Abstract
The yeast 2-micron plasmid epitomizes the evolutionary optimization of selfish extra-chromosomal genomes for stable persistence without jeopardizing their hosts’ fitness. Analyses of fluorescence-tagged single-copy reporter plasmids and/or the plasmid partitioning proteins in native and non-native hosts reveal chromosome-hitchhiking as the likely means for plasmid segregation. The contribution of the partitioning system to equal segregation is bipartite- replication-independent and replication-dependent. The former nearly eliminates ‘mother bias’ (preferential plasmid retention in the mother cell) according to binomial distribution, thus limiting equal segregation of a plasmid pair to 50%. The latter enhances equal segregation of plasmid sisters beyond this level, elevating the plasmid close to chromosome status. Host factors involved in plasmid partitioning can be functionally separated by their participation in the replication-independent and/or replication-dependent steps. In the hitchhiking model, random tethering of a pair of plasmids to chromosomes signifies the replication-independent component of segregation; the symmetric tethering of plasmid sisters to sister chromatids embodies the replication-dependent component. The 2-micron circle broadly resembles the episomes of certain mammalian viruses in its chromosome-associated propagation. This unifying feature among otherwise widely differing selfish genomes suggests their evolutionary convergence to the common logic of exploiting, albeit via distinct molecular mechanisms, host chromosome segregation machineries for self-preservation.
INTRODUCTION
Circular DNA plasmids, widespread among prokaryotes, are almost nonexistent among eukaryotes. Certain members of the budding yeast and Dictyostelium species present a rare exception by harboring circular plasmids in their nuclei (1,2). Furthermore, viruses belonging to the papilloma family and gammaherpes sub-family are propagated as episomes in infected cells during long periods of latency (3–6). Eukaryotic nuclei, however, almost ubiquitously contain non-plasmid extra-chromosomal circular DNA (eccDNA) molecules with potential roles in genome organization, dynamics and plasticity (7–10). These circles, with a wide range of sizes, are presumed to result from recombination events, which may be associated with DNA replication/repair in some instances. They have been implicated in centromere evolution, maintenance of telomere length, concerted evolution and homogenization of repeated sequences and the emergence of copy number variations. A subset of these circles provides markers for genetic instabilities associated with human diseases (11–13). In Saccharomyces cerevisiae, extra-chromosomal rDNA circles are retained with a strong preference by the mother cell during nuclear division, perhaps providing a mechanism for asymmetric segregation of a senescence factor and rejuvenation of daughters at birth (14,15). The multi-copy plasmid 2-micron circle native to S. cerevisiae, however, escapes this mother-bias, thus providing a model for the stable propagation of an extra-chromosomal DNA element in a simple eukaryotic host.
The 2-micron plasmid is a minimalized and highly optimized selfish DNA element, whose nearly chromosome-like stability is conferred by two plasmid-coded proteins Rep1 and Rep2 and a cis-acting locus STB (1,16,17). The Rep-STB system absolves the 2 micron plasmid from the strong mother bias experienced by rDNA circles and by plasmids capable of autonomous replication (ARS-plasmids) in S. cerevisiae but lack an active partitioning mechanism (18–20). The mother bias arises from the barrier to equilibration of plasmid molecules between mother and daughter compartments posed by the constricted geometry of the budding yeast nucleus, the limited duration of the mitotic cell cycle and perhaps additional constraints due to plasmid association with sub-nuclear structures (19,21). The mean loss rate of the 2-micron plasmid is as low as 10−5 to 10−4 per cell per generation. The plasmid appears to provide no advantage to the host under standard growth conditions. However, the fitness cost to the host for bearing the plasmid load, at the normal copy number of 40–60 per haploid nucleus, is also quite low (22).
The plasmid genome can be divided into two functional units, apparently devoted solely to self-serving ends. The replication origin and the partitioning system ensure, during a cell cycle, the duplication of each plasmid molecule by the host replication machinery, followed by the equal (or nearly equal) segregation of the replicated copies into mother and daughter nuclei. A drop in copy number resulting from a rare missegregation event is corrected by an amplification system, comprised of the plasmid-coded Flp site-specific recombinase and its target sites present in inverted orientation in the plasmid genome (23,24). The key to the amplification reaction is a recombination-mediated inversion of one of a pair of bi-directional replication forks. The amplified DNA spun out by the two uni-directional forks can be resolved into monomer plasmid units by homologous or Flp-mediated recombination. Intricate regulation of plasmid gene expression keeps the amplification system in check, triggers it into action promptly when required, and protects against runaway increase in plasmid copy number (25–27). The level/activity of Flp is also controlled by its post-translational modification by the host sumoylation system, thus avoiding inappropriate plasmid amplification (28,29). Furthermore sumoylation of Rep1 and Rep2 appears to promote their association with STB, and thus bolster efficient plasmid partitioning (30). The 2-micron plasmid provides an attractive system for studying self-instituted and host-imposed mechanisms by which a selfish genome and its host genome establish long-term coexistence with minimal mutual conflicts.
The remarkably high stability of the 2-micron plasmid stems from the ability of the Rep-STB system to couple plasmid partitioning to chromosome segregation (31–34). A variety of host factors that associate with centromeres and play important functional roles in faithful chromosome segregation are also detected at the STB locus, and nearly all of these appear to promote plasmid partitioning as well (32,35–38). The apparent functional resemblance between CEN and STB with respect to host factor association may suggest that the atypical, genetically defined point centromere of budding yeast and STB might have descended from a common ancestral partitioning locus (39–42). However, current evidence argues against the recruitment of kinetochore components at STB or the execution of plasmid segregation by directly utilizing spindle force (43,44).
Several lines of circumstantial evidence suggest that the 2-micron plasmid is partitioned in association with a nuclear entity that segregates without bias into mother and daughter during nuclear division. Furthermore, current results are consistent with that entity being the host chromosomes (33,34). However, direct proof for this hitchhiking model for plasmid segregation is lacking. The localization and dynamics of STB-reporter plasmids in meiotic yeast cells is suggestive of plasmid association with telomeres via a meiosis-specific nuclear envelope motor and consequent plasmid segregation as a telomere appendage during meiosis I (44). This mechanism for plasmid-chromosome tethering is not possible in mitotic cells, as at least a subset of the motor components is expressed only during meiosis (45–50). Perhaps a common hitchhiking model for 2-micron plasmid segregation may still operate during vegetative and germ-line cell divisions with mechanistic variations in how plasmid-chromosome interactions are established under the distinct cell cycle programs.
In the present study, we subjected the chromosome-hitchhiking model to incisive experimental tests in the native host, and complemented them by one set of analyses under a non-native host cell context. In addition, we revealed two distinct functional roles for the Rep-STB system in 2-micron plasmid partitioning. One effectively overcomes the mother bias in plasmid inheritance, and does not require plasmid replication. This replication-independent segregation mechanism follows the binomial rule, and limits the equal segregation of a plasmid pair to 50%. The second overcomes mother bias as well as enables efficient equal plasmid segregation (>50%), but absolutely requires plasmid replication. Host factors that interact with Rep-STB to promote plasmid can be distinguished by whether their action is dissociated from, or occurs in conjunction with, plasmid replication. Both the replication-dependent and replication-independent features of Rep-STB action fit nicely into the hitchhiking model, and can be explained by two distinct modes of plasmid-chromosome association under the two conditions.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains and the plasmids utilized in this study are listed in Supplementary Tables S1 and S2, respectively. The specific experiments in which they were employed and the figures displaying the corresponding results are summarized in Supplementary Table S3.
Fluorescence-tagged single-copy reporters excised from chromosomal integrants
The standard reporter plasmids used for studying 2 micron plasmid partitioning contain a replication origin (ORI or ARS) and the STB locus, and are complemented in trans by Rep1 and Rep2 proteins In general, these reporters are multi-copy, and their copy numbers are variable from cell to cell. For cell biological assays, the reporters are fluorescence-tagged using operator-fluorescent repressor interaction (31,32). In order to follow partitioning quantitatively, single-copy reporters have been designed. By incorporating a conditionally active centromere (CEN) sequence into the plasmid, the copy number can be lowered to nearly one (33). A site-specific recombination-based strategy has been devised for keeping the reporter copy number precisely as one (34).
The single-copy ORI-plus STB-plasmid described in a previous analysis (34) (now referred to as an STB-ARS-plasmid) and its derivatives were employed in this study. The reporters, maintained as chromosomal integrants, were excised by site-specific recombination mediated by the R recombinase. The presence of a [LacO]256 array enabled them to be visualized by their green fluorescence in host strains expressing GFP-LacI. The ORI-STB sequence was contained within the PstI-XbaI fragment (2657–3946) from the 2-micron plasmid form A. The ori-minus STB-circle (STB-arsΔ-c) was derived from this construct by deleting the sequences from coordinates 3545–3814. Two ori-minus STB-circles of identical sequence were employed in some of the assays to represent pseudo-sisters. They were excised from integrants placed on separate chromosomes (Chr IV and Chr XV). Occasionally, we noticed (by Southern analysis) small size differences between the two excised copies, presumably due to instabilities/rearrangements within their [LacO] arrays. However, these differences do not affect their functional features relevant to the experimental outcomes and interpretations. Variants of the ORI-plus or ori-minus constructs lacking STB were obtained by deleting the 2 micron circle-derived PstI-AvaI fragment (2657–3259).
Note that an STB-ARS-plasmid present in a strain without the 2 micron plasmid ([Cir0]; lacking Rep1 and Rep2) behaves effectively as an ARS-plasmid present in a [Cir+] or [Cir0] strain. As Rep1 and Rep2 act collaboratively to sustain STB function, the loss of one or both inactivates STB. Hence, in the present study, we have not tested the effects of ablating each protein individually.
Reporter plasmids harboring conditional CEN and STB
The CEN-STB-ARS reporter plasmids utilized for a subset of experiments, with a copy number of ∼1 and containing the 2 micron plasmid origin, have been described previously (33). They harbor either [LacO]256 or [TetO]112. They were visualized by [LacO]-GFP-LacI or [TetO]-RFP-TetR interaction. In a derivative of one of these plasmids, the STB sequence was deleted. The plasmid-CEN was active in glucose/raffinose grown cells, but was inactivated by galactose due to high-level transcription from the immediately upstream GAL promoter. By manipulating the host strain ([Cir+] or [Cir0]) and/or the carbon source (glucose or galactose), the same plasmid could be made to behave as a CEN-ARS-reporter, an STB-ARS-reporter or an ARS reporter (lacking both CEN and STB functions). Prior assays showed that a plasmid with an active CEN or an active CEN and STB behave similarly in segregation (33).
Consistent with the estimated copy number, a CEN-STB-ARS-reporter plasmid formed a single fluorescent focus per nucleus in >80% of the cells within a population (33; this study). Copy number variations due to missegregation events during propagation of experimental strains was avoided as follows. For each analysis, reporter plasmids were newly introduced into the recipient strains by transformation. Fresh transformants were immediately carried through G1-arrest and chromosome spread assays, or subjected to single cell cycle plasmid segregation analyses. Furthermore, for a given reporter plasmid, spreads showing more than a single focus and anaphase cells containing more than two foci, which were quite rare, were excluded from consideration.
A multi-copy STB-ARS-reporter plasmid
A multi-copy STB-plasmid (referred to as pSTB-ARS-mc) served as the reporter for assays in which plasmid and chromosome replication was blocked. This pUC19-based shuttle plasmid is a variant of pSV1 described in earlier studies (32). It contained [LacO]256, TRP1 and a 2 micron plasmid fragment harboring the replication origin and STB. The plasmid was introduced into the experimental strain by transformation and maintained by selection in medium lacking tryptophan.
Mammalian cell lines and expression vectors
COS7 (CV-1 (simian) in Origin, and carrying the SV40 genetic material) cells, a fibroblast-like cell line derived from monkey kidney tissue, were used to study the localization of the Rep1 and Rep2 proteins. The results were reproduced in HEK 293 cells (Human Embryonic Kidney) and HEK 293T cells (HEK 293 cells expressing SV40 Large T-antigen) but are not reported here. All cell lines were grown in DMEM (Dulbecco's Modified Eagle Medium) (from Cellgro) supplied with 10% FBS (Fetal Bovine Serum) (from Atlantic Biology).
Expression plasmids for mammalian cells are listed in Supplementary Table S2. Rep1 and Rep2 were expressed as carboxyl-terminal fusion proteins from pGFP-C1 and pDsRed-Express-C1 vectors (Clontech) under the control of the CMV promoter.
Antibodies
The following antibodies were used for immunofluorescence assays: mouse anti-GFP (LGB-1; Abcam), rabbit anti-RFP (ab62341; Abcam); mouse anti-Fibrillarin (38F3; Abcam), mouse anti-HA (HA.11; Covance) and rabbit anti-Rep1 (custom made; directed to a synthetic Rep1 peptide) (31). A mouse anti-Myc antibody (9E10; Covance) was used in the chromatin immunoprecipitation (ChIP) assays. Antibody dilutions ranged from 1:100 to 1:500 in individual assays, in accordance with initial standardization.
General protocols for the yeast assays
The methodologies for arresting cells in G1 and scoring the segregation of single-copy or multi-copy reporter plasmids during a single generation followed published procedures (31,33,34). For assaying the coalescence of plasmids in metaphase cells, the cell cycle stage was assessed by examining samples of a population at different time points after release from G1 arrest. The time at which ≥80% of cells were large-budded, with each cell containing a single DAPI-stained chromosome mass in the mother near the bud-neck and a spindle ∼1 μm in length, was chosen for the analysis. The protocols for preparing chromosome spreads from mitotic cells and for localizing plasmids or proteins in them by immunofluorescence have been previously described (31,32). Cell cycles were altered from the norm by overproducing or depleting protein(s) from suitably engineered expression cassettes according to published procedures: Cdc6 depletion (37,51), Mcd1-nc (non-cleavable) overexpression (32,52), monopolin assembly in mitosis (34,53), or cohesin-depletion (34,53). Additional standard protocols for growing yeast and bacterial cultures, plasmid isolation, extraction of total yeast DNA, transformation of Escherichia coli and yeast can be found on line at the web page for the Jayaram laboratory: (http://www.sbs.utexas.edu/jayaram/jayaramlab_files/Protocols.htm).
Transfection of mammalian cells
Transfections (or co-transfections) with expression vectors (∼4 μg per transfection) were carried out in cells at 70% confluence using Lipofectamine-2000 (Life Technologies) according to the supplier's instructions. The medium was replaced at 6–8 h after transfection, and cells were assayed for protein localization 40–42 h after transfer to fresh medium.
Chromosome spreads from mammalian cells
Cells, seeded on Superfrost Plus (Fisher) microscope slides, were transfected at 50% confluence (∼24 μg plasmid DNA/slide). After transfer to fresh medium at 6 h and incubation for 24 h, they were shifted to medium containing 2 mM thymidine for 12–16 h to synchronize them in G1/S phase. For releasing from thymidine block, cells were washed with 1× PBS at room temperature, twice with DMEM at 37°C, and were incubated in fresh DMEM/10% FBS for 5 h (37°C). They were then incubated in the presence of 30 ng colcemid/ml for an additional 5 h. The enrichment of mitotic cells was verified by their rounded appearance under a light microscope. Chromosome spreads were prepared according to published procedures (54).
Immunofluorescence assays in mammalian cells
Cells were fixed on microscope slides with 4% paraformaldehyde in 1× PBS for 30 min., and were permeabilized with 0.1% Triton X-100 in 1X PBS for 5 min. After washing with 1× PBS twice, cells were blocked with 1 mg/ml BSA for 1 h. Subsequent sequential steps included incubation with the primary antibody for 3 h, three washes with 1× PBS, incubation with the secondary antibody for 2 h, three washes with 1× PBS and staining with 1 μg/ml Hoechest 33342 (Sigma) in 1× PBS for 5 min. The slides were mounted, and examined by fluorescence microscopy.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described in earlier studies (35,37). Serial dilutions of template DNA were employed to determine the linear range of signal output in the PCR reactions. The signals were corrected by subtracting those from mock-immunoprecipitated (no antibody) controls. The ‘ChIP efficiency’ of a locus was obtained by normalizing the corrected signal to the corresponding signal from the input DNA.
Fluorescence microscopy
Observations were performed using an Olympus BX-60 microscope. Images were taken at room temperature at 100× (oil NA 1.30 objective) using a Photometrics Quantix camera (Roper Scientific), and then processed by MetaMorph 7.5 software (Universal Imaging Corporation) and PhotoShop CS4 (Adobe Systems, Inc.) (31,43).
Fluorescence intensities of reporter foci
The pixels constituting a fluorescent reporter focus in a G1 or anaphase cell were outlined, and the intensity of the brightest pixel within this cluster was recorded (as described under Supplementary Figure S2 in (34)). This value was corrected by subtracting the mean intensity of a set of neighboring background pixels.
RESULTS
The 2-micron plasmid is coupled to chromosomes, but not to the nuclear membrane, in its segregation
Current evidence argues against equal segregation of the 2-micron plasmid in association with the spindle pole body or by directly utilizing spindle force (43,44). However, the possibility of plasmid segregation by attachment to the nuclear membrane or to the nuclear matrix has not been ruled out. Earlier observations revealed the tendency of the 2 micron plasmid to missegregate in tandem with the bulk of the chromosomes when chromosome segregation was impaired by conditional mutations (32). However, chromosome missegregation in these mutants was almost never entirely one-sided (all chromosomes in either the mother or in the daughter). It was not possible therefore to cleanly differentiate between chromosomes and the nuclear membrane in regards to plasmid association. Furthermore, tethering an ARS-plasmid to a nuclear membrane protein or certain nuclear pore proteins has been shown to ameliorate its mother bias, and improve its stability (19,20). Strict distinction between the nuclear matrix and the nuclear envelope is not possible in budding yeast, as it lacks a nuclear lamina, typical of eukaryotic cells, with which a variety of nuclear envelope proteins become associated (55). Our experimental strategy was to induce nuclear membrane segregation into the mother and daughter cell compartments while containing chromosomes in only one of the two compartments. Plasmid-membrane association could thus be probed unambiguously without interference from chromosomes.
The analyses utilized two ‘precisely single-copy’ reporter plasmids, pSTB-ARS and pARS. They were generated in separate G1-cell populations by R recombinase-mediated excision from their chromosomally integrated forms (34) (Figure 1A; Supplementary Figure S1A–C). Except for the deletion of STB in pARS, the two plasmids were identical. The function of STB, when present within a reporter, was supported in trans by the Rep1 and Rep2 proteins supplied by a [Cir+] host strain. In a [Cir0] strain, STB was inactive. These reporters (and others used in this study), containing an operator array ([LacO]256 or [TetO]112), were fluorescence-tagged in vivo by the binding of the cognate fluorescent repressor. As they lack the Flp recombination target site (FRT), they cannot be integrated into the native 2-micron plasmid via Flp-mediated site-specific recombination. Furthermore, the limited amount of DNA sequence shared between a reporter and the native plasmid (∼1.3 kbp or less) minimizes the rate of mitotic recombination (∼2 to 3 × 10−8) (56), virtually ruling out integration via homologous recombination. Thus, the replication competence of the reporters or their segregation is not influenced in cis by the presence of the 2-micron plasmid.
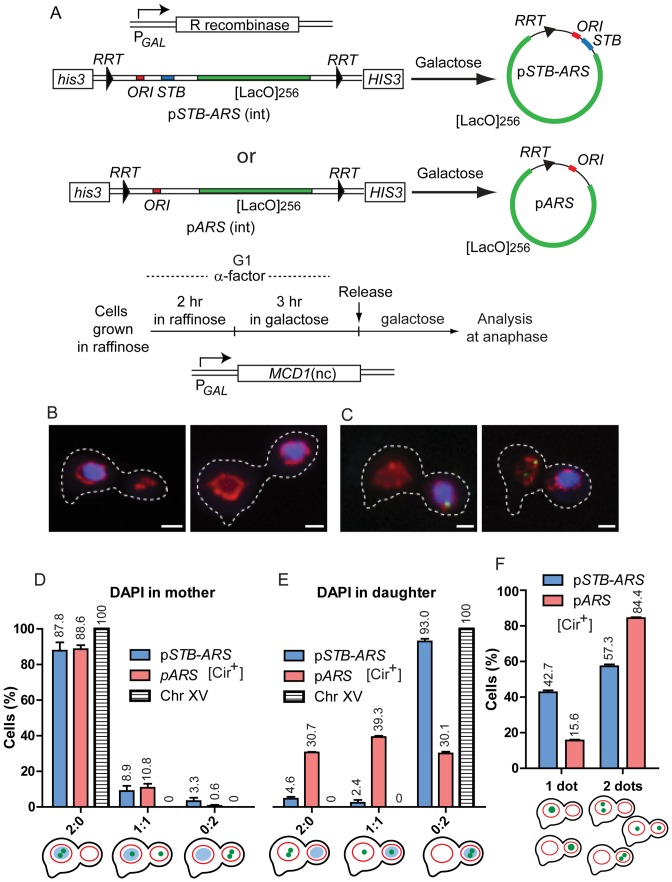
Figure 1.
Blocking cohesin disassembly reveals the coupling of an STB-ARS-plasmid to chromosomes, not to the nuclear membrane. (A) The schematic illustrates the excision of a single-copy STB-ARS- or an ARS-reporter plasmid, integrated at the HIS3 locus on chromosome XV, by expressing R recombinase from the GAL promoter (34). The arrowheads bordering the plasmid sequences represent two copies of the R recombinase target site (RRT) in direct orientation. The ORI sequence (ARS) present in these reporters, as well as others employed in this study, was derived from the 2 micron plasmid. The [LacO]256 array harbored by the reporters permits them to be visualized as green foci in host strains expressing GFP-LacI. After performing the excision reaction in G1-arrested cells, they were released into the cell cycle, and plasmid foci were scored in anaphase cells. The integrated STB-ARS-plasmid, when present in a strain lacking the R recombinase expression cassette, provided the fluorescence tag for chromosome XV. (B–E) Cohesin disassembly was blocked during a cell cycle by swamping out the native Mcd1 subunit with a non-cleavable version overexpressed from the PGAL-(MCD1-nc) expression cassette. (B) DAPI-stained chromosomes (unseparated sister chromatids) stayed in the mother (left; ∼34%) or moved into the daughter (right; ∼66%). The nuclear membrane labeled by mCherry-Nup49 was present in both mother and daughter in most cells. (C) Two cells, containing chromosomes in the daughter, exemplify replicated plasmid sisters (green) coupled to DAPI (left) or uncoupled from it (right). The fluorescence intensity of the single green dot (left) was roughly equal to the sum of the intensities of the two green dots (right) (see also Supplementary Figure S1D). (D and E) The distribution of plasmid sisters or that of Chr XV (paired sister chromatids) is plotted for anaphase cells containing DAPI in the mother (D) or in the daughter (E). (F) In this plot, well resolved individual plasmid foci were grouped separately from overlapping or coalesced foci with a doubling in fluorescence intensity. Each population assay in this figure, and subsequent ones, utilized 100–200 cells. The data are plotted as the mean ± SEM (standard error of the mean).
We uncoupled chromosome segregation from nuclear envelope segregation by over-expressing the non-cleavable form of Mcd1 (a subunit of the cohesin complex) during a single cell cycle, thus blocking cohesin disassembly at anaphase onset (32,52) (Supplementary Text S1A). Under this condition, the entire set of paired sister chromatids occupied the mother or the daughter compartment, as revealed by DAPI staining (Figure 1B and C) and by the perfect coincidence of a fluorescence-tagged ([LacO]256– (GFP-LacI)) reporter chromosome (Chr XV) with the DAPI mass present in the mother or daughter (Figure 1D and E). Daughters housed the chromosomes preferentially, roughly 2:1, over mothers. By contrast to chromosomes, the nuclear membrane outlined by mCherry-Nup49 was distributed between mother and daughter in the majority of cells (Figure 1B and C).
The STB-ARS-plasmid showed strong association with the chromosomes located in either the mother or the daughter compartment (Figure 1D and E). The presence of both plasmid sisters in the chromosome-free cell compartment was quite rare (<5%). By contrast, the strong association between the ARS-plasmid and mother-resident chromosomes (consistent with the mother bias of the plasmid) (Figure 1D) dropped sharply in the case of daughter-resident chromosomes (Figure 1E). Interestingly, there was a clear-cut increase in the 1:1 distribution of the ARS plasmid sisters when chromosomes were present in the daughter (Figure 1E). Presumably, a plasmid molecule might be randomly entrapped by the chromosome mass during its migration to the daughter.
The STB-ARS-plasmid sisters formed one coalesced focus or two separate foci more or less equally (Figure 1F). A single focus, roughly twice as intense as each of two non-coalesced foci (Supplementary Figure S1D and E), could indicate plasmid sisters paired in a cohesin-assisted fashion (33) or those associated with paired sister chromatids. The percentage of coalesced STB-ARS-plasmid sisters observed here (∼43%) was lower than that reported previously for G2/M cells (∼70%) going through a normal cell cycle (33). The difference may be due to the aberrant cell cycle regimen employed here by blocking cohesin cleavage. In contrast to the STB-ARS-plasmid sisters, sister copies of the ARS-plasmid were present predominantly as two separate foci (Figure 1F)
The behavior of the STB-ARS-plasmid under conditions that block cohesin cleavage favors a model in which the plasmid physically associates with chromosomes. The Rep1–Rep2-dependent and cohesin-assisted pairing of sister plasmids (33) may be important in how this association is established. This possibility is scrutinized in further detail in later experiments addressing the connection between plasmid replication and equal segregation. There is no evidence to suggest plasmid coupling to the nuclear envelope.
Contrary effects of plasmid-nuclear pore tethering on the segregation of STB and ARS reporter plasmids
The normally low equal segregation frequency of an ARS-plasmid can be enhanced by tethering it to the nuclear membrane by an integral membrane protein Yif1 or via a subset of the nuclear pore proteins, for example, Mlp1 or Nup2 (19,20). Segregation is presumably facilitated by the ability of inner nuclear membrane proteins and nuclear pores to traverse the nuclear envelope and enter the daughter nucleus (21,57). The findings are consistent with the physical theory of narrow escape, which predicts a diffusing particle (a nuclear plasmid in the present experimental context) to escape faster from a spherical encasement with a narrow window (the mother nucleus) if diffusion occurs along the periphery (58). The outcomes from blocking cohesin disassembly (Figure 1D and E) would suggest that nuclear pore-tethering is unlikely to help 2 micron plasmid segregation. We have now tested this notion more directly.
The reporter plasmids for these experiments were ‘nearly single-copy’ by virtue of a centromere sequence (CEN) harbored by them. The plasmid-borne CEN could be conditionally inactivated by GAL promoter-driven transcription through it (33) (Figure 2A). These plasmids harbored a [TetO]112 array, enabling their association with both TetR-GFP and Mlp1-TetR or Nup2-TetR in host stains expressing the fluorescent repressor along with one of the two nuclear pore protein-repressor fusions. Based on earlier work and the present study, the segregation properties of a reporter plasmid obtained by excision from a chromosome and an analogous plasmid whose copy number is controlled by a conditional CEN are quite similar (Supplementary Figure S2A).
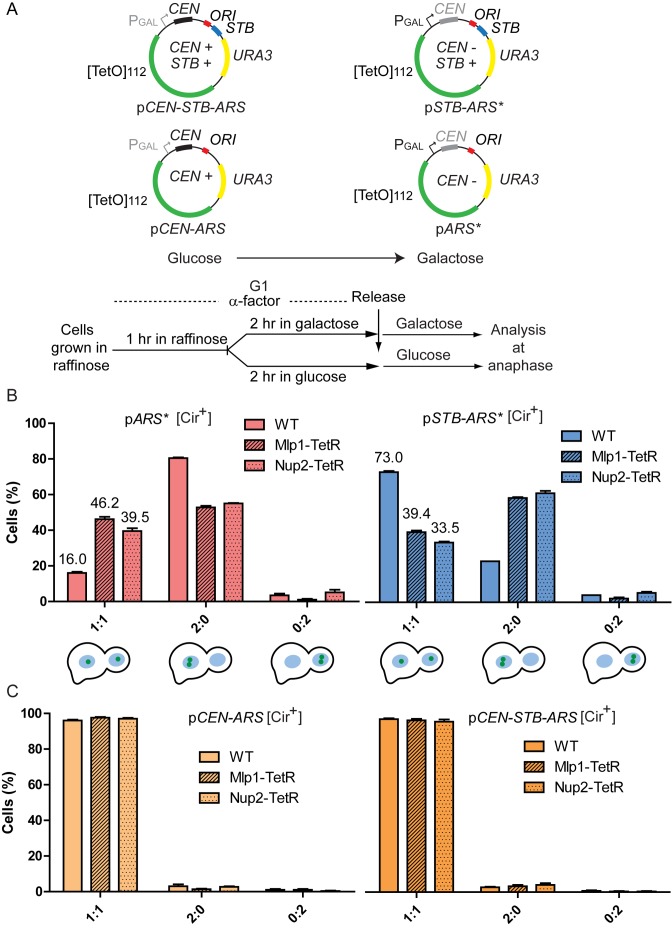
Figure 2.
Tethering to the nuclear pore via Mlp1 or Nup2 is deleterious to STB-ARS-plasmid segregation. (A) The reporter plasmids with or without STB, harboring [TetO]112 and a conditional CEN, are schematically illustrated. They were housed by a [Cir+] host strain expressing TetR-GFP. The experimental outline for following plasmid behavior in the presence or absence of an active CEN is shown. The asterisks in pSTB-ARS* and pARS* indicate the presence of a conditionally inactivated CEN in these plasmids, which are functionally analogous to plasmids pSTB-ARS and pARS, respectively, shown in Figure 1. (B and C) The percentages of anaphase cells with the indicated plasmid patterns are shown in the bar graphs. Controls denoted by ‘WT’ refer to plasmid segregation in the presence of native Mlp1 and Nup2 (not fused to TetR).
During a normal mitotic division, an STB-ARS-plasmid (pSTB-ARS*) tethered to Mlp1 or Nup2 (and presumably to the nuclear pore as a result) showed a decrease in equal segregation frequency along with an increase in mother bias (Figure 2B; right). For an ARS-plasmid (pARS*), mother bias decreased and equal segregation improved as a result of Mlp1- or Nup2-association (Figure 2B; left). In fact, when tethered to Mlp1or Nup2, the STB-ARS*- and ARS*-plasmids behaved more or less similarly with respect to segregation and mother bias. The segregation of a CEN-ARS-plasmid or a CEN-STB-ARS-plasmid in glucose-grown cells was unaffected by its association with Mlp1 or Nup2, suggesting that kinetochore-spindle association is likely dominant over nuclear pore-tethering (Figure 2C).
Taken together, the behavior of STB-ARS-plasmids upon tethering to the nuclear pore (Figure 2B) and upon preventing cohesin disassembly (Figure 1D) discredits 2 micron plasmid segregation by attaching to the nuclear envelope. Clearly, plasmid segregation mediated by the Rep-STB system is more efficient than that mediated by diffusion along the nuclear membrane. The two processes must thus be mechanistically different.
Association of single-copy reporter plasmids with yeast chromosome spreads
The strong coupling between an STB-ARS-plasmid and chromosomes (Figure 1D and E) is consistent with earlier work that revealed the association of a multi-copy STB-ARS-plasmid with yeast chromosome spreads in a Rep1–Rep2-dependent manner (32). Since the reporter plasmid was organized in the nucleus as multiple foci (roughly 3–5 per nucleus), the propensity for the chance association of a subset of the plasmid foci with the spreads could not be ignored. Furthermore, chromosome spreads are not comprised exclusively of chromosomes. To overcome these impediments, we performed the spread assays in isogenic [Cir+] and [Cir0] strains containing either one single-copy reporter plasmid or two such plasmids. Thus, the presence of a single plasmid and the simultaneous presence of a plasmid pair in chromosome spreads could be quantitated and compared. Authentic plasmid-to-chromosome tethering is expected to give comparable levels of association of one plasmid or a pair of plasmids with the spreads. Lack of such tethering should substantially diminish the probability of two-plasmid association vis a vis one-plasmid association.
Chromosome spreads were prepared from G1-arrested yeast cells housing a CEN-STB-ARS-[LacO]256-reporter or those containing, in addition, a CEN-STB-ARS-[TetO]112-reporter (Figure 3A). The copy number of each reporter estimated in previous work was ∼1 (33). Conditions were manipulated to maintain a reporter as pCEN-STB-ARS/pCEN-ARS (CEN active), pSTB-ARS* (STB active but CEN inactive) or as pARS* (neither CEN nor STB active) (Figure 3A). The isogenic [Cir+] and [Cir0] host strains were engineered to express GFP-LacI and TetR-RFP.
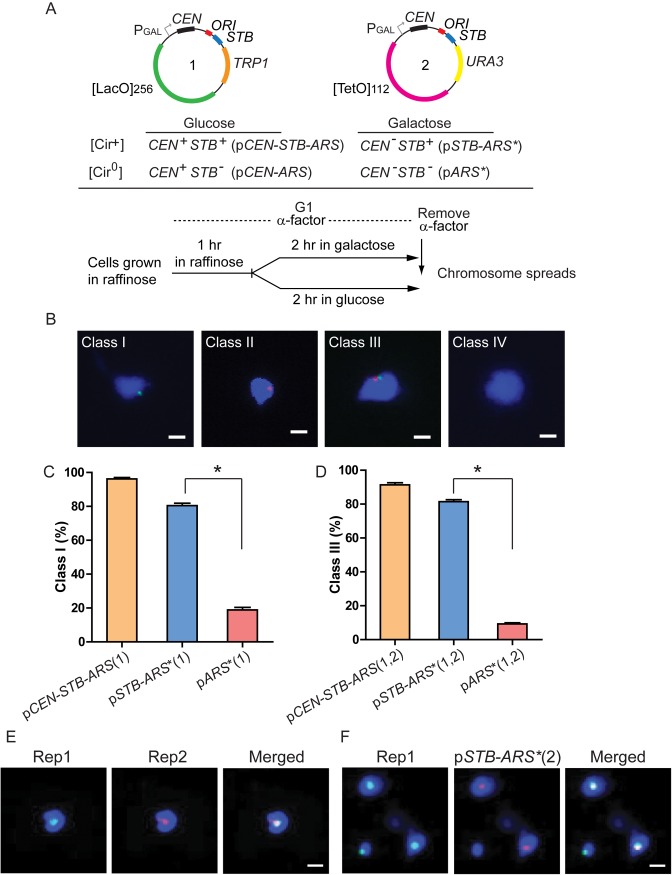
Figure 3.
Single-copy STB-ARS-reporter plasmids colocalize with the Rep proteins in yeast chromosome spreads. (A) The CEN-STB-ARS-[LacO]256-plasmid (numbered 1) was placed, either by itself or together with a companion CEN-STB-ARS-[TetO]112-plasmid (numbered 2), in [Cir+] or [Cir0] strains expressing GFP-LacI and TetR-RFP. The functional states of the plasmid-borne CEN and STB under distinct experimental conditions are tabulated. G1-cells were processed for obtaining chromosome spreads immediately after removing the α-factor employed for cell cycle arrest. (B) The spreads were designated as: Class I (containing the [LacO]256-plasmid); Class II (containing the [TetO]112-plasmid); Class III (containing both plasmids); Class IV (containing no plasmid). Cells harboring the [LacO]256-plasmid gave Class I and Class IV spreads; cells harboring, in addition, the [TetO]112-plasmid gave all four classes. (C and D) The fraction of Class I spreads from cells with the [LacO]256-plasmid (C) and that of Class III spreads from cells with the LacO]256- and [TetO]112-plasmids (D) are plotted. *P < 0.05 (two-tailed t-test). An asterisk added to a plasmid name denotes an inactivated CEN within it. (E) Native Rep1 and HA-6-tagged Rep2 were expressed from the ADH promoter in a [Cir0] strain. They were localized in chromosome spreads by indirect immunofluorescence using primary antibodies to Rep1 and to the HA-epitope, respectively. (F) These chromosome spreads were prepared from a TetR-RFP-expressing [Cir+] strain transformed with the [TetO]112-containing plasmid, pSTB-ARS*(2). Rep1 was localized as in (E). Scale bars (B, E and F) = 2 μm.
The single-plasmid analyses yielded Class I (containing the LacO-plasmid; green; numbered 1 in Figure 3A) and Class IV (plasmid-free) chromosome spreads (Figure 3B). Spreads from the two-plasmid analyses consisted of Classes I–IV (Figure 3B). Class II contained the TetO-plasmid (red; numbered 2 in Figure 3A) but not the LacO-plasmid, while Class III contained both the LacO- and TetO- plasmids. Class I in this case denoted spreads containing the LacO-plasmid but not the TetO-plasmid, while Class IV signified spreads lacking both plasmids. Note that an STB-ARS-reporter plasmid obtained by inactivating its CEN and one excised from a chromosome behave almost identically in chromosome spread assays (Supplementary Figure S2B).
The association of an STB-ARS*-plasmid (Class I; Figure 3C) and the co-association of a pair of STB-ARS*-plasmids (Class III; Figure 3D) with the spreads were high, and nearly equal (∼80%). The values were only slightly lower than the corresponding values for a CEN-STB-ARS-plasmid and a pair of CEN-STB-ARS-plasmids. The association with the spreads dropped significantly for an ARS*-plasmid, and the co-association was even lower for the ARS*-plasmid pair. The spreads also revealed the colocalization of Rep1 and Rep2 proteins with each other (Figure 3E) and with the STB plasmid (Figure 3F). The HA-6-tagged Rep2 employed for this assay was nearly as active as native Rep2 in plasmid stability assays (data not shown). The collective results suggest that Rep1-Rep2-assisted chromosome tethering of the 2 micron plasmid is responsible for its chromosome-coupled segregation.
Localization of the Rep proteins on mammalian chromosomes
Yeast chromosome spreads, in addition to containing non-chromosomal nuclear components, lack the resolution to display individual chromosomes. To circumvent these drawbacks, we examined the localization of Rep1 and Rep2 proteins expressed in mammalian cells, whose individual chromosomes can be visualized in metaphase spreads. Here, the proteins served as a proxy for an STB-ARS-plasmid with which they are normally tightly associated (31,32) (Figure 3E and F). Our use of the non-native host system is justified, as the partitioning systems of Epstein-Barr and human papilloma viruses (EBV; HPV) have been reconstituted in S. cerevisiae (Supplementary Text S1B).
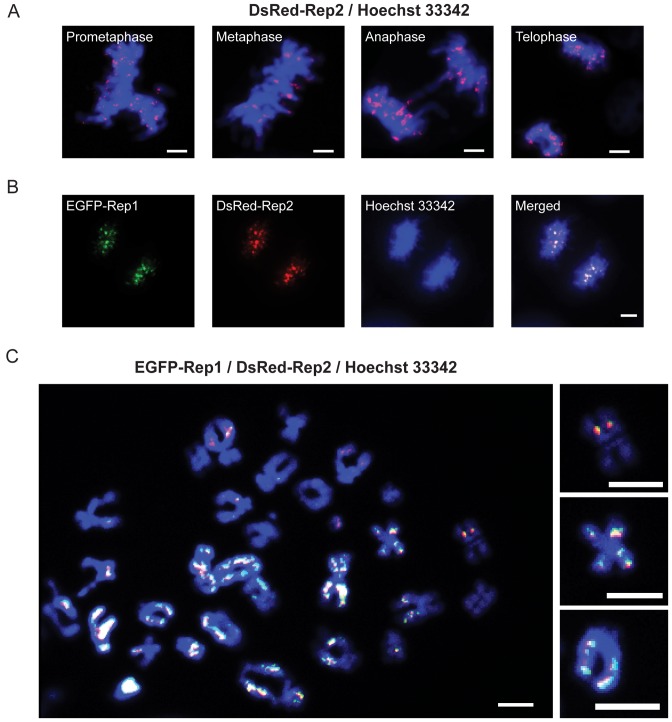
When Rep2 was expressed in COS-7 cells, the protein localized on mitotic chromosomes (Figure 4A). Rep1 was dependent on Rep2 for its nuclear localization/retention (Supplementary Figure S3A–E), and was colocalized with the latter on chromosomes (Figure 4B). Metaphase chromosome spreads, in which individual paired sister chromatids were well resolved, confirmed the physical association of Rep1–Rep2 with chromosomes (Figure 4C; Supplementary Figure S3F). Few Rep2 foci were unassociated with Rep1, and Rep2-free Rep1 foci were nearly absent (as expected from the dependence of Rep1 on Rep2). In individual spreads, most chromosomes contained at least one focus per chromatid. Strikingly, >70% of the Rep foci showed symmetric localization on sister chromatids (Figure 4C). In general, the foci were distributed non-uniformly along the chromosome arms, without an obvious preference for specialized chromosome locations such as centromeres or telomeres.
Figure 4.
Co-expressed Rep1 and Rep2 associate with mammalian chromosomes. (A) Localization of DsRed-Rep2 expressed in COS-7 cells was followed at different mitotic stages. (B) EGFP-Rep1 and DsRed-Rep2 were co-expressed in COS-7 cells, and their localization examined in mitotic cells. A cell at the late anaphase/telophase stage is shown here. (C) Chromosome spreads were prepared from colcemid-treated COS-7 cells expressing EGFP-Rep1 and DsRed-Rep2. The individual EGFP-Rep1 and DsRed-Rep2 patterns (unmerged) are shown in Supplementary Figure S3F. Examples of Rep1-Rep2 foci symmetrically located on sister chromatids are highlighted at the right. Scale bars = 5 μm.
The localization of single-copy STB-containing plasmids in association with Rep1 and Rep2 in yeast chromosome spreads, as well as the co-localization of the Rep proteins with unresolved as well as individually resolved mammalian mitotic chromosomes, suggests that the 2-micron plasmid is likely chromosome-tethered in its native biological context. The symmetric localization of the Rep proteins on sister chromatids becomes significant in the context of the experiments described below, addressing the role of DNA replication in plasmid segregation (see also ‘Discussion’).
Abrogation of mother bias by the Rep-STB system in a nearly replication-independent manner
The 2-micron circle partitioning clock is reset during each cell cycle at the G1-S window, and DNA replication appears to be an important cue for this event (32,35,37). The protein dynamics at STB (complete dissociation of late-exiting factors from a previous partitioning cycle, for example) can be delayed or blocked by delaying or blocking, respectively, the initiation of DNA replication (37). We wished to know whether overcoming mother bias, an absolute requirement for equal plasmid segregation, is dependent on DNA replication.
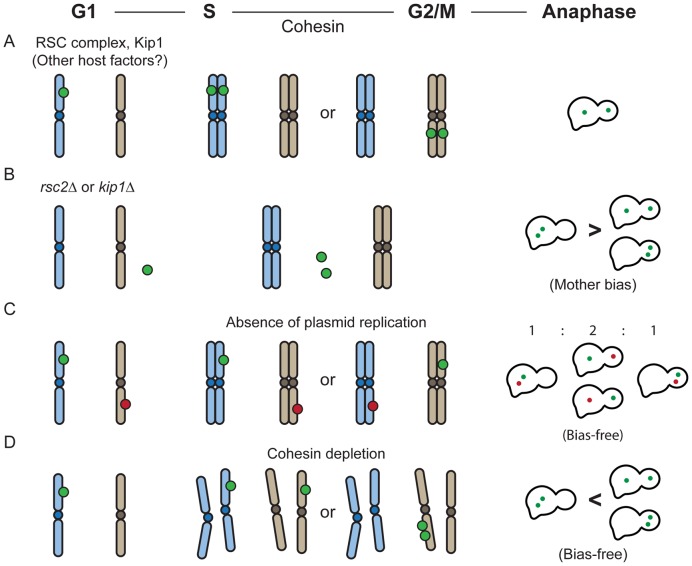
Single-copy circular reporter molecules containing STB and the [LacO}256 array, which either harbored the 2 micron circle replication origin (pSTB-ARS) or lacked the origin (STB-arsΔ-c) (Figure 5A), were excised from the chromosome in [Cir+] or [Cir0] G1-cells, and were examined in the mother and daughter nuclei of anaphase cells. Versions of theses reporters lacking STB (pARS and arsΔ-c) excised in in [Cir+] G1-cells were also included in this analysis. Whereas the replication-competent single-copy reporters reveal the segregation of plasmid sisters as 1:1 (bias-free), 2:0 (mother-biased) and 0:2 (daughter-biased), the replication-blocked reporters signify mother bias as 1:0 and daughter bias as 0:1 (Supplementary Figure S1E).
Figure 5.
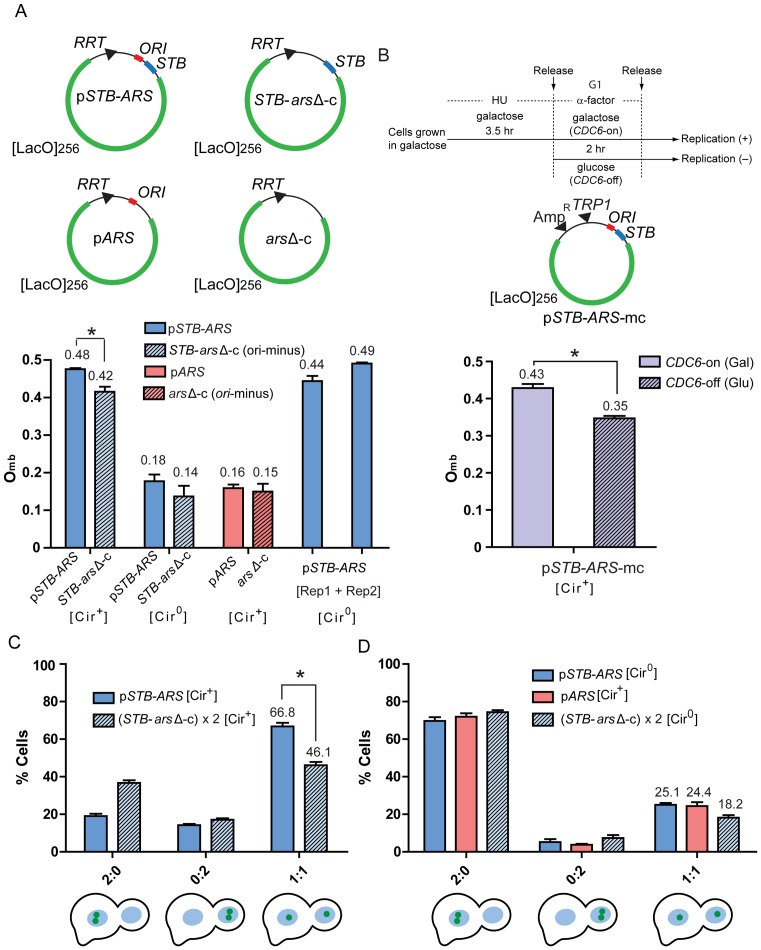
A non-replicating STB-reporter can overcome mother bias; equal segregation of plasmid sisters exceeds that of pseudo-sister circles. (A) The Omb assays were performed using a single-copy STB-plasmid (pSTB-ARS), a single-copy ARS-plasmid (pARS), an ori-minus STB-circle (STB-arsΔ-c) or an ori-minus circle (arsΔ-c) generated in G1-cells ([Cir+] or [Cir0]). The expression of the Rep proteins in a [Cir0] host strain from the ADH promoter (left) or the GAL promoter (right) is symbolized by [Rep1 + Rep2]. From the populations of plasmid/circle foci in mother and daughter nuclei (Fm and Fd, respectively) of anaphase cells, Omb was computed as Fd/[Fd + Fm]. (B) In this analysis, a multi-copy STB-ARS-reporter plasmid (pSTB-ARS-mc) was employed. The experimental protocol for depleting Cdc6 during a cell cycle is schematically diagrammed at the top. HU = hydroxyurea. (C) Two identical ori-minus STB circles, (STB-arsΔ-c) × 2, were generated in [Cir+] G1-cells, and their segregation in anaphase cells was compared to that of pSTB-ARS-sisters. *P < 0.05 (two-tailed t-test). (D) The segregation patterns of the same reporters assayed in (C) were followed in a [Cir0] strain. In addition, an ARS-plasmid was analyzed in the [Cir+] strain. The plots for pSTB-ARS in (C) and pARS in (D) represent the data sets from which their Omb values shown in (A) were derived.
We expressed the ability of a reporter to overcome mother bias Omb, as Fd/(Fd + Fm), where Fd and Fm are the number of plasmid/circle foci present in the daughter and mother nuclei, respectively. In the case of plasmid sisters, a single focus present only in the mother or the daughter nucleus, with the predicted increase in fluorescence intensity, was taken as a coalesced pair of foci (34) (Supplementary Figure S1D). The Omb values for the STB-ARS-sisters and the STB-arsΔ-circle in a [Cir+] host strain were ∼0.48 and ∼0.42, respectively (Figure 5A). The corresponding values in an isogenic [Cir0] strain were ∼0.18 and ∼0.14, respectively. Sister copies of an ARS-plasmid (STB-minus) and the ori-minus circle (arsΔ-c) failed to overcome mother bias (Omb = ∼0.16 and ∼0.15, respectively). The Omb values of the STB-ARS-plasmid in [Cir0] strains expressing REP1 and REP2 from chromosomal locales under the control of the constitutive ADH promoter or the inducible GAL1-10 bidirectional promoter were similar to its Omb in the [Cir+] strain.
The competence of the Rep-STB system to overcome mother bias is largely independent of the plasmid's ability to replicate. Conversely, the act of plasmid replication does not contribute to the bias, as the strong mother bias of an ARS plasmid is unchanged by preventing its replication. An Omb of nearly 0.5 for an STB-ARS-reporter in a [Cir0] host in the presence of the Rep proteins rules out Rep1-Rep2-mediated physical association of the reporter with the native 2-micron plasmid being responsible for the strong diminution in mother bias. Prior work showed that the Rep1 and Rep2 proteins, in the absence of the 2-micron plasmid, are sufficient to support normal segregation of a single-copy STB-ARS-plasmid (37).
Escape from mother bias by an STB-ARS-plasmid in the absence of plasmid and chromosome replication
In the STB-arsΔ-c, the consequences due to lack of replication cannot not be dissociated from those due to the absence of the origin per se. It is possible that the requirement for plasmid replication in overcoming mother bias by Rep-STB is bypassed by deleting the origin. The slight reduction in the Omb of STB-arsΔ-c (0.42) compared to that of pSTB-ARS (0.48) could be an indirect effect of removing ORI rather than the result of blocking replication. The ORI-binding Orc complex, in addition to its key role in initiating eukaryotic DNA replication (59,60), also contributes to the architecture and function of silent chromatin in S. cerevisiae (61,62) (Supplementary Text S1C). The 2-micron plasmid ORI has been shown to possess silencing activity, which is dependent on Orc (63). To rule out potential secondary effects of removing ORI, we determined Omb for a multi-copy STB reporter plasmid harboring an intact ORI when plasmid as well as chromosome replication was blocked. Because of technical limitations, this analysis could not be easily performed with the single-copy STB plasmid.
In order to program a cell cycle in the absence of DNA replication, we conditionally depleted Cdc6 (37,64) (Figure 5B; Supplementary Figure S4A), which associates with the Orc-bound replication origin to form the pre-replication complex (65). During a Cdc6-depleted cell cycle, the unreplicated chromosomes still segregate into mother and daughter roughly equally (‘reductional mitosis’). A fluorescence-tagged chromosome IV under replication-permissive (galactose) and non-permissive (glucose) conditions verified the nearly complete block in its replication as well as its bias-free distribution into the mother or daughter under Cdc6 depletion (Supplementary Figure S4B). The average number of STB-ARS-plasmid foci in G1 cells were similar in glucose and galactose (5–6 per cell) but differed in anaphase cells by a factor of 1.5 (Supplementary Figure S4C and D). While the foci number doubled with G1-to-anaphase passage in galactose, the increase was minimal in glucose. Based on the plasmid foci counts in mother and daughter, the Omb for the replicated STB-ARS-plasmids was ∼0.43 as against ∼0.35 for the unreplicated plasmids (Figure 5B).
The quantitation from a multi-copy reporter plasmid (based on foci number) is less reliable than that from a single-copy reporter plasmid. The Omb values in Figure 5B were perhaps underestimated because of the occasional tendency of foci to overlap with each other and the potential differences in plasmid count between individual foci. Nevertheless, an Omb of ∼0.35 (the smaller of the two values in Figure 5B) was still compellingly larger than Omb = ∼0.16 displayed by the ARS-plasmid (Figure 5A).
Thus, regardless of the means by which replication is blocked, the Rep-STB system diminishes mother bias to a considerable extent. However, the findings from the STB-arsΔ-circle and the Cdc6-depleted cell cycle also suggest that the efficiency of mother bias removal is further improved by the origin per se, or by replication initiated at the origin, by a small but significant margin.
Overcoming mother bias in equal plasmid segregation: necessary but not sufficient
Overcoming mother bias, in and of itself, does not ensure equal plasmid segregation. For example, equal segregation (1:1) of a pair of plasmids or their total missegregation without mother-daughter bias (2:0 = 0:2) in every anaphase cell will manifest an Omb of 0.5. It does not follow from the replication-independence of Omb that replication is not required for equal plasmid segregation. As noted before, the de novo assembly of the 2 micron plasmid partitioning complex at STB during a cell cycle is temporally coordinated with the onset of DNA replication (32,35,37). Furthermore, normal segregation occurs after the parent plasmid population has been duplicated by the cellular replication system. Finally, the bridging of plasmid sisters by the cohesin complex, assembled at STB concomitant with plasmid replication with the assistance of Rep1-Rep2 (32,33), could influence their segregation mechanism. We therefore wondered whether the Rep-STB system, capable of nearly eliminating mother bias without plasmid replication, can also bring about equal plasmid segregation in the absence of replication. To address segregation without replication, we generated two identical ori-minus STB-circles in G1 cells by recombination from two separate chromosomal locations. The excised circles are referred to as pseudo-sisters to signify their formation without duplication of a parent molecule. They were followed in anaphase cells.
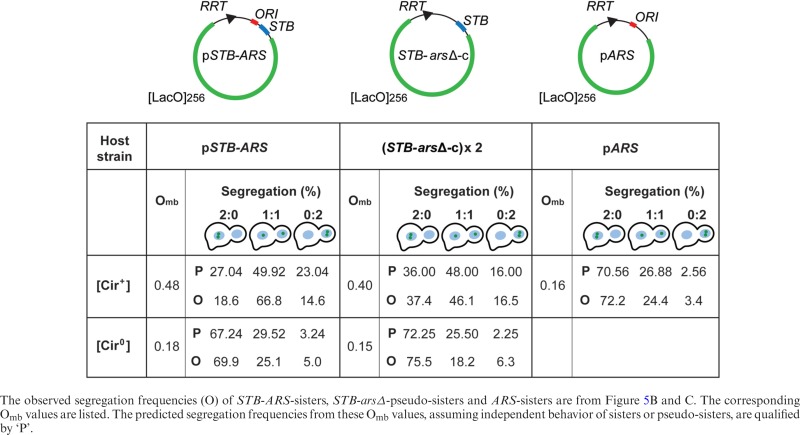
The equal segregation frequency (1:1) for the STB-arsΔ-pseudo-sisters in the presence of Rep1 and Rep2 ([Cir+] host) was ∼46%, considerably lower than ∼67% recorded for the STB-ARS-plasmid sisters (Figure 5C). The 2:0 and 0:2 frequencies for the pseudo-sisters were ∼37% and ∼17%, respectively. Their Omb, based on the combined 1:1, 2:0 and 0:2 frequencies, was ∼0.40 (Table 1). This value agreed closely with the Omb of ∼0.42 estimated for a single-copy ori-minus STB circle (STB-arsΔ-c; Figure 5A). The ARS-plasmid sisters present in a [Cir+] strain as well as the STB-ARS-sisters and STB-arsΔ-pseudo-sisters present in a [Cir0] strain showed similar, strongly mother-biased (Omb << 0.5) segregation (Figure 5D; Table 1).
Table 1. Omb values and segregation frequencies of plasmid sisters or pseudo-sisters in the presence and absence of the Rep-STB partitioning system.
|
STB-ARS-sisters clearly outperform STB-arsΔ-pseudo-sisters in 1:1 segregation, even though their Omb values are comparable. Thus, replication itself, or some step associated with replication, is responsible for enhancing equal segregation. Sister plasmids, held together by cohesin as a paired unit, may segregate to mother and daughter nuclei in a directed fashion. The potential lack of cohesion between STB-arsΔ-pseudo-sisters may limit their efficiency in 1:1 segregation without affecting their ability to overcome mother bias. In this model, the functional difference between STB-ARS-sisters and STB-arsΔ-pseudo-sisters arises from the coupled behavior of sisters versus the independent behavior of pseudo-sisters (see below).
Independent versus inter-dependent plasmid segregation
The segregation frequencies of a pair of reporters are determined by whether they behave independently or not and by the extent to which they overcome mother bias (their Omb values). For independent segregation, the binomial rule predicts the 2:0, 1:1 and 0:2 patterns to be (1-Omb)2, [2 x Omb (1-Omb)] and (Omb)2, respectively. If segregation were totally bias-free (Omb = 0.5), the expected values are 25% (2:0), 50% (1:1) and 25% (0:2). The hypothetical examples of 100% equal segregation or 100% unequal segregation without bias (50% each of 2:0 and 0:2) mentioned earlier represent the opposite extremes of dependent behavior during segregation. It is thus straightforward to deduce, from the experimentally determined Omb and segregation frequencies, whether or not a pair of reporters behave independently.
The observed 2:0, 1:1 and 0:2 frequencies of reporters lacking either STB or ARS and of an STB-ARS-reporter deprived of the Rep proteins were in agreement with the theoretical estimates for independent segregation (Table 1) (Supplementary Text S1D). The results for the STB-ARS-reporter provided with the Rep proteins (Omb = 0.48; Figure 5A) did not match the predicted values of 27% (2:0), 50.0% (1:1) and 23% (0:2) (Figure 5C) (Table 1). The absence of the Rep-STB system, or the lack of replication even in its presence, causes a pair of reporters to segregate independently. By contrast, the Rep-STB system promotes dependent segregation of sister copies of a plasmid formed by replication.
Considered in the context of Rep-STB-assisted plasmid association with chromosome spreads, the nearly complete lack of mother bias (Omb of ∼0.5) and the ∼50% equal segregation of the STB-arsΔ-pseudo-sisters can be accommodated by their independent linkage to chromosomes. The Omb = ∼0.5 of STB-ARS-sisters is also consistent with chromosome-tethering. However, the significantly higher than binomial 1:1 segregation (∼67%) of STB-ARS-sisters suggests that the Rep-STB system, in conjunction with plasmid replication, contributes more to segregation than just overcoming mother bias. The pairing of STB-ARS-plasmid sisters promoted by the Rep proteins and cohesin likely causes them to associate with chromosomes in a coupled manner, which elevates 1:1 segregation beyond 50%. While the shared feature of chromosome-association accounts for the escape of mother bias by STB-ARS-sisters and STB-arsΔ-pseudo-sisters, the difference in how sisters and pseudo-sisters establish this association may explain their disparity in equal segregation.
The segregation frequencies of plasmid sisters or pseudo-sisters lacking a functional Rep-STB system are generally consistent with the passive partitioning of a plasmid in yeast being limited by the diffusion barrier that it encounters. Omb is an indirect measure of the barrier, the two being inversely correlated.
Difference in segregation between STB-ARS-sisters and STB-arsΔ-pseudo-sisters in the presence of the monopolin complex
The monopolin complex is responsible for directing the co-segregation of sister chromatids during meiosis I (66–69). When expressed inappropriately during a mitotic cell cycle, monopolin brings about an analogous effect, though with reduced efficiency (53). In a prior study, we revealed a tight correlation between a pair of sister chromatids and a pair of STB-ARS-plasmid sisters in the extent of co-segregation during a monopolin-directed mitotic cell cycle (34). This finding is consistent with the normal segregation of STB-ARS-plasmid sisters being coupled to that of sister chromatids. In order to test whether this presumed coupling is replication-dependent, we assayed the segregation of a pair of STB-arsΔ-pseudo-sisters under the influence of monopolin (Figure 6).
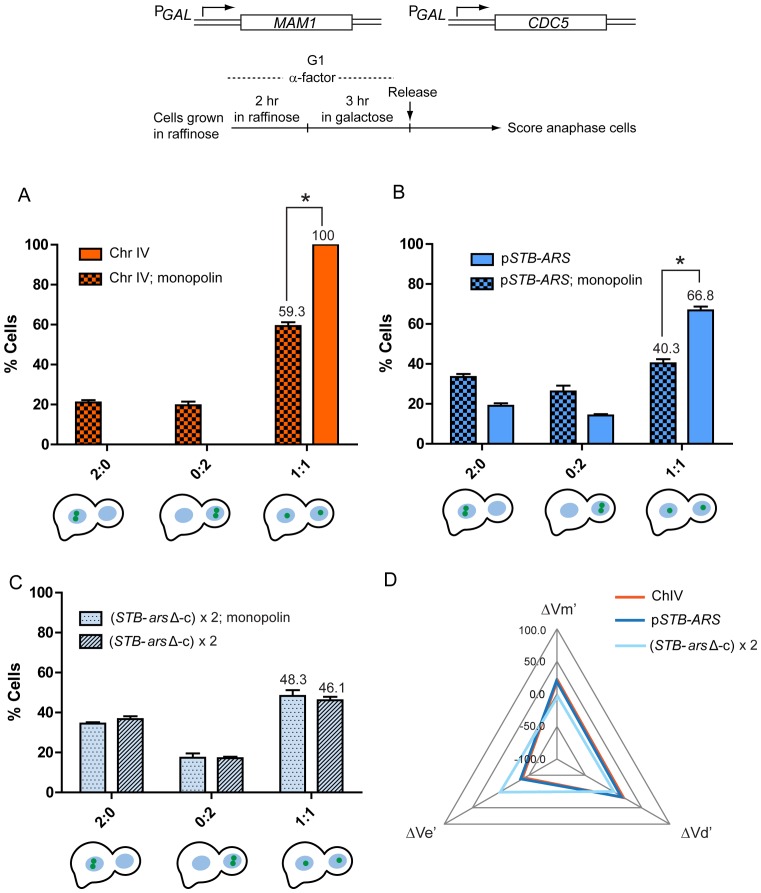
Figure 6.
STB-arsΔ-pseudo-sisters and sister chromatids are not correlated in segregation in mitotic cells expressing monopolin. The assembly of the monopolin complex was induced by turning on MAM1 and CDC5 expression from the GAL promoter in G1-arrested cells before releasing them from arrest (34,53). (A-C) The equal segregation and co-segregation frequencies of fluorescence-tagged chromosome IV (A), the STB-ARS-plasmid (B) and STB-arsΔ-pseudo-sisters (C) in the presence of monopolin or its absence (normal mitosis) are plotted. The segregation data for the STB-ARS-plasmid during normal mitosis (B) can also be seen in Figure 5C. *P < 0.05 (two-tailed t-test). (D) The data from (A–C), are represented as a radar plot, with the same scale on the three axes. The variables Ve, Vm and Vd denote equal segregation, mother-biased missegregation and daughter-biased missegregation, respectively. The corresponding ΔV values indicate their deviations from the norm due to monopolin. The normalized ΔV values (ΔV's) account for differences in the equal segregation frequencies of the individual reporters during normal mitosis (34). For example, ΔVe’ for the STB-ARS plasmid is [40.3 – 66.8]/66.8 = –39.7%; that for Chr IV is [59.3 – 100]/100 = –40.7%. The ΔVe’ for the STB-arsΔ-pseudo-sisters is [48.3 – 46.1]/46.1 = 4.8%.
Consistent with previous observations (34,53), chromosome IV sisters showed ∼41% co-segregation in the presence of monopolin, with little bias towards mother or daughter (2:0/0:2 = 1.1) (Figure 6A). The extent of co-segregation of the STB-ARS-plasmid sisters, normalized to their monopolin-free equal segregation frequency of 66.8%, was similar (∼40%), and was nearly bias-free (2:0/0:2 = 1.2) (Figure 6B). In sharp contrast, the segregation patterns of the STB-arsΔ-pseudo-sisters were nearly identical in the presence and absence of monopolin (Figure 6C).
The quantitative correlations between chromosome IV and the STB-ARS-plasmid or the STB-arsΔ-circles (STB-pseudo-sisters) in their responses to monopolin are depicted in the radar plots in Figure 6D. In this representation, ΔVe’, ΔVm’ and ΔVd’ denote monopolin-induced variations in equal segregation, mother-biased missegregation and daughter-biased missegregation, respectively, normalized for differences in the equal segregation frequencies of Chr IV, the STB-ARS-plasmid and ori-minus STB-circles during normal mitosis (34). The near congruence of the orange and dark blue triangles representing Chr IV and the STB-ARS-plasmid (Figure 6D), respectively, is consistent with previous observations on their tightly correlated behavior in the presence of monopolin (34). The STB-arsΔ-circles, characterized by the pale blue triangle, are not correlated with the chromosome and the STB-ARS-plasmid. Thus, the coupling between sister chromatids and STB-ARS-plasmid sisters in monopolin-directed missegregation is critically dependent on plasmid replication.
Plasmid replication and the contemporaneous cohesin-mediated pairing of the resulting STB-ARS-sisters provide a plausible unified explanation for their normal, better-than-binomial 1:1 segregation as well as their strongly correlated missegregation with sister chromatids clamped by monopolin. The proximity of plasmids sisters and that of sister chromatids conferred by cohesin during a normal cell cycle may promote one-to-one association between the two paired DNA entities. By contrast, STB-arsΔ-pseudo-sisters, lacking the proximity required to associate with sister chromatids, may still associate with chromosomes in a random fashion (see below).
Lack of sister plasmid cohesion from replication-independent cohesin assembly at STB
The cohesin complex, crucial for topologically bridging sister chromatids and ensuring equal chromosome segregation (70,71), is also recruited at the STB locus (32,72). Despite its presence in sub-stoichiometric amounts at STB (73), cohesin appears to function in promoting equal plasmid segregation. When cohesin assembly at STB, but not at CEN or other chromosomal loci, is postponed until after DNA replication by delaying spindle assembly, the frequency of plasmid missegregation is elevated (34,43). Equal segregation of sister chromatids paired by cohesin is not affected under this condition. Cohesin-dependent coalescence of sister STB-ARS-plasmids is impaired by this cell cycle regimen, even though STB-cohesin association is restored following spindle assembly (33,43). We now examined whether cohesin can be assembled at STB in the absence of replication, and (if so) whether such assembly can mediate the cohesion of STB-containing pseudo-sisters.
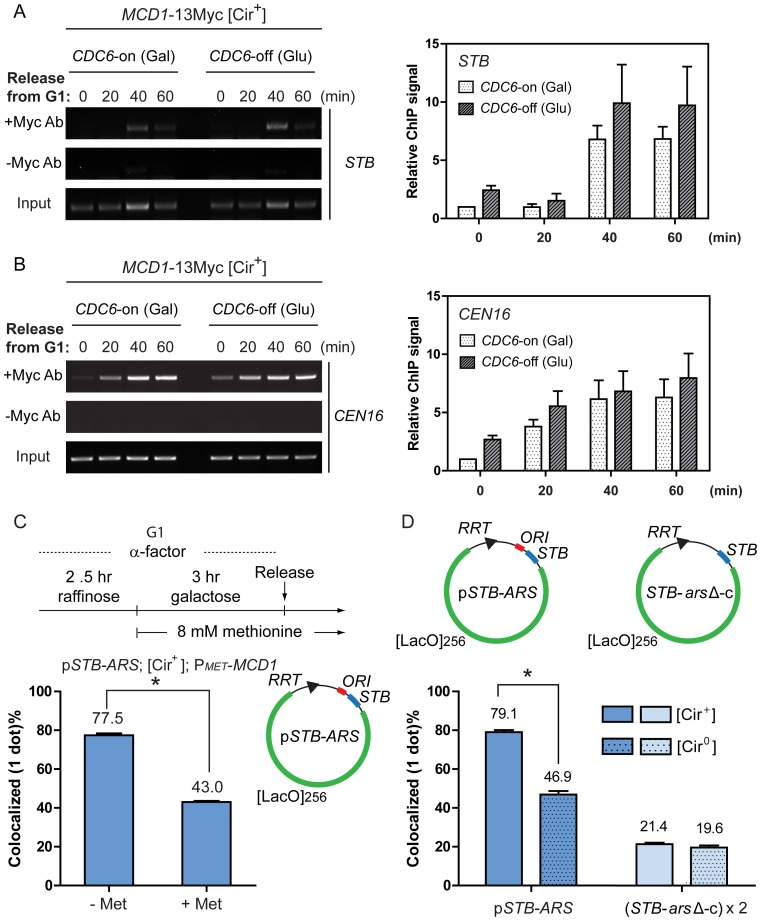
By chromatin immunoprecipitation (ChIP), no obvious difference in the association of the cohesin subunit Mcd1 at STB was detected during a replication-permissive (CDC6-on) or non-permissive (CDC6-off) cell cycle (Figure 7A). This was also the case for Mcd1 association at a centromere (CEN16) (Figure 7B). The rather delayed cohesin recruitment at STB relative to CEN may be due to subtle cell cycle perturbations from the artificial regulation of CDC6 expression (Supplementary Text S1E). The kinetics of cell cycle progression, assayed by bud emergence and morphology, were similar in the presence or absence of Cdc6 (Supplementary Figure S5).
Figure 7.
Cohesin is assembled at STB in the absence of replication but STB-arsΔ-pseudo-sisters lack cohesion. (A and B) In the chromatin immunoprecipitation (ChIP), the antibody targeted the Myc-epitope of the cohesin subunit Mcd1 expressed in the [Cir+] experimental strain. The endogenous 2-micron plasmid provided the substrate for cohesin association. The immunoprecipitate was probed by PCR-amplification using primer pairs specific for STB and CEN16. The signal at each time point was first corrected by subtracting the corresponding signal from the ‘no antibody’ controls. The corrected signals were then normalized to the corresponding ‘input’ signals. In plotting the ‘relative ChIP signals’, the signal at the 0 min time point for the ‘CDC6-on’ sample was assigned a value of 1. (C) In the [Cir+] experimental strain, MCD1 was placed under the control of the MET promoter. MCD1 expression was suppressed by the addition of methionine, as schematically outlined, to block the assembly of the cohesin complex. The STB-ARS-reporter plasmid was excised from the chromosome in G1-arrested cells. After releasing the cells from arrest, the plasmid foci in metaphase cells were classified as single (or overlapping) dots or separated dots. (D) After excising the STB-ARS-plasmid or the STB-arsΔ-circles in [Cir+] or [Cir0] cells at G1, fluorescent foci in metaphase cells were grouped into one-dot or two-dot classes as in (C). *P < 0.05 (two-tailed t-test).
The Rsc2 and Rsc8 subunits of the RSC2 chromatin remodeling complex, which is required for 2-micron plasmid segregation (37,38,74) were also detected at STB (and at CEN) by ChIP under CDC6-expressed or repressed conditions (Supplementary Figure S6A–D). Interestingly the ChIP signals for both Rsc2 and Rsc8 at STB, at the time of release from G1 arrest (time zero in Supplementary Figure S6A and C), were reproducibly higher in cells blocked in CDC6 expression.
In cells going through a normal cell cycle, the Rep1–Rep2-assisted cohesion of STB-ARS-plasmid sisters at metaphase was dependent on cohesin (Figure 7C). The decreases in coalesced plasmid foci were similar under cohesin depletion (from ∼78% to ∼43%; Figure 7C) and in the absence of the Rep proteins (from ∼79% and ∼47%; Figure 7D). For the STB-arsΔ-pseudo-sisters, the coalesced fractions were quite low, regardless of the presence or absence of the Rep proteins (∼21% and ∼20%, respectively; Figure 7D). The absence of replication apparently has a stronger adverse effect on plasmid coalescence than the absence of the Rep proteins. The topological linkage between plasmid sisters during, and immediately following, replication may help their localization at the same nuclear site.
Collectively, the above observations rule out DNA replication as a pre-requisite for cohesin or the RSC2 complex, and perhaps for other partitioning factors as well, to associate with STB. However, deviation from the normal replication program may alter the kinetics and/or dynamics of these associations. Lack of cohesin assembly at STB or cohesin assembly in the absence of replication disrupts Rep-STB function (the present study). Cohesin assembly at STB after completion of replication also has the same effect (33,43). Thus, an obligatory requirement for the cohesion of plasmid sisters and their equal segregation is cohesin assembly concomitant with replication.
As suggested earlier, the difference in the mode of chromosome-tethering between cohesed STB-ARS-sisters and STB-arsΔ-pseudo-sisters that lack cohesion is likely responsible for their distinct, though bias-free, modes of segregation.
Contributions from host factors to 2-micron plasmid segregation with or without directly impacting Omb
Host factors that associate with STB, and affect 2-micron plasmid segregation, include the RSC2 chromatin remodeling complex, the nuclear motor Kip1 and the cohesin complex (32,36–38,74). Furthermore, the integrity of the mitotic spindle is required for the assembly of cohesin at STB (43). As noted earlier, plasmid replication in the absence of the spindle, followed by spindle assembly in G2/M, results in high levels of plasmid missegregation. The present study reveals a largely replication-independent Omb component and an additional replication-dependent component to 2 micron plasmid segregation. In principle, a partitioning factor may function at the replication-independent or the replication-dependent step or at both steps. We attempted to decipher host factor contributions to the Omb and extra-Omb components of plasmid segregation by deleting or depleting individual factors.
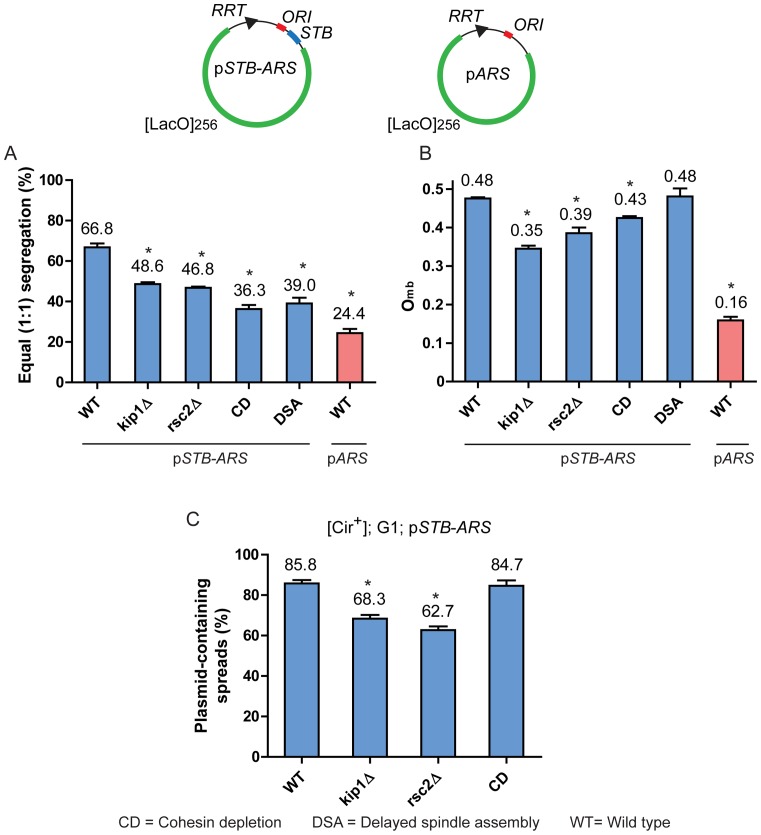
In the single-copy STB-ARS-plasmid segregation assay, kip1Δ and rsc2Δ caused similar decreases in 1:1 segregation (Figure 8A) and comparable diminutions in Omb (Figure 8B). The extent of missegregation caused by delayed spindle assembly (which also delays cohesin recruitment at STB until after plasmid replication) was nearly equal to that resulting from cohesin depletion. However, delayed spindle assembly had no effect on Omb, while the effect of cohesin depletion was quite modest. Yet, these two conditions caused higher plasmid missegregation than kip1Δ or rsc2Δ (Figure 8A), suggesting that overcoming mother bias may, in fact, disfavor equal plasmid segregation in certain contexts (see ‘Discussion’ section).
Figure 8.
Plasmid segregation factors can be distinguished by whether or not they affect Omb. (A and B) The segregation frequencies of the STB-ARS-plasmid or ARS-plasmid sisters formed by replication of the single-copy plasmids excised in G1 were scored in late anaphase nuclei. The data for the STB-ARS- and ARS-plasmids in the wild type (WT) strain are the same as those presented in Figure 4. (C) Plasmid association of the excised STB-ARS-plasmid with chromosome spreads was assayed at the G1 stage after washing off α-factor. *P < 0.05 (two-tailed t-test). For cohesin depletion (CD), the MET promoter controlling Mcd1 expression was turned off (see Figure 7C). For delayed spindle assembly (DSA), G1-arrested cells were released into the cell cycle in presence of nocodazole, and the G2/M-arrested cells were then released by washing off the drug (43).
The association of the STB-ARS-plasmid with chromosome spreads from G1-arrested cells was diminished by kip1Δ and rsc2Δ to similar extents (Figure 8C). ‘Cohesin depletion’ had no effect on this association, as expected from the efficient localization of STB-ARS-plasmids in G1-chromosome spreads (Figure 3A–C). As Mcd1 is not expressed in G1, its depletion (under our non-native experimental design) would be redundant in the context of a normal cell cycle. An earlier study showed that a sharp decline in STB-ARS-plasmid presence in chromosome spreads caused by microtubule depolymerization is reversed upon spindle restoration (43). Thus, cohesin depletion or delayed spindle assembly decreases equal plasmid segregation without apparently affecting plasmid-chromosome association per se.
In sum, these results suggest that a subset of the host factors (Kip1 and the RSC2 complex) contribute primarily to the Omb component of 2-micron plasmid segregation, perhaps by promoting plasmid-chromosome interactions. The decreases in Omb due to their absence are reflected in the reductions in STB-ARS-plasmid association with chromosome spreads (Figure 8B and C). In addition, kip1Δ and rsc2Δ appear to uncouple STB-ARS-plasmid sisters, making them behave as pseudo-sisters. The theoretical estimates for 1:1 segregation of independently behaving pseudo-sisters (as explained in describing the results depicted in Figure 5) for Omb = 0.35 (kip1Δ) and 0.39 (rsc2Δ) are 45.5% and 47.6%, respectively. They are in agreement with the observed values of 48.6% for kip1Δ and 46.8% for rsc2Δ (Figure 8A).
By contrast to kip1Δ or rsc2Δ, cohesin depletion and delayed spindle assembly (the latter likely by disrupting cohesin function at STB) (43) appear to act via a replication-dependent component of segregation that affects plasmid sisters in a coupled fashion. The experimental value for 1:1 plasmid segregation under cohesin depletion (Omb = 0.43) contradicts the expectation for pseudo-sister behavior of independent segregation, 36.3% (observed) versus 49.0% (predicted) (Figure 8A and B). Such a discrepancy is also true for delayed spindle assembly, 39.0% (observed) versus 49.9% (predicted for Omb = 0.48) (Figure 8A and B). The requirement of spindle integrity for plasmid localization in chromosome spreads (43) suggests an additional Omb-related role for the spindle in plasmid segregation. This inferred spindle contribution could not be directly tested, as a functional spindle (even one assembled late in the cell cycle) is essential for the segregation of mother and daughter nuclei.
DISCUSSION
We have unveiled the likely mechanism by which the 2-micron plasmid partitioning system promotes the equal segregation of replicated molecules to mother and daughter cells. Evidence from native and non-native host systems support the physical association of the plasmid with chromosomes, and segregation by the hitchhiking model. The Rep-STB system eliminates mother bias to a considerable extent in the absence of replication, and implements binomial segregation of a pair of pseudo-sister reporters (Supplementary Text S1F). However, the full extent of equal segregation requires overcoming mother bias to be coupled to plasmid replication. The hitchhiking model readily accommodates this relationship between bias and replication. When a pair of plasmid copies, sisters formed by replication or non-sisters, are tethered randomly to chromosomes, they are freed from bias, but are limited to only 50% equal segregation frequency (in accordance with the random assortment of chromosomes). However, if they are tethered to sister chromatids, one-to-one, they are not only released from bias but also achieve the theoretical limit for equal segregation. We suggest that the Rep-STB system takes advantage of DNA replication, as well as replication-associated (and spindle-dependent) cohesin recruitment at STB, to provide directionality/specificity to plasmid-chromosome tethering.
Chromosome-like segregation of the 2 micron plasmid without assistance from chromosomes, though formally possible, is unlikely. Although CEN and STB do display shared host factor interactions, there is no evidence for the association of kinetochore components with STB or of the Rep proteins with CEN (32; unpublished data). The normal physiology of [Cir0] strains rules out any role for the Rep proteins in chromosome segregation. By manipulating the time of spindle assembly during a cell cycle, 2-micron plasmid segregation can be disrupted without affecting chromosome segregation (43). The monopolin complex expressed artificially during mitosis associates with CEN and not STB (34), yet induces similar extents of co-segregation in sister chromatids and plasmid sisters (34; this study). The most parsimonious explanation for our collective results is that the 2-micron plasmid exploits chromosome segregation as a chromosome-attached entity.
Plasmid-chromosome association: the primary requisite of the hitchhiking model
The efficient localization of a single-copy STB-plasmid or two such plasmids in association with Rep1 and Rep2 in yeast chromosome spreads satisfies one important criterion for the validity of the hitchhiking model. The colocalization of Rep1 and Rep2 on mammalian chromosomes lends further credence to this model. The pattern of the Rep protein foci along individual chromosomes in mammalian metaphase spreads suggests plasmid sisters tethering to sister chromatids in a symmetric (non-random) fashion.
The Rep2-assisted nuclear import and chromosome localization of Rep1 in mammalian cells recapitulate several (but not all) of the cell biological attributes of these proteins in their native host system. Earlier yeast experiments utilizing GFP-fusion proteins suggested that Rep1 and Rep2 are capable of nuclear localization individually, with the nuclear-cytoplasmic contrast being sharper for Rep2 (75,76). In general agreement with these findings, independent observations indicated that Rep2 assists the nuclear import or retention of Rep1 (77). Furthermore, we found that Rep1 and Rep2 depend on each other, but not on an STB-containing plasmid, for their association with yeast chromosome spreads (32).
Neither Rep1 nor Rep2 is a strong DNA-binding protein. Rep1 does not bind STB-DNA in vitro, while the binding of Rep2 is quite weak (78). Their interaction with STB is likely promoted by a host factor or factors (79). The in vivo binding target could be STB-chromatin, rather than naked DNA. Given their poor DNA binding ability, chromosome-association of the Rep proteins is mediated presumably by a chromatin binding host protein or proteins. Such proteins may be conserved, at least in the relevant functional domain(s), between yeast and mammals. The interaction between a viral protein that associates with the viral partitioning locus and a chromatin binding host protein is a recurring theme in the tethering of viral episomes to host chromosomes (80–82) (Supplementary Text S1G).
A bi-partite mechanism for high-fidelity segregation of the 2-micron plasmid
The contribution of the Rep-STB system towards 2-micron plasmid segregation is 2-fold. First, it overcomes mother bias by promoting the random tethering of plasmid foci to chromosomes in an essentially replication-independent step. The 50% equal segregation of a pair of plasmids guaranteed by this binomial mechanism is considerably higher than that exhibited by an ARS-plasmid, but falls well short of the fidelity of chromosome segregation. Second, the Rep-STB system avails itself of plasmid replication to coordinate the association of cohesed sister plasmids with sister chromatids, thereby elevating plasmid segregation to the status of chromosome segregation. The ∼70% equal segregation frequency (less than the theoretical 100%) observed for a single-copy STB-plasmid likely reflects the limiting efficiency of the reporter system rather than the inherent efficiency of the Rep-STB system. In standard plasmid stability assays, the loss rate per generation of 2-micron derived plasmids is two to three orders of magnitude higher than that of the native 2-micron circle, yet at least an order of magnitude lower than that of ARS-plasmids.
Host factors that contribute to the replication-dependent and replication-independent components of plasmid segregation
The RSC2 chromatin remodeling complex and Kip1 help eliminate mother bias by facilitating random, replication-independent plasmid-chromosome association (Figure 9A and B). The integrity of the mitotic spindle must also contribute to this replication-independent step, as plasmid-chromosome association is strongly diminished upon microtubule depolymerization (43). By contrast, cohesin's primary effect is not on plasmid-chromosome association, or Omb per se (32,43; this study). Rather, it modulates Omb qualitatively by directing the association of plasmid sisters to sister chromatids in a strictly replication-dependent fashion. Equal segregation of sister plasmids is thus enhanced beyond 50% attainable by the binomial model (Figure 9C and D). The loss of correlation between STB-ARS-plasmid-sisters and sister chromatids during a monopolin-directed cell cycle in the absence of cohesin (34) supports this interpretation. It is also consistent with the uncorrelated segregation of STB-arsΔ-pseudo-sisters and sister chromatids even in the presence of cohesin (this study). The spindle, by its role in cohesin assembly at STB, is important for the replication-dependent partitioning step as well.
Figure 9.
Random plasmid-chromosome association overcomes mother bias; symmetric association, in addition, promotes chromosome-like equal segregation. (A) Random association of the 2-micron plasmid with chromosomes promoted by Rep-STB and a subset of the host factors is refined into association of sister plasmids with sister chromatids (symmetric tethering) in a cohesin-assisted and replication-dependent step, accomplishing equal plasmid segregation. (B) The absence of a host factor that promotes plasmid-chromosome tethering in a replication-independent fashion will increase missegregation with a clear mother bias in plasmid retention. (C) In the absence of plasmid replication, the cohesin-effect (required for symmetric tethering) is lost. The plasmid pseudo-sisters (differentially colored) overcome mother bias, but segregate randomly. (D) When replication proceeds in the absence of cohesin, plasmid sisters may tether to chromosomes randomly, or occasionally tether to the same chromosome (which is not paired with its sister). The latter mode of tethering will missegregate plasmid sisters, but without bias. Equal segregation or missegregation of plasmid sisters randomly tethered to chromosomes will also be bias-free. Cohesin may play a role in organizing plasmid sisters in a configuration that disfavors their association with the same chromosome.
During the assembly of plasmid partitioning factors, Kip1 and RSC2 components are recruited at STB earlier than cohesin (37). This temporal sequence is consistent with plasmid-chromosome tethering per se (the basic Omb component of segregation) being further refined by cohesin into tethering of plasmid sisters to sister chromatids. The early factors that directly influence Omb may facilitate plasmid replication in association with chromosomes and/or foster plasmid-chromosome reassociation following temporary detachment during replication. Lack of Kip1 or Rsc2 not only reduces the level of plasmid-chromosome tethering (decrease in Omb from ∼0.5) but also renders the residual tethering random, as revealed by the pseudo-sister (independent) behavior of plasmid sisters. This failure of plasmid sisters to attach to sister chromatids is consistent with the block in cohesin assembly at STB caused by kip1Δ or rsc2Δ (new 36, 74). However, the frequency of equal segregation of plasmid sisters under cohesin depletion (∼36%) is even lower than that expected for their random tethering (∼47%) at the observed Omb = 0.43. Cohesin may promote a configuration of plasmid sisters that favors their one-to-one attachment to sister chromatids over their co-attachment to the same sister. An increase in the latter mode of tethering in the absence of cohesin may be responsible for lowering equal segregation below that effected by random tethering (Figure 9D).
2-micron circle segregation: analogy to viral episome maintenance
The fundamental logic of chromosome tethering for stable propagation unifies the yeast plasmid and viral episomes, although the molecular mechanisms for accomplishing this goal vary among individual systems (Supplementary Text S1H). There is evidence for the symmetric patterning of the EBNA1 protein and EBV episomes along sister chromatid arms (83). The E2 proteins of papilloma viruses, and thus the viral episomes by inference, appear to associate with chromosomes in a more random fashion (84). Furthermore, at least a subset of HPVs may attach directly to the mitotic spindle via E2, and segregate in a chromosome-like, but chromosome-independent, fashion (85). Among the viral systems, EBV appears to mimic the 2-micron plasmid most closely in the symmetric tethering of episome sisters and their 1:1 segregation to daughter nuclei (83,86).
Extrapolation from the single-copy reporter plasmid to the multi-copy 2-micron plasmid
A legitimate concern is whether the sister-to-sister segregation model deduced from single-copy STB-plasmids can be extrapolated in toto to native 2-micron circle foci, each focus being comprised of several plasmid molecules. Two single copy STB-plasmids, differentially tagged by red and green fluorescence, do obey the sister-to-sister rule (33). The majority of analyses using fluorescence-tagged, single-copy plasmids has been performed in strains containing the endogenous 2-micron plasmid molecules (which remain invisible during the assays). Assuming that there is no compartmentalization of the reporter plasmid, it is expected to faithfully represent the members of the plasmid focus to which it belongs.
In their normal biological context, all 2-micron plasmids are identical molecules, as are the products of their replication. As the distinction between sisters and non-sisters is more operational than real, the partitioning system can afford to be somewhat imprecise in organizing sister foci, without sacrificing the overall efficiency of equal segregation Furthermore, the amplification system is capable of compensating for this imprecision. The rarity of plasmid amplification during steady state growth (87) suggests that the partitioning system has been optimized to maintain copy number within the range that does not require frequent correction.
Supplementary Material
Acknowledgments
We acknowledge the help from Chris Sullivan and colleagues with assays utilizing mammalian cell lines and the gifts of plasmids from Arlen Johnson and Michael Knop. We thank Alison McBride for providing technical advice and suggestions on the preparation of mammalian chromosome spreads. We are grateful to Melanie Dobson, Lorie Frappier, Alison McBride, and Chris Sullivan for their constructive criticisms of the manuscript. We also thank the reviewers for their special effort in helping us improve the clarity of presentation.
Author contributions: Y.-T.L., K.-M.C., C.-H.M. and M.J. coceived and designed experiments. Y.T.-L., K.-M.C. and C.-H.M. performed experiments. Y.T.-L., K.-M.C., C.-H.M. and M.J. analyzed data. Y.-T.L., K.-M.C. and M.J. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Robert F Welch Foundation [F-1274]; National Science Foundation [MCB-1049925]. Funding for open access charge: Robert F Welch Foundation [F-1274].
Conflict of interest statement. None declared.
REFERENCES
- 1.Volkert F.C., Wilson D.W., Broach J.R. Deoxyribonucleic acid plasmids in yeasts. Microbiol. Rev. 1989;53:299–317. doi: 10.1128/mr.53.3.299-317.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes J.E., Welker D.L. Nuclear plasmids of Dictyostelium. Genet. Eng. (NY). 1999;21:1–14. doi: 10.1007/978-1-4615-4707-5_1. [DOI] [PubMed] [Google Scholar]
- 3.McBride A.A. Replication and partitioning of papillomavirus genomes. In: Karl Maramorosch AJS, Frederick AM, editors. Advances in Virus Research. Academic Press; 2008. pp. 155–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frappier L. Viral plasmids in mammalian cells. In: Funnell BE, Phillips G, editors. Plasmid Biology. Washington, D.C.: ASM Press; 2004. pp. 325–340. [Google Scholar]
- 5.Hammerschmidt W., Sugden B. Replication of Epstein-Barr viral DNA. Cold Spring Harb. Perspect. Biol. 2013;5:a013029. doi: 10.1101/cshperspect.a013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman P.M. Keeping it quiet: chromatin control of gammaherpesvirus latency. Nat. Rev. Microbiol. 2013;11:863–875. doi: 10.1038/nrmicro3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S., Agmon N., Sobol O., Segal D. Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mob. DNA. 2010;1:1–11. doi: 10.1186/1759-8753-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesare A.J., Griffith J.D. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata Y., Kumar P., Layer R., Willcox S., Gagan J.R., Griffith J.D., Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moller H.D., Parsons L., Jorgensen T.S., Botstein D., Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc. Natl. Acad. Sci. U.S.A. 2015;112:14–22. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu N. Molecular mechanisms of the origin of micronuclei from extrachromosomal elements. Mutagenesis. 2011;26:119–123. doi: 10.1093/mutage/geq053. [DOI] [PubMed] [Google Scholar]
- 12.Autiero M., Camarca A., Ciullo M., Debily M-A., El Marhomy S., Pasquinelli R., Capasso I., D'Aiuto G., Anzisi A.M., Piatier-Tonneau D., et al. Intragenic amplification and formation of extrachromosomal small circular DNA molecules from the PIP gene on chromosome 7 in primary breast carcinomas. Int. J. Cancer. 2002;99:370–377. doi: 10.1002/ijc.10368. [DOI] [PubMed] [Google Scholar]
- 13.Cohen S., Regev A., Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14:977–985. doi: 10.1038/sj.onc.1200917. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair D.A., Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 15.Henderson K.A., Gottschling D.E. A mother's sacrifice: what is she keeping for herself. Curr. Opin. Cell Biol. 2008;20:723–728. doi: 10.1016/j.ceb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y.T., Sau S., Ma C-H., Kachroo A.H., Rowley P.A., Chang K.M., Fan H.F., Jayaram M. The partitioning and copy number control systems of the selfish yeast plasmid: an optimized molecular design for stable persistence in host cells. In: Tolmasky ME, Alonso JC, editors. Plasmids: Biology and Impact in Biotechnology and Discovery. Washington, D.C.: ASM Press; 2015. pp. 325–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang K.M., Liu Y.T., Ma C.H., Jayaram M., Sau S. The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence. Plasmid. 2013;70:2–17. doi: 10.1016/j.plasmid.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Murray A.W., Szostak J.W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 19.Gehlen L.R., Nagai S., Shimada K., Meister P., Taddei A., Gasser S.M. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Current Biol. 2011;21:25–33. doi: 10.1016/j.cub.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Khmelinskii A., Meurer M., Knop M., Schiebel E. Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr. Biol. 2011;21:R17–R18. doi: 10.1016/j.cub.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 21.Shcheprova Z., Baldi S., Frei S.B., Gonnet G., Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;45:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 22.Futcher A.B., Cox B.S. Maintenance of the 2 micron circle plasmid in populations of Saccharomyces cerevisiae. J. Bacteriol. 1983;154:612–622. doi: 10.1128/jb.154.2.612-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Futcher A.B. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 24.Volkert F.C., Broach J.R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 25.Reynolds A.E., Murray A.W., Szostak J.W. Roles of the 2 micron gene products in stable maintenance of the 2 micron plasmid of Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray J.A., Scarpa M., Rossi N., Cesareni G. Antagonistic controls regulate copy number of the yeast 2 micron plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Som T., Armstrong K.A.F., Volkert C., Broach J.R. Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 28.Chen X.L., Reindle A., Johnson E.S. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 2005;25:4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong L., Chen X.L., Silver H.R., Ahmed N.T., Johnson E.S. Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 micron circle plasmid. Mol. Biol. Cell. 2009;20:1241–1251. doi: 10.1091/mbc.E08-06-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinder J.B., McQuaid M.E, Dobson M.J. Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance. PLoS One. 2013;8:e60384. doi: 10.1371/journal.pone.0060384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velmurugan S., Yang X.M., Chan C.S., Dobson M., Jayaram M. Partitioning of the 2-micron circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta S., Yang X.M., Chan C.S., Dobson M.J., Jayaram M., Velmurugan S. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh S.K., Hajra S., Jayaram M. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13034–13039. doi: 10.1073/pnas.0702996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y.T., Ma C.H., Jayaram M. Co-segregation of yeast plasmid sisters under monopolin-directed mitosis suggests association of plasmid sisters with sister chromatids. Nucleic Acids Res. 2013;41:4144–4158. doi: 10.1093/nar/gkt096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajra S., Ghosh S.K., Jayaram M. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-micron circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui H., Ghosh S.K., Jayaram M. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J. Cell Biol. 2009;185:251–264. doi: 10.1083/jcb.200810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma C.H., Cui H., Hajra S., Rowley P.A., Fekete C., Sarkeshik A., Ghosh S.K., Yates J.R., 3rd, Jayaram M. Temporal sequence and cell cycle cues in the assembly of host factors at the yeast 2 micron plasmid partitioning locus. Nucleic Acids Res. 2013;41:2340–2353. doi: 10.1093/nar/gks1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong M.C., Scott-Drew S.R., Hayes M.J., Howard P.J., Murray J.A. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 micron plasmid maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:4218–4229. doi: 10.1128/MCB.22.12.4218-4229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik H.S, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 40.Huang C.C., Hajra S., Ghosh S.K., Jayaram M. Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: implications in centromere evolution. Mol. Cell. Biol. 2011;31:1030–1040. doi: 10.1128/MCB.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C.C., Chang K.M., Cui H., Jayaram M. Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13671–13676. doi: 10.1073/pnas.1101944108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayaram M. Association of a centromere specific nucleosom.e with the yeast plasmid partitioning locus: Implications beyond plasmid partitioning. Mob. Genet. Elements. 2011;1:203–207. doi: 10.4161/mge.1.3.17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta S., Yang X.M., Jayaram M., Velmurugan S. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2 micron plasmid-cohesin association. Mol. Cell. Biol. 2005;25:4283–4298. doi: 10.1128/MCB.25.10.4283-4298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sau S., Conrad M.N., Lee C.Y., Kaback D.B., Dresser M.E., Jayaram M. A selfish DNA element engages a meiosis-specific motor and telomeres for germ-line propagation. J. Cell Biol. 2014;205:643–661. doi: 10.1083/jcb.201312002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burns N., Grimwade B., Ross-Macdonald P.B., Choi E.Y., Finberg K., Roeder G.S., Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 46.Chua P.R., Roeder G.S. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- 47.Conrad M.N., Dominguez A.M., Dresser M.E. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1998;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 48.Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 49.Primig M., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 50.Rabitsch K.P., Toth A., Galova M., Schleiffer A., Schaffner G., Aigner E., Rupp C., Penkner A.M., Moreno-Borchart A.C., Primig M., et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 51.Severin F., Hyman A.A., Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1990;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 53.Monje-Casas F., Prabhu V.R., Lee B.H., Boselli M., Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng P.S., Brokaw J., McBride A.A. Conditional mutations in the mitotic chromosome binding function of the bovine papillomavirus type 1 E2 protein. J. Virol. 2005;79:1500–1509. doi: 10.1128/JVI.79.3.1500-1509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taddei A., Hediger F., Neumann F.R., Gasser S.M. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 56.Jinks-Robertson S., Michelitch M., Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khmelinskii A., Keller P.J., Lorenz H., Schiebel E., Knop M. Segregation of yeast nuclear pores. Nature. 2010;454:728–734. doi: 10.1038/nature09255. [DOI] [PubMed] [Google Scholar]
- 58.Singer A., Schuss Z., Holcman D., Eisenberg R. Narrow escape, Part I. J. Stat. Phys. 2006;122:437–463. [Google Scholar]
- 59.Tsakraklides V., Bell S.P. Dynamics of pre-replicative complex assembly. J. Biol. Chem. 2010;285:9437–9443. doi: 10.1074/jbc.M109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diffley J.F., Cocker J.H., Dowell S.J., Harwood J., Rowley A. Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle. J. Cell Sci. Suppl. 1995;19:67–72. doi: 10.1242/jcs.1995.supplement_19.9. [DOI] [PubMed] [Google Scholar]
- 61.Bose M.E., McConnell K.H., Gardner-Aukema K.A., Muller U., Weinreich M., Keck J.L., Fox C.A. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 2004;24:774–786. doi: 10.1128/MCB.24.2.774-786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozaydin B., Rine J. Expanded roles of the origin recognition complex in the architecture and function of silenced chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010;30:626–639. doi: 10.1128/MCB.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grunweller A., Ehrenhofer-Murray A.E. A novel yeast silencer: The 2μ origin of Saccharomyces cerevisiae has HST3-, MIG1- and SIR-dependent silencing activity. Genetics. 2002;162:59–71. doi: 10.1093/genetics/162.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piatti S., Bohm T., Cocker J.H., Diffley J.F., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a "point of no return" after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka T., Knapp D., Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 66.Corbett K.D., Yip C.K., Ee L-S., Walz T., Amon A., Harrison S.C. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petronczki M., Matos J., Mori S., Gregan J., Bogdanova A., Schwickart M., Mechtler K., Shirahige K., Zachariae W., Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 68.Rabitsch K.P., Petronczki M., Javerzat J-P., Genier S., Chwalla B., Schleiffer A., Tanaka T.U., Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 69.Tóth A., Rabitsch K.P., Gálová M., Schleiffer A., Buonomo S.B.C., Nasmyth K. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 70.Nasmyth K. Cohesin: a catenase with separate entry and exit gates. Nat. Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 71.Onn I., Heidinger-Pauli J.M., Guacci V., Ünal E., Koshland D.E. Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 72.Huang J., Hsu J.M., Laurent B.C. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 73.Ghosh S.K., Huang C.C., Hajra S., Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 2010;38:570–584. doi: 10.1093/nar/gkp993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang X.M., Mehta S., Uzri D., Jayaram M., Velmurugan S. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol. Cell. Biol. 2004;24:5290–5303. doi: 10.1128/MCB.24.12.5290-5303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahn Y.T., Wu X.L., Biswal S., Velmurugan S., Volkert F.C., Jayaram M. The 2 micron plasmid encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J. Bacteriol. 1997;179:7497–7506. doi: 10.1128/jb.179.23.7497-7506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Velmurugan S., Ahn Y.T, Yang X.M., Wu X.L., Jayaram M. The 2 micron plasmid stability system: analyses of the interactions among plasmid- and host-encoded components. Mol. Cell. Biol. 1998;18:7466–7477. doi: 10.1128/mcb.18.12.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scott-Drew S., Murray J.A. Localisation and interaction of the protein components of the yeast 2 micron circle plasmid partitioning system suggest a mechanism for plasmid inheritance. J. Cell Sci. 1998;111:1779–1789. doi: 10.1242/jcs.111.13.1779. [DOI] [PubMed] [Google Scholar]
- 78.Sengupta A., Blomqvist K., Pickett A.J., Zhang Y., Chew J.S., Dobson M.J. Functional domains of yeast plasmid-encoded Rep proteins. J. Bacteriol. 2001;183:2306–2315. doi: 10.1128/JB.183.7.2306-2315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hadfield C., Mount R.C., Cashmore A.M. Protein binding interactions at the STB locus of the yeast 2 micron plasmid. Nucleic Acids Res. 1995;23:995–1002. doi: 10.1093/nar/23.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.You J., Croyle J.L., Nishimura A., Ozato K., Howley P.M. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 81.Wu H., Ceccarelli D.F., Frappier L. The DNA segregation mechanism of Epstein–Barr virus nuclear antigen 1. EMBO Rep. 2000;1:140–144. doi: 10.1093/embo-reports/kvd026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You J., Srinivasan V., Denis G.V., Harrington W.J., Ballestas M.E., Kaye K.M., Howley P.M. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanda T., Kamiya M., Maruo S., Iwakiri D., Takada K. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J. Cell Sci. 2007;120:1529–1539. doi: 10.1242/jcs.03434. [DOI] [PubMed] [Google Scholar]
- 84.Oliveira J.G., Colf L.A., McBride A.A. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1047–1052. doi: 10.1073/pnas.0507624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Tine B.A., Dao L.D., Wu S.Y., Sonbuchner T.M., Lin B.Y., Zou N., Chiang C.M., Broker T.R., Chow L.T. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4030–4035. doi: 10.1073/pnas.0306848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nanbo A., Sugden A., Sugden B. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–4262. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zakian V.A., Brewer B.J., Fangman W.L. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979;17:923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.