code hypothesis. J. Biosci. 2002;27:165–186. doi: 10.1007/BF02703773. [DOI] [PubMed] [Google Scholar]
- 11.Voorhees R.M., Ramakrishnan V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 12.Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 13.Noller H.F. Evolution of protein synthesis from an RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:a003681. doi: 10.1101/cshperspect.a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidovich C., Belousoff M., Bashan A., Yonath A. The evolving ribosome: from non-coded peptide bond formation to sophisticated translation machinery. Res. Microbiol. 2009;160:487–492. doi: 10.1016/j.resmic.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao C., Lenz T.K., Peters J.K., Fang P.-Y., Schneider D.M., Anderson E.J., Preeprem T., Bowman J.C., O'Neill E.B., Lie L., et al. Molecular paleontology: a biochemical model of the ancestral ribosome. Nucleic Acids Res. 2013;41:3373–3385. doi: 10.1093/nar/gkt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krupkin M., Matzov D., Tang H., Metz M., Kalaora R., Belousoff M.J., Zimmerman E., Bashan A., Yonath A. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2011;366:2972–2978. doi: 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Duve C. Transfer RNAs: the second genetic code. Nature. 1988;333:117–118. doi: 10.1038/333117a0. [DOI] [PubMed] [Google Scholar]
- 18.Ribas de Pouplana L., Schimmel P. A view into the origin of life: aminoacyl-tRNA synthetases. Cell. Mol. Life Sci. CMLS. 2000;57:865–870. doi: 10.1007/PL00000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beuning P.J., Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 20.Di Giulio M. The split genes of Nanoarchaeum equitans have not originated in its lineage and have been merged in another Nanoarchaeota: a reply to Podar et al. J. Theor. Biol. 2014;349:167–169. doi: 10.1016/j.jtbi.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Schimmel P., Ribas de Pouplana L. Transfer RNA: from minihelix to genetic code. Cell. 1995;81:983–986. doi: 10.1016/s0092-8674(05)80002-9. [DOI] [PubMed] [Google Scholar]
- 22.Kanai A. Disrupted tRNA genes and tRNA fragments: a perspective on tRNA gene evolution. Life Basel Switz. 2015;5:321–331. doi: 10.3390/life5010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T., Kikuchi Y. Origin of the cloverleaf shape of transfer RNA - the double-hairpin model: Implication for the role of tRNA intro and the long extra loop. SSOEL Jpn. 2001;29:134–142. [Google Scholar]
- 24.Tamura K., Schimmel P.R. Chiral-selective aminoacylation of an RNA minihelix: Mechanistic features and chiral suppression. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13750–13752. doi: 10.1073/pnas.0606070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu M. Specific aminoacylation of C4N hairpin RNAs with the cognate aminoacyl-adenylates in the presence of a dipeptide: origin of the genetic code. J. Biochem. (Tokyo) 1995;117:23–26. doi: 10.1093/oxfordjournals.jbchem.a124715. [DOI] [PubMed] [Google Scholar]
- 26.Suga H., Hayashi G., Terasaka N. The RNA origin of transfer RNA aminoacylation and beyond. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2011;366:2959–2964. doi: 10.1098/rstb.2011.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Giulio M. The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA) J. Theor. Biol. 2006;240:343–352. doi: 10.1016/j.jtbi.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Cavarelli J., Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1993;7:79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- 29.Dong G., Nowakowski J., Hoffman D.W. Structure of small protein B: the protein component of the tmRNA-SmpB system for ribosome rescue. EMBO J. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giudice E., Gillet R. The task force that rescues stalled ribosomes in bacteria. Trends Biochem. Sci. 2013;38:403–411. doi: 10.1016/j.tibs.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Ito K., Chadani Y., Nakamori K., Chiba S., Akiyama Y., Abo T. Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One. 2011;6:e28413. doi: 10.1371/journal.pone.0028413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao C., Bhardwaj K., Sharkady S.M., Fish R.I., Driscoll T., Wower J., Zwieb C., Sobral B.W.S., Williams K.P. Variations on the tmRNA gene. RNA Biol. 2009;6:355–361. doi: 10.4161/rna.6.4.9172. [DOI] [PubMed] [Google Scholar]
- 33.Gimple O., Schön A. In vitro and in vivo processing of cyanelle tmRNA by RNase P. Biol. Chem. 2001;382:1421–1429. doi: 10.1515/BC.2001.175. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe T., Sugita M., Sugiura M. Identification of 10Sa RNA (tmRNA) homologues from the cyanobacterium Synechococcus sp. strain PCC6301 and related organisms. Biochim. Biophys. Acta. 1998;1396:97–104. doi: 10.1016/s0167-4781(97)00180-2. [DOI] [PubMed] [Google Scholar]
- 35.Williams K.P. The tmRNA website. Nucleic Acids Res. 2000;28:168. doi: 10.1093/nar/28.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowler C., Allen A.E., Badger J.H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R.P., et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 37.Hafez M., Burger G., Steinberg S.V., Lang B.F. A second eukaryotic group with mitochondrion-encoded tmRNA: in silico identification and experimental confirmation. RNA Biol. 2013;10:1117–1124. doi: 10.4161/rna.25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray M.W., Lang B.F., Burger G. Mitochondria of Protists. Annu. Rev. Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 39.Brandman O., Hegde R.S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simms C.L., Thomas E.N., Zaher H.S. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip. Rev. RNA. 2016 doi: 10.1002/wrna.1366. doi:10.1002/wrna.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atkinson G.C., Baldauf S.L., Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abelson J., Trotta C.R., Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 43.Di Giulio M. On the origin of the transfer RNA molecule. J. Theor. Biol. 1992;159:199–214. doi: 10.1016/s0022-5193(05)80702-7. [DOI] [PubMed] [Google Scholar]
- 44.Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. TIG. 1991;7:145–148. [PubMed] [Google Scholar]
- 45.Edgell D.R., Belfort M., Shub D.A. Barriers to intron promiscuity in bacteria. J. Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 47.Cech T.R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis–a review. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- 48.Costa J.-L., Paulsrud P., Lindblad P. The cyanobacterial tRNA(Leu) (UAA) intron: evolutionary patterns in a genetic marker. Mol. Biol. Evol. 2002;19:850–857. doi: 10.1093/oxfordjournals.molbev.a004142. [DOI] [PubMed] [Google Scholar]
- 49.Lacey J.C., Staves M.P. Was there a universal tRNA before specialized tRNAs came into existence. Orig. Life Evol. Biosphere J. Int. Soc. Study Orig. Life. 1990;20:303–308. doi: 10.1007/BF01808112. [DOI] [PubMed] [Google Scholar]
- 50.Martin W., Russell M.J. On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. B Biol. Sci. 2007;362:1887–1926. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller S.L. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 52.Muñoz Caro G.M., Meierhenrich U.J., Schutte W.A., Barbier B., Arcones Segovia A., Rosenbauer H., Thiemann W.H.-P., Brack A., Greenberg J.M. Amino acids from ultraviolet irradiation of interstellar ice analogues. Nature. 2002;416:403–406. doi: 10.1038/416403a. [DOI] [PubMed] [Google Scholar]
- 53.Hou Y.M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 54.Ribas de Pouplana L., Schimmel P. Operational RNA code for amino acids in relation to genetic code in evolution. J. Biol. Chem. 2001;276:6881–6884. doi: 10.1074/jbc.R000032200. [DOI] [PubMed] [Google Scholar]
- 55.Fischer A.E., Beuning P.J., Musier-Forsyth K. Identification of discriminator base atomic groups that modulate the alanine aminoacylation reaction. J. Biol. Chem. 1999;274:37093–37096. doi: 10.1074/jbc.274.52.37093. [DOI] [PubMed] [Google Scholar]
- 56.Miller M.R., Healey D.W., Robison S.G., Dewey J.D., Buskirk A.R. The role of upstream sequences in selecting the reading frame on tmRNA. BMC Biol. 2008;6:29. doi: 10.1186/1741-7007-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flynn J.M., Levchenko I., Seidel M., Wickner S.H., Sauer R.T., Baker T.A. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levchenko I., Grant R.A., Flynn J.M., Sauer R.T., Baker T.A. Versatile modes of peptide recognition by the AAA+ adaptor protein SspB. Nat. Struct. Mol. Biol. 2005;12:520–525. doi: 10.1038/nsmb934. [DOI] [PubMed] [Google Scholar]
- 59.Janssen B.D., Hayes C.S. The tmRNA ribosome-rescue system. Adv. Protein Chem. Struct. Biol. 2012;86:151–191. doi: 10.1016/B978-0-12-386497-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kieft J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katz A., Elgamal S., Rajkovic A., Ibba M. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol. Microbiol. 2016 doi: 10.1111/mmi.13419. doi:10.1111/mmi.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henkin T.M., Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 63.Di Giulio M. A Model for the Origin of the First mRNAs. J. Mol. Evol. 2015;81:10–17. doi: 10.1007/s00239-015-9691-y. [DOI] [PubMed] [Google Scholar]
- 64.Gillet R., Felden B. Transfer RNA(Ala) recognizes transfer-messenger RNA with specificity; a functional complex prior to entering the ribosome? EMBO J. 2001;20:2966–2976. doi: 10.1093/emboj/20.11.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farias S.T., Rêgo T.G., José M.V. Origin and evolution of the Peptidyl Transferase Center from proto-tRNAs. FEBS Open Biol. 2014;4:175–178. doi: 10.1016/j.fob.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Gulik P.T.S., Speijer D. How amino acids and peptides shaped the RNA world. Life Basel Switz. 2015;5:230–246. doi: 10.3390/life5010230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nameki N., Tadaki T., Muto A., Himeno H. Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in Vitro1. J. Mol. Biol. 1999;289:1–7. doi: 10.1006/jmbi.1999.2754. [DOI] [PubMed] [Google Scholar]
- 68.Meli M., Albert-Fournier B., Maurel M.C. Recent findings in the modern RNA world. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2001;4:5–11. doi: 10.1007/s101230100002. [DOI] [PubMed] [Google Scholar]
- 69.Weiner A.M., Maizels N. tRNA-like structures tag the 3’ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu X., Liu R., Sanchez I., Silva-Sanchez C., Hepowit N.L., Cao S., Chen S., Maupin-Furlow J. Ubiquitin-like proteasome system represents a eukaryotic-like pathway for targeted proteolysis in archaea. mBio. 2016;7 doi: 10.1128/mBio.00379-16. doi:10.1128/mBio.00379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat. Rev. Microbiol. 2012;10:100–111. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Giulio M. An extension of the coevolution theory of the origin of the genetic code. Biol. Direct. 2008;3:37. doi: 10.1186/1745-6150-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Giulio M. The origin of the tRNA molecule: Independent data favor a specific model of its evolution. Biochimie. 2012;94:1464–1466. doi: 10.1016/j.biochi.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 74.Giudice E., Macé K., Gillet R. Trans-translation exposed: understanding the structures and functions of tmRNA-SmpB. Front. Microbiol. 2014;5:113. doi: 10.3389/fmicb.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hudson C.M., Williams K.P. The tmRNA website. Nucleic Acids Res. 2015;43:D138–D140. doi: 10.1093/nar/gku1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Himeno H., Kurita D., Muto A. tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front. Genet. 2014;5:66. doi: 10.3389/fgene.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]